Abstract
Staphylococcus aureus is one of the major pathogens causing community—and healthcare-acquired infections. The presence of the virulence factor Panton–Valentine leukocidin (PVL) is associated with recurrent infection and clinical severity and generally regarded as a feature of community associated-methicillin-resistant Staphylococcus aureus (MRSA). To date, the focus of PVL-positive MRSA in hospitalized patients has been on outbreaks. We aimed to investigate whether PVL-positive MRSA has penetrated the community-hospital barrier by determining the prevalence of PVL in MRSA of hospitalized patients. MRSA strains isolated from patients hospitalized > 48 h in Heidelberg University Hospital between 2015 and 2018 Isolates were analysed for the presence of PVL and subjected to spa-typing. PVL-positive MRSA were then characterized by whole genome sequencing. We analysed 740 MRSA isolates in the study period and identified 6.2% (n = 46) PVL-positivity. 32.6% of PVL-positive MRSA met the criteria for nosocomial acquisition. The most frequent clones among the PVL-positive strains were ST80-t044 (21.7%, n = 10/46) and ST8-t008 (19.5%, n = 9/46). WGS identified three possible transmission clusters involving seven patients. In conclusion, we found successful epidemic PVL-positive MRSA clones entering the hospital and causing nosocomial infections. Preventive measures and constant surveillance should be maintained to prevent transmissions and clonal outbreaks.
Subject terms: Clinical microbiology, Infectious-disease epidemiology
Introduction
Staphylococcus aureus is one of the major causes of community and hospital acquired infections. Methicillin-resistance is an ongoing problem locally and globally. Although the overall prevalence of methicillin-resistant S. aureus (MRSA) in Germany is declining1, MRSA strains harboring the pathogenic marker Panton–Valentine leukocidin (PVL) are isolated more frequently2. The presence of PVL is relevant, as it is associated with a more severe clinical presentation, often with recurrences, deep and multiple lesions and frequent transmission to contact persons, irrespective of methicillin resistance3–6. In addition, PVL-positive S. aureus often displays multiple resistances to commonly used antibiotics to treat S. aureus infections3,7.
PVL has been linked to community-associated (CA-) MRSA6. Although the overall prevalence of PVL in Germany is considered low according to data acquired by Schaumburg et al.8, a study by Jappe et al.9 revealed a prevalence of 22% for PVL positivity in MRSA from a dermatological outpatient clinic from 2003 to 2005. Moreover, our recent study suggests an alarmingly high prevalence of PVL positivity (40%) in community onset (CO)-MRSA causing skin and soft-tissue infections (SSTI) in South West Germany between 2012 and 20162. Nevertheless, systematic data on the prevalence and molecular characteristics of PVL-bearing MRSA in hospitalized patients in Germany is scarce. Surveillance data and other published report suggest that a high percentage of imported epidemic MRSA clones are circulating and causing community acquired infections2–4. More importantly, patients with CO-MRSA SSTI often seek medical care in out-patient units and emergency units of tertiary hospitals10,11, providing a port of entry for these MRSA clones to the hospital setting.
In this study, we analyzed and characterized MRSA isolates from hospitalized patients with a special focus on PVL-positive MRSA by WGS retrospectively to understand the population structure and dynamics of PVL positive MRSA entering and circulating in our hospital. Clonal shift of MRSA population in the healthcare setting should be monitored closely to prevent clonal spread of highly virulent and epidemic MRSA clones.
Results
Between 2015 and 2018 we detected and isolated 756 non-duplicate MRSA from overall 756 individual patients admitted to Heidelberg University hospital with 188, 158, 210 and 184 isolates in the respective years (Table 1). 58.1% (n = 439/756) patients were male. The mean age was 52 years with a range from 3 days to 97 years.
Table 1.
Prevalence of PVL in MRSA isolated from hospitalized patient, Germany 2015–2018.
| Year | Total MRSA | PVL positive | PVL negative | ||
|---|---|---|---|---|---|
| n | n | % | n | % | |
| 2015 | 188 | 5 | 2.7 | 183 | 97.3 |
| 2016 | 158 | 17 | 10.8 | 141 | 89.2 |
| 2017 | 210 | 13 | 6.2 | 197 | 93.8 |
| 2018 | 184 | 11 | 6.0 | 173 | 94.0 |
| Total | 740 | 46 | 6.2 | 694 | 93.8 |
Molecular characteristics of MRSA from hospitalized patients
Of 756 MRSA, 22 isolates were not recoverable for further molecular characterization by spa-typing and WGS. The most common spa types (> 20 isolates) found in our MRSA collection were t003 (31.6%; n = 232/734), t002 (6.0%; n = 44/734), t127 (5.6%; n = 41/734), t008 (4.9%; n = 36/734) and t063 (2.7%; n = 20/734) (for all spa-types, see supplemental Table). The majority of characterized MRSA harboured SCCmec types II and IV (n = 326 and n = 301; 43.1% and 39.9% respectively). All MRSA isolates harboured mecA.
For 740 of 756 MRSA, data regarding the presence of PVL was available. Of 740 isolates, 46 (6.2%) were PVL-positive with 2.7% (n = 5/188), 10.8% (n = 17/158), 6.2% (n = 13/210) and 6.0% (n = 11/184) in the respective years (Table 1).
Clinical and molecular characteristics of PVL-positive MRSA
We identified 46 (6.2%) patients with PVL-positive MRSA in the study period (Table 1). Most patients were males (58.7%, n = 27/46) and mean age was 36 years with a range of 1 month to 96 years. Patients with PVL-positive MRSA were significantly younger than those with PVL-negative MRSA (mean age 36 years and 53 years, respectively, p < 0.0001). The proportion of males did not differ significantly between the two groups (p = 0.93). 58.7% (n = 27/46) of the patients had an infection with PVL-positive MRSA, of which 69.6% (n = 32/46) were simultaneously colonized while 26% (n = 12/46) were free of nasal and rectal PVL-positive MRSA on screening swabs. Two patients with PVL-positive MRSA infection were not screened for rectal or nasal MRSA colonization. 54.3% (n = 25/46) of patients had a documented history of migration. 15 patients (32.6%) met the criteria for nosocomial MRSA acquisition of being detected > 48 h after hospital admission (Fig. 1). Thus, the majority of patients (67.4%, n = 31/46) was already infected or colonized with PVL-positive MRSA on admission to hospital. Skin and soft tissue infections were the most common clinical presentation (68.8% of cases, multiple clinical presentations possible). Two cases presented with blood stream infections and two with bone and joint infections (Table 2).
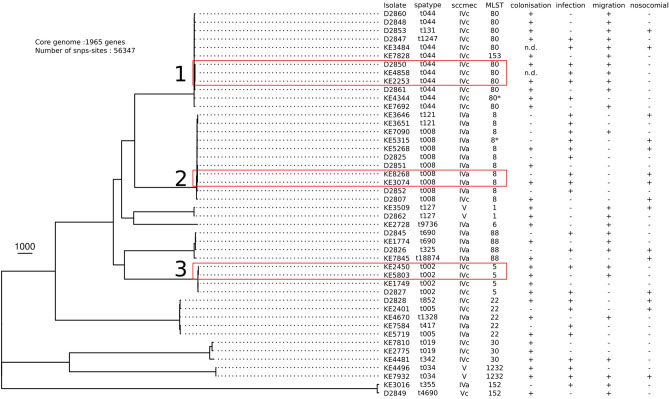
Figure 1.
Phylogenetic tree of PVL-positive MRSA. Strains with a SNP distance lower that 10 SNPs are highlighted by a red rectangle. These were potential in-hospital transmission cases between patients (named cluster 1, 2 and 3).
Table 2.
Site of infection in patients with PVL-positive MRSA
| Site of infection | n | % |
|---|---|---|
| Skin and soft tissue | 22 | 68.8 |
| Respiratory tract | 3 | 9.4 |
| Blood stream | 2 | 6.3 |
| Bone and joint | 2 | 6.3 |
| Other | 3 | 9.4 |
Multiple sites of infection possible.
Among the PVL-positive MRSA, we identified 10 clonal groups according to MLST. These were ST1, ST5, ST6, ST8, ST22, ST30, ST80, ST88, ST152 and ST1232 (Fig. 1), each representing a genetic cluster. The spa-types identified were for ST1 t127, for ST5 t002, for ST6 t9736, for ST8 t008 and t121. With ST22, spa-type t852, t005, t1328 and t417 were present. For ST30, t019 and t342 and for ST80 t044, t131 and t1247 were identified. Spa-type t690, t325 and t18874 of ST88 were detected and with ST152, t355 and t4690 were identified. With ST1232, spa-type t034 was detected. The most prevalent spa types were t044 (n = 10/46; 21.7%) and t008 (n = 9/46; 19.5%). The PVL-positive isolates were of the SCCmec type IV and V (Fig. 1). The most common spa type causing infections was t008 (n = 7/27; 25.9%), followed by t044 (n = 5/27; 18.5%).
Among the hospital onset PVL-positive MRSA (n = 15/46), ST8 (t008 and t121) MRSA was the most common, accounting for 40% (n = 6/15). The majority of these isolates (n = 5/6, 83%) belonged to the USA300 clonal group (ST8-t008-IVa/c). Other common PVL-positive MRSA clones were ST80-t044, ST88 and ST22 with two isolates each, and ST1-t127, ST5-t002 and ST1232-t034 with one isolates each (Fig. 1).
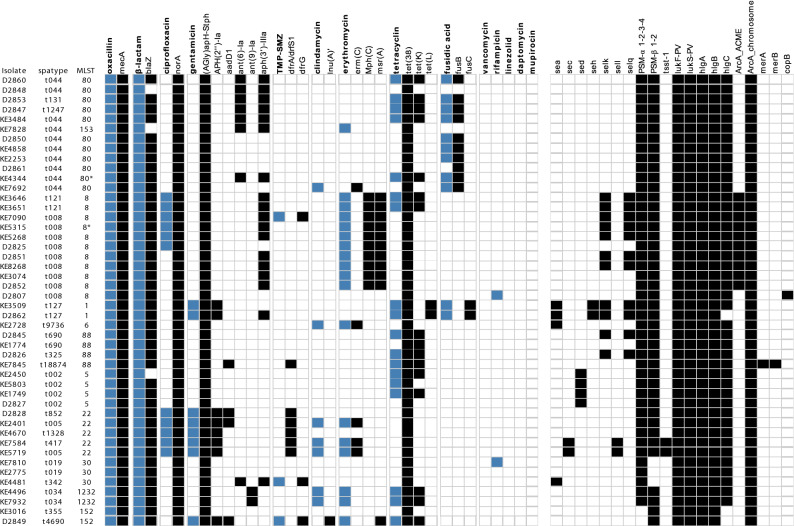
Phenotypical antimicrobial resistance and detected resistance genes in PVL-positive MRSA
Of the PVL-positive MRSA, 23.9% (n = 11/46; 3 missing data) were resistant to ciprofloxacin, belonging to ST22 (5/11) and ST8 (6/11) (Fig. 2). 17.4% (n = 8/46) were resistant to gentamicin, had all the APH(2″)-Ia gene, and belonged either to ST22, ST1 or ST152. Co-trimoxazole (TMP-SMZ) resistance was not common: only 6.5% (n = 3/46) of PVL-positive MRSA were resistant, all of which carried dfrG. Phenotypical resistance to clindamycin in 15.2% (n = 7/46) did not correlate to the presence of Inu(A)′. 39.1% (n = 18/46) were resistant to erythromycin. Erm(C) was present in 5 strains, of which 4 were phenotypically resistant. 10 strains carried Mph(C) and msr(A), were all phenotypically resistant and belonged to the ST8 (8 spa-type t008 and 2 spa-type t121). Only one strain with resistance to erythromycin harboured the msr(A) gene alone (ST152). Resistance to tetracycline was present in 39.1% (n = 18/46), 15 of these carried the tet(K) and two the tet(L) gene. Fusidic acid resistance was found in 23.9% (n = 11/46) of isolates, 9 of which carried the fusB gene; all of them belonging to the ST80. Two (4.3%) carried the fusC gene and belonged to ST1, spa-type t127. Two PVL-positive MRSA (ST8-t008 and ST30-t019) were resistant to rifampicin and carried functionally related mutations in the rpoB gene. Neither phenotypical resistance towards vancomycin, linezolid, daptomycin or mupirocin, nor any corresponding resistance gene was detected. For all detected resistance genes, see Fig. 2.
Figure 2.
Phenotypic and genotypic antimicrobial resistance of PVL-positive MRSA. Phenotypic resistance is represented by a blue square while genotypic resistance is represented by a black square. The right panel represents the virulence genes and ACME/COMER cassettes found in the strains. Strains are ordered based on the phylogenetic tree from Fig. 1.
Virulence genes present in PVL-positive MRSA
All Isolates harboured genes encoding phenol soluble modulins, 95% (n = 44/46) of them for PSM-α 1–4 and PSM-ß 1–2. Two isolates of ST152 carried only genes for PSM-ß 1–2 and three strains of ST30 only for PSM-α 1–4 (Fig. 2). Likewise, hlgA and hlgB were present in all strains, and hlgC was not detected in only three strains, both of ST152 and one of ST1 (t127). 10 of the isolates belonging to ST8 (SCCmec IVa) harboured the arginine catabolic mobile element (ACME), a hallmark of ST8-USA300 MRSA. The other, ACME-negative, ST8 t008 (SCCmec IVc) strain displayed the copB gene as part of the copper and mercury resistance mobile element (COMER), described as a key feature of the Latin American Variant (USA300-LV). In 7 of the ST8 (t008 and t121) selK and selq was detected. Two strains of ST22 were positive for tsst-1, one with spa-type t005 and the other with t417. For all genes encoding virulence factors, see Fig. 2.
Possible transmission events
SNP analysis identified three possible transmission clusters involving 7 patients (Fig. 1, red rectangles). These included three patients with ST80 (t044) in cluster 1, two patients with ST8 (t008) in cluster 2 and another two patients with ST5 (t002) in cluster 3. All three patients with t044 had overlapping hospitals stays on the same wards. The two patients with t008 and both patients with t002 had no temporal or spatial (same ward) overlap during their hospital stay.
Discussion
PVL is regarded as one of the main features of CA-MRSA and a clinically relevant virulence marker for S. aureus3. In recent years, cases of nosocomial PVL-positive MRSA transmissions and outbreaks in Europe have been accumulating5,12,13 thus suggesting that the barrier between community- and hospital-associated MRSA has already been penetrated. Although PVL is associated with the severity of infection3, the characterization of MRSA isolates in hospitalized patients in Germany in terms of PVL is not consequently pursued. To our knowledge, this is the first systematic analysis on the prevalence and molecular structure of PVL-positive MRSA in hospitalized patients in Germany.
We found that the clonal distribution as determined by MLST and spa-typing was overall concordant with existing knowledge about the molecular epidemiology of MRSA in Germany: 30.5% of isolates were spa-type t003, consistent with the predominant ST5 “Rhine-Hesse epidemic clone”14 and 5.8% t0028. Likewise ST8 t008 MRSA (4.6%,n = 35/756) is considered one of the most common MRSA clones circulating in Germany8. Unexpectedly, our sample contained 2.6% (n = 20/756) of t063 MRSA which has been rarely described in Germany, yet8. This was explained by a putative transmission cluster of PVL-negative MRSA in our hospital.
Altogether, we detected 6.2% PVL positivity in MRSA. This was considerably higher that the findings of another study on MRSA prevalence in hospitalized and non-hospitalized patients in Germany 2010–2011, which reported 2.7% being PVL-positive8. A possible explanation for this discrepancy could the premise of the analysis. The authors did not differentiate between in-patients and out-patients for their analysis, it is therefore difficult to directly compare the reported rates of PVL-positive MRSA between the two studies. Similar to our findings, a Swiss study reported 7.7% PVL-positive MRSA in a mixed population of hospitalized and non-hospitalized patients15. Similarly, a European study conducted among patients presenting with skin and soft tissue infections in the emergency room reported 8% PVL-positive MRSA for the German study centre10. Compared to the 40% prevalence of PVL-positive MRSA in patients presenting with SSTI in outpatient departments of our hospital2, these estimates and our findings support, that only a fraction of these patients actually enter the hospital. However, we also found PVL-positive MRSA in blood cultures and tissue samples from bone and joint infections, providing evidence for invasive infections in this population besides the high percentage of skin and soft tissue infections.
Over two thirds (67.4%) of patients with PVL-positive MRSA in our study were already colonized or infected on hospital admission. This was somewhat expected, since the presence of PVL has been linked to community-associated clones2,16. In line with our previous findings on the genetic background of MRSA clones in community onset SSTI, the majority of characterized isolates in this study belonged to ST8-t008 and ST80-t0442. ST80-t044 MRSA clonal lineage has been described as a frequent CA-MRSA clone circulating in Europe and the Middle East17,18. Another dominant clone was the ST8-t008 MRSA belonging to the epidemic USA300 clade (both North-American and Latin American variant). Although the prevalence of this MRSA clone in Europe is generally considered to be low8,10, we found a remarkably high proportion of USA300 MRSA clones circulating in the community2 and USA300 is indeed the most commonly imported MRSA into the European continent through intercontinental travel19. Moreover, USA300 MRSA clones can be classified as epidemic, as demonstrated by their expansive properties and dominance on the American continents20 and further highlighted by accumulating reports on nosocomial transmission events and outbreaks in Europe5,12,13.
Another interesting clone is the ST1-t127 MRSA, which is regarded as livestock-associated MRSA (LA-MRSA), made up 5.4% of our study isolates. This particular clone has been linked with bovine mastitis and colonization of farm animals. Although several reports of human infections with ST1-t127 MRSA have been published, this occurrence is still considered rare21,22. Moreover, some of the LA-MRSA clones harbor the virulence factor PVL. Although reports on the presence of PVL genes in ST1-t127 have been published, the isolates in those reports harbour the human-adapted SCCmec IV22. In contrast our PVL-positive ST1-t127 MRSA were of the SCCmec V, consistent with the porcine ST1-t127 MRSA22. The acquisition of PVL was not only restricted to the ST1 clones but several LA-MRSA isolates with ST1232 (t034), a single locus variant of the ST398 LA-MRSA, suggesting human adaptation of LA-MRSA clones22. Taken together, these finding demonstrates the relevance of the “One Health” concept for the surveillance of multi-drug resistance bacteria.
Of the three previously undetected potential transmission clusters identified by SNP analysis, one cluster most likely represents nosocomial transmission events, since these three patients had overlapping hospital stays on the same ward (t044, cluster 1, Fig. 1). The two patients with t008 MRSA (cluster 2, Fig. 1) had no temporal or spatial overlap. Comparative analysis of virulence genes revealed slight differences in genes present/absent (Fig. 2), suggesting non-identical isolates despite close clonal relationship. Therefore, patient-to-patient transmission was unlikely. In the third potential transmission cluster (t002, cluster 3, Fig. 1), both patients did not have apparent epidemiological overlap during their hospital stays. However, since both patients were housed in the same residential facility for refugees, a transmission event outside of the hospital setting is a plausible alternative explanation. Our findings underline the importance of reliable epidemiological data for investigations of transmissions and outbreaks of multi-drug resistant bacteria.
Our study has strengths and limitations. It was a single-centre study and may therefore only reflect the local epidemiological situation in a part of Germany. However, this systematic analysis provides evidence on the occurrence of PVL-positive MRSA over a 4-year period in a tertiary care hospital. Molecular typing data have indicated that epidemic PVL-positive MRSA clones have entered the hospital and may cause nosocomial infections and outbreaks. Thus, implementation of pre-emptive measures, such as systematic surveillance of MRSA population structures in hospitalized and non-hospitalized patients is needed, with special emphasis on PVL-positive clones.
Methods
Study population
We analysed and characterized MRSA isolates from hospitalized patients in our tertiary care hospital over a 4-year period between 2015 and 2018 retrospectively. The study was performed at Heidelberg University Hospital, located in the South-West of Germany. Clinical and microbiological data were acquired though the routine microbiological diagnostic, surveillance of multidrug resistant bacteria and patient care. Only hospitalized patients (> 48 h) were included in this study.
Microbiological methods
MRSA were cultured and detected from routine screening samples and clinical samples from hospitalized patients seeking medical care, as described previously2. Species identification was performed with the MALDI-TOF (Bruker GmbH, Germany), antibiotic susceptibility testing using the VITEK2 (Biomérieux GmbH, Germany) and interpreted according to current EUCAST clinical breakpoints. MRSA was confirmed by the presence of nuc and mecA as described earlier2. Isolates were cryopreserved for surveillance purposes.
Spa typing, SCCmec typing and detection of PVL
PVL detection was performed by PCR, as previously described3. Spa typing was performed by Sanger sequencing as published elsewhere23. SCCmec typing for the cassette types I to V were determined by multiplex PCR as described earlier24. All PVL positive strains were subject to WGS for a more detailed analysis.
WGS
Genomic DNA was extracted from overnight bacterial culture using the DNeasy Blood and Tissue Minikit (Qiagen GmbH, Germany) according to manufacturer’s protocol with the addition of a prior lysis step with lysostaphin (Genaxxon GmbH, Germany). Standard genomic library was prepared from the DNA and sequenced with the Illumina MiSeq platform (2 × 250 bp paired end). Quality control and assembly was performed as described previously25.
Briefly, raw sequences were trimmed for quality using Sickle 1.33 (parameters, q > 30; 1 > 45)6. The cleaned sequences were then assembled using SPAdes 3.13.07. Contigs obtained from the assembly were curated for length (> 500 bp) and coverage (> 10 ×) to ensure no errors and contamination in the draft genome. Annotation was performed using Prokka 1.14.1 (based on Genetic Code Table 11)8 and the NCBI Prokaryotic Genome Annotation Pipeline. Resistance and virulence genes were found using Abricate 0.8.13 (https://github.com/tseemann/abricate with respectively the antibiotic database from ResFinder 3.0; ARG-ANNOT; CARD; NCBI-BARRGD and the virulence database VFDB. MLST was perform using the MLST 2.0 pipeline from the centre for Genomic Epidemiology26. Sequences will be made publically available upon publication of the manuscript.
Cluster analysis by SNP and allelic difference
Genomes were compared by calculating the core genomes with Roary and only genes present in all isolates were considered (1965 genes, 74.7 ± 1.7% of the genomes). Phylogenetic distance was calculated with Gubbins 2.3.4 to take in account recombination events and not over-estimate the SNP polymorphism27. Clonal groups were defined as genomes distant from less than 10 SNPs.
Data and statistical analysis
Patient data was extracted from the medical record. A history of migration was defined as a patient with home address outside of Germany or living in a community facility for refugees. Descriptive statistics was analyzed using the STATA 13 software (StataCorp, USA). Two-sample t test or chi square test was used calculating the p value for age and sex, p < 0.05 was considered statistically significant.
Ethical considerations
Ongoing molecular characterization of MRSA isolates is performed as part of obligatory surveillance and infection control measures mandated by the German Infection Protection Act. All methods were carried out in accordance with relevant guidelines and regulations. The Ethical review board of the University of Heidelberg approved the study protocol (S-474/2018) and waived informed consent.
Supplementary information
Acknowledgements
Open access funding provided by Projekt DEAL.
Author contributions
D.N., S.K., K.H. and P.Z. designed the study, J.H., D.N., S.K. and S.B. performed the molecular typing and acquired the data, D.N., S.K. and S.B. analysed the dataset, S.K. and D.N. drafted the manuscript, all authors critically read, revised and approved the final version of the manuscript.
Data availability
Sequences were uploaded to https://www.ncbi.nlm.nih.gov/biosample with the BioProject Number PRJNA637212. Accession numbers are listed in Supplementary Table 3.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-70112-z.
References
- 1.Walter J, Noll I, Feig M, Weiss B, Claus H, Werner G, et al. Decline in the proportion of methicillin resistance among Staphylococcus aureus isolates from non-invasive samples and in outpatient settings, and changes in the co-resistance profiles: an analysis of data collected within the antimicrobial resistance surveillance network, Germany 2010 to 2015. BMC Infect. Dis. 2017;17(1):169. doi: 10.1186/s12879-017-2271-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klein S, Menz MD, Zanger P, Heeg K, Nurjadi D. Increase in the prevalence of Panton–Valentine leukocidin and clonal shift in community-onset methicillin-resistant Staphylococcus aureus causing skin and soft-tissue infections in the Rhine-Neckar Region, Germany, 2012–2016. Int. J. Antimicrob. Agents. 2019;53(3):261–267. doi: 10.1016/j.ijantimicag.2018.10.026. [DOI] [PubMed] [Google Scholar]
- 3.Nurjadi D, Friedrich-Janicke B, Schafer J, Van Genderen PJ, Goorhuis A, Perignon A, et al. Skin and soft tissue infections in intercontinental travellers and the import of multi-resistant Staphylococcus aureus to Europe. Clin. Microbiol. Infect. 2015;21(6):567e1-10. doi: 10.1016/j.cmi.2015.01.016. [DOI] [PubMed] [Google Scholar]
- 4.Zanger P, Nurjadi D, Schleucher R, Scherbaum H, Wolz C, Kremsner PG, et al. Import and spread of Panton–Valentine leukocidin-positive Staphylococcus aureus through nasal carriage and skin infections in travelers returning from the tropics and subtropics. Clin. Infect. Dis. 2012;54(4):483–492. doi: 10.1093/cid/cir822. [DOI] [PubMed] [Google Scholar]
- 5.Nurjadi D, Klein S, Zimmermann S, Heeg K, Zanger P. Transmission of ST8-USA300 Latin American variant methicillin-resistant Staphylococcus aureus on a neonatal intensive care unit: recurrent skin and soft-tissue infections as a marker for epidemic community-associated-MRSA colonization. Infect. Control Hosp. Epidemiol. 2017;38(7):883–885. doi: 10.1017/ice.2017.88. [DOI] [PubMed] [Google Scholar]
- 6.Boyle-Vavra S, Daum RS. Community-acquired methicillin-resistant Staphylococcus aureus: the role of Panton–Valentine leukocidin. Lab Invest. 2007;87(1):3–9. doi: 10.1038/labinvest.3700501. [DOI] [PubMed] [Google Scholar]
- 7.Nurjadi D, Olalekan AO, Layer F, Shittu AO, Alabi A, Ghebremedhin B, et al. Emergence of trimethoprim resistance gene dfrG in Staphylococcus aureus causing human infection and colonization in sub-Saharan Africa and its import to Europe. J. Antimicrob. Chemother. 2014;69(9):2361–2368. doi: 10.1093/jac/dku174. [DOI] [PubMed] [Google Scholar]
- 8.Schaumburg F, Kock R, Mellmann A, Richter L, Hasenberg F, Kriegeskorte A, et al. Population dynamics among methicillin-resistant Staphylococcus aureus isolates in Germany during a 6-year period. J. Clin. Microbiol. 2012;50(10):3186–3192. doi: 10.1128/JCM.01174-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jappe U, Heuck D, Strommenger B, Wendt C, Werner G, Altmann D, et al. Staphylococcus aureus in dermatology outpatients with special emphasis on community-associated methicillin-resistant strains. J. Invest. Dermatol. 2008;128(11):2655–2664. doi: 10.1038/jid.2008.133. [DOI] [PubMed] [Google Scholar]
- 10.Bouchiat C, Curtis S, Spiliopoulou I, Bes M, Cocuzza C, Codita I, et al. MRSA infections among patients in the emergency department: a European multicentre study. J. Antimicrob. Chemother. 2017;72(2):372–375. doi: 10.1093/jac/dkw431. [DOI] [PubMed] [Google Scholar]
- 11.Kossow A, Stuhmer B, Schaumburg F, Becker K, Glatz B, Mollers M, et al. High prevalence of MRSA and multi-resistant gram-negative bacteria in refugees admitted to the hospital-but no hint of transmission. PLoS ONE. 2018;13(5):e0198103. doi: 10.1371/journal.pone.0198103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kossow A, Kampmeier S, Schaumburg F, Knaack D, Moellers M, Mellmann A. Whole genome sequencing reveals a prolonged and spatially spread nosocomial outbreak of Panton–Valentine leucocidin-positive meticillin-resistant Staphylococcus aureus (USA300) J. Hosp. Infect. 2019;101(3):327–332. doi: 10.1016/j.jhin.2018.09.007. [DOI] [PubMed] [Google Scholar]
- 13.Kairet K, Ho E, Van Kerkhoven D, Boes J, Van Calenbergh S, Pattyn L, et al. USA300, A strain of community-associated methicillin-resistant Staphylococcus aureus, crossing Belgium's borders: outbreak of skin and soft tissue infections in a hospital in Belgium. Eur. J. Clin. Microbiol. Infect. Dis. 2017;36(5):905–909. doi: 10.1007/s10096-016-2883-6. [DOI] [PubMed] [Google Scholar]
- 14.Kock R, Mellmann A, Schaumburg F, Friedrich AW, Kipp F, Becker K. The epidemiology of methicillin-resistant Staphylococcus aureus (MRSA) in Germany. Dtsch. Arztebl. Int. 2011;108(45):761–767. doi: 10.3238/arztebl.2011.0761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Von Dach E, Diene SM, Fankhauser C, Schrenzel J, Harbarth S, Francois P. Comparative genomics of community-associated methicillin-resistant Staphylococcus aureus shows the emergence of clone ST8-USA300 in Geneva Switzerland. J. Infect. Dis. 2016;213(9):1370–1379. doi: 10.1093/infdis/jiv489. [DOI] [PubMed] [Google Scholar]
- 16.Miller LG, Perdreau-Remington F, Rieg G, Mehdi S, Perlroth J, Bayer AS, et al. Necrotizing fasciitis caused by community-associated methicillin-resistant Staphylococcus aureus in Los Angeles. N. Engl. J. Med. 2005;352(14):1445–1453. doi: 10.1056/NEJMoa042683. [DOI] [PubMed] [Google Scholar]
- 17.Tristan A, Bes M, Meugnier H, Lina G, Bozdogan B, Courvalin P, et al. Global distribution of Panton–Valentine leukocidin-positive methicillin-resistant Staphylococcus aureus, 2006. Emerg. Infect. Dis. 2007;13(4):594–600. doi: 10.3201/eid1304.061316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harastani HH, Tokajian ST. Community-associated methicillin-resistant Staphylococcus aureus clonal complex 80 type IV (CC80-MRSA-IV) isolated from the Middle East: a heterogeneous expanding clonal lineage. PLoS ONE. 2014;9(7):e103715. doi: 10.1371/journal.pone.0103715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nurjadi D, Fleck R, Lindner A, Schafer J, Gertler M, Mueller A, et al. Import of community-associated, methicillin-resistant Staphylococcus aureus to Europe through skin and soft-tissue infection in intercontinental travellers, 2011–2016. Clin. Microbiol. Infect. 2019;25(6):739–746. doi: 10.1016/j.cmi.2018.09.023. [DOI] [PubMed] [Google Scholar]
- 20.Planet PJ. Life after USA300: the rise and fall of a superbug. J. Infect. Dis. 2017;215(suppl_1):S71–S77. doi: 10.1093/infdis/jiw444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aspiroz C, Lozano C, Vindel A, Lasarte JJ, Zarazaga M, Torres C. Skin lesion caused by ST398 and ST1 MRSA Spain . Emerg. Infect. Dis. 2010;16(1):157–159. doi: 10.3201/eid1601.090694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Earls MR, Kinnevey PM, Brennan GI, Lazaris A, Skally M, O'Connell B, et al. The recent emergence in hospitals of multidrug-resistant community-associated sequence type 1 and spa type t127 methicillin-resistant Staphylococcus aureus investigated by whole-genome sequencing: Implications for screening. PLoS ONE. 2017;12(4):e0175542. doi: 10.1371/journal.pone.0175542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harmsen D, Claus H, Witte W, Rothganger J, Claus H, Turnwald D, et al. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 2003;41(12):5442–5448. doi: 10.1128/JCM.41.12.5442-5448.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghaznavi-Rad E, Nor Shamsudin M, Sekawi Z, van Belkum A, Neela V. A simplified multiplex PCR assay for fast and easy discrimination of globally distributed staphylococcal cassette chromosome mec types in meticillin-resistant Staphylococcus aureus. J. Med. Microbiol. 2010;59(Pt 10):1135–1139. doi: 10.1099/jmm.0.021956-0. [DOI] [PubMed] [Google Scholar]
- 25.Kocer K, Boutin S, Dalpke AH, Heeg K, Mutters NT, Nurjadi D. Comparative genomic analysis reveals a high prevalence of inter-species in vivo transfer of carbapenem-resistance plasmids in patients with haematological malignancies. Clin. Microbiol. Infect. 2019;26:780e-1. doi: 10.1016/j.cmi.2019.10.014. [DOI] [PubMed] [Google Scholar]
- 26.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, et al. Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 2012;50(4):1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43(3):e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequences were uploaded to https://www.ncbi.nlm.nih.gov/biosample with the BioProject Number PRJNA637212. Accession numbers are listed in Supplementary Table 3.