SUMMARY
Dynein is a microtubule motor that transports myriad cargos in various cell types and contexts. How dynein is regulated to perform these activities with spatial and temporal precision remains unclear. Human dynein is regulated by autoinhibition, whereby intermolecular contacts limit motor activity. Whether this mechanism is conserved throughout evolution, whether it can be affected by extrinsic factors, and its role in regulating dynein function remain unclear. Here, we use a combination of negative stain EM, single molecule assays, genetic, and cell biological techniques to show that autoinhibition is conserved in budding yeast, and plays a key role in coordinating in vivo dynein function. Moreover, we find that the Lissencephaly-related protein, LIS1 (Pac1 in yeast) plays an important role in regulating dynein autoinhibition. Our studies demonstrate that rather than inhibiting dynein motility, Pac1/LIS1 promotes dynein activity by stabilizing the uninhibited conformation, which ensures appropriate dynein localization and activity in cells.
INTRODUCTION
Cytoplasmic dynein relies on a complex network of regulators to support its myriad functions. For instance, processive motility of human dynein requires the dynactin complex and an adaptor that links them together1,2. The reliance on cargo-specific adaptors (e.g., BicD2, Hook31,2) ensures that dynein-dynactin are linked together at the right time and place for appropriate motility. Dynactin supports dynein motility by promoting microtubule binding3, and by orienting the motor domains in a parallel manner that is conducive for motility4. In addition to its regulation by extrinsic factors, studies have shown that human dynein-1 and dynein-2 are auto-regulated by intra-complex interactions. Specifically, intermolecular interactions between the motor domains stabilize an autoinhibited conformation of human dynein called the phi particle (for its resemblance to the Greek letter)4–7. For dynein-2 (responsible for intraflagellar transport), the autoinhibited conformation – which has been observed in its native context7 – reduces its velocity, ATPase activity, and microtubule landing rate6. Similarly, the autoinhibited dynein-1 conformation has been shown to reduce its microtubule landing rate and motility properties4,8. Autoinhibition of dynein-1 also precludes its interaction with dynactin and BicD24.
Although it is well established that human dynein adopts the autoinhibited conformation, the extent of its evolutionary conservation is unclear. Yeast dynein is of particular interest due to two notable in vitro discrepancies with human dynein: (1) yeast dynein is processive in single molecule assays without the need for other factors, such as dynactin9; and, (2) yeast dynein has been reported to interact with dynactin in the absence of adaptors10. The reasons for these differences are unclear, but one possibility is that yeast dynein does not adopt the autoinhibited conformation, which could potentially account for its ability to walk in the absence of dynactin. This is supported by studies showing that artificially separating the motor domains of human dynein-1 with a rigid linker (thus preventing intermolecular contacts) is sufficient to convert it to a processive motor8.
Another important effector of dynein activity is the lissencephaly protein LIS1, which is required for numerous dynein functions in cells11–13, including promoting dynein recruitment to cellular sites14,15, and assisting in dynein transport functions, including nuclear migration in neurons16,17, and high-load vesicular transport18–21. However, the mechanism by which LIS1 affects dynein activity remains controversial. For instance, in vitro studies have shown that LIS1 reduces the velocity of dynein alone21–23, but increases the velocity of dynein-dynactin-BicD223,24. In addition to promoting dynein force production21, studies have shown that LIS1 promotes initiation of dynein-dynactin-BicD2 motility from the plus ends of dynamic microtubules23,25. Studies with the budding yeast homolog of LIS1 – Pac1 – have shown that it reduces the velocity of dynein motility26–29, presumably by uncoupling the ATPase cycle from the conformational changes in the motor and microtubule-binding domains that elicit microtubule release27,28. Thus, the precise role for Pac1/LIS1 in dynein function remains confounded by these contrasting results. Although a role for Pac1/LIS1 in regulating the autoinhibited conformation has not yet been reported, two studies found that LIS1 can indeed promote dynein-dynactin interaction30,31, which is an expected consequence of relieving dynein autoinhibition4.
We set out to address the question of whether yeast dynein adopts an autoinhibited conformation, and if so, what role it plays in regulating dynein activity. Our recent findings suggested that yeast dynein indeed adopts such a conformation32. Specifically, we found that engineering a neurological disease-correlated mutation into yeast dynein leads to increased run lengths of single molecules of dynein, and a localization pattern in cells that is indicative of an enhanced dynein-dynactin interaction. This mutation was at a residue recently shown to be important for maintenance of the autoinhibited conformation of human dynein4. Here, we use a combination of in vitro and in vivo approaches to show that yeast dynein indeed adopts an autoinhibited conformation that restricts its in vitro processivity, and coordinates it localization and activity in cells. Moreover, we find that Pac1 is an important effector of this conformational state: rather than inhibiting dynein motility, Pac1 promotes its activity by stabilizing the uninhibited conformation.
RESULTS
Yeast dynein adopts an autoinhibited ‘phi’ particle conformation
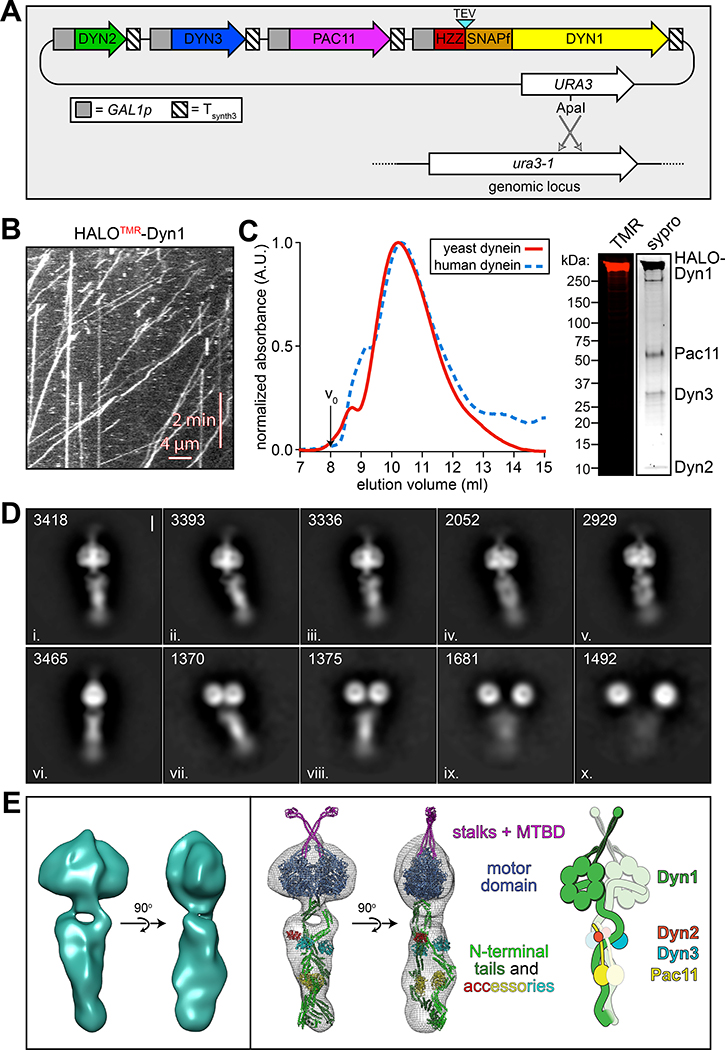
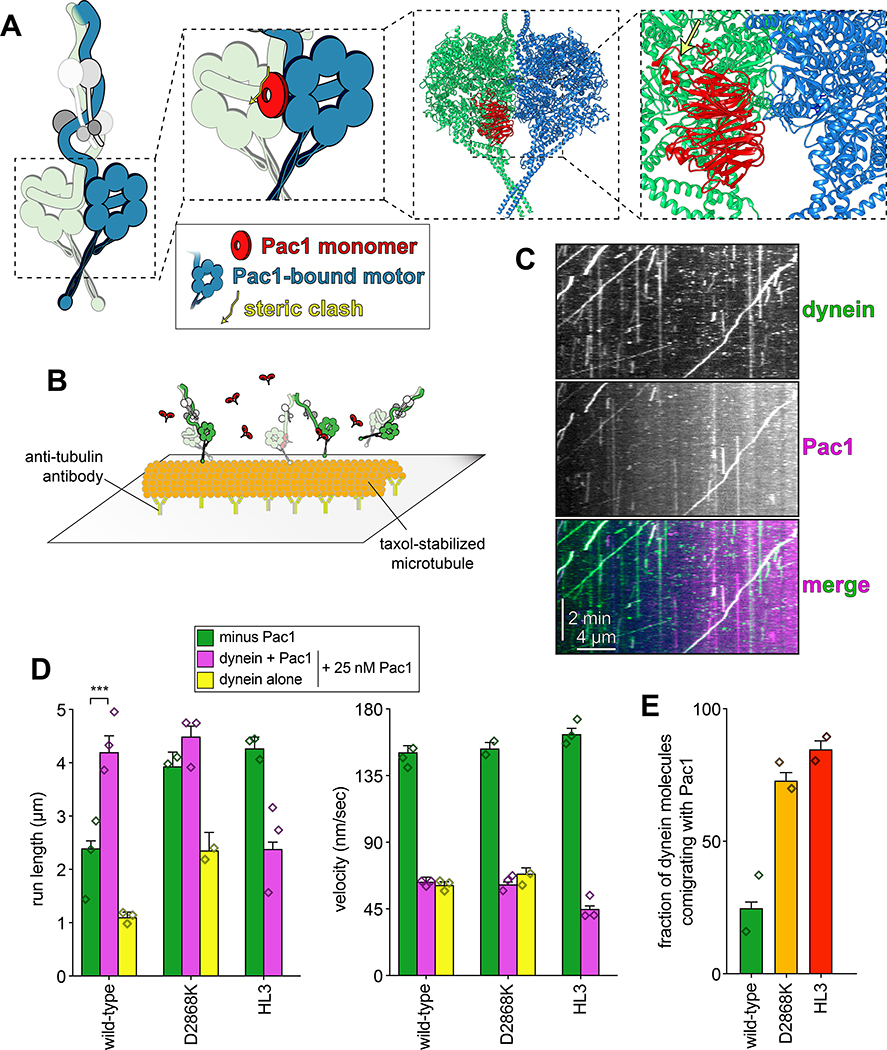
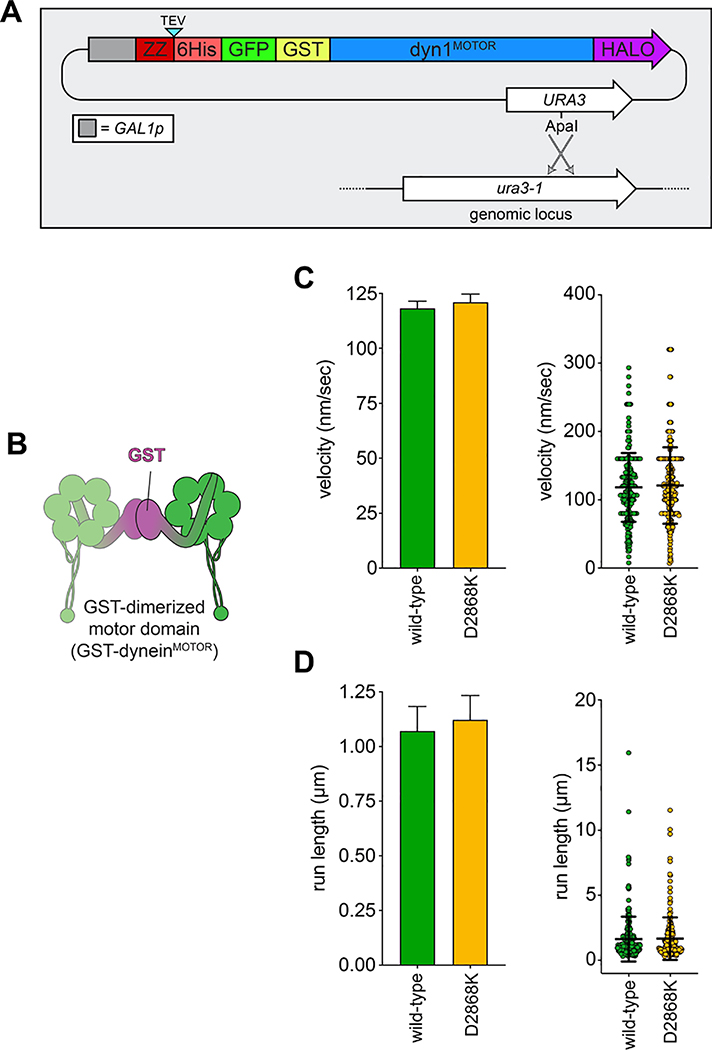
To determine whether yeast dynein adopts an autoinhibited conformation5, we developed a strategy to isolate biochemical quantities of the intact yeast dynein complex to a high degree of purity. The yeast dynein complex is comprised of light (Dyn2), light-intermediate (Dyn3), intermediate (Pac11), and heavy chains (Dyn1)33, the latter of which is the catalytic subunit responsible for motility. We generated a polycistronic plasmid containing all dynein complex subunits each under the control of the galactose-inducible promoter (GAL1p). We included a tandem affinity tag (8His-ZZ, or “HZZ”) followed by either a SNAP or HALO tag on the N-terminus of Dyn1 for purification and fluorescent labeling of the complex, respectively (Fig. 1A). Single molecule motility assays with the purified complex confirmed its activity was nearly identical to the non-overexpressed complex (Fig. 1B; also see below). The increased yield permitted us to isolate the complex to a high degree of purity using size exclusion chromatography, which revealed an elution profile nearly identical to the recombinant human dynein complex (Fig. 1C).
Figure 1. The yeast dynein complex adopts an autoinhibited phi particle conformation.
(A) Schematic of the polycistronic plasmid used to produce the intact yeast dynein complex (GAL1p, galactose-inducible promoter; Tsynth3, terminator sequence56). Restriction digest with ApaI (cuts within URA3 gene) targets the plasmid for homologous recombination into the ura3–1 locus, as depicted. (B) Representative kymograph depicting single molecule motility of the purified overexpressed yeast dynein complex. (C) Representative elution profiles of yeast and human dynein complexes from Superose 6 resin (left), and scans of the same polyacrylamide gel depicting fluorescently labeled Dyn1 (via HaloTag-TMR) and the entire complex (via Sypro Ruby staining; right; 3 independent preparations yielded very similar results). (D) Representative negative stain EM class averages of the intact yeast dynein complex (2D classes were generated from one preparation; however, independent preparations provided very similar raw images, as shown in Extended Data Fig. 1; scale bar, 10 nm). Number of particles used to generate each class indicated in each panel. Classes i – vi depict dynein in the autoinhibited, phi particle conformation, whereas vii – x depict dynein in various open, uninhibited states. (E) 3D models of dynein in the autoinhibited state generated from 2D class averages with (right) and without (left) a high resolution 3D structure of human dynein-1 in the phi particle conformation (pdb 5NVU4) manually docked into it. Note that the structures of the two tail domains have been slightly rotated with respect to the motor domains to better fit the 3D model, and that the structures of both TcTEX and Robl have been eliminated due to their absence from the yeast dynein complex. Also see Extended Data Figure 1 and Supplementary Video 1.
We used negative stain electron microscopy to obtain the first high magnification view of the intact yeast dynein complex. This revealed the presence of dynein complexes in both an open, uninhibited state, and those in an apparent autoinhibited conformation, with the large majority being in the latter state (Extended Data Fig. 1). Class averages appeared strikingly similar to the intact human dynein-1 complex1,4, and to an artificially dimerized motor domain truncation of dynein-2 in the autoinhibited conformation6 (Fig. 1D). The N-terminal tail domains – which exhibit flexibility with respect to the motor domains – appear twisted around one another in 2D and 3D averages (Fig. 1E, and Supplementary Video 1). Intermolecular contacts appear to extend to the motor domains and the coiled-coil stalks (the latter of which cross in an “X”-like configuration), which connect the AAA rings to the microtubule-binding domains (MTBDs). We confirmed a high degree of similarity between the human and yeast dynein phi particles by manually docking a high resolution structure of human dynein4 into our 3D model (Fig. 1E, and Supplementary Video 1).
Previous observations of an artificially-dimerized, truncated yeast dynein motor domain fragment (lacking the N-terminal tails) revealed no phi particles (as noted by negative stain EM26, and within the crystal lattice34,35). Thus, in contrast to human dynein-2, for which the motor domain is sufficient to adopt the phi particle6,36, yeast dynein requires the tail to do so. We also noted several class averages depicting conformations in which the motor domains are closely apposed, but unbound, and the tails are wrapped around one another (Fig. 1D; classes vii and viii), indicating that intermolecular contacts within the tails can stabilize motor domain-independent tail-tail interactions. This also suggests that the transition to the autoinhibited conformation may be initiated by tail-tail contacts, which appear to be required to stabilize the motor domains in the autoinhibited conformation.
The autoinhibited conformation restricts in vitro processivity
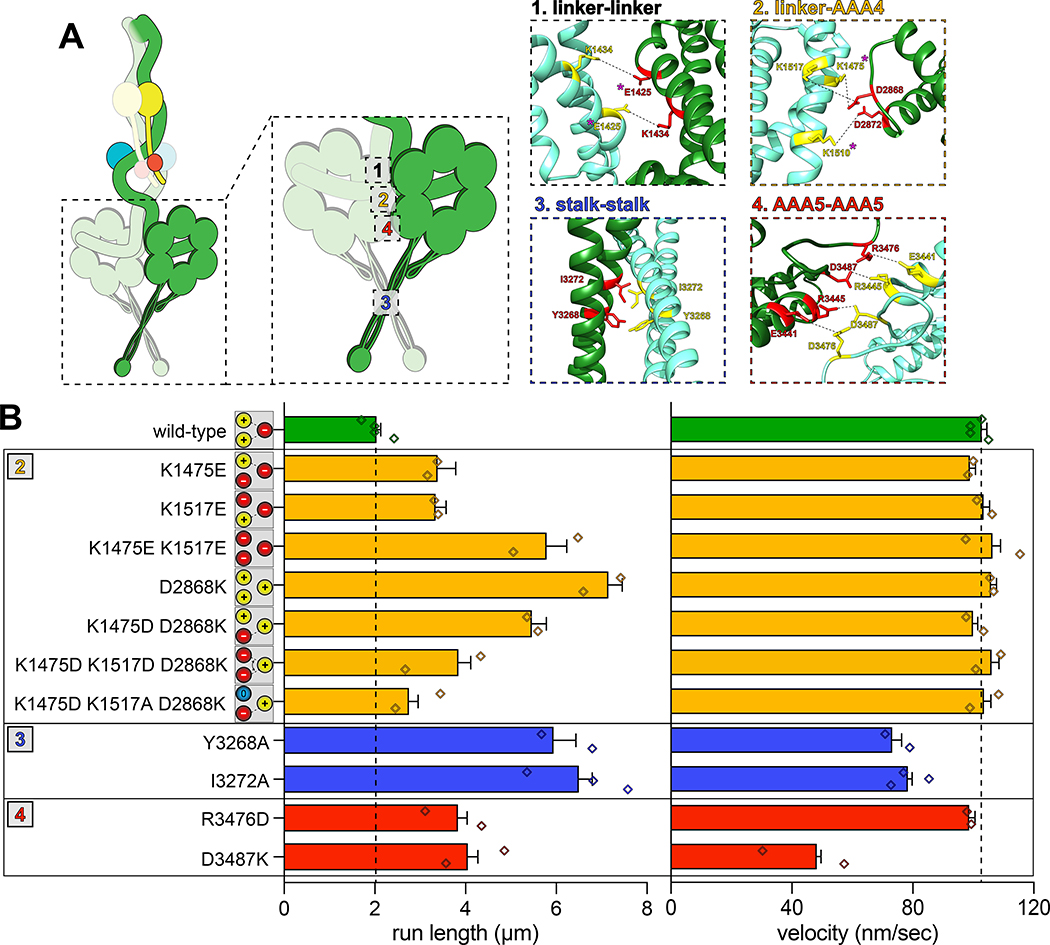
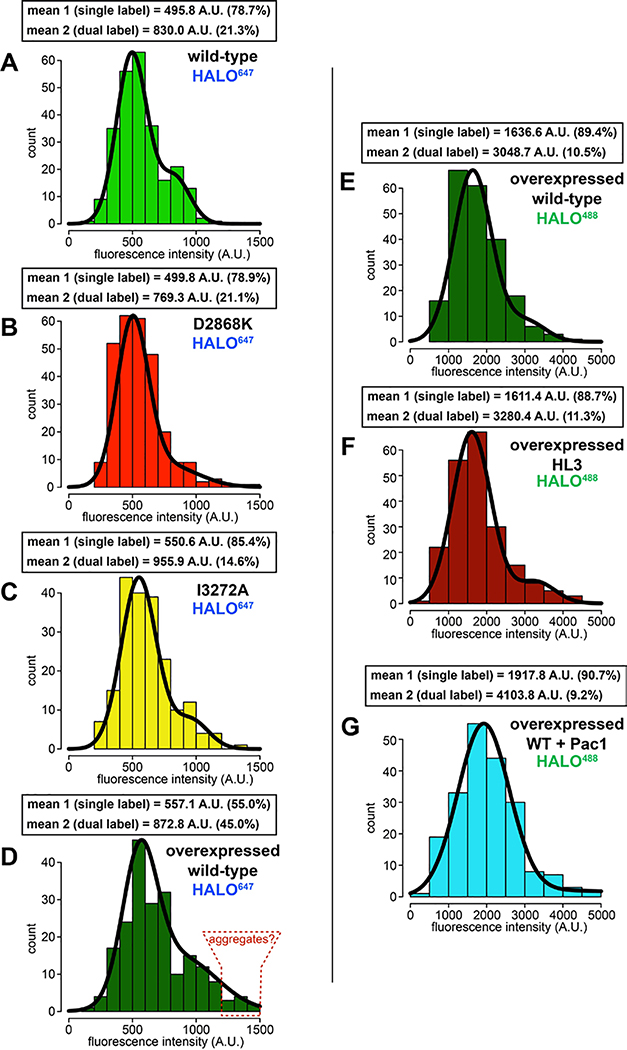
To determine if the increased processivity we previously noted for the neurological disease-correlated K1475Q32 is a consequence of disrupted phi particle formation, we sought to assess the consequences of mutating other intermolecular surfaces. The human dynein phi particle structure identified four key intermolecular surfaces (Fig. 2A)4. Sequence alignment and homology modeling of Dyn1 into this structure revealed a high degree of conservation at all four surfaces (Fig. 2A). We found that increasing degrees of disruption of a predicted tripartite salt bridge at surface 2 increased processivity in a step-wise manner that we were able to reduce by repairing the broken electrostatic pairing (Fig. 2B). Consistent with the apparent stalk-stalk contact (Fig. 1D, classes i - v), we noted a large increase in run length for surface 3 mutants. Unlike surface 2 mutants, these latter mutants moved slower, which may be due to disrupted kinetics of helix sliding in the coiled coil stalk, which is responsible for communicating nucleotide-dependent conformational changes within the motor domain to the MTBD36–39. Processivity also increased for surface 4 mutants, with one of them reducing velocity to ~50% that of wild-type. Finally, fluorescence intensity measurements confirmed that the run length increases for the most processive mutants (D2868K and I3272A) are not a consequence of protein aggregation (Extended Data Fig. 2A – C). These results indicate that the three surfaces tested here are important for formation of the autoinhibited conformation of yeast dynein, which limits its processivity (see Supplementary Video 3).
Figure 2. Disrupting phi particle contact points extends single molecule run lengths.
(A) Cartoon depicting four predicted intermolecular contact surfaces within the motor domains that stabilize the phi particle conformation. Four insets show respective regions of yeast dynein modeled into the human dynein phi particle structure. Structural models were generated using one-to-one threading of the yeast DYN1 sequence into 5NVU4 on the Phyre2 server57. Residues with magenta asterisks are mutated in patients suffering from neurological disease58–60, suggesting that autoinhibition is critical for normal cellular dynein function. (B) Single molecule run length and velocity values for wild-type and mutant dyneins with phi particle disrupting mutations (at surfaces 2, 3 and 4, as indicated). Cartoons along vertical axis depict electrostatic interactions (or lack thereof) among residues 1517, 1475 (left circles) and 2868 (right circle) at linker-AAA4 surface. Note that the degree of processivity enhancement is inversely proportional to the number of charge interactions at this surface. Diamonds represent mean values obtained from each independent replicate experiment (n = 528 [4], 400 [2], 361 [2], 213 [2], 409 [2], 352 [2], 233 [2], 323 [2], 170 [2], 462 [3], 362 [2], 319 [2] motors [independent replicates], left to right; error bars indicate standard error). Also note that we generated and tested the motility of two other point mutants at interface 4, E3441K and R3445D, both of which were inactive in single molecule assays (not shown). Also see Extended Data Figures 2, 3 and 4, and Supplementary Video 3.
As noted above, the GST-dimerized yeast dynein motor domain (GST-dyneinMOTOR; Extended Data Fig. 3B) likely does not adopt the autoinhibited conformation. Consistent with this notion, GST-dyneinMOTOR with D2868K did not exhibit increased processivity (Extended Data Fig. 3), further indicating that the reason for the increased run lengths for full-length dynein is disruption of the autoinhibited conformation, and not altered mechanochemistry. It is interesting to note that GST-dyneinMOTOR exhibits run lengths (1.1 – 1.6 μm; see Extended Data Figs. 3 and 7F) much lower than that of the uninhibited dynein mutants (≤ 7.2 μm, or 4.5-fold higher), in spite of this fragment not adopting the autoinhibited conformation. This indicates that the tail domain permits a motor domain arrangement that is more conducive to processive motility than the GST, which is consistent with the higher force generation capacity40 of the intact dynein complex with respect to GST-dyneinMOTOR.
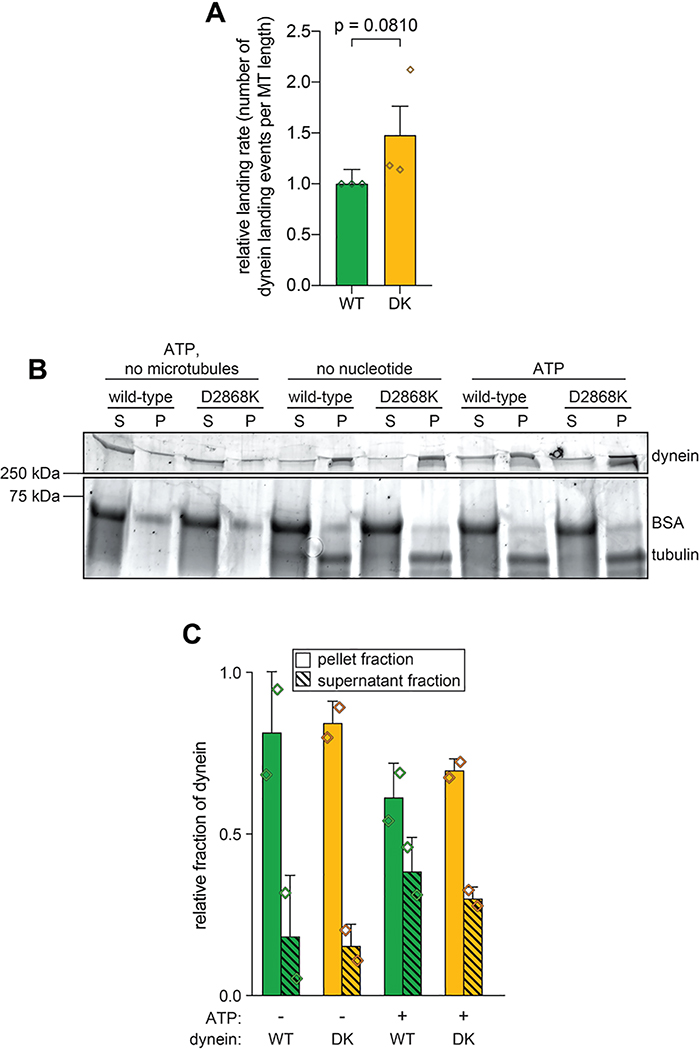
Although disruption of the human dynein phi particle increases its microtubule association4, we observed only a small increase in landing rate for the uninhibited mutant (Extended Data Fig. 4A). We noted a similarly small increase in microtubule affinity for the uninhibited mutant using a microtubule cosedimentation assay (Extended Data Fig. 4B and C). Although it is unclear why uninhibited yeast dynein does not exhibit increased microtubule binding affinity, our findings indicate that microtubule association of wild-type yeast dynein is likely not restricted by the autoinhibited conformation.
Dynein autoinhibition restricts cortical localization and dynactin association
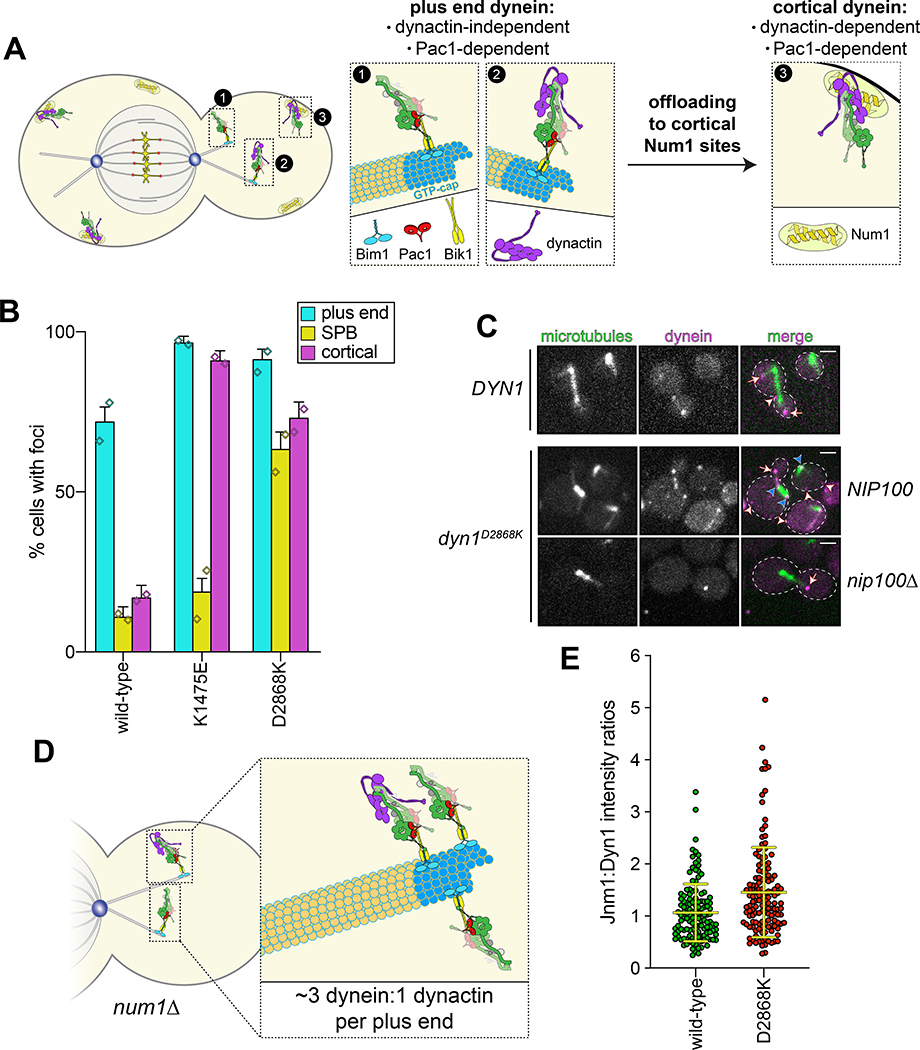
Although uninhibited human dynein mutants exhibited unchanged processivity, they were noted to have higher affinity for dynactin and BicD24. In yeast, dynein-mediated recruitment of dynactin to microtubule plus ends is required for their localization to cortical Num1 receptor sites41,42 (Fig. 3A). Thus, uninhibited dynein mutant cells would be expected to show enhanced dynactin recruitment to plus ends, and an increased frequency of dynein cortical localization. As expected, cortical dynein foci were more frequently observed in mutant cells in a manner that required dynactin (Fig. 3B and C). The dyn1D2868K cells also had a greater frequency of spindle pole body-associated dynein foci (i.e., at minus ends), where active dyneins were shown to accumulate43. Finally, ratiometric fluorescence intensity measurements revealed a significant increase in the relative ratio of dynactin to dynein at microtubule plus ends in the D2868K mutant (1.06 to 1.45; Fig. 3D and E).
Figure 3. The autoinhibited conformation restricts cellular localization of dynein and its interaction with dynactin.
(A) Cartoon depicting the two main sites of dynein localization (microtubule plus ends, and cell cortex), and the molecular requirements for each. Dynein plus end localization (1) requires Bik150 and Pac144, with Bim1 potentially playing some role in this process, but does not require dynactin41. Rather, dynactin plus end localization (2) relies on dynein41. Subsequent to plus end targeting, dynein-dynactin complexes are offloaded to cortical Num1 sites29,61 (3). (B) Plot depicting the fraction of cells (weighted mean) with indicated mutant or wild-type Dyn1–3GFP foci cells (n = 100, 90, 82 mitotic cells from two independent experiments; error bars indicate weighted standard error of proportion; diamonds represent mean values obtained from each independent replicate experiment). (C) Representative images of wild-type or mutant dynein (D2868K) localizing in otherwise wild-type or nip100Δ (dynactin component) cells. Note the lack of cortical localization of dyneinD2868K in nip100Δ cells (white arrowheads, cortical foci; white arrows, plus end foci; blue arrowheads, SPB foci; similar results were obtained from 2 independent replicates; scale bars, 2 μm). (D) Cartoon depicting the relative ratio of dynein to dynactin at microtubule plus ends (~3 dynein:1 dynactin), based on previous quantitative ratiometric imaging49. (E) Scatter plot (shown with bars depicting mean and standard deviation values) of the ratios of fluorescence intensity values for Jnm1–3mCherry (dynactin component p50/dynamitin):Dyn1–3GFP at individual microtubule plus ends and SPBs (n = 132 and 145 plus ends or SPBs, left to right, from two independent experiments). Background corrected intensity values of colocalizing Jnm1–3mCherry and Dyn1–3GFP foci were each divided to obtain individual ratio values. Measurements were taken from num1Δ cells to prevent offloading of assembled dynein-dynactin complexes from plus ends.
Peptide insertion between motor and tail domains ablates autoinhibited conformation
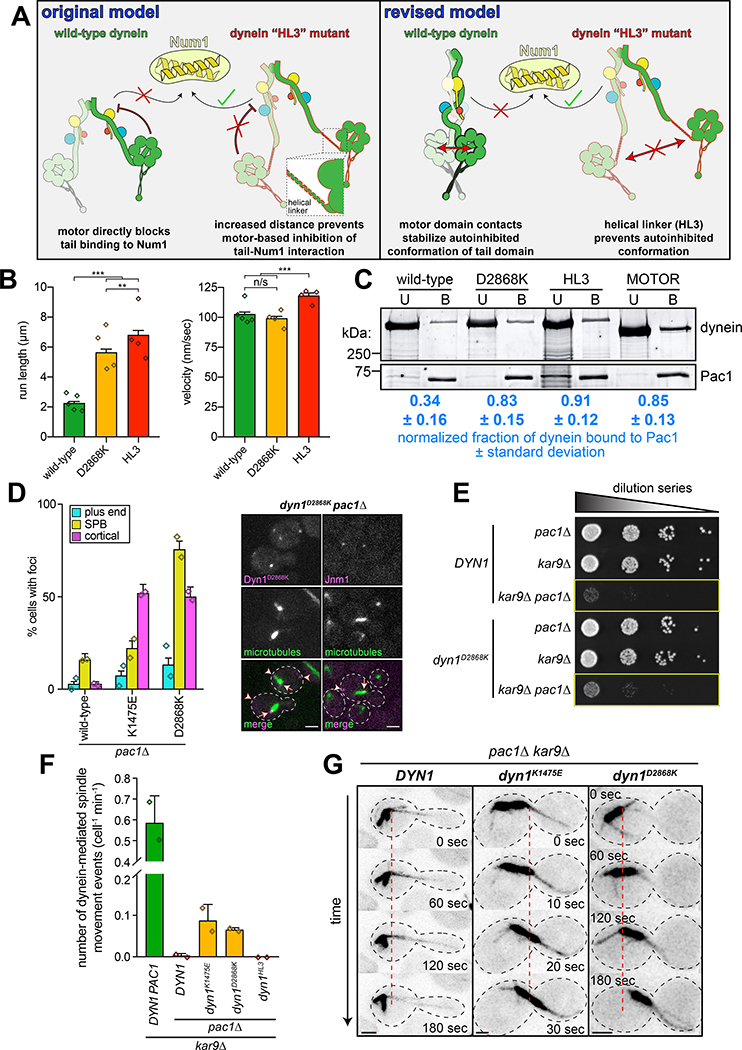
We noted that the uninhibited mutants also localized more frequently to plus ends (Fig. 3B), a pattern similar to that of a truncated dynein motor fragment (dyneinMOTOR)42, and a dynein mutant with a helical linker peptide (helical linker 3, HL3) inserted between the tail and motor domains (dyneinHL3; Fig. 4A)29. Of note, dyneinHL3 also localizes to the cell cortex more frequently than wild-type29, much like a tail domain fragment42. We originally generated the dyneinHL3 mutant29 to test the hypothesis42 that the motor domain restricts the Num1-binding capacity of the dynein tail. We predicted that the HL3 peptide would sufficiently separate the tail and motor domains such that the motor domain would no longer have this ability (i.e., would “unmask” the tail domain; Fig. 4A, “original model”). Although localization data29 supported this hypothesis, there has been no additional evidence to favor this mechanism. We wondered whether HL3 simply disrupts the autoinhibited conformation (Fig. 4A, “revised model”), which would result in an enhanced interaction between dynein, dynactin and Num1.
Figure 4. Preventing dynein autoinhibition permits Pac1/LIS1-independent localization and function.
(A) Cartoons depicting models accounting for dyneinHL3 phenotypes29. The tail domain of full-length, wild-type dynein cannot associate with Num1 in the absence of plus end-targeting; however, addition of HL3 between tail and motor domains permits plus end-targeting-independent dynein-Num1 interaction29. Our original model posited that this was due to the motor precluding the tail-Num1 interaction. Our revised model posits that motor domain contacts stabilize the autoinhibited conformation, in which the tail domains adopt a twisted state that is unable to interact with Num1. We propose that HL3 insertion prevents adoption of the autoinhibited conformation. (B) Motility assay parameters, measured from single molecules of motors purified using the strategy described in Figure 1A (n = 840 [5], 586 [4], 642 [4] motors [independent replicates], left to right; error bars indicate standard error; n/s, p = 0.1563; **, p = 0.0092; ***, p < 0.0001; diamonds, mean values obtained from independent replicates). (C) Binding assay illustrating increased affinity of Pac1 for uninhibited dyneins (see Methods). Values represent the mean corrected band intensities (n = 3, 3, 2, 3 independent experiments, left to right). (D; left) Fraction of cells (weighted mean) with mutant or wild-type Dyn1–3GFP foci in pac1Δ cells (n = 113, 108, 90 mitotic cells from two independent experiments, left to right; error bars indicate weighted standard error of proportion; diamonds represent mean values obtained from independent replicates). (right) Representative images depicting the presence of cortical dynein and dynactin (Jnm1) in dyn1D2868K pac1Δ cells (arrowheads, cortical foci; arrows, SPB foci; scale bars, 2 μm). (E) Serial dilutions of cells with indicated genotype (note differences in cell growth in yellow boxes; representative assay shown; similar results were obtained from 2 independent replicates). (F and G) Relative in vivo dynein activity with representative time-lapse images (see Methods; n = 35, 30, 30, 30 and 32 cells from two independent experiments; diamonds, mean values obtained from independent replicates; scale bars in panel G, 2 μm; dashed line provides point of reference). Also see Extended Data Figure 5 and Supplementary Video 2.
Single molecule analysis of dyneinHL3 revealed that this mutant exhibits longer run lengths than dyneinD2868K (Fig. 4B; both produced as in Fig. 1A), and velocities similar to GST-dyneinMOTOR (see Extended Data Fig. 3C), suggesting that HL3 indeed disrupts autoinhibition. We noted that the overexpressed wild-type dynein complex exhibited slightly longer runs than the non-overexpressed complex (2.3 μm versus 2.0 μm, p = 0.0049). This is likely due to a subset of the motors aggregating, as apparent from the presence of a small fraction of particles with higher fluorescence intensities (Extended Data Fig. 2D). Importantly, aggregation cannot account for the increased processivity of dyneinHL3 (Extended Data Fig. 2E and F).
We previously noted that, like dyneinMOTOR, dyneinHL3 exhibits higher affinity for Pac1 (LIS1 homolog) than wild-type dynein9,29,42. We wondered whether this is a consequence of disrupted autoinhibition. Using a co-pelleting assay, we found that Pac1 indeed preferentially binds to dyneinD2868K, dyneinHL3 and GST-dyneinMOTOR (Fig. 4C). This also indicates that the likely reason for the altered localization29 and single molecule behavior of dyneinHL3 is that it is in a constitutively uninhibited state.
Disruption of autoinhibition permits Pac1-independent cellular dynein activity
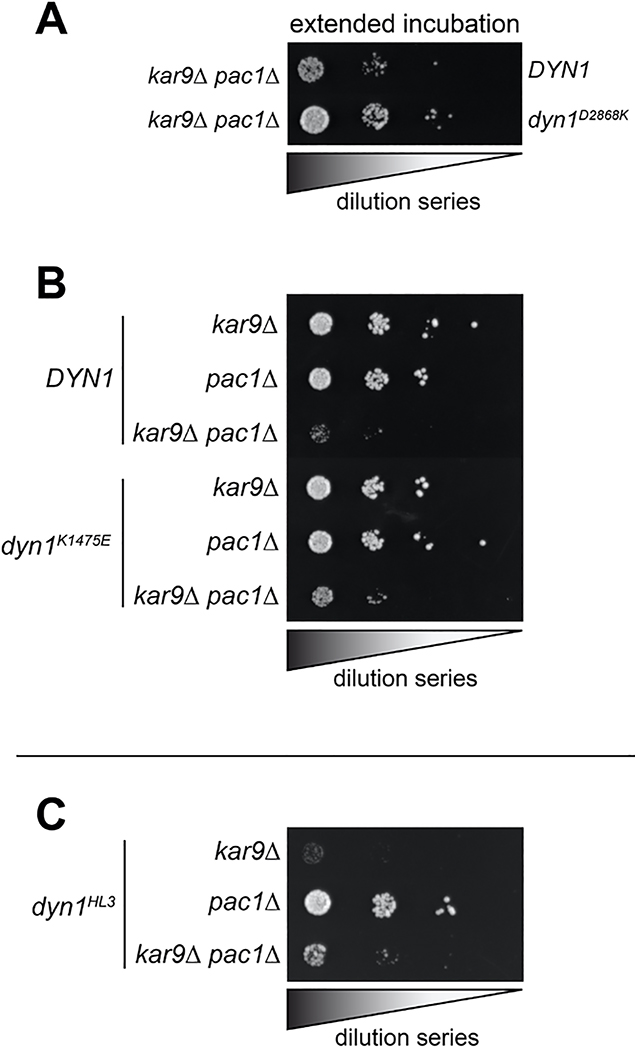
Given the similarities between the uninhibited mutants and dyneinHL3, we wondered if the Pac1-independent cortical localization of dyneinHL3 is also a consequence of uninhibition29. Pac1 is required for normal plus end and cortical localization of wild-type dynein42,44 (Fig. 4D), which is consistent with the offloading model for dynein function, in which dynein is offloaded from plus ends to cortical Num1 sites29. However, dyneinK1475E and dyneinD2868K were found at the cortex in pac1Δ cells (Fig. 4D). We also found that the uninhibited mutants partly rescued pac1Δ-dependent synthetic growth defects with kar9Δ (which effects a parallel mechanism of spindle orientation45–47), suggesting that the uninhibited dynein mutants can partly compensate for loss of Pac1 (Fig. 4E and Extended Data Fig. 5A and B). We did not observe a similar rescue for cells expressing dyneinHL3 (Extended Data Fig. 5C), suggesting that, although this mutant bypasses Pac1 for cortical localization29, and is highly processive in vitro, it is unable to move the spindle in cells.
As expected, we observed almost no dynein-mediated spindle movements in pac1Δ cells expressing either wild-type or dyneinHL3 (Fig. 4F and G). However, we did observe dynein-mediated spindle movements in pac1Δ cells expressing either dyneinK1475E or dyneinD2868K, indicating that the Pac1-independent cortical populations of these mutants are active (Fig. 4F and G, and Supplementary Video 2). Given the ability of the uninhibited dynein mutants to rescue loss of Pac1, this suggests that one function of Pac1 is to relieve dynein autoinhibition.
Pac1 stabilizes the uninhibited conformation of motile dynein complexes
To gain additional insight into the potential mechanism by which Pac1 may be affecting dynein autoinhibition we studied available structures. Docking of a Pac1-bound motor domain structure28 into one of the two motors in the autoinhibited state4 revealed an apparent clash between Pac1 and the motor domain to which Pac1 is not bound (Fig. 5A), suggesting that when Pac1 is bound to dynein, the autoinhibited conformation is sterically precluded. This accounts for the enhanced affinity of Pac1 for the uninhibited mutants, and also for dyneinMOTOR.
Figure 5. Pac1 promotes release of the autoinhibited conformation of dynein.
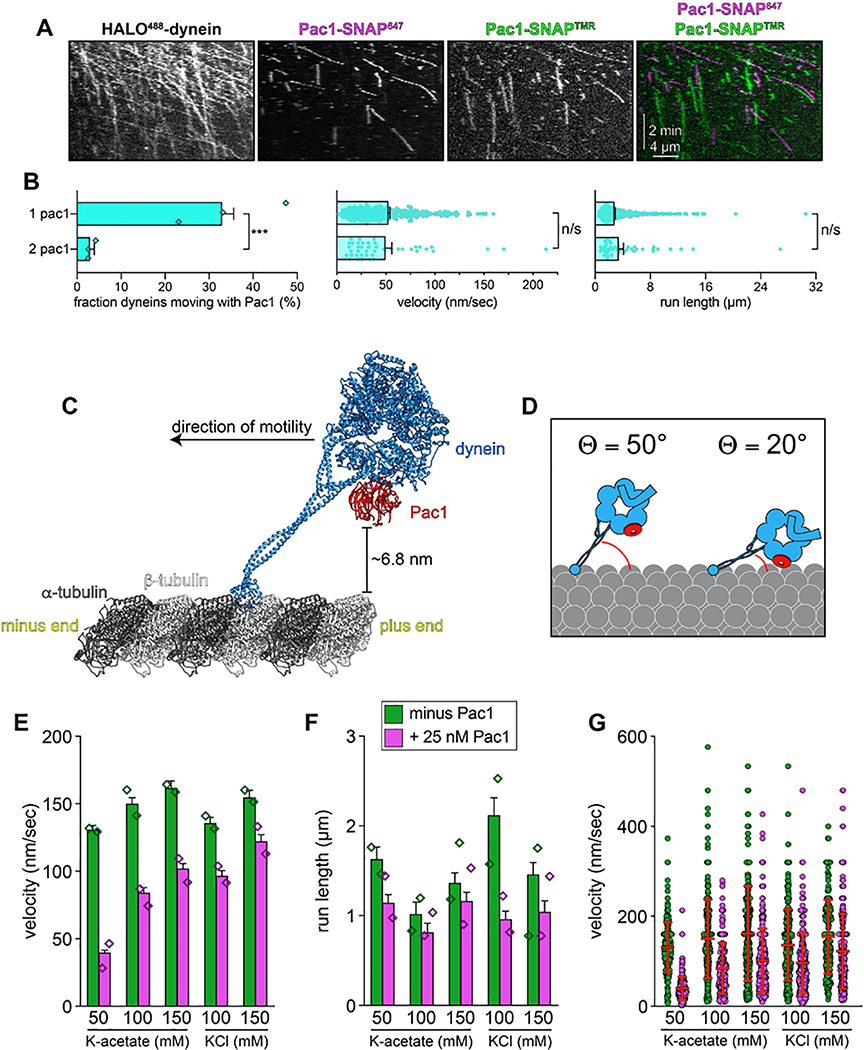
(A) Cartoon and structural model depicting steric clash between autoinhibited dynein and Pac1. Structural model was generated by aligning the Pac1-bound dynein monomer structure (pdb 5VH928) into one of the heavy chains in the phi particle structure (pdb 5NVU4). Note the steric clash (depicted with jagged yellow arrow) between Pac1 (in red) with the non-Pac1-bound heavy chain (in green). (B) Cartoon depicting experimental setup for dynein-Pac1 single molecule assay. (C) Representative kymograph illustrating comigrating dynein-Pac1 complexes in motility buffer supplemented with 150 mM potassium acetate (see panels D and E for n values). (D) Plots depicting motility parameters (left, mean run length values, from fitting of raw data to one-phase decay; right, velocity; error bars indicate standard error) of indicated dyneins moving in the absence (i.e., those not pre-incubated with Pac1, green) or presence of 25 nM Pac1 (dimer concentration). For those experiments in which Pac1 and dynein were pre-incubated, we separately scored those dyneins comigrating with Pac1 (magenta), or migrating without Pac1 (yellow; n values: wild-type, 437, 353, 553 dyneins ± Pac1 from 3 independent replicates; D2868K, 329, 664, 134 dyneins ± Pac1 from 2, 3, and 2 independent replicates, respectively; HL3, 595 and 416 dyneins ± Pac1 from 2 independent replicates; all from left to right; diamonds represent mean values obtained from each independent replicate experiment). To acquire movies of dynein alone, 1-second durations were used; however, for two-color dynein + Pac1 movies, we used 3 second durations due to the speed limitations of our microscope. Statistical significance was determined using a two-tailed Mann-Whitney test. (E) The fraction of dynein molecules migrating with Pac1 is plotted for the indicated dynein. Error bars depict standard error of proportion (n = 665, 398, and 376 dynein molecules from two independent experiments, left to right; diamonds represent mean values obtained from each independent replicate experiment). Also see Extended Data Figures 1 and 7.
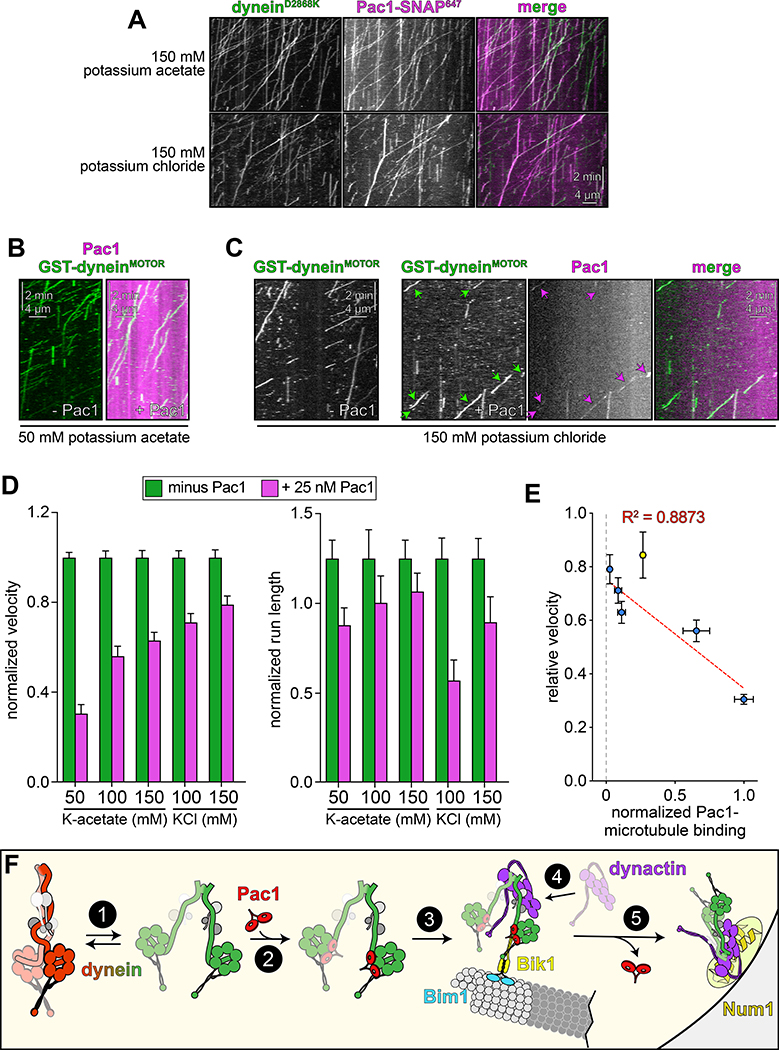
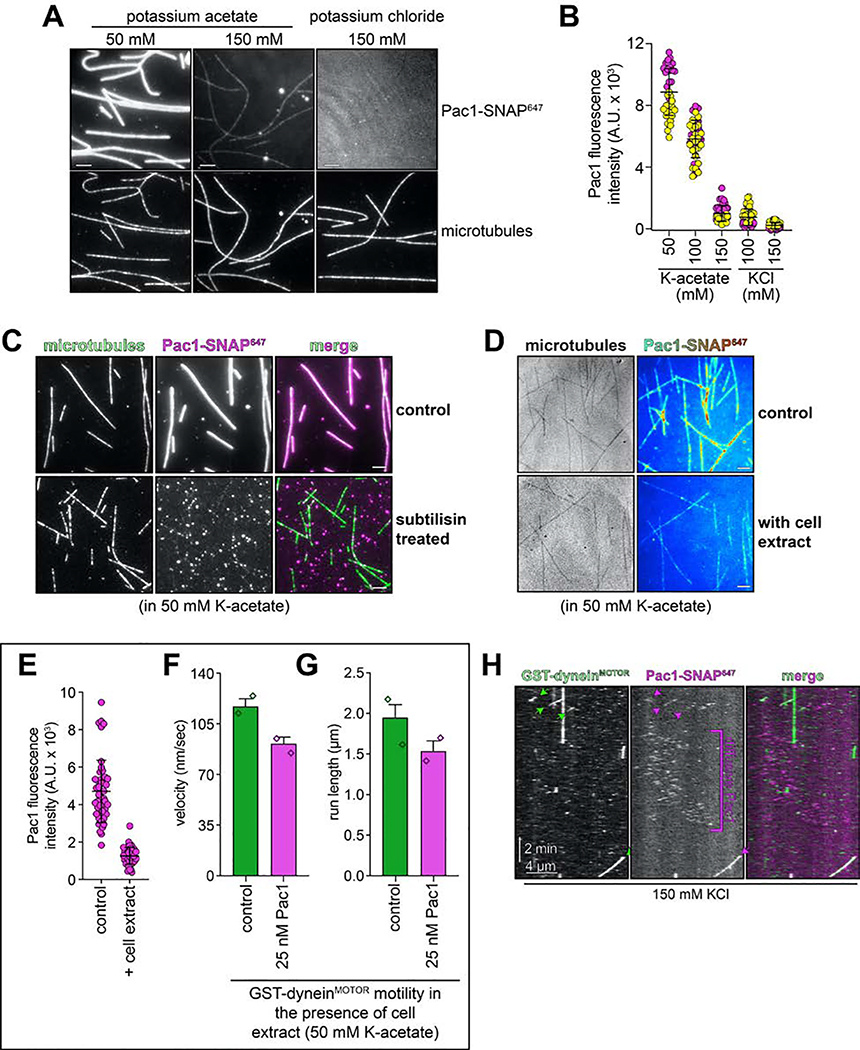
To test whether Pac1 could affect dynein autoinhibition, we sought to reassess the effect of Pac1 on dynein motility. We predicted that if Pac1 prevents autoinhibition, then it would increase dynein processivity. Studies describing the effect of Pac1 on dynein26,27,29 contrast with recent studies using human LIS123–25. Whereas Pac1 reduces dynein velocity, and promotes a microtubule-bound state26–28, LIS1 either increases the velocity of human dynein-dynactin complexes23,24, or has no effect on velocity25. In these latter studies, LIS1 was observed comigrating with dynein-dynactin at varying degrees. Thus, to clearly define how Pac1 affects dynein, we sought to assess comigrating dynein-Pac1 complexes. However, we noted that even at nanomolar concentrations, Pac1 bound extensively along microtubules in our motility buffer (Extended Data Fig. 6A). This is in stark contrast to LIS1, which does not bind microtubules as assessed by various means21,23–25. We found that the Pac1-microtubule interaction was sensitive to ionic strength: Pac1 strongly binds microtubules in buffer supplemented with 50 mM potassium acetate (conditions that have been used to assess the effect of Pac1 on dynein previously26,29), but binds to a much lesser extent in 150 mM potassium acetate (161.5 mM ionic strength; Extended Data Fig. 6A and B). We also found that Pac1-microtubule binding requires the tubulin C-terminal tails (Extended Data Fig. 6C), and can be reduced by the presence of other cellular factors (from the addition of cellular extracts; Extended Data Fig. 6D and E). To clearly assess the effect of Pac1 on dynein motility, we thus used conditions in which Pac1-microtubule binding is strongly reduced (Fig. 5B).
Two-color imaging of dynein preincubated with Pac1 revealed many instances of their comigration (Fig. 5C and E). We noted that comigrating dynein-Pac1 complexes (Fig. 5D, magenta bars) moved with significantly longer run lengths than those dyneins not comigrating with Pac1 in the same imaging chamber (yellow bars), and than those not preincubated with Pac1 (green bars). Fluorescence intensity analysis revealed that the increased dynein-Pac1 run lengths are not a consequence of dynein aggregation (Extended Data Fig. 2E and G). Consistent with the enhanced affinity of Pac1 for the uninhibited mutants, we observed a greater frequency of Pac1 comigrating with dyneinD2868K and dyneinHL3 (Fig. 5E). However, we noted only a small (D2868K) or no (HL3) Pac1-dependent run length increase for either of these mutants, further supporting the notion that they are already uninhibited, and thus refractory to Pac1 activity. These data indicate that Pac1 indeed stabilizes an uninhibited conformation of motile dynein complexes.
By incubating HALO488-dynein with an equimolar mixture of Pac1-SNAP647 and Pac1-SNAP561, we found that the large majority of dynein-Pac1 complexes possessed only one Pac1 dimer, and only a small number were bound to two (5.7% after correcting for those dynein complexes that are migrating with two Pac1s of the same color; Extended Data Fig. 7A and B). Thus, although dynein can bind two Pac1 dimers, this is a low probability event. We noted no significant difference in either run length or velocity values for 1 Pac1 versus 2 Pac1-bound dyneins (Extended Data Fig. 7B).
Pac1-mediated dynein velocity reduction requires non-specific Pac1-microtubule interaction
We next sought to clarify the prevailing model for Pac1 in dynein velocity reduction26,27,29. We hypothesized that Pac1 reduces dynein velocity as a direct consequence of its ability to bind both dynein and microtubules (as with the MAP She148). Consistent with this possibility, structural analysis revealed that Pac1 binds dynein at a region that is proximal to the microtubule (Extended Data Fig. 7C and D). Our first clue that this may be the case came from separately analyzing dynein complexes that comigrated with Pac1 versus those that did not (Fig. 5D, yellow versus magenta bars). Dynein complexes that comigrated with Pac1 moved with the same reduction in velocity as those that did not (Fig. 5D). Moreover, those dyneins moving with two Pac1 dimers did not move at a lower velocity than those bound by one (Extended Data Fig. 7B), suggesting that Pac1 binding does not directly affect dynein mechanochemistry. These data indicate that while processivity enhancement of dynein by Pac1 occurs in a manner that requires a stable dynein-Pac1 complex, velocity reduction does not.
Given the low but detectable Pac1-microtubule binding in these conditions (Extended Data Fig. 6A and B) we sought to further minimize Pac1-microtubule binding. Supplementing the buffer with 150 mM potassium chloride reduced Pac1-microtubule binding almost to background levels (Extended Data Fig. 6A, B, and H), yet did not negatively impact the Pac1-dynein interaction, as assessed from two-color imaging of dyneinD2868K and Pac1 (Fig. 6A; an identical ionic strength buffer was also used previously to assess dynein-Pac1 interaction26,27). By performing the motility assay in varying buffer conditions, we found that the degree to which Pac1 reduces dynein velocity directly correlates with the extent of Pac1-microtubule binding (Fig. 6B – D, and Extended Data Fig. 7E and G), which we further confirmed by plotting and fitting the relationship to a linear regression (Fig. 6E, blue points; R2 = 0.8873). Finally, inclusion of cell extracts in the motility chamber, which reduces Pac1-microtubule binding even in low ionic strength buffers (Extended Data Fig. 6D and E) also severely attenuated the velocity reduction effect (Fig. 6E, yellow point, and Extended Data 6F and G). Thus, in contrast to processivity enhancement by Pac1 – which occurs in a manner that is independent of Pac1-microtubule binding – velocity reduction of dynein by Pac1 occurs only when Pac1 is bound to microtubules. Taken together, these findings support a model in which Pac1 is not an inhibitor, but rather an activator of dynein.
Figure 6. Reducing Pac1-microtubule binding minimizes Pac1-mediated dynein velocity reduction.
(A) Representative kymographs depicting comigrating dyneinD2868K-Pac1 complexes in motility buffers with increased ionic strength (from 3 and 2 independent replicates, top to bottom). Note that Pac1 and dynein still interact robustly in these conditions, as apparent from the high degree of persistent colocalization. (B and C) Representative kymographs depicting different motility characteristics of GST-dyneinMOTOR (used extensively in previous Pac1 studies26–28) in the presence of Pac1 when the latter is either extensively bound to the microtubule (B), or to a much less extent (C; see panel D for n values). (D) Mean normalized motility parameters (see Extended Data Fig. 7E – G for means from independent replicates, and scatter plots) of GST-dyneinMOTOR in the absence (green) or presence (magenta) of 25 nM Pac1 (dimer concentration; n = 348, 268, 396, 226, 447, 359, 385, 315, 251, 320 motors, from two independent experiments each; left to right). Error bars indicate standard error. (E) Relative degree of Pac1-microtubule binding (mean values normalized to 1; see Extended Data Fig. 6B and E for scatter plot of intensity values, and n values for each) versus mean relative velocity of GST-dyneinMOTOR in the presence of Pac1 (mean GST-dyneinMOTOR velocity in the absence of Pac1 equals 1; see panel D, and Extended Data Figs. 6F and G, and 7E – G for relative and absolute velocity values, and n values). Blue and yellow points (error bars represent standard error) are from increasing ionic strength buffer experiment (see panel D), and the cell extract experiment (see Extended Data Fig. 6D – G), respectively. The blue points were fit to a linear regression with R2 value shown. (F) Model for dynein and Pac1 activity in cells: (1) dynein stochastically switches between closed and open states, the latter of which is stabilized by Pac1 binding (2); dynein-Pac1 associates with plus ends via direct interactions with Bik1 (3), which may rely partly on Bim1. (4) Plus end dynein-Pac1 associates with dynactin, which is then offloaded to cortical Num1 (5). Given the lack of apparent Pac1 cortical foci, Pac1 likely dissociates either concomitant with, or subsequent to dynein-dynactin offloading. Also see Extended Data Figure 7.
DISCUSSION
We propose the following model for dynein function (Fig. 6F): (1) dynein stochastically switches between autoinhibited and uninhibited states; (2) one or two Pac1 dimers bind dynein when it is in the uninhibited state, consequently preventing it from switching to the autoinhibited conformation; (3) Pac1-dynein associate with plus ends49 via Bik1/CLIP-17050 (in a manner that does not require the dynein MTBD43, as depicted); (4) uninhibited, plus end-bound dynein interacts with dynactin, where they remain inactive, possibly due to the lack of an adaptor at this site23,25,43; (5) upon encountering cortical Num1, the dynein-dynactin complex is offloaded29 and activated for motility43, possibly due to the arrangement of the motor heads in a parallel manner that is conducive for motility4. It is interesting to note that dyneinHL3 is inactive in cells, in spite of it exhibiting in vitro and localization phenotypes similar to other uninhibited mutants29. This could be a consequence of the helical linker disrupting the adoption of the parallel head configuration that may be needed for in vivo activity4.
Our work indicates that Pac1 promotes dynein activity by stabilizing the uninhibited state, and that prior observations of a velocity reduction phenotype26–29 are likely due to the low ionic strength buffers used in these assays. Although we observe a small effect of Pac1 on dynein velocity even at higher ionic strengths, this is likely due to residual Pac1-microtubule-binding (e.g., Extended Data Fig. 6H). These data challenge the current model for Pac1 activity, whereby its binding to the motor domain sterically blocks dynein’s mechanochemical cycle27, thereby promoting its association with microtubule plus ends26–28 (the latter of which is also contradicted by the dispensable nature of the dynein MTBD for plus end localization43). Our data indicate that Pac1 reduces dynein velocity in low ionic strength buffers by exerting drag on dynein via simultaneous contacts with dynein and microtubules, similar to observations with the MAP She148. This raises the question of whether microtubule-binding by Pac1 is a relevant activity in cells. Several lines of evidence indicate this is not the case. Pac1 only localizes to plus ends in cells, and not along microtubules44,49,51. Its plus end binding is indirect, as it relies on dynein49 and Bik1/CLIP-17049,51. Finally, studies have observed no microtubule-binding activity of LIS121,23–25. In fact, in contrast to an inhibitory function, two of these studies observed a LIS1-dependent dynein-dynactin velocity increase23,24, for reasons now determined by two new studies published in this issue52,53: by stabilizing uninhibited dynein, LIS1 promotes assembly of faster 2 dynein:1 dynactin complexes54. Several studies demonstrate the conserved nature of the autoinhibitory mechanism6,7, including a recent study in Aspergillus nidulans55.
Our findings raise interesting questions regarding the distinct motility capabilities of yeast versus human dynein, the latter of which requires dynactin and an adaptor for processive motility1,2. In particular, why does yeast dynein not need such factors in spite of its high propensity to adopt the autoinhibited state? Perhaps the yeast dynein motor domains are more likely to orient in a parallel configuration4 in the absence of dynactin binding? Higher resolution structural data will be required to determine if this is indeed the case.
METHODS
Media and strain construction
Strains are derived from either W303 or YEF473A and are available upon request. We transformed yeast strains using the lithium acetate method. Strains carrying mutations were constructed by PCR product-mediated transformation or by mating followed by tetrad dissection. Proper tagging and mutagenesis was confirmed by PCR, and in most cases sequencing (all point mutations were confirmed via sequencing). Fluorescent tubulin-expressing yeast strains were generated using common strategies. Strains overexpressing the yeast dynein complex (or the GST-dyneinMOTOR fragment) were generated by transforming p8His-ZZ-SNAPf-Dynein, p8His-ZZ-HALO-Dynein, or pZZ-dyneinMOTOR-HALO (wild-type or mutants; see below) linearized by digestion with ApaI (cuts within the URA3 gene; see Fig. 1A). Integration was confirmed by PCR. Yeast synthetic defined (SD) media was obtained from Sunrise Science Products (San Diego, CA).
Plasmid generation
For overexpression and purification of the yeast dynein complex (wild-type or mutants), we generated a polycistronic plasmid expressing all four dynein complex subunits using strategies analogous to the biGBAC assembly62. We first made a yeast expression “library” vector – pLIBy – which enables generation of a gene expression cassettes (GEC) with a strong, inducible GAL1 promoter (GAL1p) on the 5’ end, and a synthetic terminator sequence (Tsynth356) on the 3’ end. A PCR product encompassing GAL1p, and an oligonucleotide containing Tsynth3 and a multicloning site (XbaI-NotI-SpeI-BamHI) were assembled into pRS305 digested with BamHI and NotI using Gibson assembly, yielding pLIBy. We also generated a yeast genomic-integration vector with optimized linker sequences for Gibson assembly62 flanked by PmeI restriction sites (equivalent to pbiG1a and pbiG1b). These plasmids – pbiG1ay and pbiG1by– were generated by using Gibson assembly to insert a PCR product encompassing these elements from pbiG1a and pbiG1b62 into pRS306. PCR products encompassing the DYN2 (without the native intron), DYN3 or PAC11 open reading frames were assembled into pLIBy digested with BamHI and NotI. Subsequently, these GECs were amplified from each respective pLIBy vector using oligonucleotides that include regions for priming preceded on the 5’ end by predefined “Cas” sequences62: the DYN2 GEC was amplified with Casα-forward and Casβ-reverse; the DYN3 GEC was amplified with Casβ-forward and Casγ-reverse; and, the PAC11 GEC was amplified with Casγ-forward and Casω-reverse. These three PCR products were assembled into pbiG1by digested with SwaI to generate pbiG1by:GAL1p:Dyn2::GAL1p:DYN3::GAL1p:PAC11.
We generated pLIBy:6His-StrepII-SNAPf-DYN1 using Gibson assembly. However, due to complications generating a PCR product from this vector, we chose to clone everything into this vector. We first substituted the LEU2 expression cassette in the pLIBy backbone with a URA3 marker by assembling a PCR product encompassing the URA3 cassette from pRS306 into pLIBy:6His-StrepII-SNAPf-DYN1 digested with KasI and AatII, yielding pLIBy:6His-StrepII-SNAPf-DYN1::URA3. To enable assembly of the DYN2/DYN3/PAC11 polygene cassette into pLIBy:6His-StrepII-SNAPf-DYN1::URA3, we inserted the optimized “B” and “C” linker sequences for Gibson assembly62 into this plasmid by assembling a PCR product encompassing “B”-PmeI site-“C” into pLIBy:6His-StrepII-SNAPf-DYN1::URA3 digested with KpnI and SalI. Subsequent to digestion with PmeI, this plasmid was assembled with the PmeI restriction digest product from pbiG1by:GAL1p:Dyn2::GAL1p:DYN3::GAL1p:PAC11 (encompassing GAL1p:Dyn2::GAL1p:DYN3::GAL1p:PAC11), yielding pLIBy: GAL1p:Dyn2::GAL1p:DYN3::GAL1p:PAC11::GAL1p:6His-StrepII-SNAPf-Dyn1::URA3, hereafter referred to as p6His-StrepII-SNAPf-Dynein. Prior to using this plasmid for pilot tests, we decided to swap the 6His-StrepII affinity tag for an 8His-ZZ tag (followed by a tandem TEV protease recognition site). We did this by assembling a PCR product encompassing 8His-ZZ into p6His-StrepII-SNAPf-Dynein digested with AatII and XhoI, yielding p8His-ZZ-SNAPf-Dynein. We replaced the SNAPf tag with a HALO tag using a similar strategy, yielding p8His-ZZ-HALO-Dynein. All mutations were engineered into these plasmids using common strategies.
Due to difficulties in engineering the D2868K mutation into the GST-dyneinMOTOR-expressing yeast strain (SMY1008), we generated a plasmid that overexpresses this fragment in yeast (under control of the GAL1 promoter; see Extended Data Figure 3A). Like the full-length dynein complex expression plasmids described above, this plasmid can also be integrated into the ura3–1 locus. PCR products encompassing the entire GAL1p:ZZ-2TEV-6His-GFP-GST-dyn1MOTOR-HALO cassette (comprised of 4 individual pieces, each with 23 nt of overlapping sequences; amplified from SMY1008) were assembled into pRS305 digested with KpnI and BamHI using Gibson assembly, yielding pZZ-dyneinMOTOR-HALO. The D2868K mutation was engineered into this plasmid using common strategies.
Protein purification
Purification of yeast dynein (ZZ-TEV-Dyn1-HALO, under the native DYN1 promoter; or, ZZ-TEV-HALO-(or SNAPf)-Dynein, with all genes under control of the GAL1p promoter; or, ZZ-TEV-6His-GFP-3HA-GST-dyneinMOTOR-HALO, under the control of the GAL1p promoter) was performed as previously described with minor modifications used for the overexpressed complex48. Briefly, yeast cultures were grown in YPA supplemented with either 2% glucose (for non-overexpressed full-length dynein) or 2% galactose (for the GAL1p-inducible strains; overexpression of the full-length dynein complex was induced for no more than 3 hours), harvested, washed with cold water, and then resuspended in a small volume of water. The resuspended cell pellet was drop frozen into liquid nitrogen and then lysed in a coffee grinder (Hamilton Beach). For most purifications (with exception of those used for negative stain/EM imaging) we used the following procedure: after lysis, 0.25 volume of 4X dynein lysis buffer (1X buffer: 30 mM HEPES, pH 7.2, 50 mM potassium acetate, 2 mM magnesium acetate, 0.2 mM EGTA) supplemented with 1 mM DTT, 0.1 mM Mg-ATP, 0.5 mM Pefabloc SC (concentrations for 1X buffer) was added, and the lysate was clarified at 22,000 × g for 20 min. The supernatant was then bound to IgG sepharose 6 fast flow resin (GE) for 1–1.5 hours at 4°C, which was subsequently washed three times in 5 ml lysis buffer, and twice in 5 ml TEV buffer (50 mM Tris, pH 8.0, 150 mM potassium acetate, 2 mM magnesium acetate, 1 mM EGTA, 10% glycerol) supplemented with 0.005% Triton X-100, 1 mM DTT, 0.1 mM Mg-ATP, and 0.5 mM Pefabloc SC. To fluorescently label the motors for single molecule analyses, the bead-bound protein was incubated with either 6.7 μM HaloTag-AlexaFluor660, HalotTag-AlexaFluor488, or HaloTag-TMR (Promega), or SNAP-Surface Alex Fluor 647 (NEB), as appropriate, for 10–20 minutes at room temperature. The resin was then washed four more times in TEV buffer supplemented with 1 mM DTT, 0.005% Triton X-100, and 0.1 mM Mg-ATP, and then incubated with TEV protease for 1–1.5 hours at 16°C. Following TEV digest, the beads were pelleted, and the resulting supernatant was collected, aliquoted, flash frozen in liquid nitrogen, and stored at −80°C. Protein preparations used for negative stain/EM imaging were subject to tandem affinity purification. To do so, subsequent to lysis, 0.25 volume of 4X NiNTA dynein lysis buffer (1X buffer: 30 mM HEPES, pH 7.2, 200 mM potassium acetate, 2 mM magnesium acetate, 10% glycerol) supplemented with 1 mM beta-mercaptoethanol, 0.1 mM Mg-ATP, 0.5 mM Pefabloc SC (concentrations for 1X buffer) was added, and the lysate was clarified as above. The supernatant was then bound to NiNTA agarose for 1 hour at 4°C, which was subsequently washed three times in 5 ml NiNTA lysis buffer. The protein was eluted in NiNTA lysis buffer supplemented with 250 mM imidazole by incubation for 10 minutes on ice. The eluate was then diluted with an equal volume of dynein lysis buffer, which was then incubated with IgG sepharose 6 fast flow resin for 1 hour at 4°C. The beads were washed and the protein was eluted as described above. Eluted protein was either applied to a size exclusion resin (Superose 6; GE), or snap frozen. The gel filtration resin was equilibrated in GF150 buffer (25 mM HEPES pH 7.4, 150 mM KCl, 1 mM MgCl2, 5 mM DTT, 0.1 mM Mg-ATP) using an AKTA Pure. Peak fractions (determined by UV 260 nm absorbance and SDS-PAGE) were pooled, concentrated, aliquoted, flash frozen, then stored at −80°C. We noted that prolonged periods of storage in the −80°C (≥ 3–5 days) led to an increase in the prevalence of dynein aggregates (as apparent from fluorescence intensity analysis), which exhibited longer run lengths in single molecule assays. We also noted that aggregation was much more prevalent when the motor was labeled with the HALO-AlexaFluor660 than it was with the HALO-AlexaFluor488.
Purification of Pac1-FLAG-SNAP was performed as previously described26, with the addition of a gel filtration step to remove any residual, unbound fluorescent dye. Specifically, TEV protease eluted protein was applied to a size exclusion resin (Superose 6; GE), that was equilibrated in TEV buffer supplemented with 1 mM DTT using an AKTA Pure. Peak fractions (determined by UV 260 nm absorbance and SDS-PAGE) were pooled, concentrated, aliquoted, flash frozen, then stored at −80°C.
For comparison of elution profiles between yeast and human dynein complexes, the human dynein complex was expressed and purified from insect cells (ExpiSf9 cells; Life Technologies) as previously described with minor modifications1. Briefly, 4 ml of ExpiSf9 cells at 2.5 × 106 cells/ml, which were maintained in ExpiSf CD Medium (Life Technologies), were transfected with 1 μg of bacmid DNA (see above) using ExpiFectamine (Life Technologies) according to the manufacturer’s instructions. 5 days following transfection, the cells were pelleted, and 1 ml of the resulting supernatant (P1) was used to infect 300 ml of ExpiSf9 cells (5 × 106 cells/ml). 72 hours later, the cells were harvested (2000 × g, 20 min), washed with phosphate buffered saline (pH 7.2), pelleted again (1810 × g, 20 min), and resuspended in an equal volume of human dynein lysis buffer (50 mM HEPES, pH 7.4, 100 mM NaCl, 10% glycerol, 1 mM DTT, 0.1 mM Mg-ATP, 1 mM PMSF). The resulting cell suspension was drop frozen in liquid nitrogen and stored at −80°C. For protein purification, 30 ml of additional human dynein lysis buffer supplemented with cOmplete protease inhibitor cocktail (Roche) was added to the frozen cell pellet, which was then rapidly thawed in a 37°C water bath prior to incubation on ice. Cells were lysed in a dounce-type tissue grinder (Wheaton) using ≥ 150 strokes (lysis was monitored by microscopy). Subsequent to clarification at 22,000 × g, 45 min, the supernatant was applied to 2 ml of IgG sepharose fast flow resin pre-equilibrated in human dynein lysis buffer, and incubated at 4°C for 2–4 hours. Beads were then washed with 50 ml of human dynein lysis buffer, and 50 ml of human dynein TEV buffer (50 mM Tris pH 7.4, 150 mM potassium acetate, 2 mM magnesium acetate, 1 mM EGTA, 10% glycerol, 1 mM DTT, 0.1 mM Mg-ATP). The bead-bound protein was eluted with by incubation with TEV protease overnight at 4°C. The next morning, the recovered supernatant was applied to a Superose 6 gel filtration column as above.
Single molecule motility assays
The yeast dynein single-molecule motility assay was performed as previously described with minor modifications48. Briefly, flow chambers constructed using slides and plasma cleaned and silanized coverslips attached with double-sided adhesive tape were coated with anti-tubulin antibody (8 μg/ml, YL1/2; Accurate Chemical & Scientific Corporation) then blocked with 1% Pluronic F-127 (Fisher Scientific). Taxol-stabilized microtubules assembled from unlabeled and fluorescently-labeled porcine tubulin (10:1 ratio; Cytoskeleton) were introduced into the chamber. Following a 5–10 minute incubation, the chamber was washed with dynein lysis buffer (see above) supplemented with 20 μM taxol. Subsequently, purified dynein motors diluted in motility buffer (30 mM HEPES pH 7.2, 2 mM magnesium acetate, 1 mM EGTA, 1 mM DTT, 1 mM Mg-ATP, 0.05% Pluronic F-127, 20 μM taxol, and an oxygen-scavenging system consisting of 1.5% glucose, 1 U/ml glucose oxidase, 125 U/ml catalase) supplemented with either 50 mM potassium acetate, or as indicated in figure legend, were introduced in the chamber, and imaged.
To image comigrating Pac1-dynein complexes, 500 nM Pac1-SNAP647 (dimer concentration) and 10–50 nM HALOTMR-Dynein were preincubated on ice for 10–15 minutes prior to a 20-fold dilution into modified motility buffer (30 mM HEPES pH 7.2, 2 mM magnesium acetate, 1 mM EGTA, 1 mM DTT, 1 mM Mg-ATP) supplemented with potassium acetate or potassium chloride as indicated in figure legends, 0.05% Pluronic F-127, 20 μM taxol, and an oxygen-scavenging system (as above). The higher yield overexpressed dynein complex was needed for these assays given the low landing rate of dynein in the higher ionic strength buffers. We ensured that comigrating Pac1-SNAP647 spots were not due to bleed-through from the HALOTMR-dynein channel by performing two-color imaging with HALOTMR-dynein alone (no spots were apparent in the far-red channel in these cases). To measure Pac1 fluorescence intensity on microtubules, 50 nM of Pac1-SNAP647 (dimer concentration) diluted in motility buffer was added to chambers containing taxol-stabilized microtubules.
To remove the unstructured E-hooks from microtubules, taxol-stabilized microtubules were digested with a 1 mg/ml subtilisin (Sigma) for 90 min at 37°C prior to preparation of flow chambers. For experiments in which cellular extracts were included in the imaging chamber (Extended Data Fig. 6D and E), log phase cultures of SMY2532 (cells not expressing dynein; see Supplementary Table 1) were pelleted, resuspended in a small volume of motility buffer (with 50 mM potassium acetate), drop frozen in liquid nitrogen, lysed by mortar and pestle, and then clarified at 21,000 × g for 15 minutes. To assess Pac1-microtubule binding in the presence of cell extracts, 50 nM Pac1-SNAP647 (dimer concentration) was diluted in motility buffer supplemented with clarified extract (0.96 mg/ml final) and introduced into an imaging chamber. Similar conditions were used to assess the effect of Pac1 on dynein motility in the presence of cell extract, except 25nM Pac1 and ~50 pM GST-dyneinMOTOR were also included in the imaging chamber; moreover, ATP levels were increased to 10 mM (compared with 1 mM, used for the other single molecule assays).
Total internal reflection fluorescence microscopy (TIRFM) images were collected using a 1.49 NA 100X TIRF objective on a Nikon Ti-E inverted microscope equipped with a Ti-S-E motorized stage, piezo Z-control (Physik Instrumente), and an iXon X3 DU897 cooled EM-CCD camera (Andor). 488 nm, 561 nm, and 640 nm lasers (Coherent) were used along with a multi-pass quad filter cube set (C-TIRF for 405/488/561/638 nm; Chroma) and emission filters mounted in a filter wheel (525/50 nm, 600/50 nm and 700/75 nm; Chroma). To image nonfluorescent microtubules (e.g., Extended Data Fig. 6D), we employed interference reflection microscopy, as recently described63. For time-lapse movies, we acquired images at 1, 2, or 3 second intervals for 8–10 min. Velocity and run length values were determined from kymographs generated using the MultipleKymograph plugin for ImageJ (http://www.embl.de/eamnet/html/body_kymograph.html). Those motors that moved for ≥ 3 time points were measured. Reported run lengths were determined from fitting raw values to a one-phase decay.
Negative stain electron microscopy and image analysis
EM grids were prepared with a standard negative stain protocol by applying fresh dynein samples to glow discharged carbon coated 200 mesh copper grids. After ~1 minute incubation, 2% uranyl acetate was added. 1600 micrographs were collected on a FEI Tecnai F20 200kV TEM equipped with a Gatan US4000 CCD (model 984), at a nominal magnification of 90,000X with the digital pixel size 6.19 angstroms. All image analysis was performed in Relion 3.0 on the University of Colorado Boulder High Performance Computer Cluster, Summit. Particles were manually picked from ~20 micrographs (~200 particles), which were used to generate a low resolution 2D class average. Using these 2D averages as a starting point, we then used an iterative process to autopick particles that were used to generate our final 2D averages, and for 3D model building (in total, 42,611 particles were used for final averages shown in Figure 1D).
Calculation of ionic strength
We calculated the ionic strength of our buffers as previously described64. In brief, we empirically determined the amount of KOH required to pH a 30 mM HEPES buffer to 7.2. We estimated the proportion of protonated to unprotonated HEPES at pH 7.2 to be 2:1 (using the Henderson-Hasselbalch equation). Using the formulation for ionic strength (I = 1/2 Σ ci zi2), we determined the ionic strength of the 50 mM K-acetate motility buffer to be 61.5 mM, the 100 mM K-acetate and KCl buffers to be 111.5 mM, and the 150 mM K-acetate and KCl buffers to be 161.5 mM. Note we did not include the proportion of zwitterionic HEPES species (20 mM) in the calculation of ionic strength as it has been demonstrated previously to have no effect on the ionic strength of a solution, despite contributing to electrostatic interactions65.
Dynein-Pac1 binding experiments
Purified, gel filtered Pac1-FLAG-SNAP (0.5 – 0.75 μg per binding experiment) was bound to FLAG M2 magnetic beads (Sigma Aldrich) by incubation in TEV supplemented with 0.005% Triton X-100, 1 mM DTT, and 0.1 mM Mg-ATP at 4°C. After 1 hour, unbound Pac1-FLAG-SNAP was removed by washing the beads four times with in the same buffer. Subsequently, roughly equal amounts of purified dynein proteins (wild-type, mutant, or truncation) were incubated with Pac1-FLAG-SNAP-decorated beads (diluted in same buffer; reaction volume ranged from 50 – 120 μl among replicates). After 1 hour, the unbound fraction was removed by pipetting, while the bound fraction was eluted with 0.25 mg/ml 3XFLAG peptide (Sigma Aldrich) by incubation for 20 minutes on ice. Bound and unbound fractions were resolved by SDS-PAGE, and the normalized, relative bound and unbound fractions were determined by measuring background corrected band intensities. Note that we observed no binding of dynein to FLAG M2 magnetic beads in the absence of Pac1-FLAG-SNAP (not shown).
Microtubule copelleting experiments
To perform the microtubule copelleting assay, 1 μM microtubules were incubated for 10 minutes at room temperature with 2 nM of either wild-type or mutant dynein in motility buffer supplemented with 0.1 mg/ml bovine serum albumin (Bio-Rad, cat. # 5000206) with or without 1 mM Mg-ATP. Reactions were subsequently pelleted at 21,130 × g for 20 minutes. The supernatant and pellet were separated, resuspended in sample buffer, and run on a 4–15% gradient acrylamide gel. Gels were stained with Sypro Ruby, and then imaged on a Typhoon gel imaging system (FLA 9500).
Live cell imaging experiments
For the spindle dynamics assay, cells were arrested with hydroxyurea (HU) for 2.5 hours, and then mounted on agarose pads containing HU for fluorescence microscopy32. Full Z-stacks (15 planes with 0.2 μm spacing) of GFP-labeled microtubules (GFP-Tub1) were acquired every 10 seconds for 9.66 minutes (58 time points) on a stage prewarmed to 30°C. To eliminate any dynein-independent contributions to spindle movements, these assays were performed in cells lacking Kar9, a protein that is required for an actin/myosin-mediated spindle orientation pathway45–47. To image dynein localization in live cells, cells were grown to mid-log phase in SD media supplemented with 2% glucose, and mounted on agarose pads. Images were collected on a Nikon Ti-E microscope equipped with a 1.49 NA 100X TIRF objective, a Ti-S-E motorized stage, piezo Z-control (Physik Instrumente), an iXon DU888 cooled EM-CCD camera (Andor), a stage-top incubation system (Okolab), and a spinning disc confocal scanner unit (CSUX1; Yokogawa) with an emission filter wheel (ET480/40m for mTurquoise2, ET525/50M for GFP, and ET632/60M for mRuby2; Chroma). Lasers (445 nm, 488 nm and 561 nm) housed in a LU-NV laser unit equipped with AOTF control (Nikon) were used to excite mTurquoise2, GFP and mRuby2, respectively. The microscope was controlled with NIS Elements software (Nikon).
Statistics and Reproducibility
All data were collected from at least two independent replicates (independent protein preparations, or cell cultures, for in vitro and in vivo experiments, respectively). The values from each independent replicate – which are indicated on each plot (see diamonds on relevant plots) – showed similar results. T-tests were performed using Graphpad Prism. Statistical significance was determined using a two-tailed Mann-Whitney test (for single molecule run length values), or with an unpaired two-tailed Welch’s t test (for single molecule velocity values). Z scores were calculated using the following formula:
where:
Z scores were converted to two-tailed P values using an online calculator.
Data availability
All yeast strains, datasets, and raw movie files generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Extended Data
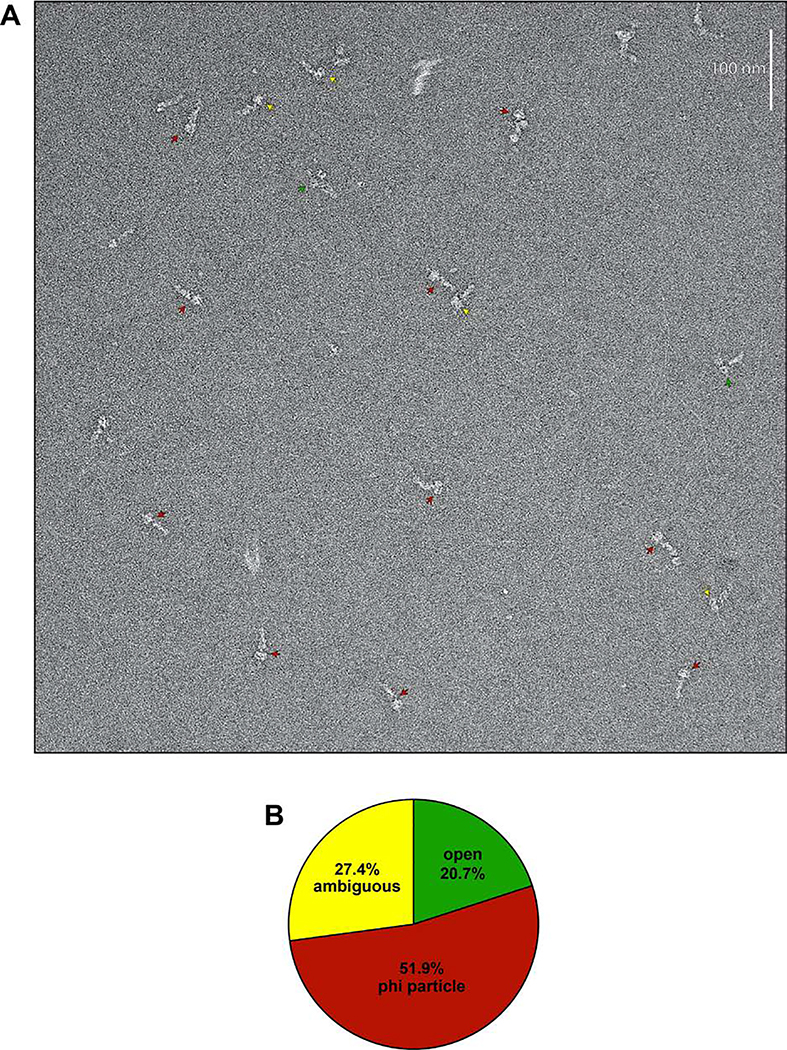
Extended Data Figure 1. Representative raw EM image and quantitation of conformational states.
(A) Representative EM image of negative stained yeast dynein complex (red arrow, phi particle conformation; green arrow, open, uninhibited conformation; yellow arrow, ambiguous). (B) Quantitation of indicated conformational states from raw images (n = 435 particles).
Extended Data Figure 2. Fluorescent intensity analysis of native and overexpressed single molecules of dynein.
Histogram of fluorescence intensity values for single molecules of motile dyneins, as indicated, along with accompanying Gaussian fits and modeled parameters (determined using the model-based clustering algorithm Mclust66). The percentages reflect the relative proportion of molecules that fall within each component (i.e., for mean 1, and mean 2). The two mean values for each likely represent single-labeled (mean 1) and dual-labeled (mean 2) dynein dimers, respectively. Red outlined region in panel D (“aggregates”), delineate particles with ~3-fold higher fluorescence intensity values than the single labeled complexes.
Extended Data Figure 3. D2868K mutation has no effect on GST-dyneinMOTOR motility.
(A) Schematic of the plasmid used to produce GST-dyneinMOTOR (wild-type and D2868K mutant). Restriction digest with ApaI (cuts within URA3 gene) targets the plasmid for homologous recombination into the ura3–1 locus as depicted. (B) Cartoon representation of the minimal GST-dimerized dynein motor domain (amino acids 1219–4092 of the dynein heavy chain, Dyn1). (C and D) Plots depicting mean values (left) and all values (right) for velocity (C) and run length (D) of wild-type and D2868K GST-dyneinMOTOR, along with the standard error (n = 217 and 238 motors for each).
Extended Data Figure 4. Uninhibited dynein mutant exhibits only moderate increase in microtubule landing activity.
(A) Plots depicting relative microtubule landing rate of full length wild-type (WT) and D2868K (DK) dynein, as measured from single molecule motility experiments (mean ± standard deviation; n = 554 wild-type motors from 1532 μm of microtubules, and 553 D2868K motors from 1177 μm of microtubules; 3 independent experiments were quantitated for each). Diamonds represent mean normalized values obtained from each independent replicate experiment. Briefly, equivalent concentrations of full length wild-type or D2868K dynein were added to imaging chambers (after taking into account relative differences in labeling efficiencies, as determined from fluorescent scans of protein gels), and the number of moving motors were quantitated. Statistical significance was determined using a two-tailed Welch’s t test. (B and C) Representative gel (B; Sypro Ruby-stained) and quantitation (C) of microtubule co-sedimentation assay with full length wild-type (WT) and D2868K (DK) dynein done in the absence and presence of ATP (mean ± standard deviation; n = 2 independent experiments; diamonds represent values obtained from each replicate). Relative microtubule binding was determined by measuring background-corrected band intensities of each, and subtracting any non-specific microtubule-independent pelleting (as determined from experiment performed in the absence of microtubules).
Extended Data Figure 5. Synthetic interactions between dynein mutants and Kar9.
(A-C) Serial dilutions of cells with indicated genotype were plated on rich media (YPA supplemented with 2% glucose) and grown at 30°C for 2–4 days (A, extended incubation of plates shown in Figure 4E were grown for 4 days; all others were incubated for 2 days). Note the severe growth defects in dyn1HL3 kar9Δ cells (in panel C), suggesting that dyneinHL3 is not active in cells. Note that similar results were obtained from 2 independent replicates.
Extended Data Figure 6. Pac1-microtubule binding behavior, and its contribution to dynein activity.
(A and B) Representative images (A) and intensity scatter plots (B; bars depict mean ± standard deviation) of microtubule-bound Pac1 in different buffers. Pac1-SNAP647 diluted in motility buffer (50 nM, dimer concentration) with indicated salts was introduced into a chambers with coverglass-adhered microtubules, and images were acquired (yellow and magenta circles represent data acquired from each independent experiments; n = 38, 49, 41, 49, and 45 microtubules that span 911 μm, 1074 μm, 1077 μm, 906 μm, 1017 μm in length for each condition, left to right). (C) Pac1-microtubule binding is reduced after enzymatic removal of the unstructured tubulin carboxy-terminal tails (see Methods; similar results were obtained from 2 independent experiments). (D - G) Addition of cell extracts reduces Pac1-microtubule binding, and attenuates Pac1-mediated dynein velocity reduction. Representative fluorescence images of Pac1-SNAP647 on microtubules (D; Pac1-SNAP647 shown as a heat map) and scatter plots depicting intensity values (E; bars depict mean ± standard deviation; n= 48 and 47 microtubules that span 890 μm and 841 μm in length for each condition, left to right; similar results were obtained from 2 independent experiments). (F and G) Plots depicting the motility properties for GST-dyneinMOTOR in the absence and presence of 25 nM Pac1 (dimer concentration) in low ionic strength buffer (50 mM potassium acetate) in the presence of cell extracts (0.96 mg/ml final; 275 and 258 motors, left to right, from 2 independent experiments were quantitated). Note the small Pac1-mediated GST-dyneinMOTOR velocity reduction in the presence of cell extracts (22.1%, compared to 69.5% in the absence of extracts). (H) Additional representative kymograph of GST-dyneinMOTOR comigrating with Pac1 in buffer with 150 mM KCl (see Fig. 6D and Extended Data Fig. 7E – G for quantitation and statistics). Note the diffusive behavior of Pac1 on microtubules. Scale bars in panels A, C and D, 4 μm.
Extended Data Figure 7. Assessment of dynein-Pac1 stoichiometry, structural analysis of the Pac1-dynein-microtubule complex, and additional motility plots.
(A and B) Representative kymographs (A) and quantitation (B) of dynein-Pac1/2xPac1 complex motility (in motility buffer supplemented with 120 mM potassium acetate; n = 870 dynein molecules from 3 independent experiments; mean values with standard error are shown, along all datapoints for middle and right plots; similar results were obtained from each replicate). Statistical significance was determined by calculating Z scores (left; ***, p < 0.0001), using a two-tailed Mann-Whitney test (middle; p = 0.6068), or with a two-tailed Welch’s t test (right; p = 0.6581). Note the estimated fraction of dynein-2xPac1 complexes (5.7%; see main text) is less than what would be expected if there was no cooperativity for Pac1-dynein binding (i.e., the product of the probabilities of two single, independent binding events, 10.8%). (C and D) Structural and cartoon model of a microtubule and Pac1-bound dynein monomer (generated with pdbs 4RH736, 3J1T67, 5VH928, and 3J6G68). Note the close proximity of Pac1 to the microtubule surface, the latter of which is lacking the unstructured E-hooks. (D) Cryo-EM data reveals the dynein-microtubule angle varies due to a hinge point within the MTBD, and can be much steeper than that shown in panel A69,70 (Θ ≥ 15–20°, with average = 55°). Cartoons depict range of angles sampled by dynein on microtubules, and thus the distances between Pac1 and the microtubule. (E - G) Non-normalized plots of mean values (E and F) and all data points (G; see Figure 6D for n values) showing the relationship between Pac1-mediated dynein velocity reduction and Pac1-microtubule binding (for panels B left, E, and F, diamonds represent mean values obtained from each independent replicate experiment; for panel G, mean values and standard deviations are depicted with red lines).
Supplementary Material
Supplementary Video 2. Uninhibited dynein mutants can bypass the need for Pac1 to move the mitotic spindle. The mitotic spindle (via GFP-Tub1) is observed in pac1Δ kar9Δ cells expressing either wild-type (top), K1475E (middle), or D2868K (bottom) dynein. Each frame is a maximum-intensity projection of a 2.4 μm z stack of confocal images collected at 10 second intervals. The appearance of green circles in the top left corner of each movie corresponds to frames in which dynein-mediated spindle movements are apparent. Note the presence of two spindles in the wild-type cell. See Figure 4F for quantitation and n values.
Supplementary Video 3. Proposed mechanism for autoinhibition restricting dynein processivity. In combination with the tail-dependence of the autoinhibited conformation, our observation of 2D classes with apparent tail-tail contacts, but not motor-motor contacts (Figure 1, classes vii and viii) indicate that these contacts may initiate the transition to the autoinhibited state. Subsequent to adopting the autoinhibited conformation, we hypothesize that dynein dissociates from the microtubule.
Supplementary Video 1. Three-dimensional classification of the full-length yeast dynein phi particle reveals similarities to human dynein. Movie of the 3D class average of the autoinhibited yeast dynein conformation. The high resolution cryoEM structure of the human dynein phi particle (PDB 5NVU1) was manually docked into the EM density map. Note that the structures of the two tail domains have been slightly rotated with respect to the motor domains to better fit the 3D model, and that the structures of both TcTEX and Robl have been eliminated due to their absence from the yeast dynein complex.
ACKNOWLEDGEMENTS
We are grateful to Samara Reck-Peterson for the 8His-ZZ-Pac1-SNAP-expressing yeast strain, and members of the Markus and DeLuca laboratories for valuable discussions. Electron microscopy was done at the University of Colorado, Boulder EM Services Core Facility in the MCDB Department, with the technical assistance of facility staff. This work utilized the RMACC Summit supercomputer, which is supported by the National Science Foundation (awards ACI-1532235 and ACI-1532236), the University of Colorado Boulder and Colorado State University. The RMACC Summit supercomputer is a joint effort of the University of Colorado Boulder and Colorado State University. We are also extremely grateful to Erin Osborne-Nishimura, David King, and Samuel Bowerman for their invaluable assistance with using software on Summit. This work was funded by the NIH/NIGMS (GM118492 to S.M.M.).
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES CITED
- 1.Schlager MA, Hoang HT, Urnavicius L, Bullock SL & Carter AP In vitro reconstitution of a highly processive recombinant human dynein complex. EMBO J 33, 1855–1868, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McKenney RJ, Huynh W, Tanenbaum ME, Bhabha G & Vale RD Activation of cytoplasmic dynein motility by dynactin-cargo adapter complexes. Science 345, 337–341, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McKenney RJ, Huynh W, Vale RD & Sirajuddin M Tyrosination of alpha-tubulin controls the initiation of processive dynein-dynactin motility. EMBO J 35, 1175–1185, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang K, Foster HE, Rondelet A, Lacey SE, Bahi-Buisson N, Bird AW & Carter AP Cryo-EM Reveals How Human Cytoplasmic Dynein Is Auto-inhibited and Activated. Cell 169, 1303–1314 e1318, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amos LA Brain dynein crossbridges microtubules into bundles. J Cell Sci 93 ( Pt 1), 19–28, (1989). [DOI] [PubMed] [Google Scholar]
- 6.Toropova K, Mladenov M & Roberts AJ Intraflagellar transport dynein is autoinhibited by trapping of its mechanical and track-binding elements. Nat Struct Mol Biol 24, 461–468, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jordan MA, Diener DR, Stepanek L & Pigino G The cryo-EM structure of intraflagellar transport trains reveals how dynein is inactivated to ensure unidirectional anterograde movement in cilia. Nat Cell Biol 20, 1250–1255, (2018). [DOI] [PubMed] [Google Scholar]
- 8.Torisawa T, Ichikawa M, Furuta A, Saito K, Oiwa K, Kojima H, Toyoshima YY & Furuta K Autoinhibition and cooperative activation mechanisms of cytoplasmic dynein. Nat Cell Biol 16, 1118–1124, (2014). [DOI] [PubMed] [Google Scholar]
- 9.Reck-Peterson SL, Yildiz A, Carter AP, Gennerich A, Zhang N & Vale RD Single-molecule analysis of dynein processivity and stepping behavior. Cell 126, 335–348, (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kardon JR, Reck-Peterson SL & Vale RD Regulation of the processivity and intracellular localization of Saccharomyces cerevisiae dynein by dynactin. Proc Natl Acad Sci U S A 106, 5669–5674, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sasaki S, Shionoya A, Ishida M, Gambello MJ, Yingling J, Wynshaw-Boris A & Hirotsune S A LIS1/NUDEL/cytoplasmic dynein heavy chain complex in the developing and adult nervous system. Neuron 28, 681–696, (2000). [DOI] [PubMed] [Google Scholar]
- 12.Wynshaw-Boris A & Gambello MJ LIS1 and dynein motor function in neuronal migration and development. Genes & development 15, 639–651, (2001). [DOI] [PubMed] [Google Scholar]
- 13.Moughamian AJ, Osborn GE, Lazarus JE, Maday S & Holzbaur EL Ordered recruitment of dynactin to the microtubule plus-end is required for efficient initiation of retrograde axonal transport. J Neurosci 33, 13190–13203, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raaijmakers JA, Tanenbaum ME & Medema RH Systematic dissection of dynein regulators in mitosis. J Cell Biol 201, 201–215, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coquelle FM, Caspi M, Cordelieres FP, Dompierre JP, Dujardin DL, Koifman C, Martin P, Hoogenraad CC, Akhmanova A, Galjart N, De Mey JR & Reiner O LIS1, CLIP-170’s key to the dynein/dynactin pathway. Mol Cell Biol 22, 3089–3102, (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsai JW, Bremner KH & Vallee RB Dual subcellular roles for LIS1 and dynein in radial neuronal migration in live brain tissue. Nat Neurosci 10, 970–979, (2007). [DOI] [PubMed] [Google Scholar]
- 17.Tsai JW, Chen Y, Kriegstein AR & Vallee RB LIS1 RNA interference blocks neural stem cell division, morphogenesis, and motility at multiple stages. J Cell Biol 170, 935–945, (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yi JY, Ori-McKenney KM, McKenney RJ, Vershinin M, Gross SP & Vallee RB High-resolution imaging reveals indirect coordination of opposite motors and a role for LIS1 in high-load axonal transport. J Cell Biol 195, 193–201, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chapman DE, Reddy BJN, Huy B, Bovyn MJ, Cruz SJS, Al-Shammari ZM, Han H, Wang W, Smith DS & Gross SP Regulation of in vivo dynein force production by CDK5 and 14–3-3epsilon and KIAA0528. Nature communications 10, 228, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reddy BJ, Mattson M, Wynne CL, Vadpey O, Durra A, Chapman D, Vallee RB & Gross SP Load-induced enhancement of Dynein force production by LIS1-NudE in vivo and in vitro. Nature communications 7, 12259, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKenney RJ, Vershinin M, Kunwar A, Vallee RB & Gross SP LIS1 and NudE induce a persistent dynein force-producing state. Cell 141, 304–314, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamada M, Toba S, Yoshida Y, Haratani K, Mori D, Yano Y, Mimori-Kiyosue Y, Nakamura T, Itoh K, Fushiki S, Setou M, Wynshaw-Boris A, Torisawa T, Toyoshima YY & Hirotsune S LIS1 and NDEL1 coordinate the plus-end-directed transport of cytoplasmic dynein. EMBO J 27, 2471–2483, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baumbach J, Murthy A, McClintock MA, Dix CI, Zalyte R, Hoang HT & Bullock SL Lissencephaly-1 is a context-dependent regulator of the human dynein complex. eLife 6, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gutierrez PA, Ackermann BE, Vershinin M & McKenney RJ Differential effects of the dynein-regulatory factor Lissencephaly-1 on processive dynein-dynactin motility. J Biol Chem 292, 12245–12255, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jha R, Roostalu J, Cade NI, Trokter M & Surrey T Combinatorial regulation of the balance between dynein microtubule end accumulation and initiation of directed motility. EMBO J 36, 3387–3404, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang J, Roberts AJ, Leschziner AE & Reck-Peterson SL Lis1 acts as a “clutch” between the ATPase and microtubule-binding domains of the dynein motor. Cell 150, 975–986, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toropova K, Zou S, Roberts AJ, Redwine WB, Goodman BS, Reck-Peterson SL & Leschziner AE Lis1 regulates dynein by sterically blocking its mechanochemical cycle. eLife 3, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeSantis ME, Cianfrocco MA, Htet ZM, Tran PT, Reck-Peterson SL & Leschziner AE Lis1 Has Two Opposing Modes of Regulating Cytoplasmic Dynein. Cell 170, 1197–1208 e1112, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Markus SM & Lee WL Regulated offloading of cytoplasmic dynein from microtubule plus ends to the cortex. Dev Cell 20, 639–651, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dix CI, Soundararajan HC, Dzhindzhev NS, Begum F, Suter B, Ohkura H, Stephens E & Bullock SL Lissencephaly-1 promotes the recruitment of dynein and dynactin to transported mRNAs. J Cell Biol 202, 479–494, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang S, Ketcham SA, Schon A, Goodman B, Wang Y, Yates J 3rd, Freire E, Schroer TA & Zheng Y Nudel/NudE and Lis1 promote dynein and dynactin interaction in the context of spindle morphogenesis. Mol Biol Cell 24, 3522–3533, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marzo MG, Griswold JM, Ruff KM, Buchmeier RE, Fees CP & Markus SM Molecular basis for dyneinopathies reveals insight into dynein regulation and dysfunction. eLife 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Markus SM & Lee WL Microtubule-dependent path to the cell cortex for cytoplasmic dynein in mitotic spindle orientation. Bioarchitecture 1, 209–215, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmidt H, Gleave ES & Carter AP Insights into dynein motor domain function from a 3.3-A crystal structure. Nat Struct Mol Biol 19, 492–497, S491, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carter AP, Cho C, Jin L & Vale RD Crystal structure of the dynein motor domain. Science 331, 1159–1165, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmidt H, Zalyte R, Urnavicius L & Carter AP Structure of human cytoplasmic dynein-2 primed for its power stroke. Nature 518, 435–438, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gibbons IR, Garbarino JE, Tan CE, Reck-Peterson SL, Vale RD & Carter AP The affinity of the dynein microtubule-binding domain is modulated by the conformation of its coiled-coil stalk. J Biol Chem 280, 23960–23965, (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kon T, Imamula K, Roberts AJ, Ohkura R, Knight PJ, Gibbons IR, Burgess SA & Sutoh K Helix sliding in the stalk coiled coil of dynein couples ATPase and microtubule binding. Nat Struct Mol Biol 16, 325–333, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rao L, Berger F, Nicholas MP & Gennerich A Molecular mechanism of cytoplasmic dynein tension sensing. Nature communications 10, 3332, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gennerich A, Carter AP, Reck-Peterson SL & Vale RD Force-induced bidirectional stepping of cytoplasmic dynein. Cell 131, 952–965, (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moore JK, Li J & Cooper JA Dynactin function in mitotic spindle positioning. Traffic 9, 510–527, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Markus SM, Punch JJ & Lee WL Motor- and tail-dependent targeting of dynein to microtubule plus ends and the cell cortex. Curr Biol 19, 196–205, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lammers LG & Markus SM The dynein cortical anchor Num1 activates dynein motility by relieving Pac1/LIS1-mediated inhibition. J Cell Biol 211, 309–322, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee WL, Oberle JR & Cooper JA The role of the lissencephaly protein Pac1 during nuclear migration in budding yeast. J Cell Biol 160, 355–364, (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yin H, Pruyne D, Huffaker TC & Bretscher A Myosin V orientates the mitotic spindle in yeast. Nature 406, 1013–1015, (2000). [DOI] [PubMed] [Google Scholar]
- 46.Hwang E, Kusch J, Barral Y & Huffaker TC Spindle orientation in Saccharomyces cerevisiae depends on the transport of microtubule ends along polarized actin cables. J Cell Biol 161, 483–488, (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liakopoulos D, Kusch J, Grava S, Vogel J & Barral Y Asymmetric loading of Kar9 onto spindle poles and microtubules ensures proper spindle alignment. Cell 112, 561–574, (2003). [DOI] [PubMed] [Google Scholar]
- 48.Ecklund KH, Morisaki T, Lammers LG, Marzo MG, Stasevich TJ & Markus SM She1 affects dynein through direct interactions with the microtubule and the dynein microtubule-binding domain. Nature communications 8, 2151, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Markus SM, Plevock KM, St Germain BJ, Punch JJ, Meaden CW & Lee WL Quantitative analysis of Pac1/LIS1-mediated dynein targeting: Implications for regulation of dynein activity in budding yeast. Cytoskeleton (Hoboken) 68, 157–174, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sheeman B, Carvalho P, Sagot I, Geiser J, Kho D, Hoyt MA & Pellman D Determinants of S. cerevisiae dynein localization and activation: implications for the mechanism of spindle positioning. Curr Biol 13, 364–372, (2003). [DOI] [PubMed] [Google Scholar]
- 51.Li J, Lee WL & Cooper JA NudEL targets dynein to microtubule ends through LIS1. Nat Cell Biol 7, 686–690, (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Elshenawy MM, Kusakci E, Volz S, Baumbach J, Bullock SL, Yildiz A Lis1 activates dynein motility by pairing it with dynactin. Nat Cell Biol (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Htet ZM, Gillies JP, Baker RW, Leschziner AE, DeSantis ME, Reck-Peterson SL Lis1 promotes the formation of activated cytoplasmic dynein-1 complexes. Nat Cell Biol (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Urnavicius L, Lau CK, Elshenawy MM, Morales-Rios E, Motz C, Yildiz A & Carter AP Cryo-EM shows how dynactin recruits two dyneins for faster movement. Nature 554, 202–206, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qiu R, Zhang J & Xiang X LIS1 regulates cargo-adapter-mediated activation of dynein by overcoming its autoinhibition in vivo. J Cell Biol, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Curran KA, Morse NJ, Markham KA, Wagman AM, Gupta A & Alper HS Short Synthetic Terminators for Improved Heterologous Gene Expression in Yeast. ACS Synth Biol 4, 824–832, (2015). [DOI] [PubMed] [Google Scholar]
- 57.Kelley LA, Mezulis S, Yates CM, Wass MN & Sternberg MJ The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10, 845–858, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Poirier K, Lebrun N, Broix L, Tian G, Saillour Y, Boscheron C, Parrini E, Valence S, Pierre BS, Oger M, Lacombe D, Genevieve D, Fontana E, Darra F, Cances C, Barth M, Bonneau D, Bernadina BD, N’Guyen S, Gitiaux C, Parent P, des Portes V, Pedespan JM, Legrez V, Castelnau-Ptakine L, Nitschke P, Hieu T, Masson C, Zelenika D, Andrieux A, Francis F, Guerrini R, Cowan NJ, Bahi-Buisson N & Chelly J Mutations in TUBG1, DYNC1H1, KIF5C and KIF2A cause malformations of cortical development and microcephaly. Nature genetics 45, 639–647, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scoto M, Rossor AM, Harms MB, Cirak S, Calissano M, Robb S, Manzur AY, Martinez Arroyo A, Rodriguez Sanz A, Mansour S, Fallon P, Hadjikoumi I, Klein A, Yang M, De Visser M, Overweg-Plandsoen WC, Baas F, Taylor JP, Benatar M, Connolly AM, Al-Lozi MT, Nixon J, de Goede CG, Foley AR, McWilliam C, Pitt M, Sewry C, Phadke R, Hafezparast M, Chong WK, Mercuri E, Baloh RH, Reilly MM & Muntoni F Novel mutations expand the clinical spectrum of DYNC1H1-associated spinal muscular atrophy. Neurology, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Willemsen MH, Vissers LE, Willemsen MA, van Bon BW, Kroes T, de Ligt J, de Vries BB, Schoots J, Lugtenberg D, Hamel BC, van Bokhoven H, Brunner HG, Veltman JA & Kleefstra T Mutations in DYNC1H1 cause severe intellectual disability with neuronal migration defects. Journal of medical genetics 49, 179–183, (2012). [DOI] [PubMed] [Google Scholar]
- 61.Heil-Chapdelaine RA, Oberle JR & Cooper JA The cortical protein Num1p is essential for dynein-dependent interactions of microtubules with the cortex. J Cell Biol 151, 1337–1344, (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
ADDITIONAL REFERENCES CITED
- 62.Weissmann F et al. biGBac enables rapid gene assembly for the expression of large multisubunit protein complexes. Proc Natl Acad Sci U S A 113, E2564–2569, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mahamdeh M, Simmert S, Luchniak A, Schaffer E & Howard J Label-free high-speed wide-field imaging of single microtubules using interference reflection microscopy. J Microsc 272, 60–66, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thiede C, Lakamper S, Wessel AD, Kramer S & Schmidt CF A chimeric kinesin-1 head/kinesin-5 tail motor switches between diffusive and processive motility. Biophys J 104, 432–441, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stellwagen E, Prantner JD & Stellwagen NC Do zwitterions contribute to the ionic strength of a solution? Anal Biochem 373, 407–409, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fraley C & Raftery AE Bayesian Regularization for Normal Mixture Estimation and Model-Based Clustering Journal of Classification 24, 155–181, (2007). [Google Scholar]
- 67.Redwine WB et al. Structural basis for microtubule binding and release by dynein. Science 337, 1532–1536, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alushin GM et al. High-resolution microtubule structures reveal the structural transitions in alphabeta-tubulin upon GTP hydrolysis. Cell 157, 1117–1129, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Imai H et al. Direct observation shows superposition and large scale flexibility within cytoplasmic dynein motors moving along microtubules. Nature communications 6, 8179, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Can S, Lacey S, Gur M, Carter AP & Yildiz A Directionality of dynein is controlled by the angle and length of its stalk. Nature 566, 407–410, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Video 2. Uninhibited dynein mutants can bypass the need for Pac1 to move the mitotic spindle. The mitotic spindle (via GFP-Tub1) is observed in pac1Δ kar9Δ cells expressing either wild-type (top), K1475E (middle), or D2868K (bottom) dynein. Each frame is a maximum-intensity projection of a 2.4 μm z stack of confocal images collected at 10 second intervals. The appearance of green circles in the top left corner of each movie corresponds to frames in which dynein-mediated spindle movements are apparent. Note the presence of two spindles in the wild-type cell. See Figure 4F for quantitation and n values.
Supplementary Video 3. Proposed mechanism for autoinhibition restricting dynein processivity. In combination with the tail-dependence of the autoinhibited conformation, our observation of 2D classes with apparent tail-tail contacts, but not motor-motor contacts (Figure 1, classes vii and viii) indicate that these contacts may initiate the transition to the autoinhibited state. Subsequent to adopting the autoinhibited conformation, we hypothesize that dynein dissociates from the microtubule.
Supplementary Video 1. Three-dimensional classification of the full-length yeast dynein phi particle reveals similarities to human dynein. Movie of the 3D class average of the autoinhibited yeast dynein conformation. The high resolution cryoEM structure of the human dynein phi particle (PDB 5NVU1) was manually docked into the EM density map. Note that the structures of the two tail domains have been slightly rotated with respect to the motor domains to better fit the 3D model, and that the structures of both TcTEX and Robl have been eliminated due to their absence from the yeast dynein complex.
Data Availability Statement
All yeast strains, datasets, and raw movie files generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.