Abstract
Populations of the common chimpanzee (Pan troglodytes) are in an impending risk of going extinct in the wild as a consequence of damaging anthropogenic impact on their natural habitat and illegal pet and bushmeat trade. Conservation management programmes for the chimpanzee have been established outside their natural range (ex situ), and chimpanzees from these programmes could potentially be used to supplement future conservation initiatives in the wild (in situ). However, these programmes have often suffered from inadequate information about the geographical origin and subspecies ancestry of the founders. Here, we present a newly designed capture array with ~60,000 ancestry informative markers used to infer ancestry of individual chimpanzees in ex situ populations and determine geographical origin of confiscated sanctuary individuals. From a test panel of 167 chimpanzees with unknown origins or subspecies labels, we identify 90 suitable non-admixed individuals in the European Association of Zoos and Aquaria (EAZA) Ex situ Programme (EEP). Equally important, another 46 individuals have been identified with admixed subspecies ancestries, which therefore over time, should be naturally phased out of the breeding populations. With potential for future re-introduction to the wild, we determine the geographical origin of 31 individuals that were confiscated from the illegal trade and demonstrate the promises of using non-invasive sampling in future conservation action plans. Collectively, our genomic approach provides an exemplar for ex situ management of endangered species and offers an efficient tool in future in situ efforts to combat the illegal wildlife trade.
Subject terms: Genetic variation, Conservation biology, Conservation biology
Introduction
In an era of human-induced acceleration of species loss, often referred to as the sixth mass extinction era (Ceballos et al. 2015), conservation efforts to save endangered species are calling for novel approaches to mitigate the ongoing extinction crisis.
Since the discovery of the common chimpanzee (Pan troglodytes), humans have been drawn to this charismatic species. Despite our fascination, human activities have led to a drastic decline in the population size of the chimpanzee. In the last two decades, chimpanzees have been listed as ‘Endangered’ at the species level on the IUCN Red List, with one of the four recognised subspecies, the western chimpanzee (P. t. verus) being listed as ‘Critically Endangered’ in the latest assessment (Humle et al. 2016). Human encroachment on the natural range of the chimpanzee has further caused an intensified conflict between humans and chimpanzees (Hockings et al. 2015). One by-product of the human wildlife conflicts has been a rise in opportunistic trafficking of chimpanzees, which, in recent years has become more organised and systematic (Stiles et al. 2013). Besides wildlife trade, other continuous threats including habitat destruction, poaching for local consumption, and human linked disease outbreaks has led to a drastic decline in the wild chimpanzee populations (Humle et al. 2016). Together, these threats emphasise the importance of a ‘One Plan Approach’ conservation programme linking in situ and ex situ efforts (Traylor-Holzer et al. 2019) to prevent the predicted extinction of chimpanzees within the current century (Estrada et al. 2017).
Outside Africa, several regional chimpanzee conservation programmes exist, with the largest being the European Association of Zoos and Aquaria (EAZA) Ex situ Programme (henceforth EEP). The EEP targets the subspecies level and today, breeding programmes for two of the four recognised subspecies, the western chimpanzee (P. t. verus) and the central chimpanzee (P. t. troglodytes) have been established (Carlsen and de Jongh 2019). The primary aim of the EEP is to safeguard the survival of healthy self-sustaining populations targeting the taxonomical level of subspecies (Carlsen and de Jongh 2019). The extant EEP populations consist of wild founders and descendants thereof. However, in times before high resolution genetic technologies were available and even in its early development, knowledge of subspecies labels and relatedness between founders were inaccurate and has led to admixture of subspecies in the captive population (Hvilsom et al. 2013). Early attempts to add a genetic layer to the EEP management has confirmed that knowledge of subspecies ancestries, inbreeding and relatedness estimates are instrumental to preserve genetic diversity in captive populations (Hvilsom et al. 2013). Yet, most recent attempts based on microsatellite markers (Hvilsom et al. 2013), did not have the necessary resolution or predictive power to disentangle several generations of hybridisations in the EEP breeding population. Although we still do not know its full extent, hybridisation between neighbouring subspecies of chimpanzees has been shown to occur in the wild (Hvilsom et al. 2013; Prado-Martinez et al. 2013; de Manuel et al. 2016) and therefore, it is not unlikely that some founders in the EEP harbour shared ancestries from more than one subspecies. The current strategy in the EEP targets un-admixed breeding individuals and with the current methods, it is impossible to tell if small admixture proportions arose from an early ex situ hybridisation event followed by several generations of backcrossing or from a naturally admixed founder. Therefore, founders are potentially being wrongfully excluded from the breeding programme due to their admixed ancestry.
The scenario outlined above, is by no means exclusive to captive management of chimpanzees but extends to practically any ex situ management programme of populations based on wild born founders with a taxonomical subdivision. When morphology alone is insufficient in taxonomical delimitation between subspecies or the targeted conservation units, genetic resources becomes increasingly important. Yet, the choice of genetic resource is not always trivial. In response to a growing availability of different types of genetic resources with widely different applications, several studies have tried to develop guidelines based on the management requirements (see e.g. Grueber et al. 2019; Norman et al. 2019).
As described, the complexities in EEP management of chimpanzees requires a new rigorous solution as previous attempts using either mitochondrial DNA, or microsatellites have proven insufficient. With a genome-wide set of ancestry informative markers, we predict that it will be possible to obtain the desired depth of predictive power to infer ancestries in the present and previous generations and classify individuals with shared ancestries as either descendants of admixed founders or ex situ hybrids. This could provide the foundation of a possible reassessment of the current management strategies under the EEP and in turn, allow for inclusion of wild born hybrids in the breeding programme if these are found to resemble the diversity of the species in the wild.
In their natural range, chimpanzees have become a commodity and organised illegal trade poses a serious threat to the species. Over the period from 2005 to 2011 a reported minimum of 643 chimpanzees were harvested from the wild for illegal trade activities (Stiles et al. 2013). However, extrapolations suggest that 20 times as many individuals have become victims of the illegal wildlife trade in that relatively short time span (Stiles et al. 2013). While most of the captured individuals are sold as bushmeat, a considerable number of mostly juvenile chimpanzees end up in the illegal pet trade. When conservation authorities confiscate illegally kept chimpanzees, they are placed at wildlife sanctuaries, often arbitrarily based on availability of space and proximity to the confiscation site. Whilst some of the rescued chimpanzees require specialised lifetime care, others may be successfully reintroduced into their natural habitats after extensive preparation (Beck et al. 2007). For chimpanzees destined to lifetime care, proper management planning requires knowledge about relatedness among sanctuary chimpanzees in order to set up family groups. In cases, were chimpanzees are suitable for reintroduction, knowledge of geographical origin is essential as several studies have shown lineage-specific adaptations in all four subspecies in their respective geographical ranges (e.g. Nye et al. 2018). In the first complete geo-referenced genomic map of the chimpanzee, de Manuel et al. (2016) portrayed a strong correlation between geographical origin and genetic diversity, where the former can be inferred solely based on the latter. Employing genetic testing at the site of confiscation (e.g. airports and transport hubs) would enable conservation authorities to infer geographical origin of confiscated individuals and with time, strive to facilitate a return of these individuals to a protected area in the region where they were captured. Alternatively, confiscated chimpanzees can be sent to a neighbouring sanctuary with housing capacity, where specialised care and rehabilitation can be provided, and if possible, future reintroduction can be planned. Genetic testing at an early stage of confiscation also has the potential to understand and help break trafficking routes and enable CITES authorities to track and enforce law control in situations where chimpanzees are housed in disreputable zoos and entertainment facilities. However, to be a practical tool in conservation, the genetic test needs to maximise the inference accuracy, require very little investment, and pose as little risk to animal health as possible. These requirements limit our choice of applicable data types. With a novel SNP array design where the level of genetic information is only surpassed by costly whole genome sequencing, we argue that our approach constitutes the most cost-efficient option for conservation management in situations where funding is often scarce and demands for rigorous solutions are high.
Using a selected panel of 59,800 targeted ancestry informative markers, we demonstrate the ability to infer robust estimates of ancestry in several generations of the EEP chimpanzee breeding population. We further show how this set of ancestry informative markers can be used to determine geographical origin of confiscated individuals and demonstrate how these methodologies can readily be applied to using non-invasive sampling. In combination, these methods harbour great potential for future global management plans for the chimpanzee and provides an important exemplar for management of endangered species in general.
Materials and methods
Samples
A total of 179 chimpanzee samples were collected and analysed in the present study (Supplementary File S1 SequencingStatistics.xlsx). For the purpose of cross-validation between sequencing batches and to test our methodology on non-invasive hair sampling, a number of individuals were sequenced in duplicates and triplicates, which lead to 167 unique individuals. 136 from the EEP population housed in 47 different European zoos and primate rescue and rehabilitation centres (Table S2), and 31 from eight sanctuaries across Africa (Table S3). To form a reference panel, we complemented the genotypes of EEP and sanctuary chimpanzees with whole genome data from 58 geo-referenced wild-born chimpanzees, representing the four chimpanzee subspecies, and additionally, one known admixed individual (Ptv-Donald) and one known descendant of wild born individuals (Ptv-Clint) (Prado-Martinez et al. 2013; de Manuel et al. 2016).
DNA extraction and library preparation
DNA was extracted using a standard phenol-chloroform protocol. Samples were quantified with a Qubit 2.0 fluorometer, Qubit® dsDNA BR Assay Kit (Thermo Fisher Scientific). DNA library preparation was carried out in three batches. For the first batch (24 samples) and the second batch (63 samples), extracted DNA was sheared with a Covaris S2 ultrasonicator using the recommended fragmentation settings to obtain a 350 bp insert size. For the third batch (92 samples) DNA was sheared using the recommended settings of Covaris S2 to obtain 200 bp insert size. The first batch of 24 libraries (with 6 more samples not used in this study) were prepared using 1.5 μg of DNA and the TruSeq DNA HT Sample Prep Kit (Illumina), following manufacturer’s instructions and 14 cycles of polymerase chain reaction (PCR) amplification. The second batch of 63 samples (with 17 more samples not used in this study) were processed using 500 ng of starting DNA and following the custom dual-indexed protocol described by Kircher et al. (2012) and 12 cycles of PCR were done for indexing and amplification. The remaining 92 samples (with two more samples not used in this study) were processed using 200 ng of starting DNA following the BEST protocol (Carøe et al. 2018) with minor modifications (initial reaction volume was incremented up to 50 μl to accommodate a larger amount of starting DNA and 10 cycles of PCR amplification). For this third batch, we used inline barcoded short adaptors with the same seven nucleotide barcodes at the P5 and P7 adaptors. Clean-ups were done using homemade SPRI beads (Rohland and Reich 2012). Libraries were eluted in 25 μl of ddH2O and quantified with an Agilent 2100 Bioanalyzer using a DNA 1000 assay kit.
Target capture design
We performed a target capture enrichment experiment using baits synthesised by Agilent Technologies. We targeted 59 800 autosomal sites that were ancestry informative markers and designed using the panTro4 genome. Marker selection was done using published chimpanzee genomes (Prado-Martinez et al. 2013) and by applying a sparse PCA method on 10 Mbp bins of the genomes (Lee et al. 2012). Variant sites were then weighted to identify the most informative markers for the first two principal components (PCs) and 200 AIMs were extracted per segment. The genome was binned to have an unbiased and evenly distributed sampling of the genome and to have enough resolution to provide estimates of ancestry in highly admixed individuals.
For target enrichment hybridisation, libraries were pooled equimolarly based on a library prep method to obtain a total of 19 pools (see Supporting Information for a detailed description of the targeted enrichment hybridisation). PCR amplification product was cleaned up using our homemade SPRI beads (Rohland and Reich 2012). Each enriched sample was then quantified on a NanoDrop, BioAnalyzer and then sequenced.
Fastq filtering and mapping
Libraries were sequenced on five lanes of a HiSeq 2500 ultra-high-throughput sequencing system, one lane for 24 chimpanzee samples, 2 lanes for 63 chimpanzee samples and 2 lanes for the remaining 92 samples. Inline barcoded libraries captured in the same pool (92 from Batch 3) were de-multiplexed using Sabre software v. 1.0 (https://github.com/najoshi/sabre).
Prior to mapping, paired-end reads were filtered to remove PCR duplicates using FASTUNIQ v. 1.1 (Xu et al. 2012) and adaptors (Illuminaclip) and low quality first five bases in a read (Slidingwindow:5:20) were trimmed using TRIMMOMATIC v. 0.36 (Bolger et al. 2014). Overlapping reads were merged with a minimum overlap of 10 bp and minimum length of final read to 50 bp, using PEAR v. 0.9.6 (Zhang et al. 2014). Then, reads were mapped using BWA v. 0.7.12 (Li and Durbin 2009) to the Hg19 reference genome (GRCh37, Feb.2009 (GCA_000001405.1)). PCR duplicates were removed using PICARDTOOLS v. 1.95 (http://broadinstitute.github.io/picard/) with the MarkDuplicates option. Further filtering of the reads was done to discard secondary alignments and reads with mapping quality lower than 30 using SAMTOOLS v. 1.5 (Li et al. 2009). We then filtered for the targeted space (4 bp around the selected SNP) using BEDTOOLS intersect v. 2.16.2 (Quinlan and Hall 2010).
The total aligned reads were calculated by dividing the number of uniquely mapped reads (the remaining reads after removing duplicates) by the number of production reads. The on-target aligned reads were calculated by dividing the target filtered reads by the production reads. Then, the total coverage was calculated by dividing aligned bases by the length of the assembly (Hg19) and the target effective coverage dividing the on-target bases by the targeted genomic space. Finally, the enrichment factor of the capture performance was calculated by taking the ratio between the on-target reads by total mapped reads over the target size by genome size.
Variant calling
Variant discovery was performed using GATK ‘Unified Genotyper’ (DePristo et al. 2011) for each sample independently with the following parameters -out_mode EMIT_ALL_SITES -stand_call_conf 5.0 -stand_emit_conf 5.0 -A BaseCounts -A GCContent -A RMSMappingQuality -A BaseQualityRankSumTest. Genotypes from each sample were combined in a single VCF using GATK ‘CombineVariants’ (DePristo et al. 2011) with -genotypeMergeOptions UNIQUIFY –excludeNonVariant parameters. We also included the genotype information of available whole genome data of aforementioned 58 wild-born geo-referenced chimpanzees and Ptv-Donald and Ptv-Clint (Prado-Martinez et al. 2013; de Manuel et al. 2016). Unless differently stated in separate analysis, the variants with a depth of coverage less than 3, a quality score less than 30 (QUAL < 30), minor allele frequency (MAF) of 0.005 and a missingness rate of >60 % were removed using VCFTOOLS v. 0.1.12 (Danecek et al. 2011). We only kept the genotypes that were inside the target space by using the -bed option in VCFTOOLS v. 0.1.12 (Danecek et al. 2011).
Ancestry inference and inbreeding
We inferred proportions of shared ancestries in two approaches. First, to detect underlying genetic structure with a reduction of the dimensionality in the data, we performed a principle component analysis (PCA) using EIGENSOFT v. 6.1.3. (Price et al. 2006). All samples were included without pruning of sites in linkage disequilibrium or MAF, in order to avoid exclusion of fixed sites between populations. Analyses on shared ancestry in ex situ and sanctuary populations were done with reference to the genetic structure in the wild born individuals with ADMIXTURE v. 1.2 (Alexander et al. 2009). To avoid any bias introduced from a joint analysis with related individuals, each of the 167 unique individuals from the EEP and sanctuary populations were analysed separately one by one against a reference panel of all wild born individuals. After applying a MAF filter (--maf 0.05) in PLINK v. 1.07 (Purcell et al. 2007) to exclude sites polymorphic in only one individual, a set of 45,542 sites where kept for analysis. Each analysis of ADMIXTURE v. 1.2 (Alexander et al. 2009) was iterated 100 times under an EM optimisation algorithm and termination criteria of a log-likelihood increase of 10−5 between iterations. A value of K = 4 was chosen to obtain clusters in line with the four recognised subspecies of chimpanzees. To assess convergence, the 100 iterations were evaluated to ensure that iterations did not differ by more than 1 log-likelihood value.
For each of the individuals with admixture coefficients >0.99, we applied NGSRELATEv2 (Hanghøj et al. 2019) to estimate pairwise relatedness and individual inbreeding coefficients based on population allele frequencies from each of the inferred admixture clusters, after excluding MAF < 0.05 (see Supplementary Information for details along with per population and global estimates of FIS).
Hybrid classification
To further explore the ancestry sharing in the EEP and sanctuary individuals and to be able to differentiate shared ancestry originating from the founding individuals and EEP hybrids, we developed a hidden Markov model (available on GitHub http://gihub.com/svendvn/ImmediateAncestry) to allow for an inference of the posterior proportion of ancestries in the three immediate previous generations. In addition, we estimate where these immediate ancestors belong in the pedigree. For full documentation of the model, see Supplementary Information.
Re-assignment of geographical origin
We applied the methodology of ORIGEN (CRAN R package https://cran.r-project.org/web/packages/OriGen/index.html) as described by Rañola et al. (2014), to re-assign the geographical origin of confiscated sanctuary individuals. We applied the FitOriGenModelFindUnknowns parameter to the 1690 highest ranked informative markers to assign individual geographical origin onto the allele frequency surface, inferred from the wild born reference panel.
Non-invasive sampling
To test our targeted capture approach on non-invasively collected hair samples, we sequenced three individuals where we had both blood samples, whole genome reference data and hair samples. Hair samples were capture sequenced using the same methodology as described above for blood samples, except we added a pre-treatment step in the DNA extraction of hair samples to enhance lysis of keratin. Shared ancestry and geo-graphical origin was analysed as described above.
Results
Capture sequencing and variant calling
First we quantified and assessed the performance of our capture methodology in the selected targeted space. We wanted to ensure sufficient representation of the targeted genomic regions to reliably call the selected variants. In a total of five lanes of HiSeq2500 we obtained ~1000 million production reads, and on average, each sample received five million reads. After removing PCR duplicates and considering only primary alignments with a mapping quality higher than 30, we obtained an average of 3.6 million mapped reads (74.31%) per sample (Supplementary File S1). The average effective target coverage on the 59,800 autosomal SNPs was 21.69X with 12.91% of on-target reads (four base pairs around the targeted SNP, Supplementary File S1) which fulfilled our theoretical prediction of 20X. In terms of capture performance, this last statistic is an underestimate since the full length of the capture bait is 120 base pairs and in this analysis, we only considered the four base pairs around the targeted SNP. Still, we considered it to be more accurate since it is the true space where the informative SNP falls. Lastly, to summarise the performance of the capture methodology, we computed the enrichment factor that relates the number of aligned reads on the target space divided by the production reads, with the size of the target space to the size of the whole genome. The resulting enrichment factor of 89.31X reasserts the advantages of capture to ensure enough coverage for genotyping purposes (Supplementary File S1).
Considering all samples without overlap, we obtained a total of ~150,000 genotypes. However the average number of SNPs called per sample was 30,337 sites passing the filtering steps (MAF 0.05 and max-missing 0.6, after we excluded samples ‘12103’ and ‘12349’ due to low coverage). The maximum number of SNPs called in one individual was 51,952 and the minimum was 10,783 (Fig. S1). Among the variation found in western chimpanzees, only a third of these were polymorphic in the western chimpanzee (Table S1), yet, of the 46,260 polymorphic sites, 15,738 were private in the western chimpanzee (Fig. S2). For fixed sites, the western chimpanzee also had the highest number of private sites (Fig. S2). Among the four subspecies, the eastern chimpanzee had the highest total number of polymorphic sites, followed by the central chimpanzee, Nigerian-Cameroon chimpanzee, and western chimpanzee, respectively (Table S1).
Population structure, ancestry, and inbreeding
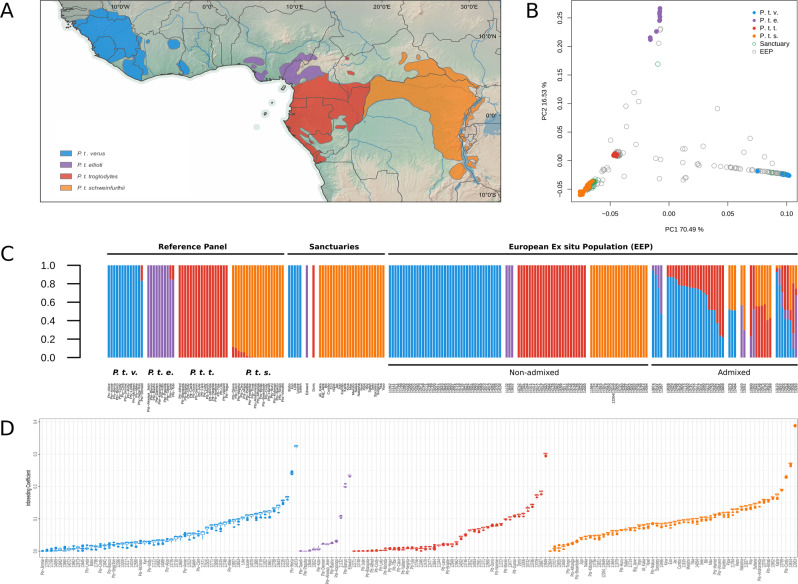
The major axes of variance in EEP and sanctuary individuals were explored with a PC analysis with reference to the panel of geo-referenced individuals with known subspecies label from Prado-Martinez et al. (2013) and de Manuel et al. (2016). The first PC (PC1) explained 70.49% of the variance in our data, separating the western chimpanzees from the three other subspecies in the reference panel (Fig. 1b). With 16.53 % of explained variance, PC2 separated the Nigerian-Cameroon chimpanzee, central chimpanzee, and eastern chimpanzee.
Fig. 1. Subspecies ancestry and inbreeding in wild and captive populations of chimpanzees.
a Geographical distribution ranges of the four chimpanzee subspecies (IUCN 2015; QGIS 2018). b Population structure by principal component decomposition of sanctuary and the EAZA Ex situ Programme (EEP) populations with reference to wild born individuals. c Shared ancestry inferences of sanctuary and EEP individuals summarised from individual ADMIXTURE analysis against the reference panel of wild born individuals. Individuals from the reference panel are labelled with a subspecies ancestry prefix and known sample name in previous literature (Prado-Martinez et al. 2013; de Manuel et al. 2016), sanctuary individuals are labelled with common sample name identifiers, and individuals from the EEP are labelled by studbook number (Tables S2 and S3). d Individual inbreeding coefficients for all individuals with admixture proportions >0.99 in either of the four inferred clusters. Inbreeding estimates were estimated within each cluster independently. Clusters are colour labelled in accordance to (a–c).
The majority of the 167 tested individuals from the EEP and sanctuary populations, clustered with either of the four reference populations, while a minor part of the individuals scattered in between the defined populations (Fig. 1b). The inferred ancestries from the ADMIXTURE analysis conveyed the same patterns of genetic population structure separating the geo-referenced individuals into four distinct clusters with varying degree of ancestry sharing between geographically neighbouring subspecies (Fig. 1c). With this as a reference, we assigned the EEP and sanctuary individuals into groupings in terms of their ancestry patterns of either non-admixed or hybrids with multiple components of ancestry. Of the 167 tested individuals, 121 could be confidently assigned as non-admixed (admixture proportion from one subspecies ≥ 0.99). All 31 sanctuary individuals were assigned to subspecies level without evidence of admixture, where five clustered with the western chimpanzee, one with the Nigerian-Cameroon chimpanzee, one with the central chimpanzee, and 24 with the eastern chimpanzee. In the EEP population, we inferred the majority of the 90 non-admixed individuals to belong to the western chimpanzee (41), three with the Nigerian-Cameroon chimpanzee, 25 with the central chimpanzee, and 21 with the eastern chimpanzee. Of the remaining 46 EEP individuals, 38 were inferred to be hybrids with two ancestry components while the last eight had three ancestry components.
Of all the individuals from the EEP, sanctuary, and the reference panel with admixture coefficients >0.99, relatedness estimates were low (Figs. S3–S6) while we identified eight individuals with inbreeding coefficients above 0.2 (Fig. 1d). Within these eight individuals, all four subspecies were represented, as were wild and captive born chimpanzees.
Hybrid classification
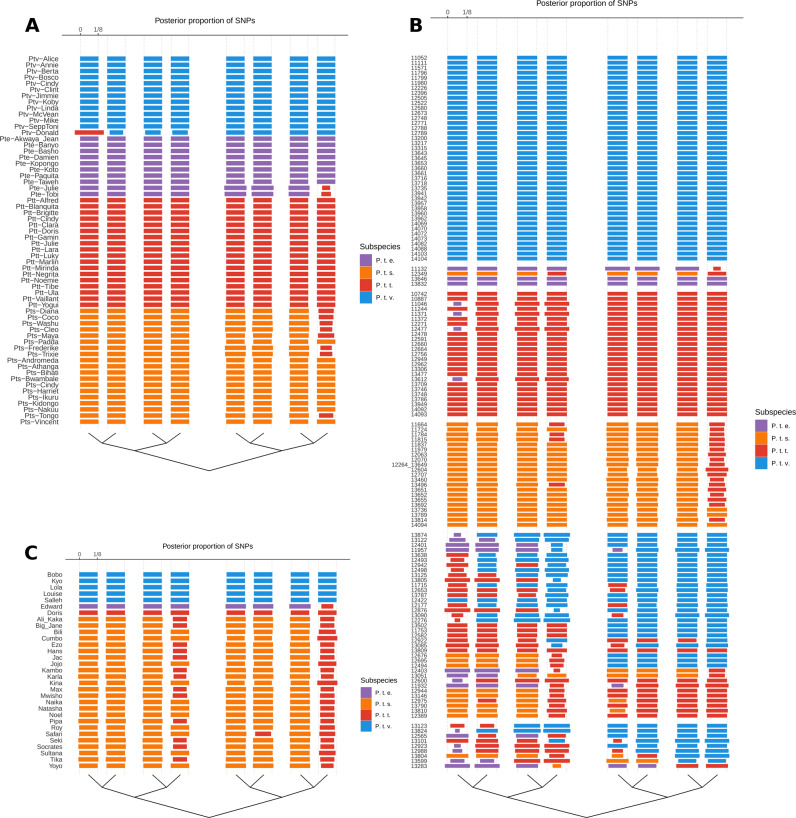
To explore ancestry patterns in the previous three generations, we ran our ancestry classification model going back k = 3 generations and visualised the number of loci each ancestor in generation k contributed to the ancestral informative part of the genome (see Supplementary Information). In general, our method correctly estimated the expected ancestries of our reference panel individuals (Fig. 2a). Several eastern and Nigerian-Cameroonian chimpanzee individuals were estimated to contain substantial ancestry components from the mutually neighbouring central subspecies. The known hybrid Ptv-Donald (Prado-Martinez et al. 2013) was estimated by the method to be at least one-eighth central chimpanzee, yet the large proportion of loci that were assigned to the central chimpanzee in the posterior distribution might suggest that Ptv-Donald could be as much as one-fourth central chimpanzee.
Fig. 2. Hybrid classification.
Hybrid ancestry in a the reference panel, b the EEP population, and c the sanctuary population. The estimated posterior ancestries, θ is shown for the eight ancestors k = 3 generations back in time, for each individual in the three populations. The ancestors are orderedaccording to the “unphased” pedigree in the bottom of the plot. The width of each rectangle indicate the expected proportion of loci that are assigned to thatancestor (conditioned on the estimate of θ). Small widths suggest deviations from the model and features that could be improved by posterior correction.
Similar to the ancestries inferred with ADMIXTURE, our method classified a large fraction of the EEP and sanctuary individuals to have ancestors from only one subspecies in the last three generations (Figs. 1c, 2b, c). In general, individuals inferred to belong to the eastern chimpanzee had third generation ancestors of central chimpanzee ancestry (Fig. 2b, c). Similarly, four inferred central chimpanzees in the EEP population, showed small proportions of ancestry from the Nigeria-Cameroon chimpanzee. Comparably, one sanctuary individual, Edward, was inferred here as a Nigeria-Cameroon chimpanzee with a small proportion of central chimpanzee ancestry. However, performing posterior correction by replacing the low central chimpanzee ancestor with another high posterior Nigeria-Cameroon ancestor, would likely make a more accurate estimate. Among the admixed EEP individuals, our model showed similar results to those obtained with ADMIXTURE but as ancestry patterns became increasingly complex (more than two ancestral subspecies) our inferred posterior proportions became increasingly uncertain (Figs. 2b, S14). We further observed that in some cases, small deviating (possibly deep coalescing) segments could have let the model to prefer configurations in the ancestry patterns to switch halves (Fig. 2c), while the correct configuration would probably be a simple case of hybridisation in the parent generation.
Geo-localisation
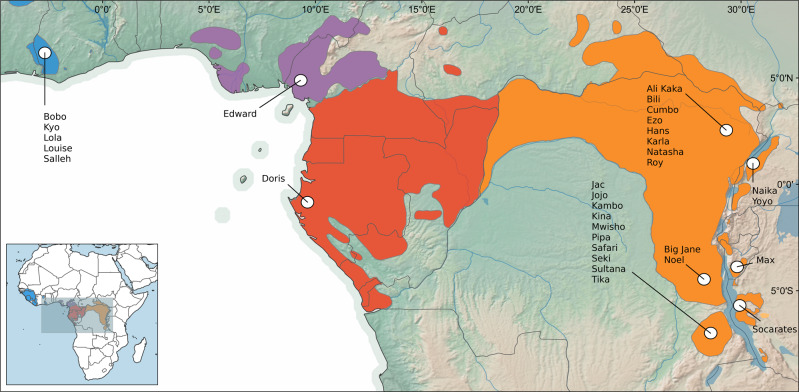
Based on an allele frequency surface map, built from our reference panel of wild born individuals, we determined the geographical origin of all 31 sanctuary individuals. Generally, the inferred probabilities of geographical origin gave accurate estimates (i.e. high probabilities assigned to just one or a few adjacent grid cells) for all sanctuary individuals (Fig. 3). Also, all individuals assigned to the natural range of their inferred subspecies label. The majority of our tested sanctuary individuals belonged to the eastern chimpanzee where the geographical origins were inferred to six provinces along the eastern part of the natural range of the subspecies. Seven of the eastern individuals had low probability estimates divided over a cluster of adjacent grid cells, with the highest ranking cell assigned probability of less than 0.1. All five western chimpanzee individuals were assigned to the same grid cell in the eastern limits of their range. The single individual from the Nigeria-Cameroon chimpanzee was assigned to a locality in Cameroon while the one central chimpanzee was assigned to the coastal region of Gabon.
Fig. 3. Geographical origin estimates for sanctuary individuals.
Based on the allele frequency surface map of the reference panel, sanctuary individuals are assigned probabilities of geographical origin, here summarised from individual estimates.
Non-invasive sampling
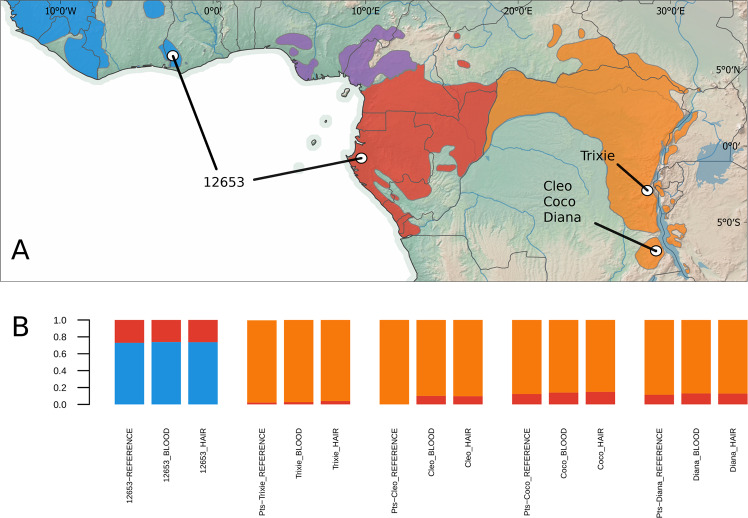
Expanding our targeted capture approach to non-invasively collected hair samples, corroborated the results obtained with blood samples. ADMIXTURE estimates converged to the same result in the two sample types for all tested individuals and geographical origin was assigned to the same locality between samples (Figs. 4, S15–S19). Compared to the reference, ancestry estimates in our capture array approach did not always reveal the minor components of shared ancestries found when including all variant sites in the genome (Fig. 4).
Fig. 4. Ancestry and geographical origin estimates from non-invasive samples.
a Geographical origin estimates from hair samples based on the allele frequency surface map of the reference panel, tested individuals are assigned probabilities of geographical origin, here summarised from individual estimates with comparison to blood samples (Figs. S15–S19). b Shared ancestry estimates for hair samples compared to whole genome reference data and capture sequenced data from blood.
Discussion
As an exemplar for conservation genetics of endangered species, we have designed a novel capture array that targets identified ancestry informative markers across the genomes of 24 wild born chimpanzees (Prado et al. 2013) and the PanTro4 reference genome. Acknowledging that the selected ancestry markers were derived from a relatively limited set of genomes, which could potentially introduce an ascertainment bias towards specific subspecies, we confirmed that our design has the power to correctly identify the subspecies of an extended panel of newly sequenced chimpanzee genomes (de Manuel et al. 2016) (Fig. 1). Based on this proof of concept, we sequenced 167 chimpanzees from the EEP and sanctuary populations and analysed subspecies ancestries and geographical origin. We further show how this approach can be extended to non-invasive samples with robust results.
Ancestry of the ex situ population
In our test panel of 167 chimpanzees, 136 were from the EEP population housed at 47 European zoos and rehabilitation centres. Based on information on disembarkation or place of capture, we know that the majority of chimpanzees who founded the current EEP population came from West Africa. In accordance to this, a majority of the 90 non-admixed individuals could be assigned to the western chimpanzee (Fig. 1c). Our findings confirm that for the western chimpanzee, early efforts of the EEP that sought to identify a core group of non-admixed western chimpanzees using mitochondrial DNA (Jepsen and Carlsen unpublished) and microsatellites (Hvilsom et al. 2013), have been momentarily successful. Yet, using similar methodologies, previous attempts have only managed to identify a small group of central chimpanzees since the breeding effort for this subspecies was established (Carlsen and de Jongh 2019). Here, we identify 25 central chimpanzee individuals in the EEP population that show no evidence of shared ancestry with other subspecies (Fig. 1c), and hence from a genetic viewpoint, would qualify as a suitable bolster to the current breeding population. Similarly, the 21 inferred non-admixed eastern chimpanzee individuals could form the crucial starting point from where a separate breeding effort could be established under the EEP. In contrast to this, of our tested 136 EEP individuals, only three could be assigned to the Nigerian-Cameroonian subspecies (Fig. 1c) and in general, of the four subspecies, the Nigeria-Cameroon chimpanzee is by far the least represented in the EEP population (Carlsen and de Jongh 2019). Yet, with our targeted capture approach, it will now be feasible to scan the remaining EEP population (~1000 housed individuals) for additional non-admixed chimpanzee individuals in order to explore the possibilities of creating separate breeding populations for the two remaining subspecies.
Still, with a presumed small EEP population of eastern and Nigerian-Cameroonian chimpanzees, it might prove difficult to avoid inbreeding, although our estimates suggests, that high inbreeding coefficients are not exclusive to these particular subspecies. In fact, individuals with inbreeding coefficients in the range of 0.2–0.4 were found in each of the four subspecies and includes both wild and captive born individuals (Fig. 1d). It is therefore difficult to establish whether the amount of inbreeding in EEP individuals are a consequence of breeding among closely related individuals or whether it stems from inbred founders. In a few cases, like individual ‘14073’, we know from reliable pedigree information, that this individual is the offspring of two full-siblings (Carlsen and de Jongh 2019). For the large majority of the EEP population, this knowledge is not available or is associated with high levels of uncertainties. Together with accurate ancestry inferences, genetically-based inbreeding estimates will be of high importance in management of the breeding population as will other factors such as age, fecundity, behaviour and housing capacities.
Of our 136 tested EEP individuals, 46 were inferred to be of hybrid origin (Fig. 1c). In terms of distinguishing founder individuals with shared ancestry components (wild born hybrids) from ex situ hybrids, our ancestry analyses show that the majority of our inferred hybrids are between non-neighbouring populations in the wild (e.g. between the western chimpanzee and either of the three other subspecies) and are therefore most likely the result of hybridisation in the EEP breeding population. From a management standpoint, these should eventually be phased out of the breeding programme. Yet, some known hybrids have been allowed to breed under the current management. This has been done with the purpose to maintain population numbers in an interim period while the populations reach their target size and also to allow experienced females to pass on up-bringing behaviour to young individuals in the housed groups. To explore the extent of wild born hybrids in the EEP and the possibility of including these in the breeding efforts, we developed a new method for hybrid classification that can trace ancestry patterns three generations back. This could possibly allow us to distinguish between hybrids bred in captivity and wild born hybrids, where the latter could be included in breeding programmes, as they represent natural processes in the wild. However, two key requirements to such an inclusion are a better understanding of the extent of hybridisation in the wild and an EEP management decision on what a suitable admixture threshold would be.
As validation for the hybrid classification model (see also Supplementary Information), our method infers the known hybrid background of Ptv-Donald to have received at least 12.5% of its ancestry from the central chimpanzee, which is in the range of what was previously estimated using whole-genome sequencing data (Prado-Martinez et al. 2013). Yet, in the EEP population, only a few of the inferred hybrids fit with the expectations of ancestry patterns in wild born hybrids. The majority of the inferred hybrids include a western chimpanzee ancestry component (Fig. 2b), which is highly unlikely to occur in the wild due to the vast geographical distance to any neighbouring subspecies (Fig. 1a). Of the eight inferred hybrids with adjacent distribution ranges, one central/Nigerian-Cameroonian and seven central/eastern hybrids (Fig. 2b), we know from studbook information that all eight individuals were captive born (Carlsen and de Jongh 2019) (Table S2). The only cases where our model might have picked up remnants from natural hybridisations are the ancestry components of central chimpanzee in what we inferred to be non-admixed eastern chimpanzees using ADMIXTURE (Fig. 1c, Fig. 2b). However, this could likely be due to a general limitation of our model to separate these two subspecies due to their evolutionary close relationship and history of allele sharing (Prado-Martinez et al. 2013; de Manuel et al. 2016). Although we did not identify any wild born hybrids in the tested set of individuals, our model predictions will be highly useful in terms of pinpointing the timing of admixture and help to illuminate blanks in the studbook regarding possible sires.
Sanctuary ancestry and geographical origin
In contrast to the predominance of western chimpanzee individuals in the EEP population, the majority of the tested sanctuary individuals are inferred to belong to the eastern chimpanzee. Of the 31 tested individuals, we only find four that can be assigned to the western chimpanzee and a single individual from each of the Nigeria-Cameroon chimpanzee and the central chimpanzee (Fig. 1c). When exploring ancestry patterns in the last three generations, we obtained similar results as in the EEP population, where small posterior proportions of central chimpanzee were found in individuals of the eastern chimpanzee (Fig. 2c). This is most likely due to the limitations of our model when it comes to distinguishing shared alleles between these two subspecies, and we do not infer any geographical origin close to possible contact zones between the two subspecies (Fig. 3).
For western and Nigerian-Cameroonian chimpanzees, we obtained high probabilities in the assigned origins but with little spatial resolution. Essentially, all five western chimpanzee individuals assign to the same grid cell. As de Manuel et al. (2016) have previously shown, population structure inferred in the western and Nigerian-Cameroonian populations, may not offer enough resolution to provide fine scale determination of geographical origin. To improve origin estimates in these populations, it is crucial to obtain a better representation of georeferenced samples across their distribution ranges. This has been achieved for most of the central and eastern chimpanzee ranges, but with only one central chimpanzee individual (Doris), we cannot fully evaluate the prediction power and resolution for this subspecies. Nevertheless, the estimated geographical origin of Doris is very close to the reported confiscation site (Table S3), which gives us some assurance that future efforts to determine origins in the central chimpanzee will be possible. With a larger set of individuals from the eastern chimpanzee, we can start to appreciate the full potential of the method. The 24 analysed individuals can be assigned to geographical origins in six localities along the eastern edge of the distribution range of the eastern chimpanzee, where the majority originates from two locations in the northern and southern regions of the Democratic Republic of Congo (DRC) (Fig. 3). First of all, this might tell us that these regions are heavily affected by poaching and illegal trafficking, although the abundance of confiscation sites might also be biased by the locality of contributing sanctuaries. Only further testing of individuals from sanctuaries across the species range will allow us to assess regional threat levels. However, with the inferred origins of the eastern chimpanzee individuals all along the eastern edge of the range, we can conclude that the threats are not confined to a few regions for this subspecies but are distributed across the eastern boarders of the DRC.
When comparing the inferred geographical origins with the reported confiscation sites for all our tested sanctuary individuals (Table S3), it becomes apparent that the trafficking routes generally operate within a relatively local scale. Overall, we see that most of the tested individuals originate from locations that are within close proximity to where they have been confiscated, though with two notable exceptions, Louise and Edward. Louise was confiscated in Moscow, Russia and inferred to have originated from West Africa, while Edward was confiscated in Nairobi Airport, Kenya with inferred origin in Cameroon. This confirms that the illegal trade of wild chimpanzees spans beyond country borders and the African continent as reported in Stiles et al. (2013). Both individuals are now housed in sanctuaries where specialised care can be provided, yet, in these cases, both individuals have been placed in sanctuaries far from their geographical origin and possibly within mixed subspecies groups (other individuals from these sanctuaries have been assigned to different subspecies). Without proper knowledge of their ancestry, sanctuaries might face the same challenges as we have seen in the EEP population, with admixture of subspecies as a result of (unintended) breeding. Genetic testing at an early stage could help to ameliorate these challenges and as we have shown, our genomic approach extents to non-invasive sampling (Fig. 4), making these methods both an accurate and practical tool in conservation efforts to help combat the illegal trade of chimpanzees.
We further predict that this approach will be self-empowering as sampling gaps in the distribution range of the chimpanzee are continuously covered and DNA extraction methods for non-invasive samples improve. This will significantly advance our predictive power of geographical origin and provide valuable insight to shared ancestries in natural populations with positive knock-on effects to hybrid assessment in the ex situ populations.
Our capture array approach of targeting ancestry informative markers offers a standardised and cost-effective method that accurately guides ex situ and in situ conservation management programmes. At the current rate of decline, chimpanzees are predicted to go extinct within the current century (Estrada et al. 2017). Conservation efforts might therefore, in a foreseeable future, be obligated to supplement wild populations with individuals from the ex situ populations as a last resort to prevent them from going extinct. Should it come to this, our approach facilitates the safeguarding of genetically self-sustainable populations that will have preserved a genetic profile that resembles their wild counterparts.
The current extinction crisis however, extends well beyond chimpanzees and the demand for molecular genetics to help guide future population management programmes is immense, ranging across the taxonomical scale of birds, reptiles, amphibians, and mammals. For the latter alone, more than ten EEP genetic projects are underway and globally, regional zoo associations are undertaking molecular genetic studies for which the present study serves as an important blueprint for linking in situ and ex situ conservation efforts.
Data archiving
The genetic data used in the present study is a publicly accessible through the Dryad Digital Repository, 10.5061/dryad.31zcrjdh7.
Supplementary information
Acknowledgements
The authors would like to thank all institutions who provided samples for this study: The EAZA Biobank, Stichting AAP Amersfoort Zoo, Antwerp Zoo, Zoo Delle Star ad Aprilia, Royal Burgers’ Zoo Arnhem, Monde Sauvage Safari, Badoca Safari Park, Barcelona Zoo, Zoo Parc de Beauval, Bioparc Valencia, Borås Zoo, Parco Natura Viva, Edinburgh Zoo, Ölands Zoo, Plättli Zoo, Givskud Zoo/Zootopia, Safaripark Beekse Bergen, Hodonin Zoo, Xanthus Zoo, Kolmården Zoo, Krakow Zoo, Kristiansand Zoo, Lagos Zoo, Le Pal Zoo, Leipzig Zoo, Liberec Zoo, Lisbon Zoo, Madrid Zoo, Magdeburg Zoo, Olmense Zoo, Zoo Osnabrück, African Safari, Plaisance du Touch, Plzen Zoo, Centro de Rescate de Primates Rainfer, Zoological Center Tel Aviv—Ramat Gan, Safari Ravenna, Zoo di Roma, Leintal Zoo, Serengeti-park Hodenhagen, Reserve Africaine de Sigean, Wilhelma Zoo, Loro Parque Zoo, Touroparc, Twycross Zoo, AAP Primadomus, Warsaw Zoo, Schwabenpark, Zagreb Zoo, Cente International de Recherches Médicales de Franceville, Chimfunshi Wildlife Orphanage, Jeunes Animaux Confisques au Katanga, Ngamaba Island Chimpanzee Sanctuary, Tacugama Chimpanzee Sanctuary, The Chimpanzee Conservation Center. We further wish to thank Tom de Jongh and Lisbeth Borbye for valuable language editing and comments to the manuscript and Abigail Ramsøe for early developmental stages of the hybrid classification model. PF is supported by the Innovation Fund Denmark doctoral fellowship programme and the Candys Foundation. CF is supported by “la Caixa” doctoral fellowship programme. TSK is funded by Carlsberg grant CF19-0712 prepared within the framework of the HSE University Basic Research Program. TMB is supported by BFU2017-86471-P (MINECO/FEDER, UE), U01 MH106874 grant, Howard Hughes International Early Career, Obra Social “La Caixa” and Secretaria d’Universitats i Recerca and CERCA Programme del Departament d’Economia i Coneixement de la Generalitat de Catalunya (GRC 2017 SGR 880). EL is supported by CGL2017-82654-P (MINECO/FEDER, UE).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Associate Editor: Xiangjiang Zhan
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Peter Frandsen, Claudia Fontsere
These authors jointly supervised this work: Tomas Marques-Bonet, Christina Hvilsom
Contributor Information
Peter Frandsen, Email: pef@zoo.dk.
Claudia Fontsere, Email: claudia.fontsere@upf.edu.
Supplementary information
The online version of this article (10.1038/s41437-020-0313-0) contains supplementary material, which is available to authorised users.
References
- Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck B, Walkup K, Rodriques M, Unwin S, Travis D, Stoinski T (2007) Best practice guidelines for the re-introduction of Great Apes. Gland, Switzerland
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsen F, de Jongh T (2019) European studbook for the chimpanzee Pan troglodytes. Copenhagen
- Carøe C, Gopalakrishnan S, Vinner L, Mak SST, Sinding MHS, Samaniego JA, et al. Single-tube library preparation for degraded DNA. Methods Ecol Evol. 2018;9:410–419. doi: 10.1111/2041-210X.12871. [DOI] [Google Scholar]
- Ceballos G, Ehrlich PR, Barnosky AD, García A, Pringle RM, Palmer TM. Accelerated modern human–induced species losses: entering the sixth mass extinction. Sci Adv. 2015;1:e1400253. doi: 10.1126/sciadv.1400253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Manuel M, Kuhlwilm M, Frandsen P, Sousa VC, Desai T, Prado-Martinez J, et al. Chimpanzee genomic diversity reveals ancient admixture with bonobos. Science. 2016;354:477–481. doi: 10.1126/science.aag2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada A, Garber PA, Rylands AB, Roos C, Fernandez-duque E, Fiore A Di et al. (2017) Impending extinction crisis of the world’s primates: why primates matter. Sci Adv 3:e1600946 [DOI] [PMC free article] [PubMed]
- Grueber CE, Fox S, McLennan EA, Gooley RM, Pemberton D, Hogg CJ, et al. Complex problems need detailed solutions: harnessing multiple data types to inform genetic management in the wild. Evol Appl. 2019;12:280–291. doi: 10.1111/eva.12715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanghøj K, Moltke I, Andersen PA, Manica A, Korneliussen TS. Fast and accurate relatedness estimation from high-throughput sequencing data in the presence of inbreeding. Gigascience. 2019;8:1–9. doi: 10.1093/gigascience/giz034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockings KJ, McLennan MR, Carvalho S, Ancrenaz M, Bobe R, Byrne RW, et al. Apes in the anthropocene: flexibility and survival. Trends Ecol Evol. 2015;30:215–222. doi: 10.1016/j.tree.2015.02.002. [DOI] [PubMed] [Google Scholar]
- Humle T, Maisels F, Oates JF, Plumtre A, Williamson EA (2016) Pan troglodytes. IUCN Red List Threat Species
- Hvilsom C, Frandsen P, Børsting C, Carlsen F, Sallé B, Simonsen BT, et al. Understanding geographic origins and history of admixture among chimpanzees in European zoos, with implications for future breeding programmes. Heredity. 2013;110:586–593. doi: 10.1038/hdy.2013.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IUCN (2015) IUCN Red List of Threatened Species. Version 20153: www.iucnredlist.org
- Jepsen BI, Carlsen F Genetic identification of West African Chimpanzee, Pan troglodytes verus, based on mitochondrial DNA analysis. Unpublished
- Kircher M, Sawyer S, Meyer M. Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res. 2012;40:e3. doi: 10.1093/nar/gkr771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Epstein MP, Duncan R, Lin X. Sparse principal component analysis for identifying ancestry-informative markers in genome-wide association studies. Genet Epidemiol. 2012;36:293–302. doi: 10.1002/gepi.21621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman AJ, Putnam AS, Ivy JA. Use of molecular data in zoo and aquarium collection management: benefits, challenges, and best practices. Zoo Biol. 2019;38:106–118. doi: 10.1002/zoo.21451. [DOI] [PubMed] [Google Scholar]
- Nye J, Laayouni H, Kuhlwilm M, Mondal M, Marques-Bonet T, Bertranpetit J. Selection in the introgressed regions of the chimpanzee genome. Genome Biol Evol. 2018;10:1132–1138. doi: 10.1093/gbe/evy077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado-Martinez J, Sudmant PH, Kidd JM, Li H, Kelley JL, Lorente-Galdos B, et al. Great ape genetic diversity and population history. Nature. 2013;499:471–475. doi: 10.1038/nature12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- QGIS (2018) QGIS Geographic Information System. Open Source Geospatial Foundation Project. http://qgis.osgeo.org
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rañola JM, Novembre J, Lange K. Fast spatial ancestry via flexible allele frequency surfaces. Bioinformatics. 2014;30:2915–2922. doi: 10.1093/bioinformatics/btu418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohland N, Reich D. Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Res. 2012;22:939–946. doi: 10.1101/gr.128124.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiles D, Redmond I, Cress D, Nellemann C, Formo RK (2013) Stolen apes—the illicit trade in chimpanzees, Gorillas, Bonobos and Orangutans. Birkeland Trykkeri AS, Norway
- Traylor-Holzer K, Leus K, Bauman K. Integrated Collection Assessment and Planning (ICAP) workshop: helping zoos move toward the One Plan Approach. Zoo Biol. 2019;38:95–105. doi: 10.1002/zoo.21478. [DOI] [PubMed] [Google Scholar]
- Xu H, Luo X, Qian J, Pang X, Song J, Qian G, et al. FastUniq: a fast de novo duplicates removal tool for paired short reads. PLoS ONE. 2012;7:e52249. doi: 10.1371/journal.pone.0052249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Kobert K, Flouri T, Stamatakis A. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics. 2014;30:614–620. doi: 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genetic data used in the present study is a publicly accessible through the Dryad Digital Repository, 10.5061/dryad.31zcrjdh7.