Abstract
The availability of MHC-binding prediction tools has been useful in guiding studies aimed at identifying candidate target Ags to generate reactive T cells and to characterize viral and tumor-reactive T cells. Nevertheless, prediction algorithms appear to function poorly for epitopes containing cysteine (Cys) residues, which can oxidize and form disulfide bonds with other Cys residues under oxidizing conditions, thus potentially interfering with their ability to bind to MHC molecules. Analysis of the results of HLA-A*02:01 class I binding assays carried out in the presence and absence of the reducing agent 2-ME indicated that the predicted affinity for 25% of Cys-containing epitopes was underestimated by a factor of 3 or more. Additional analyses were undertaken to evaluate the responses of human CD8+ tumor-reactive T cells against 10 Cys-containing HLA class I–restricted minimal determinants containing substitutions of α-aminobutyric acid (AABA), a cysteine analogue containing a methyl group in place of the sulfhydryl group present in Cys, for the native Cys residues. Substitutions of AABA for Cys at putative MHC anchor positions often significantly enhanced T cell recognition, whereas substitutions at non-MHC anchor positions were neutral, except for one epitope where this modification abolished T cell recognition. These findings demonstrate the need to evaluate MHC binding and T cell recognition of Cys-containing peptides under conditions that prevent Cys oxidation, and to adjust current prediction binding algorithms for HLA-A*02:01 and potentially additional class I alleles to more accurately rank peptides containing Cys anchor residues.
Extensive analysis of peptide binding to individual MHC class I molecules has led to the development of tools for identifying Ags recognized by class I-restricted T cells. Combined, in vitro peptide-binding assays and analysis of peptides eluted from cell surface MHC molecules (1–4) have led to the development of MHC binding algorithms that can facilitate the identification of minimal T cell determinants (5, 6). Nevertheless, studies of tumor-reactive T cells and T cells that respond to viral immunization have demonstrated limitations of using peptide/MHC binding prediction tools to query proteome and transcriptome databases. These analyses have led to the identification of T cell determinants that arise from multiple mechanisms that include protein splicing (7–10) and the translation of alternative open reading frames (11) or intronic sequences (12).
The influence of peptide modifications on T cell recognition has also been demonstrated in studies of peptides containing cysteine (Cys), a category of peptides that can be difficult to investigate in tissue culture assay systems because of the ability of Cys residues to oxidize and form disulfide linkages and disulfide-linked peptide multimers. Human T cells have been identified that preferentially recognize either a cysteinylated or an unmodified form of the minor-histocompatibility Ag H-Y peptide FIDSYICQV (13). Substitution of serine (Ser) or alanine (Ala) for the native Cys residues enhanced the binding and in vitro and in vivo immune response of murine T cells to Cys-containing influenza nucleoprotein epitopes, and the addition of the reducing agent tris(2-carboxyethyl) phosphine or DTT to tissue culture media enhanced in vitro T cell responses to those peptides (14). Responses against additional cysteine-containing peptides that include the NY-ESO-1 (CTAG2) cancer germline HLA-A*02:01-restricted epitope SLLMWITQC (14, 15) and the tyrosinase HLA-A*01:01-restricted epitope KCDICTDEY (16), however, appear to be limited to the reduced form of the peptide. The NY-ESO-1 peptide contains a C-terminal Cys residue, one of the primary anchor positions responsible for binding to HLA-A*02:01 (1), and the tyrosinase peptide contains a Cys at position 2, corresponding to an auxiliary anchor for binding to HLA-A*01:01 (17). The inclusion of reducing agents in cell culture media significantly enhanced recognition of the NY-ESO-1 peptide. Furthermore, binding affinity measurements provided evidence for a more stable interaction of the NY-ESO-1 peptide containing a substitution of valine (Val) for Cys to HLA-A*02:01, and of a soluble NY-ESO-1-reactive TCR to these complexes, than formed by the native NY-ESO-1 peptide (18). Peptides containing conservative substitutions of either Ala or Ser for the Cys at position 2 in a tyrosinase epitope were recognized by human tumor-reactive T cells at between 10- and 100-fold lower concentrations than the native peptide (16). Similarly, conservative substitution of Ser or Ala for Cys residues in viral epitopes significantly enhance their ability to stimulate in vitro and in vivo murine T cell responses (14). Substitution of the isosteric amino acid α-aminobutyric acid (AABA) for the three Cys residues present in a murine class I-restricted Moloney Murine Sarcoma virus GagL epitope was needed to generate MHC tetramers because of the formation of multimers that were incapable of binding to the H-2Db restriction element when the native peptide was used (19).
The problematic nature of analyzing Cys-containing peptides, particularly with respect to determining their HLA binding affinities, has consequences for the development of accurate peptide-binding algorithms, further exacerbating the problem of identifying immunogenic determinants containing Cys residues when those algorithms are in turn used as the basis for identifying candidate Ags. In addition, conventional methods used for identifying peptides eluted from cell surface MHC molecules appear to lead to a significant loss of Cys-containing peptides, as demonstrated in two recent studies that appear to at least partially correct for these losses using a bio-informatic (20) or chemical approach (21). The observation that modifications of the MHC anchor residue positions in the HLA-A*02:01-restricted NY-ESO-1 peptide enhanced MHC binding provided an opportunity to further explore the nature of these interactions, as- HLA-A*02:01, which is one of the most common class I alleles in the general worldwide population (22), was the first HLA class I allele whose structure was solved by x-ray crystallography (23), and to date remains one of the most well-studied and characterized class I alleles. The binding specificity of A*02:01 is largely dictated by a canonical configuration of main anchor B and F pockets (24) in its peptide-binding groove, although secondary interactions are also important for high affinity binding (25). The relatively narrow and nonpolar structure of the B pocket cavity confers a preference for aliphatic hydrophobic residues (i.e., L, I, M, and V) in position 2, although a tolerance for small polar residues (S and T) has also been noted. The F pocket, which engages the peptide C-terminal residue, is more polar than the B pocket, but is also associated with a preference for aliphatic hydrophobic residues, and a tolerance for small polar residues. The tolerance for small polar residues in the B and F pockets of A*02:01 makes study of unoxidized Cys particularly interesting. These findings presumably extend to additional alleles that can accommodate aliphatic residues in their B and F pocket cavities.
In this report, the binding affinities of a panel of nine known Cys-containing HLA-A*02:01-restricted determinants and modified peptides containing substitutions of AABA for Cys were evaluated in the presence and absence of a reducing agent. Immune reactivity was assessed against six of the nine A*02:01-restricted and four additional non-A*02:01-restricted peptides containing Cys residues in putative anchor and nonanchor positions. Significant differences were noted between the predicted and measured binding affinities of peptides containing cysteine anchors, and AABA substitutions were found to have different effects on T cell recognition, depending upon the position of the substitution.
Materials and Methods
MHC-peptide-binding assays
Quantitative assays to measure the binding of peptides to HLA A*02:01 class I molecules based on the inhibition of binding of a radiolabeled standard peptide (hepatitis B virus [HBV] core 18–27 analogue, FLPSDYFPSV). MHC molecules were purified by affinity chromatography from the Epstein-Barr virus transformed homozygous cell line JY, and assays were performed, as described previously (26, 27). Briefly, radiolabeled peptide is coincubated at room temperature with varying concentrations (~1 μM-0.1 nM) of purified MHC in the presence of a mixture of protease inhibitors and 1 μM B2-microglobulin. Following a 2-d incubation, MHC-bound radioactivity was determined following capture of MHC/peptide complexes on W6/32 (anti-class I) Ab-coated assay plates. Peptides were tested at six different concentrations covering a 100,000-fold dose range in three or more independent assays, and the concentration of peptide yielding IC50 of the binding of the radiolabeled probe peptide was calculated. Under the conditions used, where [radiolabeled probe] < [MHC] and IC50 ≥ [MHC] (bracketed values correspond to concentrations), the measured IC50 values are reasonable approximations of the true Kd values (28, 29). For assays under reducing conditions, 1 mM 2-ME was added.
Positional scanning combinatorial libraries and bioinformatic analyses
Nine- and ten-mer positional scanning combinatorial libraries (PSCL) were synthesized as described previously (30). Each library contains randomized 9- or 10-mer peptides with one fixed residue at a single position. With each of the 20 naturally occurring residues represented at each position along a 9-mer (or 10-mer) backbone, the entire library consisted of 180 (or 200) peptide mixtures. A library completely randomized at each position is also tested.
PSCLs were tested in a standard competition assays, or in the presence of 2-ME as described above, and data analyzed as described previously (31–33). Briefly, the IC50 nM value of each mixture is standardized as a ratio to the geometric mean IC50 nM value of the entire set of 180 mixtures and then normalized at each position so that the value associated with optimal binding at each position corresponds to 1. For each position, an average (geometric) relative binding affinity (ARB) is calculated, and then the ratio of the ARB for the entire library to the ARB for each position is derived.
Predicted binding values using the PSCL were generated by summing the corresponding log ARB value for each residue at each position. Percentile scores were generated by indexing against a set of 10,000 randomly generated peptide sequences (ExPASy, RandSeq) scored with the same matrix. Predicted IC50 nM values were derived by indexing against NetMHCpan-generated IC50 nM with the same set of random peptide sequences.
HPLC analysis
Selected Cys-containing peptides were incubated for 2 d at room temperature in the presence of varying concentrations of 2-ME, a chemical compound commonly used to reduce disulfide bonds and, because of its capacity to scavenge hydroxyl radicals, as a biological antioxidant. The 2-d room temperature incubation replicates the conditions each peptide is subjected to during a standard in vitro peptide-binding assay. As a control, each peptide was similarly incubated, but without 2-ME present. For a second control, the same peptides were air-oxidized with no 2-ME.
Two 9-mer peptides with free acid C termini (peptide 3186.0013, sequence AAICTHLEV-OH, and peptide 3186.0017, sequence GALASCMGL-OH), each containing a single internal Cys, were dissolved from lyophilized material in 100% DMSO at an approximate concentration of 10 mg/ml. In separate trials, ~10 nmol (1 μl) of each peptide was subsequently diluted to a final volume of 1 ml in a sterile PBS solution (pH 7.2), containing 2-ME at one of the following concentrations: 0 mM, 100, 500 μM, 1, 10, 20, 50, or 100 mM. In addition to the no-DMSO control, additional controls of 48-h air oxidation (i.e., air bubbled into the sample) and 48-h air oxidation in the presence of 20% aqueous DMSO were also performed. To minimize experimental variables and simplify liquid chromatography postrun analysis, protease inhibitors were not included in the reaction mix.
After 48 h, each sample was analyzed by reversed phase HPLC. HPLC analysis was carried out on a Prominence LC-20AT module equipped with an SIL-20A autosampler and in-line SPD-20A UV-Vis detector (Shimadzu Scientific Instruments, Columbia, MD). Separation was performed on a Hypersil Gold ODS column (3 μm, 125 mm × 2.1 mm; Thermo Fisher Scientific, Hanover Park, IL) equipped with a C18 guard column at 40°C. Pump control and peak integration was carried out with LabSolutions Software (version 5.96 SP3; Shimadzu Corporation, Tokyo, Japan). Mobile phase consisted of buffer A, 0.22 μm Milli-Q filtered water containing 0.1% TFA, and buffer B containing acetonitrile in 0.1% TFA. Reduced and oxidized peptides were eluted from the column with the following gradient: 0–2 min 5% buffer B, 2–37 min 5–52.5% buffer B, 37–42 min 52.58–0% buffer B, 42–45 min 80–5% buffer B, and 45–50 min 5% buffer B under a constant flow rate of 0.3 ml/min. For each 2-ME concentration sample, 1 nmol (100 μl of the treated sample) was injected. The minimum 2-ME concentration required to produce and maintain a reduced state of cysteine (free sulfhydryl) was defined as the minimum amount of 2-ME between the two trials at which the predominant peak eluting from reversed phase indicated reduced peptide.
Evaluation of T cell responses
Assays of HLA-A*02:01-restricted T cells were carried out by initially reconstituting lyophilized peptides in 100% DMSO, carrying out 10-fold serial dilutions in media at concentrations ranging between 1 mg/ml and 1 ng/ml, and immediately pulsing diluted peptides onto K562 cells that had been stably transduced with the gene encoding HLA-A*02:01. After 2 h, peptide-pulsed target cells were washed and incubated with relevant T cells populations. Recombinant constructs encoding tumor Ag-specific TCRs were generated by cloning paired TCR α– and β-chains in the MSGV1 retroviral expression vector (34). A TCR identified from in vitro-cultured tumor infiltrating lymphocytes (TIL) generated from a patient with colorectal cancer were used to evaluated reactivity against the TP53(168–176) p.R175H epitope (35), and T cells transduced with a TCR isolated from a melanoma patient were used to evaluate responses against the CTAG2 epitope (36). Cells transduced with TCRs isolated from melanoma patients 1200, 4000, and 4155 were used to evaluate responses against the PMEL(639–647), HIVEP2(167–1682) P1682L, and RSL1D1(175–183) R183C epitopes, respectively. Similarly, cells transduced with TCRs isolated from in vitro-cultured TIL generated from colorectal cancer patients 4032, 4266, 4266, and 4235 were used to evaluate responses against the PHLPP1(563–571) G556E, TP53(241–249) R248W, ECI2(350–358) N352I, and GALK1(242–250) epitopes, respectively. Responses against the CYFIP1(116–124) I121V neoantigen were assessed using in vitro-cultured TIL generated from a colorectal tumor fragment. Following an 18-h coculture of 5 × 104 T cells and 5 × 104 APC, culture supernatants were tested for the presence of IFN-γ using a standard ELISA Ab capture assay (Thermo Fisher Scientific).
Because of the diversity of HLA-restriction elements in this group of epitopes, a variety of different APCs were used for T cell recognition assays. The peptides encoding GALK1 A249V were tested against a tumor infiltrating lymphocyte fragment culture known to contain mutation-specific cells. These peptides were pulsed onto dendritic cells (DCs) autologous to the patient from whom the TIL were originally identified. The RSL1D1 R183C were pulsed onto DCs autologous to the patient from whom the mutation-specific TCR was originally isolated. The peptides encoding TP53 R248W and ECI2 N352I were pulsed onto Cos7 cells transiently transfected with HLA-A*68:02 and HLA-A*68:01, respectively, because of a lack of available DC samples from the patient in whom those reactivities were identified.
Results
Decreased accuracy of HLA binding predictions for cysteinecontaining peptides
As part of a recent study (37), we assembled a set of peptides consisting of 32 epitopes encoded by nonsynonymous somatic mutations (neoepitopes) from the Cancer Immunity Epitope database (38) plus an additional 45 neoepitopes from the published literature curated by the Immune Epitope Database (IEDB) (39) (Supplemental Table I). Approximately 90% (69/77) of the neoepitopes could be captured using a NetMHCpan predicted affinity threshold of 500 nM (Supplemental Fig. 1), a finding in agreement with previous studies that had identified both predicted and measured affinities of 500 nM as a suitable threshold to identify ~85% of pathogenic class I-restricted T cell epitopes (26, 40, 41).
Review of the data set, particularly the eight epitopes predicted to bind with an IC50 >500 nM, suggested that prediction of Cys-containing epitopes may be less accurate than non-Cys-containing epitopes. The 500 nM threshold only identified 10 (71%) of the 14 Cys-containing epitopes (Supplemental Fig. 1). Also, although Cys-containing epitopes constituted 18% of the entire epitope set, they composed 50% (4/8) of the outliers (p < 0.014, Fisher exact test) (Table I top), all of which were restricted by A*02:01 (p < 0.0001) (Table I middle), whereas none of the non-A*02:01 outlier epitopes contained Cys (p = 0.375, Table I bottom). These findings indicate that the affinities of class I-restricted Cys-containing candidate epitopes may be significantly underestimated by current class I binding algorithms and that this may more frequently occur for peptides predicted to bind to class I alleles such as A*0201, where Cys residues may be tolerated at both the position 2 and C-terminal primary anchor positions.
Table I.
Influence of the presence of Cys residues within an epitope on predicted MHC class I binding affinity
| No. of Peptides |
|||
|---|---|---|---|
| Epitope Status | With Cys | No Cys | Total |
| AU epitopes | |||
| Predicted nonbinders | 4 | 4 | 8 |
| Predicted binders | 10 | 59 | 69 |
| Total (p = 0.014) | 14 | 63 | 77 |
| A*02:01 epitopes | |||
| Predicted nonbinders | 4 | 0 | 4 |
| Predicted binders | 3 | 24 | 27 |
| Total (p = 0.0001) | 7 | 24 | 31 |
| Non-A*02:01 epitopes | |||
| Predicted nonbinders | 0 | 4 | 4 |
| Predicted binders | 7 | 35 | 42 |
| Total (p = 0.375) | 7 | 39 | 46 |
Determination of effective concentrations of 2-ME to protect Cys-containing peptides from oxidation in HLA binding assays
MHC binding predictions are largely based on in vitro peptide-binding assays performed in solution over a 24–48-h time span (6, 42). As Cys residues have a propensity to oxidize in solution, it is possible that oxidation interferes with or attenuates the binding capacity of Cys-containing peptides. As a result, in vitro binding assays may underestimate the binding capacity of Cys-containing peptides that may be naturally processed and presented within reducing cellular compartments.
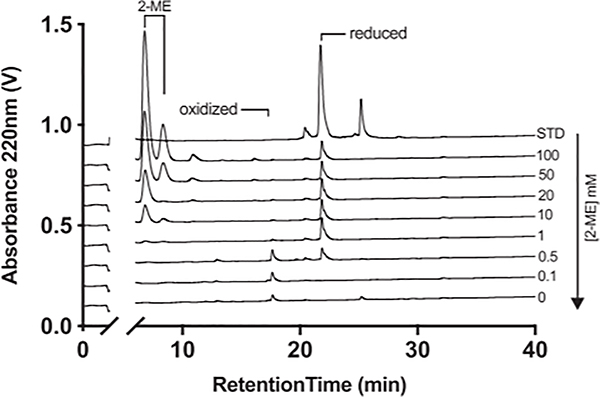
To identify conditions that would allow the binding affinity of Cys-containing monomeric peptides to be determined, a Cys-containing peptide (AAICTHLEV-OH) was incubated for 2 d at room temperature in the presence or absence of varying concentrations of the reducing reagent 2-ME. A different aliquot of the peptide was oxidized by bubbling air into the sample for 2 d in the absence of 2-ME, and the minimum 2-ME concentration required to generate a predominantly reduced cysteine peak was determined by reverse phased HPLC analysis (Fig. 1). For this peptide, increasing concentrations of 2-ME resulted in a shift from a predominant peak eluting at the position of the oxidized peptide to a predominant peak eluting at the position of the reduced species at concentrations of 1 mM or higher.
FIGURE 1.
Representative HPLC elution profile of oxidized and reduced peptide under reducing conditions. Assays were performed as described in the text.
Functional tolerance of purified HLA A*02:01 to varying concentrations of 2-ME
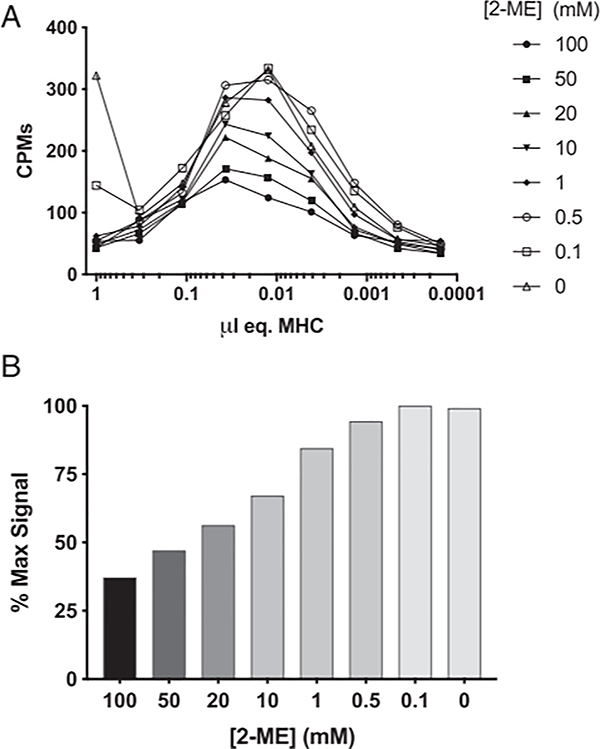
Next, we sought to define optimal conditions for testing peptide binding capacity to purified HLA-A*02:01 MHC molecules in the presence of 2-ME. Accordingly, a radiolabeled version of the HLA-A*02:01-restricted HBV core 18–27 (sequence FLPSDYFPSV) T cell epitope was tested in direct binding assays with varying concentrations of 2-ME and purified MHC, and the amount of bound cpm was determined (Fig. 2). As summarized in Fig. 2A, maximal binding of the radiolabeled ligand was observed with between 33 and 10 nM MHC across all 2-ME conditions. Furthermore, detectable signal was obtained regardless of 2-ME concentration, showing that the assay is generally tolerant of the presence of 2-ME. However, binding was reduced at concentrations above 500 μM 2-ME, a concentration that was only partially effective at reducing Cys-containing peptides. Approximately 85% of the peak signal intensity was seen at 1 mM, a concentration shown above to be capable of maintaining the tested peptides predominantly in their reduced state (Fig. 2B). Thus, 1 mM 2-ME was used for subsequent assays performed in reducing conditions.
FIGURE 2.
Influence of the presence of varying amounts of 2-ME on the binding activity of HLA A*02:01. Various concentrations of A*02:01 were titrated for binding activity using radiolabeled HBV core 18–27 as a probe ligand, as described in Materials and Methods. Activity was assessed as a function of concentration of added 2-ME (A). Maximal counts of >85% could be obtained in the presence of 1 mM 2-ME (B).
HLA peptide binding capacity influenced by cysteine oxidation
A set of nine known HLA-A*02:01-restricted epitopes recognized by tumor-reactive T cells containing one or two Cys residues were chosen to investigate the potential effects of peptide oxidation on MHC binding and T cell recognition. The panel included four nonmutant epitopes and five neoepitopes and their corresponding wild-type (WT) peptides, all containing either one or two Cys residues except for the CDK4 WT peptide. In addition, four non-HLA-A*02:01-restricted Cys-containing neoepitopes were chosen to evaluate the effects of peptide oxidation on T cell recognition (Table II)
Table II.
Sequences of epitope evaluated for binding to HLA-A*02:01 and recognition by T cells
| Gene (Residues) | Mutant/Nonmutant | Epitope | Mutation | HLA Restriction Element | No. of Cys Residues | Position of Mutation in Epitope | Position of Cys |
|---|---|---|---|---|---|---|---|
| CYFIP1016-124) | Mutant | SCSRAVPQV | 1121V | A*02:01 | 1 | 6 | 2 |
| CTAG2(157–165) | Nonmutant | SLLMWITQC | N/A | A*02:01 | 1 | N/A | 9 |
| TP53(168–176) | Mutant | HMTEVVRHC | R175H | A*02:01 | 1 | 8 | 9 |
| PMEL(639–647) | Nonmutant | RLPRIFCSC | N/A | A*02:01 | 2 | N/A | 7,9 |
| PHLPP1(563–571) | Mutant | ILCETCLIV | G566E | A*02:01 | 2 | 4 | 3,6 |
| HIVEP2(1674–1682) | Mutant | CISSCNPNL | P1682L | A*02:01 | 2 | 9 | 1, 5 |
| BING4(altemative. orf) | Nonmutant | CQWGRLWQL | N/A | A*02:01 | 1 | N/A | 1 |
| GnTV(intronic) | Nonmutant | VLPDVFIRC | N/A | A*02:01 | 1 | N/A | 9 |
| CDK4(23–32) | Mutant | ACDPHSGHFV | R23C | A*02:01 | 1 | 2 | 2 |
| TP53(241–249) | Mutant | SCMGGMNWR | R248W | A*68:01 | 1 | 8 | 2 |
| ECI2(350–358) | Mutant | CTIAVVNFL | N352I | A*68:02 | 1 | 3 | 1 |
| GALK1(242–250) | Mutant | qceevarvl | A249V | B* 35:03 | 1 | 8 | 2 |
| RSL1D1(175–183) | Mutant | KSPNPSTPC | R183C | C*01:02 | 1 | 9 | 9 |
N/A, not applicable.
Competitive binding assays were then carried out under standard conditions, in conditions excluding protease inhibitors that can potentially modify Cys residues, or in the presence of the reducing agent 2-ME, as described in Materials and Methods. Furthermore, binding assays were carried out using modified peptide variants in which the native Cys residues were replaced with AABA. The binding data, including predicted affinities, are summarized in Table III.
Table III.
Measurement of HLA-A*02:01 binding affinities of native and modified peptides in the presence or absence of 2-ME
| IC50 (ng/ml) |
||||||
|---|---|---|---|---|---|---|
| Ag | Sequence | Peptide Modification | Predicted | Normal | No Protease Inhibitors | 2-ME |
| CTAG2(157–165) | SLLMWITQC | Native | 390 | 13,487 | 820 | 157 |
| CTAG2(157–165) | SLLMWITQBa | P9 C to AABA | —b | 64 | 94 | 99 |
| HIVEP2(1674–1682) P1682L | CISSCNPNL | Native | 1,593 | >20,(KK) | >20,(KK) | 4,034 |
| HIVEP2(1674–1682) P1682L | BISSCNPNL | PI C to AABA | — | 212 | 573 | 250 |
| HIVEP2(1674–1682) P1682L | CISSBNPNL | P5 C to AABA | — | 845 | 1,047 | 618 |
| HIVEP2(1674–1682) P1682L | BISSBNPNL | PI, P5 C to AABA | — | 575 | 604 | 1,260 |
| HIVEP2(1674–1682) WT | CISSCNPNP | Native | >20,000 | >20,(KK) | >20,(KK) | >20,000 |
| HIVEP2(1674–1682) WT | BISSCNPNP | PI C to AABA | — | >20,(KK) | >20,(KK) | >20,000 |
| HIVEP2(1674–1682) WT | CISSBNPNP | P5 C to AABA | — | >20,(KK) | >20,(KK) | >20,000 |
| HIVEP2(1674–1682) WT | BISSBNPNP | PI, P5 C to AABA | — | 13,800 | 17,100 | 13,400 |
| BING4(alt orf) | CQWGRLWQL | Native | 20 | 1,425 | 1,132 | 155 |
| BING4(alt orf) | BQWGRLWQL | PI C to AABA | — | 143 | 121 | 93 |
| PHLPP 1(563–571) G556E | ILCETCLIV | Native | 60 | 461 | 325 | 306 |
| PHLPP 1(563–571) G556E | ILBETCLIV | P3 C to AABA | — | 63 | 58 | 82 |
| PHLPP 1(563–571) G556E | ILCETBLIV | P6 C to AABA | — | 162 | 104 | 93 |
| PHLPP 1(563–571) G556E | 1LBETBLIV | P3, P6 C to AABA | — | 18 | 14 | 24 |
| PHLPP 1(563–571) WT | ILCGTCLIV | Native | 110 | 192 | 544 | 228 |
| PHLPP 1(563–571) WT | ILBGTCLIV | P3 C to AABA | — | 419 | 516 | 808 |
| PHLPP 1(563–571) WT | ILCGTBLIV | P6 C to AABA | — | 1,395 | 839 | 885 |
| PHLPP 1(563–571) WT | ILBGTBLIV | P3, P6 C to AABA | — | 1,103 | 733 | 1,547 |
| CYFIP1016–124) 1121V | SCSRAVPQV | Native | 12,117 | 11,649 | 6,952 | 2,344 |
| CYFIP1016–124) 1121V | SBSRAVPQV | P2 C to AABA | — | 1,251 | 807 | 1,684 |
| CYFIP1016–124) WT | SCSRAIPQV | Native | 9,850 | 1,469 | 529 | 199 |
| CYFIP1016–124) WT | SBSRAIPQV | P2 C to AABA | — | 1,120 | 1,018 | 867 |
| CDK4(23–32) R24C | ACDPHSGHFV | Native | 18,383 | >20,(KK) | 3,302 | 13,477 |
| CDK4(23–32) R24C | ABDPHSGHFV | P2 C to AABA | — | 2,272 | 1,972 | 2,053 |
| CDK4(23–32) WT | ARDPHSGHFV | Native | >20,000 | >20,(KK) | >20,(KK) | >20,000 |
| GnTV(intron) | VLPDVFIRC | Native | 439 | 482 | 128 | 32 |
| GnTV(intron) | VLPDVFIRB | P9 C to AABA | — | 0.6 | 3.3 | 2.5 |
| TP53(168–176) R175H | HMTEVVRHC | Native | 4,984 | 4,210 | 1,850 | 1,440 |
| TP53(168–176) R175H | HMTEVVRHB | P9 C to AABA | — | 318 | 301 | 187 |
| TP53(168–176) WT | HMTEVVRRC | Native | 7,043 | 10,186 | 1,147 | 1,025 |
| TP53(168–176) WT | HMTEVVRRB | P9 C to AABA | — | 337 | 261 | 272 |
| PMEL(639–647) | RLPR1FCSC | Native | 1,492 | 1,975 | 512 | 68 |
| PMEL(639–647) | RLPR1FBSC | P7 C to AABA | — | 508 | 63 | 25 |
| PMEL(639–647) | RLPRIFCSB | P9 C to AABA | — | 11 | 11 | 27 |
| PMEL(639–647) | RLPRIFBSB | P7, P9 C to AABA | — | 4.7 | 7.2 | 14 |
B indicates AABA.
Dashes indicate that the binding affinities of these peptides could not be predicted as they contain amino acids that are not naturally incorporated into proteins.
Using standard assay conditions, the measured IC50s for the BING4(alt orf), CTAG2(157–165), HIVEP2(1674–1682) P1682L, and PHLPP1(563–571) G556E peptides were 71, 35, 13, and 8-fold higher, respectively, than predicted values. For the remaining peptides, except for the CYFIP WT peptides, there was a less than 2-fold variation between the predicted and measured IC50s (Table III).
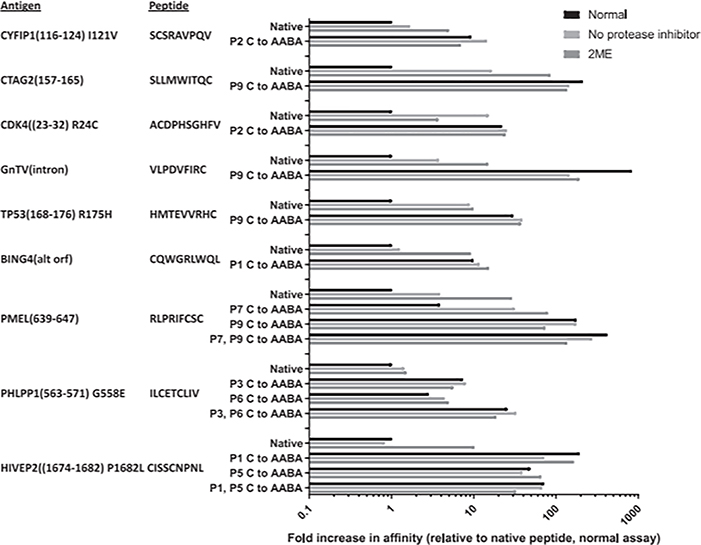
The effects of varying the assay conditions by removing protease inhibitors, adding 2-ME to the assay buffer, testing peptides containing AABA substitutions, or testing AABA-substituted peptides in the presence of 2-ME on binding of the peptide panel to HLA-A*02:01 were then evaluated. The affinity increases were noted for all the peptides whose binding capacities could be determined when assays were carried out either in the absence of protease inhibitors or in the presence of 2-ME, except for the PHLPP1(563–571) WT peptide (Table III), which was associated with negligible change. The ratio between the IC50 values measured under standard conditions and in the absence of protease inhibitors ranged between 16 and 0.35, with a median ratio of 2.5, and the ratio between the IC50 values measured under standard conditions and in the presence of 2-ME ranged between 86 and 0.84, with a median ratio of 8.3 (Fig. 3). The ratio between the IC50 values measured under standard conditions with the native Cys-containing peptides and peptides where all the native Cys residues were replaced by AABA ranged between 840 and 0.17, with a median ratio of 19, and the ratio observed with AABA-substituted peptides in the presence of 2-ME ranged between 192 and 0.12, with a median ratio of 21 (Fig. 3). The addition of 2-ME to assays carried out with ABBA-containing peptides resulted in a median 0.73-fold change in the measured IC50 values relative to those obtained with AABA-containing peptides without 2-ME (Fig. 3), indicating that 2-ME did not exert a nonspecific effect on binding assay results. The PHLPP1(563–571) WT peptide was the only peptide for which AABA substitutions appeared to decrease measured binding affinity relative to standard conditions (Fig. 3). Although these results could indicate that there is a preference for Cys over ABBA at position 3, a potential secondary anchor in the PHLPP1(563–571) WT peptide, binding of the ABBA-substituted PHLPP1(563–571) G566E peptide was enhanced relative to the native peptide, indicating that the mutant residue at position 4 may also influence peptide binding (Fig. 3).
FIGURE 3.
Relative binding capacity of various Cys-containing CTL epitopes to A*02:01 under conditions to mitigate potential effects of oxidation. Native peptides, or analogs with Cys residues substituted to AABA, were tested under normal conditions, in the absence of protease inhibitors, or in the presence of 2-ME. The top bar in each group represents normal, the middle bar in each group represents no protease inhibitor, and the bottom bar in each group represents 2-ME.
For the seven peptides used for these comparisons containing Cys residues in a primary anchor position, the ratio between the predicted IC50 values and those measured in the presence of 2-ME ranged between 50 and 2.5, whereas for the three peptides containing one or two Cys residues in potential secondary anchor positions, the ratio ranged between 0.78 and 0.20. Similarly, the ratio between the predicted IC50 values for the same group of seven peptides and those measured using AABA-substituted peptides ranged between 763 and 6.1, whereas for the three peptides containing Cys residues in nonanchor positions, the ratio ranged between 3.3 and 0.1, results consistent with the dominant role of primary anchor positions on peptide/MHC binding.
Although the binding capacity of the CDK4(23–32) R24C and HIVEP2(1674–1682) P1682L peptides could not be measured under standard conditions, both AABA-substituted peptides demonstrated relatively weak but significant binding, and significant binding of the HIVEP2(1674–1682) P1682L peptide could be detected in the presence of 2-ME (Fig. 3).
Taken together, these results indicate that current MHC binding algorithms underestimate the binding affinities of most peptides containing Cys residues in primary anchor positions and that their measured binding affinity can be significantly enhanced by inhibiting peptide oxidation.
Generation of improved combinatorial PSCL matrices
We and others have previously demonstrated the use of PSCL, consisting of combinatorial mixtures of peptides sharing a single residue at one position and random residues at all other positions, to derive quantitative peptide binding motifs for human, nonhuman primate, and mouse MHC class I molecules (31, 32, 43–46). Using a set of 180 or 200 mixtures containing the 20 naturally occurring residues at each position along 9-mers or 10-mers backbones, respectively, residue-by-residue and position-by-position matrices detailing binding specificity of class I MHC molecules can be derived.
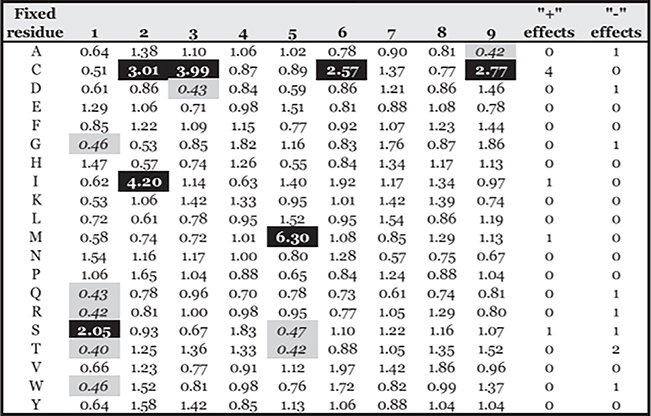
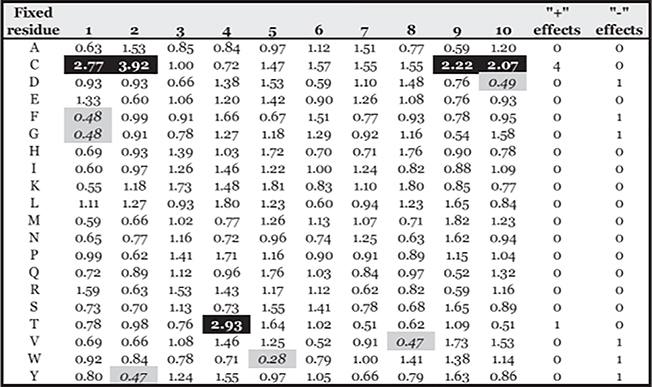
Accordingly, to examine the potential effect of Cys oxidation on peptide binding, 9- and 10-mer PSCL were tested in HLA-A*02:01 binding assays. Corresponding 9-mer and 10-mer matrices (Supplemental Table II) were derived, and the average (geometric) relative binding capacity (ARB) of peptides containing each amino acid at each position determined, as detailed in Materials and Methods. Assays were performed both in the presence or absence of 2-ME, and the ratio between the ARB for each position and residue measured under normal assay conditions and in the presence of 2-ME were determined (Figs. 4, 5). Greater or less than 2-fold differences, corresponding to ~1 SD, are highlighted in green or red, respectively.
FIGURE 4.
Relative changes in binding capacity associated with specific residues in specific positions in the presence of 2-ME for 9-mer peptides. Values represent the ratio of the ARB associated with each residue and position in the 2-ME assay to the ARB in normal conditions (2-ME ARB/normal ARB). Positive (+) and negative (−) effects show the number of positions a residue is associated with (>2-fold increases [black] or decreases [gray]) in ARB.
FIGURE 5.
Relative changes in binding capacity associated with specific residues in specific positions in the presence of 2-ME for 10-mer peptides. Values represent the ratio of the ARB associated with each residue and position in the 2-ME assay to the ARB in normal conditions (2-ME ARB/normal ARB). Positive (+) and negative (−) effects show the number of positions a residue is associated with (>2-fold increases [black] or decreases [gray]) in ARB.
As expected, most of the significant positive effects resulting from the addition of 2-ME were associated with the peptides containing Cys, particularly at the C terminus and at positions 2 or 3 in 9-mers and positions 1 or 2 in 10-mers; moderately negative or no effects were noted for most of the other residues and positions.
Validation of the new matrices in an additional broader set of epitopes from the IEDB
The ability of the new matrices to facilitate identification of Cys-containing epitopes was next examined. A search of the IEDB identified 703 nonredundant HLA-A*02:01-restricted 9–10-mer epitopes that had been validated using ELISPOT, intracellular cytokine staining, or MHC multimer assays (47). This set included 81 epitopes that contained Cys. A*02:01 binding affinities were predicted as described in Materials and Methods for all the peptides using both the control (normal) and 2-ME matrices (Supplemental Table III).
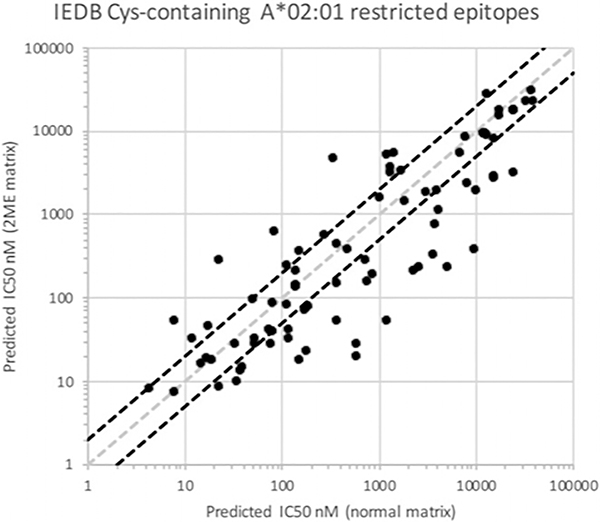
The corresponding affinities of the Cys-containing peptides are illustrated in Fig. 6, where affinities from the control matrix are plotted on the x-axis and the corresponding affinities from the 2-ME matrix on the y-axis. In the figure, the diagonal dotted gray line indicates unity, where the predicted affinity for a peptide from the two matrices is identical, and diagonal dotted red lines demarcate 2-fold or greater differences in predicted affinity.
FIGURE 6.
Comparison of predicted binding capacity of known Cys-containing epitopes using matrices derived from PSCL tested under normal conditions, or in the presence of 2-ME. The gray diagonal line demarcates equivalency in the predictions. Points below the diagonal indicate increases in predicted affinity using the 2-ME-derived matrix. Diagonal black lines highlight 2-fold differences in affinity.
As shown in Fig. 6, 33 of the 81 Cys peptides (41%) were associated with 2-fold or greater increases in predicted affinity using the 2-ME matrix (lower right), compared with 13 (16%) using the control matrix (upper left); for non-Cys peptides, the corresponding rates were 28 and 22%, respectively (Supplemental Table II). In addition, 22 (27%) of the Cys peptides were predicted to bind with 3-fold or higher affinities, compared with just six peptides (7%) predicted to bind with a 3-fold or lower affinity, when comparing the values obtained with the 2-ME and control matrices.
Overall, the average affinity predicted for Cys-containing peptides was nearly 2-fold higher (1.7 geometric mean) when using the 2-ME matrix, compared with the control matrix. An average increase of almost 3-fold higher (2.7 geometric mean) was noted when the Cys residue was either in position 2 or at the C terminus (Supplemental Table II), the main anchors for A*02:01 binding; by comparison, the difference was 1.1-fold for non-Cys peptides.
Prediction of HLA binding affinity is widely used to identify candidate T cell epitopes, and an affinity of 500 nM is routinely used as a threshold for peptide selection (26). With the present set of epitopes, it is noted that this threshold identifies 40 of the Cys-containing epitopes when using the control matrix, although the 2-ME matrix accounts for 48, representing a 20% increase in the number of epitopes identified. Taken together, these analyses show that the capacity for predicting the binding affinity of Cys-containing peptides is enhanced when algorithms account for potential effects of oxidation of Cys, with potential implications for epitope identification studies.
Evaluating potential role of peptide oxidation on T cell responses to cysteine-containing epitopes
The effects of peptide oxidation on T cell recognition were then examined by assaying responses to titrated doses of native and AABA-substituted Cys-containing neoepitopes corresponding to six of the HLA-A*02:01 restricted epitopes tested in the MHC binding assays detailed above plus four additional Cys-containing neoepitopes recognized in the context of A*68:01, A*68:02, B*35:03, or C*01:02 (Table II).
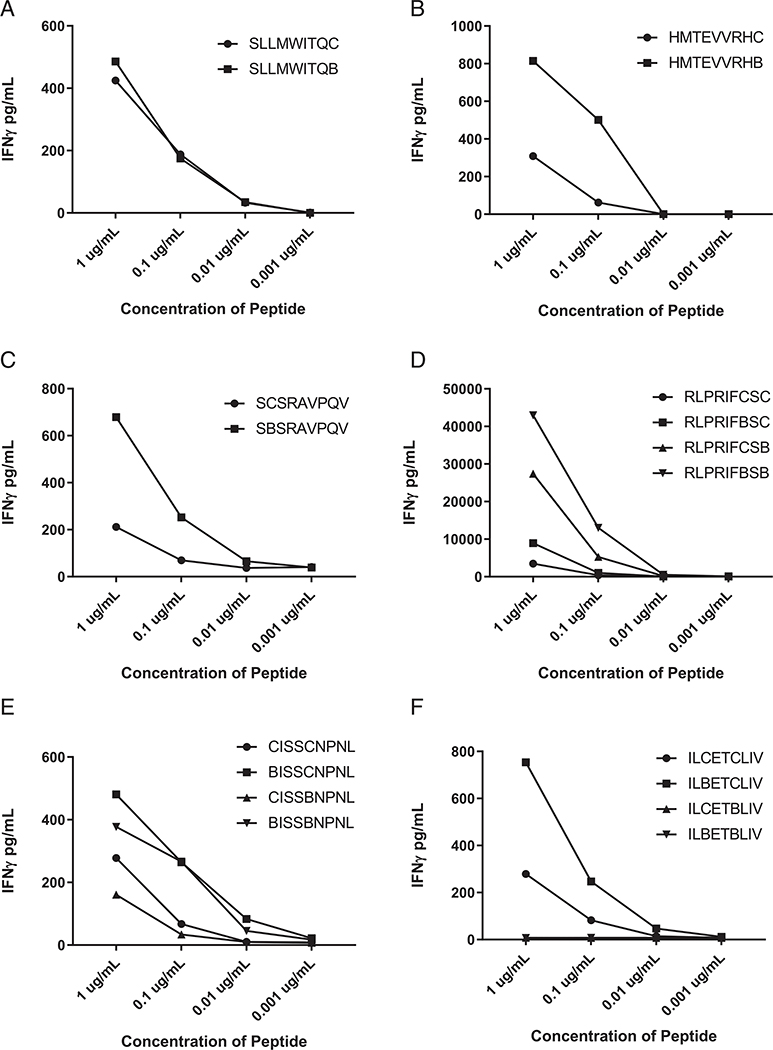
Similar dose titration curves were observed for the native and AABA-substituted CTAG2 peptides, whereas AABA-substituted TP53(168–176) R175H, CYFIP1(116–124) I121V peptides were recognized at a 10–100-fold lower concentration than the corresponding native peptides (Fig. 7A–C). Substitutions of AABA at position 9 alone or positions 7 and 9 in the PMEL peptide resulted in enhanced T cell recognition, whereas minimal enhancement was seen in response to the PMEL peptide containing a single AABA substitution at position 7 (Fig. 7D), indicating that oxidation of the C-terminal primary anchor position may have interfered with binding to HLA-A*02:01. Similarly, recognition of the HIVEP2(1674–1682) P1682L peptide appeared to be enhanced by modification of position 1, a potential secondary MHC anchor, whereas modification of position 5 alone did not appear to lead to enhanced recognition (Fig. 7E). Modification of the PHLPP1(563–571) G556E peptide at position 3 also appeared to enhanced T cell recognition; however, peptides containing AABA substitutions at position 6 alone or both positions 3 and 6 in the PHLPP1(563–571) G556E peptide were not recognized at 1 μg/ml, the highest concentration tested, indicating that the cysteine at position 6 may represent a critical TCR contact residue (Fig. 7F).
FIGURE 7.
T cell recognition of Cys-containing HLA-A*02:01-restricted peptides and the corresponding AABA-substituted peptides. Curves indicate IFN-γ secretion by relevant T cells in response to coculture with APCs pulsed with titrated concentrations of (A) CTAG2(157–165), (B) TP53(168–176) R175H, (C) CYFIP1(116–124) I121V, (D) HIVEP2(1674–1682) P1682L, (E) PMEL(639–647), and (F) PHLPP1(563–571) G556E peptides.
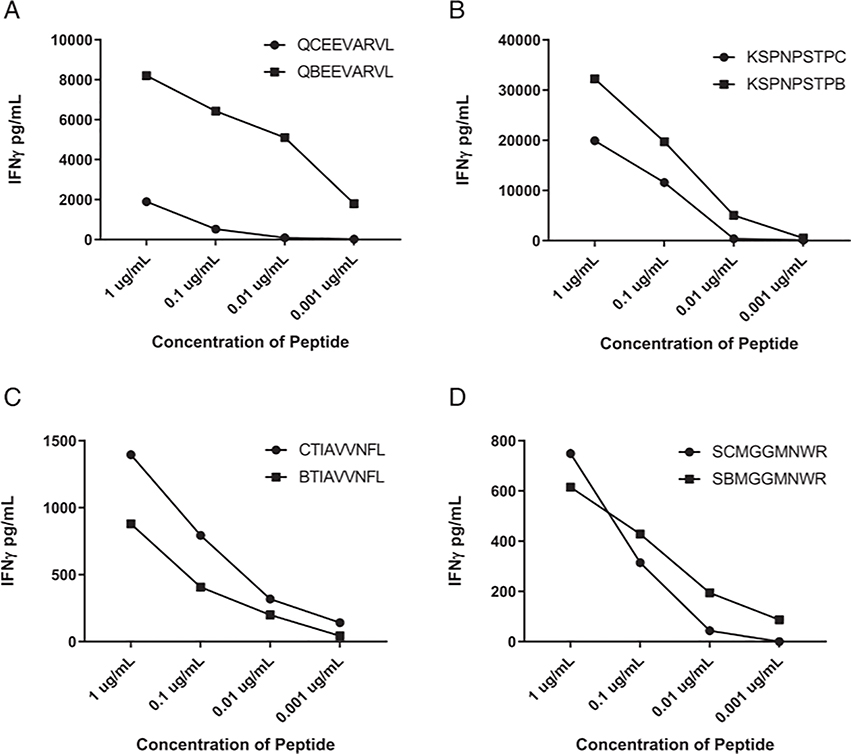
To further investigate the generality of these findings, T cell responses were investigated against four additional Cys-containing peptides recognized in the context of additional class I alleles. Substitution of AABA for the single Cys at position 2 in the HLA-A*35:03-restricted GALK1(242–250) A249V neoepitope enhanced T cell reactivity by a factor of ~1000-fold relative to the native peptide (Fig. 8A), although substitution of the single C-terminal Cys residue in the HLA-C*01:02-restricted RSL1D1(175–183) R183C neoepitope and the single position 1 Cys residue in the HLA*A68:02-restricted ECI2(350–358) N352I neoepitope enhanced reactivity by a factor of ~10-fold (Fig. 8B, 8C). In contrast, responses against the HLA-A*68:01-restricted TP53(241–249) R248W epitope containing a single position 2 Cys residue did not appear to be significantly altered by AABA substitution (Fig. 8D), as observed with the CTAG2(157–165) epitope (Fig. 7A).
FIGURE 8.
T cell recognition of Cys-containing peptides restricted by HLA alleles other than HLA-A*02:01 and the corresponding AABA-substituted peptides. Curves indicate IFN-γ secretion by relevant T cells in response to coculture with APCs pulsed with titrated concentrations of (A) HLA-A*35:03-restricted GALK1(242–250) A249V, (B) HLA-C*01:02-restricted RSL1D1(175–183) R183C, (C) HLA*A68:02-restricted ECI2(350–358) N352I, and (D) HLA-A*68:01-restricted TP53(241–249) R248W peptides.
Discussion
The evaluation of immune responses against Cys-containing peptides has been hampered by the fact that peptide oxidation can interfere with accurate measurement of peptide binding affinity and can significantly diminish T cell reactivity. Previous studies analyzing responses to individual Cys-containing peptides have demonstrated that MHC binding and T cell recognition can be substantially enhanced by substitution of Cys residues with Val, Ser or Ala, or the nonnatural amino acid AABA. Binding affinity measurements, however, have typically been carried out under conditions that allow peptide oxidation and in the presence of protease inhibitors, some of which can modify sulfhydryl groups, reducing the peptide monomer concentration and inhibiting MHC binding.
In many in vitro sensitization assays and vaccination protocols that are dependent upon accurate predictions of MHC binding due to the need to narrow down candidate epitopes, these issues can lead to a bias against the selection of Cys-containing peptides. The analysis of peptides that have been eluted from MHC molecules has provided an opportunity to enhance the accuracy of prediction data, although oxidation during sample processing can lead to an underestimate of the frequency of Cys-containing peptide in these samples. Overall, Cys residues compose ~2% of all residues in the proteome, indicating that ~18% of 9-mer peptides would be expected to contain a Cys residue, but these frequencies vary significantly among proteins and within protein regions. The DNA binding domain of the human Tp53 protein, generally considered to comprise residues 101–290 (48), encompasses the most frequently mutated or hot-spot mutations and all 10 of the Cys residues, representing 5% of all residues in this region of the Tp53 protein. In addition, Cys-containing peptides within the DNA binding region of the Tp53 protein appear to represent frequent targets of tumor infiltrating lymphocytes in patients with ovarian and gastrointestinal cancer (49), providing opportunities for therapeutic interventions in patients with these common epithelial malignancies.
Further analysis demonstrated the influence of the position of the Cys residue within the peptide sequence on MHC binding and T cell recognition. For the nine peptides tested for binding to HLA*A*02:01, addition of the reducing agent 2-ME increased the measured affinity by between 1.5-fold and 86-fold. Substitutions of Cys residues with AABA, an amino acid that was isosteric with Cys but incapable of forming disulfide bonds, led to between 9- and 838-fold increases in measured binding affinities. The addition of 2-ME did not appear to affect the binding affinities of AABA-containing peptides, providing evidence for the specificity of these effects, as 2-ME addition should not affect the binding of AABA-containing peptides. Furthermore, the measured affinities of the PMEL(639–647) and PHLPP1(563–571) G556E peptides, two of the three peptides containing two Cys residues, were higher for the peptides containing two AABA substitutions than those containing only one substitution. For four of the peptides containing Cys residues at presumed MHC anchor positions CYFIP1(116–124) I121V, TP53(168–176) R175H, PMEL(639–647) and GALK1(242–250) A249V, ABBA substitution significantly enhanced T cell recognition. Substitution of non-MHC anchor residue positions in the HIVEP2(1674–1682) P1682L and PHLPP1(563–571) G566E peptides with AABA, however, also significantly enhanced T cell recognition. These results are consistent with the hypothesis that peptide oxidation can diminish T cell recognition either by inhibiting MHC binding or by altering the interface of T cell contact residues. The fact that ABBA substitution enhanced recognition of some but not all of the evaluated Cys-containing peptides may be related to the influence of flanking residues on the susceptibility of the native peptides to oxidation (50). It is also notable that the HLA-B*35:03-restricted GALK1(242–250) A249V, HLA-C*01:02-restricted RSL1D1(175–183) R183C, and HLA-A*68:01-restricted TP53(241–249) A249V neoepitopes contain Cys residues at primary anchor positions, presumably reflecting the known preferences in these alleles for aliphatic or small polar anchor residues, as noted above for HLA-A*02:01 epitopes (51).
The increased binding affinities measured for peptides containing Cys substitutions appear to result primarily from preventing peptide oxidation but also can result from the fact that interactions between Cys residues in anchor positions and the MHC binding pockets are suboptimal. The higher affinity measured for the HLA-A*02:01 shown for the CTAG2 peptide containing a Val substitution for the C-terminal Cys residue appeared to result from additional side-chain interactions not present in the native CTAG2 peptide, demonstrated in x-ray crystallographic studies (52); however, our results demonstrated that the addition of 2-ME or the substitution of AABA for Cys could significantly enhance MHC binding. These findings are consistent with the hypothesis that the major effects of these maneuvers are to prevent peptide oxidation, but x-ray crystallographic studies of binding of native and AABA-substituted peptides would be informative.
These findings demonstrate that MHC binding and T cell recognition of Cys-containing peptides can be enhanced either by the inclusion of a reducing agent or the substitution of AABA for the native Cys residue, which should facilitate a more accurate assessment of the immunological potency of these peptides. In addition, the results point to the need to develop improved binding prediction algorithms that accurately score peptides containing Cys in anchor residues for multiple alleles that contain aliphatic residues at these positions.
Supplementary Material
Acknowledgments
This work was supported by the National Cancer Institute, National Institutes of Health (NIH) and by NIH Grant R21 AI134127-01 awarded to A.S. and B.P.
Abbreviations used in this article
- AABA
α-aminobutyric acid
- ARB
average (geometric) relative binding affinity
- DC
dendritic cell
- HBV
hepatitis B virus
- IEDB
Immune Epitope Database
- PSCL
positional scanning combinatorial library
- WT
wild-type
Footnotes
Disclosures
The authors have no financial conflicts of interest.
The online version of this article contains supplemental material.
References
- 1.Falk K, Rötzschke O, Stevanović S, Jung G, and Rammensee HG. 1991. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature 351: 290–296. [DOI] [PubMed] [Google Scholar]
- 2.Pearson H, Daouda T, Granados DP, Durette C, Bonneil E, Courcelles M, Rodenbrock A, Laverdure JP, Côté C, Mader S, et al. 2016. MHC class I-associated peptides derive from selective regions of the human genome. J. Clin. Invest. 126: 4690–4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murphy JP, Konda P, Kowalewski DJ, Schuster H, Clements D, Kim Y, Cohen AM, Sharif T, Nielsen M, Stevanovic S, et al. 2017. MHC-I ligand discovery using targeted database searches of mass spectrometry data: implications for T-cell immunotherapies. J. Proteome Res. 16: 1806–1816. [DOI] [PubMed] [Google Scholar]
- 4.Berlin C, Kowalewski DJ, Schuster H, Mirza N, Walz S, Handel M, Schmid-Horch B, Salih HR, Kanz L, Rammensee HG, et al. 2015. Mapping the HLA ligandome landscape of acute myeloid leukemia: a targeted approach toward peptide-based immunotherapy. [Published erratum appears in 2016 Leukemia 30: 1003–1004.] Leukemia 29: 647–659. [DOI] [PubMed] [Google Scholar]
- 5.Rammensee H, Bachmann J, Emmerich NP, Bachor OA, and Stevanović S. 1999. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics 50: 213–219. [DOI] [PubMed] [Google Scholar]
- 6.Kim Y, Sidney J, Buus S, Sette A, Nielsen M, and Peters B. 2014. Dataset size and composition impact the reliability of performance benchmarks for peptide-MHC binding predictions. BMC Bioinformatics 15: 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanada K, Yewdell JW, and Yang JC. 2004. Immune recognition of a human renal cancer antigen through post-translational protein splicing. Nature 427: 252–256. [DOI] [PubMed] [Google Scholar]
- 8.Vigneron N, Stroobant V, Chapiro J, Ooms A, Degiovanni G, Morel S, van der Bruggen P, Boon T, and Van den Eynde BJ. 2004. An antigenic peptide produced by peptide splicing in the proteasome. Science 304: 587–590. [DOI] [PubMed] [Google Scholar]
- 9.Warren EH, Vigneron NJ, Gavin MA, Coulie PG, Stroobant V, Dalet A, Tykodi SS, Xuereb SM, Mito JK, Riddell SR, and Van den Eynde BJ. 2006. An antigen produced by splicing of noncontiguous peptides in the reverse order. Science 313: 1444–1447. [DOI] [PubMed] [Google Scholar]
- 10.Dalet A, Robbins PF, Stroobant V, Vigneron N, Li YF, El-Gamil M, Hanada K, Yang JC, Rosenberg SA, and Van den Eynde BJ. 2011. An antigenic peptide produced by reverse splicing and double asparagine deamidation. Proc. Natl. Acad. Sci. USA 108: E323–E331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang RF, Parkhurst MR, Kawakami Y, Robbins PF, and Rosenberg SA. 1996. Utilization of an alternative open reading frame of a normal gene in generating a novel human cancer antigen. J. Exp. Med. 183: 1131–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coulie PG, Lehmann F, Lethé B, Herman J, Lurquin C, Andrawiss M, and Boon T. 1995. A mutated intron sequence codes for an antigenic peptide recognized by cytolytic T lymphocytes on a human melanoma. Proc. Natl. Acad. Sci. USA 92: 7976–7980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pierce RA, Field ED, den Haan JM, Caldwell JA, White FM, Marto JA, Wang W, Frost LM, Blokland E, Reinhardus C, et al. 1999. Cutting edge: the HLA-A*0101-restricted HY minor histocompatibility antigen originates from DFFRY and contains a cysteinylated cysteine residue as identified by a novel mass spectrometric technique. J. Immunol. 163: 6360–6364. [PubMed] [Google Scholar]
- 14.Chen W, Yewdell JW, Levine RL, and Bennink JR. 1999. Modification of cysteine residues in vitro and in vivo affects the immunogenicity and antigenicity of major histocompatibility complex class I-restricted viral determinants. J. Exp. Med. 189: 1757–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jäger E, Chen YT, Drijfhout JW, Karbach J, Ringhoffer M, Jäger D, Arand M, Wada H, Noguchi Y, Stockert E, et al. 1998. Simultaneous humoral and cellular immune response against cancer-testis antigen NY-ESO-1: definition of human histocompatibility leukocyte antigen (HLA)-A2-binding peptide epitopes. J. Exp. Med. 187: 265–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kittlesen DJ, Thompson LW, Gulden PH, Skipper JC, Colella TA, Shabanowitz J, Hunt DF, Engelhard VH, and Slingluff CL Jr. 1998. Human melanoma patients recognize an HLA-A1-restricted CTL epitope from tyrosinase containing two cysteine residues: implications for tumor vaccine development. [Published erratum appears in 1999 J. Immunol. 162: 3103–3106.] J. Immunol. 160: 2099–2106.9498746 [Google Scholar]
- 17.Falk K, Rötzschke O, Takiguchi M, Grahovac B, Gnau V, Stevanović S, Jung G, and Rammensee HG. 1994. Peptide motifs of HLA-A1, -A11, -A31, and -A33 molecules. Immunogenetics 40: 238–241. [DOI] [PubMed] [Google Scholar]
- 18.Chen JL, Stewart-Jones G, Bossi G, Lissin NM, Wooldridge L, Choi EM, Held G, Dunbar PR, Esnouf RM, Sami M, et al. 2005. Structural and kinetic basis for heightened immunogenicity of T cell vaccines. J. Exp. Med. 201: 1243–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schepers K, Toebes M, Sotthewes G, Vyth-Dreese FA, Dellemijn TA, Melief CJ, Ossendorp F, and Schumacher TN. 2002. Differential kinetics of antigen-specific CD4+ and CD8+ T cell responses in the regression of retrovirus-induced sarcomas. J. Immunol. 169: 3191–3199. [DOI] [PubMed] [Google Scholar]
- 20.Abelin JG, Keskin DB, Sarkizova S, Hartigan CR, Zhang W, Sidney J, Stevens J, Lane W, Zhang GL, Eisenhaure TM, et al. 2017. Mass spectrometry profiling of HLA-associated peptidomes in mono-allelic cells enables more accurate epitope prediction. Immunity 46: 315–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abelin JG, Harjanto D, Malloy M, Suri P, Colson T, Goulding SP, Creech AL, Serrano LR, Nasir G, Nasrullah Y, et al. 2019. Defining HLA-II ligand processing and binding rules with mass spectrometry enhances cancer epitope prediction. Immunity 51: 766–779.e17. [DOI] [PubMed] [Google Scholar]
- 22.González-Galarza FF, Takeshita LY, Santos EJ, Kempson F, Maia MH, da Silva AL, Teles e Silva AL, Ghattaoraya GS, Alfirevic A, Jones AR, and Middleton D. 2015. Allele frequency net 2015 update: new features for HLA epitopes, KIR and disease and HLA adverse drug reaction associations. Nucleic Acids Res. 43(D1): D784–D788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, and Wiley DC. 1987. Structure of the human class I histocompatibility antigen, HLA-A2. Nature 329: 506–512. [DOI] [PubMed] [Google Scholar]
- 24.Madden DR 1995. The three-dimensional structure of peptide-MHC complexes. Annu. Rev. Immunol. 13: 587–622. [DOI] [PubMed] [Google Scholar]
- 25.Ruppert J, Sidney J, Celis E, Kubo RT, Grey HM, and Sette A. 1993. Prominent role of secondary anchor residues in peptide binding to HLA-A2.1 molecules. Cell 74: 929–937. [DOI] [PubMed] [Google Scholar]
- 26.Paul S, Weiskopf D, Angelo MA, Sidney J, Peters B, and Sette A. 2013. HLA class I alleles are associated with peptide-binding repertoires of different size, affinity, and immunogenicity. J. Immunol. 191: 5831–5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sidney J, Southwood S, Moore C, Oseroff C, Pinilla C, Grey HM, and Sette A. 2013. Measurement of MHC/peptide interactions by gel filtration or monoclonal antibody capture. Curr. Protoc. Immunol. Chapter 18: Unit 18.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gulukota K, Sidney J, Sette A, and DeLisi C. 1997. Two complementary methods for predicting peptides binding major histocompatibility complex molecules. J. Mol. Biol. 267: 1258–1267. [DOI] [PubMed] [Google Scholar]
- 29.Cheng Y, and Prusoff WH. 1973. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 22: 3099–3108. [DOI] [PubMed] [Google Scholar]
- 30.Pinilla C, Appel JR, Blanc P, and Houghten RA. 1992. Rapid identification of high affinity peptide ligands using positional scanning synthetic peptide combinatorial libraries. Biotechniques 13: 901–905. [PubMed] [Google Scholar]
- 31.Sidney J, Assarsson E, Moore C, Ngo S, Pinilla C, Sette A, and Peters B. 2008. Quantitative peptide binding motifs for 19 human and mouse MHC class I molecules derived using positional scanning combinatorial peptide libraries. Immunome Res. 4: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reed JS, Sidney J, Piaskowski SM, Glidden CE, León EJ, Burwitz BJ, Kolar HL, Eernisse CM, Furlott JR, et al. 2011. The role of MHC class I allele Mamu-A*07 during SIV(mac)239 infection. Immunogenetics 63: 789–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loffredo JT, Sidney J, Bean AT, Beal DR, Bardet W, Wahl A, Hawkins OE, Piaskowski S, Wilson NA, et al. 2009. Two MHC class I molecules associated with elite control of immunodeficiency virus replication, Mamu-B*08 and HLA-B*2705, bind peptides with sequence similarity. J. Immunol. 182: 7763–7775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao Y, Zheng Z, Robbins PF, Khong HT, Rosenberg SA, and Morgan RA. 2005. Primary human lymphocytes transduced with NY-ESO-1 antigen-specific TCR genes recognize and kill diverse human tumor cell lines. J. Immunol. 174: 4415–4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lo W, Parkhurst M, Robbins PF, Tran E, Lu YC, Jia L,Gartner JJ, Pasetto A, Deniger D, et al. 2019. Immunologic recognition of a shared p53 mutated neoantigen in a patient with metastatic colorectal cancer. Cancer Immunol. Res. 7: 534–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robbins PF, Li YF, El-Gamil M, Zhao Y, Wargo JA, Zheng Z, Xu H, Morgan RA, Feldman SA, et al. 2008. Single and dual amino acid substitutions in TCR CDRs can enhance antigen-specific T cell functions. J. Immunol. 180: 6116–6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koşaloğlu-Yalçln Z, Lanka M, Frentzen A, Logandha Ramamoorthy Premlal A, Sidney J, Vaughan K, Greenbaum J, Robbins P, Gartner J, Sette A, and Peters B. 2018. Predicting T cell recognition of MHC class I restricted neoepitopes. OncoImmunology 7: e1492508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vigneron N, Stroobant V, Van den Eynde BJ, and van der Bruggen P. 2013. Database of T cell-defined human tumor antigens: the 2013 update. Cancer Immun. 13: 15. [PMC free article] [PubMed] [Google Scholar]
- 39.Vita R, Overton JA, Greenbaum JA, Ponomarenko J, Clark JD, Cantrell JR, Wheeler DK, Gabbard JL, Hix D, Sette A, and Peters B. 2015. The immune epitope database (IEDB) 3.0. Nucleic Acids Res. 43(D1): D405–D412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sette A, Vitiello A, Reherman B, Fowler P, Nayersina R, Kast WM, Melief CJ, Oseroff C, Yuan L, Ruppert J, et al. 1994. The relationship between class I binding affinity and immunogenicity of potential cytotoxic T cell epitopes. J. Immunol. 153: 5586–5592. [PubMed] [Google Scholar]
- 41.Assarsson E, Sidney J, Oseroff C, Pasquetto V, Bui HH, Frahm N, Brander C, Peters B, Grey H, and Sette A. 2007. A quantitative analysis of the variables affecting the repertoire of T cell specificities recognized after vaccinia virus infection. J. Immunol. 178: 7890–7901. [DOI] [PubMed] [Google Scholar]
- 42.Trolle T, Metushi IG, Greenbaum JA, Kim Y, Sidney J, Lund O, Sette A, Peters B, and Nielsen M. 2015. Automated benchmarking of peptide-MHC class I binding predictions. Bioinformatics 31: 2174–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stryhn A, Pedersen LO, Romme T, Holm CB, Holm A, and Buus S. 1996. Peptide binding specificity of major histocompatibility complex class I resolved into an array of apparently independent subspecificities: quantitation by peptide libraries and improved prediction of binding. Eur. J. Immunol. 26: 1911–1918. [DOI] [PubMed] [Google Scholar]
- 44.Lauemøller SL, Holm A, Hilden J, Brunak S, Holst Nissen M, Stryhn A, Østergaard Pedersen L, and Buus S. 2001. Quantitative predictions of peptide binding to MHC class I molecules using specificity matrices and anchor-stratified calibrations. Tissue Antigens 57: 405–414. [DOI] [PubMed] [Google Scholar]
- 45.Sidney J, Peters B, Moore C, Pencille TJ, Ngo S, Masterman KA, Asabe S, Pinilla C, Chisari FV, and Sette A. 2007. Characterization of the peptide-binding specificity of the chimpanzee class I alleles A 0301 and A 0401 using a combinatorial peptide library. Immunogenetics 59: 745–751. [DOI] [PubMed] [Google Scholar]
- 46.Udaka K, Wiesmüller KH, Kienle S, Jung G, Tamamura H, Yamagishi H, Okumura K, Walden P, Suto T, and Kawasaki T. 2000. An automated prediction of MHC class I-binding peptides based on positional scanning with peptide libraries. Immunogenetics 51: 816–828. [DOI] [PubMed] [Google Scholar]
- 47.Carrasco Pro S, Sidney J, Paul S, Lindestam Arlehamn C, Weiskopf D, Peters B, and Sette A. 2015. Automatic generation of validated specific epitope sets. J. Immunol. Res. 2015: 763461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pavletich NP, Chambers KA, and Pabo CO. 1993. The DNA-binding domain of p53 contains the four conserved regions and the major mutation hot spots. Genes Dev. 7(12B): 2556–2564. [DOI] [PubMed] [Google Scholar]
- 49.Malekzadeh P, Pasetto A, Robbins PF, Parkhurst MR, Paria BC, Jia L, Gartner JJ, Hill V, Yu Z, Restifo NP, et al. 2019. Neoantigen screening identifies broad TP53 mutant immunogenicity in patients with epithelial cancers. J. Clin. Invest. 129: 1109–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alcock LJ, Perkins MV, and Chalker JM. 2018. Chemical methods for mapping cysteine oxidation. Chem. Soc. Rev. 47: 231–268. [DOI] [PubMed] [Google Scholar]
- 51.Rapin N, Hoof I, Lund O, and Nielsen M. 2010. The MHC motif viewer: a visualization tool for MHC binding motifs. Curr. Protoc. Immunol. Chapter 18: Unit 18.17. [DOI] [PubMed] [Google Scholar]
- 52.Chen JL, Dunbar PR, Gileadi U, Jäger E, Gnjatic S, Nagata Y, Stockert E, Panicali DL, Chen YT, Knuth A, et al. 2000. Identification of NY-ESO-1 peptide analogues capable of improved stimulation of tumor-reactive CTL. J. Immunol. 165: 948–955. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.