A synthetic switch shows tunable transgene expression, regulatable genome engineering, and programmable biocomputation in mammals.
Abstract
Dynamically adjustable gene- and cell-based therapies are recognized as next-generation medicine. However, the translation of precision therapies into clinics is limited by lack of specific switches controlled by inducers that are safe and ready for clinical use. Ferulic acid (FA) is a phytochemical with a wide range of therapeutic effects, and its salt sodium ferulate (SF) is used as an antithrombotic drug in clinics. Here, we describe an FA/SF-adjustable transcriptional switch controlled by the clinically licensed drug SF. We demonstrated that SF-responsive switches can be engineered to control CRISPR-Cas9 systems for on-command genome/epigenome engineering. In addition, we integrated FA-controlled switches into programmable biocomputers to process logic operations. We further demonstrated the dose-dependent SF-inducible transgene expression in mice by oral administration of SF tablets. Engineered switches responsive to small-molecule clinically licensed drugs to achieve adjustable transgene expression profiles provide new opportunities for dynamic interventions in gene- and cell-based precision medicine.
INTRODUCTION
Gene- and cell-based therapies are rapidly emerging innovation drivers in modern medical sciences (1, 2). Mammalian synthetic biology is boosting this innovation by providing the necessary molecular tools and design strategies for cell-based precision medicine (3). A particular focus in cell-based precision therapy is on externally programmable gene switches to align target gene activities with complex therapeutic requirements in living cells and organisms (4). This is of particular interest with regard to improving therapeutic outcome while reducing possible side effects. For example, there is a high demand for on-command switchable CRISPR (clustered regularly interspaced short palindromic repeats)–Cas9 systems to achieve desired genome/epigenome editing while avoiding off-target effects due to prolonged, unspecific Cas9 activity (5, 6). Furthermore, despite the observation that genetic engineered chimeric antigen receptor T (CAR-T) cells are achieving unprecedented response rates against leukemia (7), there is the urgent need to improve treatment safety and reliability by integrating sophisticated control switches to avoid side effects such as the cytokine release syndrome (8). Obviously, the further advancement of such promising therapeutic technologies is currently severely limited by a lack of safe and specific control systems to tune transgene activity and regulate cell behavior in a dynamic, patient-tailored fashion.
Initial efforts in the design of dynamically controlled genetic switches relied on antibiotics [e.g., tetracycline (9), streptogramins (10), or macrolides (11)] as triggers to control cell behaviors. Later on, genetic switches were engineered to be activated by cosmetic compounds [e.g., phloretin (12) and paraben (13)], food additives [e.g., vanillic acid (14) and benzoate (BA) (15)], and even food components [e.g., caffeine (16), spearmint (17), and protocatechuic acid (18)]. However, these inducers either can lead to antibiotic resistance, are not yet clinically approved for long-term use in patients, or are present in common food leading to undesired switch activation. Physical inducers, such as light (19, 20), magnetic force (21), and temperature (22), have recently been developed to control the expression and release of therapeutic outputs. However, specialized hardware and software are often required for administration of these stimuli. Furthermore, fluctuations of the physical inducers in everyday life make them less orthogonal and likely lead to unwanted interference. For these reasons, ideal inducers would act in a specific manner, would have no toxic effects, would be safe and clinically approved for human use, and would be easily obtained from natural sources (23). Genetic switches responsive to such ideal inducers would show substantially increased chances for translation into clinical practice (24).
Ferulic acid (FA) is a widely distributed hydroxycinnamic acid, an abundant polyphenolic compound found in various vegetables, fruits, and grain, exhibiting potent antioxidant and various therapeutic activities (25). Because of its strong antioxidant activity by scavenging free radicals and activating antioxidant enzymes, it has been widely applied to prevent reactive oxygen species–related diseases (26), such as cancer (27), cardiovascular diseases (28), and diabetes mellitus (29). It is also approved as a food additive in some countries (30). Its sodium salt, sodium ferulate (SF), is a compound widely used in traditional Chinese medicine for several decades and is approved by the National Medical Products Administration (NMPA) for treating cardiovascular and cerebrovascular diseases and to prevent thrombosis (31–33). Overall, FA/SF has all it takes to be a safe and physiologically inert inducer for genetic switches designed for future therapeutic applications.
It has been reported that FA can trigger dissociation of the phenolic acid decarboxylase regulator (PadR) repressor, a transcriptional regulator of the padA gene in Bacillus subtilis 168, from its specific DNA binding sequence OPadR (34, 35). On the basis of this switchable molecular interaction, we propose to construct a novel genetic switch in which PadR is fused to eukaryotic epigenetic effector domains. This synthetic transcription factor would specifically modulate the activity of synthetic promoters containing the PadR-specific OPadR operator in response to varying concentrations of FA/SF. We here present the FA/SF-controlled, orthogonal genetic switches termed FAR (FA regulator) that are responsive to the clinical drug SF to fine-tune transgene expression in various mammalian cell lines and in mice. We further demonstrate applicability of the designed FAR switches for multiple applications: (i) in combination with CRISPR-Cas9 for inducible gene editing and epigenome engineering, (ii) as modular building block for engineering programmable biocomputation to perform five logic operations in single mammalian cells, and (iii) the ability to program transgene expression in mice in response to oral administration of clinically approved SF tablets. This versatility combined with robust switch characteristics make the FAR switches a potent instrument for cybergenetics and expand the molecular intervention strategies that may foster novel advances in gene- and cell-based precision therapies.
RESULTS
Design and validation of an FAROFF switch
To engineer a mammalian cell–compatible FA-controlled OFF switch, where gene expression is switched off in the presence of FA, we constructed a synthetic mammalian transcription activator, the activation version of phenolic acid decarboxylase regulator (aPadR), by fusing a VP64 transactivation domain (four copies of the Herpes simplex virus protein 16) to the N terminus of PadR (VP64-PadR). The transactivator aPadR binds and initiates transcription from synthetic promoters termed PaPadR harboring a hexameric OPadR operator upstream of a minimal human cytomegalovirus immediate early promoter (PhCMVmin; Fig. 1A). However, in the presence of FA, aPadR dissociates from its promoter PaPadR, and transgene expression is deactivated.
Fig. 1. Design and characterization of the FAROFF switch.
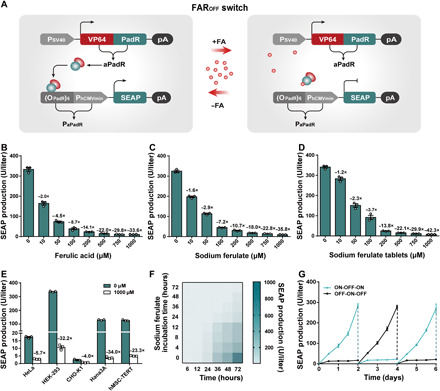
(A) Schematic design of the FAROFF switch. The binding domain PadR was fused to the transactivation domain VP64 to create an FA-dependent transactivator aPadR (VP64-PadR), which is driven by the simian virus 40 promoter. In the absence of FA (−FA), aPadR binds to a chimeric target promoter PaPadR [(OPadR)6-PhCMVmin] and activates the expression of reporter gene SEAP. In the presence of FA (+FA), aPadR is released from PaPadR, and SEAP expression is switched off. (B to D) Dose-dependent SEAP production profile of the FAROFF switch. SEAP expression of HEK-293 cells cotransfected with pLS125 and pLS74 was assessed after cultivation with different concentrations of FA (B), SF (C), or SF tablets (D). (E) FA-induced SEAP expression in different mammalian cell lines. (F) Time-dependent SEAP expression kinetics of the FAROFF switch. SEAP levels of stable cell line HEKFAR-OFF-SEAP, exposed to 500 μM SF for different time periods (0 to 72 hours), were profiled at specific time points (X axis, 6 to 72 hours). (G) Reversibility of the FAROFF switch. SEAP expression of HEKFAR-OFF-SEAP cells, alternating the SF concentrations (500 μM: OFF; 0 μM: ON) every 2 days, was profiled every 12 hours. Data are means ± SD; n = 3 independent experiments.
To optimize FA/SF-inducible OFF switch (FAROFF switch) characteristics with regard to maximal transgene expression in the absence of FA and minimal basal expression in its presence, we constructed and tested four different configurations of the hybrid transactivator aPadR (fig. S1), four different linkers (La×) between VP64 and PadR (fig. S2), and six different tandem repeat variants of OPadR in the synthetic promoters (fig. S3). We found that a combination of aPadR (VP64-La3-PadR) and PaPadR6 [(OPadR)6-PhCMVmin] showed the highest fold induction (~44.1-fold) of the reporter gene SEAP (secreted alkaline phosphatase) when cultivated in the presence or absence of FA (fig. S3F). To exclude that the observed change in gene expression was due to pleiotropic effects of the inducer, we transfected human embryonic kidney (HEK) 293 cells with a constitutive SEAP expression plasmid (pSEAP2-control, PSV40-SEAP-pA) and cultivated the cells in the presence of increasing concentrations (0 to 2000 μM) of different inducers (FA, SF, and commercial SF tablets) for 48 hours. Subsequent scoring of SEAP production and cell viability demonstrate that the tested inducers do not have negative effects neither on overall gene expression capacity of the transfected cells (fig. S4, A to C) nor on cell viability within the tested concentration range (fig. S4, D to F).
Next, we investigated gene expression kinetics as function of the inducer concentration. Cotransfection of the constitutive aPadR expression vector pLS125 (PhCMV-VP64-La3-PadR-pA) and the response plasmid pLS74 (PaPadR6-SEAP-pA) encoding a PaPadR6-driven SEAP expression unit resulted in high SEAP production (332.98 ± 12.96 U/liter) (Fig. 1B). Cultivation in the presence of increasing concentrations (0 to 1000 μM) of FA, SF, or SF tablets, however, resulted in a dose-dependent decrease in SEAP production up to complete deactivation (Fig. 1, B to D). To assess cross-cell compatibility of FAROFF, we tested the switch in various commonly used mammalian cell lines. The data showed that FAROFF-controlled transgene expression was functional in all cell lines tested (Fig. 1E). Differences in expression level between the different cell lines can be attributed to cell-specific factors such as differences in transfection efficiency as also observed in previous studies (18, 19).
To study long-term performance of the FAROFF switch, we generated and randomly selected eight FAROFF-transgenic stable cell lines (HEKFAR-OFF-SEAP) by transfection of pLS168 (ITR-PaPadR6-SEAP-pA::PhCMV-VP64-La3-PadR-P2A-puromycin-pA-ITR) into HEK-293 cells and clonal selection using the Sleeping Beauty transposon system (36). All the selected cell lines showed SF-induced SEAP repression (fig. S5A), but with different regulation performance, probably as a result of differences in transgene copy number and integration sites that remains beyond control using traditional transient transfection method (19). The cell line with the highest regulation performance was used in follow-up experiments. This cell line HEKFAR-OFF-SEAP showed excellently adjustable gene expression time courses and full reversibility of transgene expression (Fig. 1, F and G). Furthermore, FA-dependent expression performance was observed to be stable for 1 week in cell culture plate (fig. S5B). This comprehensive characterization demonstrating excellent FA-adjustable regulation of gene expression, cross-cell line compatibility, and long-term stability and reversibility strongly suggests that this control switch has a broad application potential.
Design and validation of an FA/SF-inducible ON switch (FARON switch)
In certain configurations such as in whole animals, FA/SF-inducible gene expression might be preferable compared to the OFF-type switch characteristics as demonstrated above. To obtain an FA-controlled ON switch (FARON switch) in mammalian cells, we constructed a synthetic mammalian transcription repressor iPadR (KRAB-PadR) by fusing the human Krüppel-associated box (KRAB) domain to the N terminus of PadR. The transrepressor iPadR binds and silences constitutive gene expression from synthetic promoters (PiPadR) comprising the simian virus 40 promoter (PSV40) fused to different configurations of OPadR-binding sites. However, in the presence of FA, iPadR dissociates from PiPadR, resulting in the derepression of SEAP production (Fig. 2A). To engineer an optimal FARON switch with maximal transgene expression in the presence of FA and minimal basal expression in its absence, we optimized the FARON switch by testing multiple configurations of OPadR motifs in the promoter (fig. S6) and different linkers (Li×) between KRAB and PadR (fig. S7). We found that a combination of the transrepressor iPadR (KRAB-Li6-PadR) and the promoter PiPadR5 [OPadR-PSV40-OPadR(RC)] showed the highest fold induction (~43.0-fold) in the presence of FA (Fig. 2B).
Fig. 2. Design and characterization of the FARON switch.
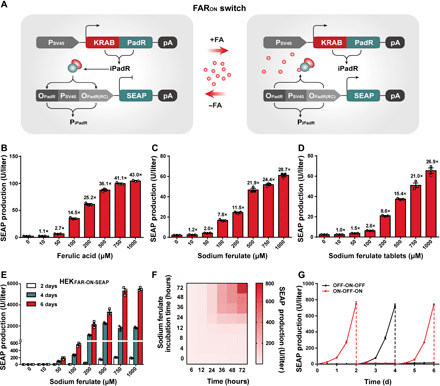
(A) Schematic design of the FARON switch. PadR was fused to a trans-silencing domain KRAB to obtain the FA-dependent transrepressor iPadR (KRAB-PadR) driven by the constitutive simian virus 40 promoter. In the absence of FA (−FA), iPadR binds to a chimeric target promoter PiPadR [OPadR-PSV40-OPadR(RC)] and represses PSV40-driven SEAP expression. In the presence of FA (+FA), iPadR is dissociated from the target promoter PiPadR and derepresses SEAP expression. (B to D) Dose-dependent SEAP expression profile of the FARON switch. SEAP expression of HEK-293 cells cotransfected with pLS163 and pLS64 was assessed after cultivation with different concentrations of FA (B), SF (C) or SF tablets (D). (E) Dose-dependent SEAP expression kinetics of the stable cell line HEKFAR-ON-SEAP. (F) Time-dependent SEAP expression kinetics of the FARON switch. SEAP levels of HEKFAR-ON-SEAP cells, incubated with 500 μM SF for different time periods (0 to 72 hours), were profiled at different time points (X axis, 6 to 72 hours). (G) Reversibility of the FARON switch. SEAP expression of HEKFAR-ON-SEAP cells, alternating the SF concentrations (500 μM: ON; 0 μM: OFF) every 2 days, was profiled every 12 hours. Data are means ± SD; n = 3 independent experiments.
Next, we characterized the FARON switch in detail. We cotransfected the plasmids pLS163 (PSV40-KRAB-Li6-PadR-pA) and pLS64 (PiPadR5-SEAP-pA) into HEK-293 cells and incubated the cells in the presence of different concentrations (0 to 1000 μM) of FA, SF, or SF tablets. Scoring SEAP production after 48 hours revealed tight repression in the absence of inducers and a dose-dependent increase in transgene expression correlating with higher inducer concentrations (Fig. 2, B to D). Similarly to the FAROFF (Fig. 1E), the FARON switch switch showed excellent switching properties in various mammalian cell lines, indicating its broad application potential (fig. S8A).
To evaluate long-term expression, adjustability, and reversibility of the FARON switch, we created a HEK-293–based stable cell line for FARON-mediated SEAP expression (HEKFAR-ON-SEAP) using the Sleeping Beauty transposon system (36) and subsequent clonal selection. All the selected clones showed significant SF-induced transgene expression with different maximum SEAP production levels (fig. S8B). One of the clones with the best performance (~240-fold induction) out of the eight evaluated ones was used for the following studies (fig. S8B). Characterizing the stable cell line revealed that HEKFAR-ON-SEAP cell line revealed that SEAP expression could be dose-dependently activated for 6 days (Fig. 2E). Furthermore, fine-tunable, inducer-dependent dynamic expression characteristics (Fig. 2F) and excellent reversibility (Fig. 2G) qualify the FARON system as a precise, robust, and adjustable FA/SF-triggered control.
SF-controlled CRISPR-Cas9/dCas9 activity for genome/epigenome engineering
The CRISPR technology is well known for gene editing and genome/epigenome engineering (37). The ability to control the activity of CRISPR with side effect–free inducers will enable tight regulation of arbitrary endogenous genes for precision gene therapy and or for studying the dynamics of transcriptional regulation. Capitalizing on the strong activation and finely adjustable regulation potential of the FARON switch, we engineered three SF-controlled CRISPR-Cas9 systems for activation (PadRa), inhibition (PadRi), and deletion (PadRdel) of endogenous genes in human cells.
We first explored epigenetic remodeling–based activation of endogenous genes (PadRa) (Fig. 3A). In this design, we used the PiPadR6 promoter to promote SF-inducible expression of an engineered activation mediator complex MS2-P65-HSF1 (38). This complex harbors a transcription activation domain (P65) and further an MS2-binding site, which allows its recruitment to dCas9 via single guide RNAs (sgRNAs) fused to MS2-specific RNA aptamers. Thus, the SF-inducible expression of MS2-P65-HSF1 triggers transcription of dCas9-targeted exogenous and endogenous genes. Functionality of this system was successfully validated by the SF-mediated dose-dependent expression of exogenous SEAP gene (Fig. 3B) and endogenous ASCL1, IL1RN, and RHOXF genes (Fig. 3C).
Fig. 3. FA-controlled CRISPR-Cas9 devices for genome and epigenome editing.
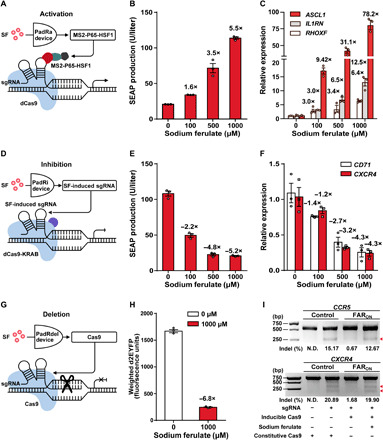
(A) Schematic design of FA-controlled PadR-mediated activation (PadRa). In the presence of SF, the transactivator MS2-P65-HSF1 is produced and recruited by constitutively expressed dCas9 and sgRNAMS2 (sgRNA with MS2 loop) to form a transcriptional activation complex (sgRNAMS2-dCas9-MS2-P65-HSF1). (B) Dose-dependent SF-inducible SEAP expression for PadRa. (C) Dose-dependent PadRa-mediated activation of different endogenous genes, including ASCL1, IL1RN, and RHOXF. (D) Schematic design of SF-controlled PadR-mediated inhibition (PadRi). In the presence of SF, the expression of sgRNA is induced to allow assembly of a repressive complex (sgRNA-dCas9-KRAB) targeting sgRNA-specific DNA sites to inhibit gene expression. (E) Dose-dependent SF-repressible SEAP expression by PadRi. (F) PadRi-mediated repression of different endogenous genes, including CD71 and CXCR4. (G) Schematic design of FA-controlled PadR-mediated gene deletion (PadRdel). Cas9 expression is induced by SF and enables Cas9-mediated target gene deletion. (H) FA-induced gene editing of exogenous gene d2EYFP. The data [(B), (C), (E), (F), and (H)] represent the means ± SD; n = 3 independent experiments. (I) PadRdel-mediated genome CCR5 and CXCR4 editing. Red arrowheads indicate the expected cleavage bands. N.D.: not detectable; bp: base pair. See table S1 for detailed description of genetic components and table S6 for detailed transfection mixtures.
We next applied the FARON switch for the SF-dependent inhibition of target genes (PadRi) by controlling sgRNA expression to induce the transcriptional repressor dCas9-KRAB to the desired genome region. To this aim, OPadR-binding sites were sandwiched upstream and downstream of the U6 (constitutive) promoter [termed OPadR-PU6-OPadR(RC)] that drives sgRNA expression. When cotransfecting this system with an expression plasmid for iPadR, sgRNA expression is silenced, whereas the addition of SF derepresses U6-driven sgRNA expression by disrupting iPadR binding (Fig. 3D). The produced sgRNA is assembled into a repressive complex (sgRNA-dCas9-KRAB) to target specific DNA sites and to inhibit gene expression. To test whether the PadRi system can achieve sgRNA-mediated target gene inhibition, we cotransfected HEK-293 cells with the PadRi components together with the sgRNAs targeting defined genes. With this system, we used SF to control the expression of exogenous SEAP (Fig. 3E) and d2EYFP (fig. S9A) or of the endogenous CD71 and CXCR4 genes (Fig. 3F).
Last, we repurposed the SF-controlled CRISPR-Cas9 system for controllable gene editing (PadRdel). Cas9-mediated cleavage of an endogenous genomic locus induces indel mutations via nonhomologous end joining (NHEJ), thus disrupting functional gene expression. We used the FARON system with the PiPadR6 promoter to express Cas9 in an SF-inducible manner (Fig. 3G). We first validated functionality of the system by disrupting expression of the exogenous d2EYFP gene in HEK-293 as demonstrated by flow cytometry analysis and by fluorescence microscopy (Fig. 3H and fig. S9B). We next demonstrated the applicability of the PadRdel system to edit the gene of the endogenous CCR5 chemokine receptor that acts as co-receptor for HIV uptake and the c-x-c motif chemokine receptor 4. As detected by the mismatch-sensitive T7 endonuclease I (T7E1) assay, the PadRdel system successfully edited the CCR5 and CXCR4 genes (Fig. 3I).
SF- and food additive–controlled programmable biocomputers in mammalian cells
Mammalian cells engineered with programmable control networks performing higher-order arithmetic calculations may enable the assembly biocomputers to allow the design of complex human-machine interfaces and provide patient-tailored therapeutic interventions in future gene- and cell-based precision therapies (39, 40). By rational combination of mammalian synthetic gene switches, Boolean logic gates could be constructed to program complex intracellular computation that allows cells to process with a computer-like algorithm (15, 18). One of the long-term goals of these efforts is to develop programmable logic computations controlled by side effect-free inputs. To seek a generalizable biocomputing platform controlled by safe, food-derived, and clinically approved molecules, we combined our FARON system with a previously reported system responsive to the food additive BA (15). To demonstrate the general applicability of this approach, we have constructed five examples of logic gates to achieve a variety of Boolean operations controlled by SF and BA. Of special interest are the negation of the OR gate (NOR) and the complement of an AND gate (NAND) operations that are functionally complete, i.e., these operations can be used to express all possible truth tables by combining members of the set into a Boolean expression (41).
First, two BA-controlled genetic switches (BAON and BAOFF) were constructed and validated as reported (15). An AND logic gate is exclusively induced in the presence of both inputs. To engineer an AND gate, we constructed a synthetic promoter PBF1 [PSV40-OPadR-(OCbaR)2] comprising an SF-repressible promoter (PSV40-OPadR) fused to a BA-inducible CbaR-binding site (OCbaR)2. Only in the presence of both SF and BA, the transrepressors KRAB-PadR and KRAB-CbaR are released and derepress expression of the reporter d2EYFP (Fig. 4A). As a complementary phase to the AND gate, a NAND gate was designed. To this aim, we constructed a chimeric promoter PBF2 [(OCbaR)2-OPadR-PhCMVmin] comprising an SF-inducible promoter (OPadR-PhCMVmin) fused to a CbaR-binding site (OCbaR)2 in front of the OPadR site. Only in the presence of both SF and BA, the transactivators VP16-CbaR and VP64-PadR dissociate from their cognate operators, resulting in effective deactivation of d2EYFP expression (Fig. 4B).
Fig. 4. SF- and BA-controlled programmable biocomputers in human cells.
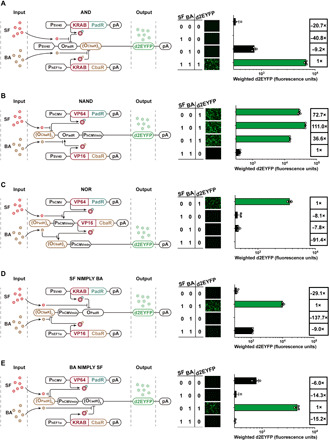
Processing performance of the (A) AND, (B) NAND, (C) NOR, (D) SF NIMPLY BA, and (E) BA NIMPLY SF logic gates in HEK-293 cells. The schematic design of the five logic gates is displayed on the left panels. The processing output performance of five logic gates is shown on the right panels. HEK-293 cells (8 × 104) were cotransfected with (A) pLS25, pMX34, and pDL55; (B) pLS56, pMX43, and pDL58; (C) pLS125, pDL41, and pDL68; (D) pMX43, pLS25, and pDL73; and (E) pLS125, pMX34, and pDL77 and cultivated with different combinations of the two input signals SF (1 mM) and BA (0.75 mM) in accordance with the truth tables. d2EYFP expression in the cells was quantified by fluorescence microscopy and flow cytometric analysis 48 hours after the addition of input signals. Data are means ± SD; n = 3 independent experiments. See table S1 for detailed description of genetic components and table S7 for detailed transfection mixtures for each logic gate.
A NOR gate is turned on only when no input signal is present. A NOR gate was constructed by wiring the FAROFF and BAOFF switches in a cascade configuration. In the absence of both inducers, SF and BA, the constitutively expressed transactivator VP64-PadR binds to the SF-responsive minimal promoter PaPadR6 [(OPadR)6-PhCMVmin] to initiate expression of the BA-responsive transactivator CbaR-VP16, which, in turn, binds to the BA-responsive minimal promoter PBF3 [(OCbaR)2-PhCMVmin] to induce expression of the output gene d2EYFP (Fig. 4C). An A NIMPLY B gate is exclusively induced in the presence of A while B is absent (42). For example, in an SF NIMPLY BA gate, the output is expected to be only induced if SF is present while BA is absent. To engineer the SF NIMPLY BA logic gate, we assembled a chimeric promoter PBF4 [(OCbaR)2-PhCMVmin-OPadR] comprising a BA-deactivatable promoter [(OCbaR)2-PhCMVmin] flanked in 3′ by a PadR-binding site OPadR. Only in the presence of SF and the absence of BA, the transrepressor KRAB-PadR dissociates from PBF4, while the transactivator CbaR-VP16 binds to PBF4 to activate d2EYFP expression (Fig. 4D). To obtain a reversed BA NIMPLY SF logic gate, we engineered a chimeric promoter PBF5 [(OPadR)6-PhCMVmin-(OCbaR)2] comprising an SF-deactivatable promoter [(OPadR)6-PhCMVmin] flanked in 3′ by a CbaR-binding site (OCbaR)2. Only in the presence of BA and the absence of SF, the transrepressor KRAB-CbaR dissociates from PBF5, while the transactivator VP64-PadR binds to PBF5 to activate d2EYFP expression (Fig. 4E). These exciting proof-of-concept results for biocomputing show that complex genetic programs could precisely be controlled by food additives and clinically licensed drugs, indicating the application potential for complex engineered cell-based therapeutic dosing regimens.
Control of transgene expression in mice by the clinically licensed drug SF
For future applications in precision gene- and cell-based therapies, it is essential that state-of-the-art gene regulation systems are functional within whole organisms (43). To validate SF-controlled transgene gene expression in animals, we microencapsulated HEKFAR-ON-SEAP cells into coherent, immunoprotective alginate-poly-(l-lysine)-alginate beads. After confirming SF-inducible SEAP production in microencapsulated cells in vitro (fig. S10), we next implanted the cell-containing capsules intraperitoneally into mice (Fig. 5A). The mice were given a dose of SF within the range of 1 to 1000 mg/kg each day. Notably, the acute LD50 (median lethal dose) of 3200 mg/kg was calculated in mice (35). SEAP production quantified in the blood showed SF-dependent dose-response kinetics for up to 15 days (Fig. 5B). When the mice were implanted with HEKFAR-ON-SEAP and orally administrated with clinically approved solubilized SF tablets, we observed significantly induced SEAP levels in the bloodstream for 15 days as well (Fig. 5C). However, we observed SEAP levels in vivo decreased at day 15, which may be attributed to the cell encapsulation technologies containing xenogeneic molecules that may escape from capsules and trigger immune responses destroying the encapsulated cells or their surrounding tissues to hinder the long-term performance of HEKFAR-ON-SEAP cells (22). Despite the limitations of transplant technology, these results demonstrate that SF tablets can be used as a trigger for the tight and precise control of implanted cells in future clinical investigations.
Fig. 5. Performance of the FARON switch in mice.
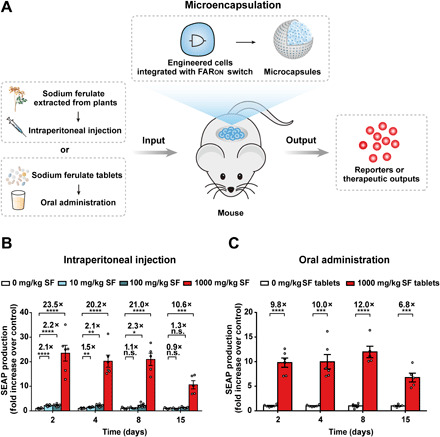
(A) Schematic showing the mouse experimental design and procedure for assessing SF-controlled transgene expression in vivo. HEK-293 cells engineered for FARON-inducible SEAP production were microencapsulated into alginate-poly-(l-lysine)-alginate beads allowing free diffusion of oxygen, nutrients, and secreted proteins across the membrane while simultaneously shielding encapsulated cells from the immune system. SEAP production in microencapsulated HEKFAR-ON-SEAP cells implanted in the peritoneum of mice could be controlled either via injection of SF or via oral administration of SF tablets. (B and C) Dose-dependent SF-inducible SEAP expression in mice. Eight-week-old male C57BL/6J mice were intraperitoneally implanted with 2 × 106 microencapsulated HEKFAR-ON-SEAP cells (200 cells per capsule) and received intraperitoneal administration of SF (B) or oral administration of clinically licensed SF tablets (C) three times per day (0 to 1000 mg/kg per day). SEAP levels in the bloodstream of mice were quantified on days 2, 4, 8, and 15 after implantation. Data are expressed as means ± SEM; n = 5 to 6 mice. n.s., not significant. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 versus control.
DISCUSSION
In this work, we have developed an FA/SF-controlled transcription switch (FAR switch), which can be applied to control transgene expression, genome/epigenome engineering, and complex biological logic operations in mammalian cells. One of the great advantages of the FAR switch is that it is responsive to an off-the-shelf clinical drug that was shown to be safe and does not trigger inadvertent side reactions (31). SF has been used to treat patients with cardiovascular and cerebrovascular diseases for decades in China (31), indicating that SF is very safe for long-term therapeutic usage. Such beneficial properties of the inducer might facilitate the clinical approval for the corresponding gene switch. Another potential benefit of SF being clinically licensed is the perspective of developing a combined therapy. In such a perspective, SF not only would coordinate both the expression of desired therapeutic genes but would also elicit its original therapeutic effect. For example, the system could be deployed to control both expression of antithrombotic peptides expression, such as hirudin (44) or antithrombotic bioactive peptides (45), and the intrinsic antithrombotic activity of SF. Such scenarios would represent an attractive therapeutic option as the original activity of a drug could be invigorated and improved by a complementing therapeutic gene network.
Combining the PadR repressor from B. subtilis 168 (35) and mammalian epigenetic regulators, we have constructed the novel mammalian transgene expression systems FARON and FAROFF, which respond to the clinically approved drug SF. The optimized FARON/FAROFF switch demonstrated very low background, high induction folds, robust induction kinetics, precise adjustability, and full reversibility. Because of the high modularity of the switch, we were able to construct SF-controlled Cas9/dCas9 systems for controllable genome editing and epigenome remodeling. Moreover, the FAR switch showed interference-free regulation characteristics when used with the BA-inducible switch, thus enabling to perform logic computations in mammalian cells, indicating that this switch is likely an ideal building block for the design of complex and high-order synthetic gene networks. The suitability of the FARON to regulate gene expression in mice opens the perspective of transferring such complex regulation networks into a therapeutic setting functional in living mammals.
Despite the strong regulation performance in vitro and in vivo, there is room for improvement with regard to a translational perspective. For example, the sensitivity of the FAR switch to its inducer could further be improved to obtain a higher dynamic range in vivo in response to SF tablets. We anticipate that the sensitivity of the FAR switch could be optimized by engineering the transcription factor and the operator sequence in the promoter. For example, by using directed evolution approaches (46) or rational protein design based on bioinformatics (47), it may be possible to obtain a more sensitive mutated version of the transcriptional repressor PadR.
Collectively, our work demonstrates how genetic switches responsive to the clinically approved drug SF can expand the synthetic biology tool box and provide a new device to cybergenetics. This work shows a safe and robust strategy for dynamic control of transgene expression and epigenetic engineering, which provides an alternative strategy to achieve dynamic interventions in future gene- and cell-based precision medicine.
MATERIALS AND METHODS
Plasmid construction
Comprehensive design and construction details for all expression vectors are listed in table S1. The expression vectors were constructed by T4 DNA ligation (Takara Bio, catalog no. 2011B) or Gibson assembly according to the manufacturer’s instructions [Seamless Assembly Cloning Kit, Obio Technology Inc., catalog no. BACR(C) 20144001]. All constructions have been confirmed by sequencing (Genewiz Inc., China).
Cell culture
Human cervical adenocarcinoma cells [HeLa, American Type Culture Collection (ATCC): CCL-2], HEK cells (HEK-293, ATCC: CRL-11268), HEK-293–derived Hana3A cells that were engineered for the stable expression of GαολΦ and chaperones RTP1/RTP2/REEP1, and telomerase-immortalized human mesenchymal stem cells (hMSC-TERT, ATCC: SCRC4000) were cultivated in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, lot no. 31600-083) supplemented with 10% (v/v) fetal bovine serum (FBS; Biological Industries, lot no. 04-001-1C) and 1% (v/v) penicillin/streptomycin solution. Chinese hamster ovary cells (CHO-K1, ATCC: CCL-61) were cultivated in its specific culture medium (Procell, lot no. CM-0062) supplemented with 5% (v/v) FBS and 1% (v/v) penicillin/streptomycin solution. All cell types were cultured at 37°C in a humidified atmosphere containing 5% CO2.
Cell transfection
All cell lines were transfected with a standard polyethyleneimine (PEI)–based protocol. Briefly, 5.0 × 104 HEK-293 cells were seeded in a 24-well cell culture plate. After 20 hours of cultivation, 200 ng of plasmid DNA diluted in 50 μl of FBS-free DMEM was mixed with 0.6 μl of PEI (PolyScience, catalog no. 24765; 1 μg/μl). The transfection mixture was incubated at room temperature for 15 min and subsequently dropwise added to the cells. After 6 hours, the culture medium was replaced by fresh cell culture medium optionally supplemented with different concentrations of FA (Sigma-Aldrich, catalog no. 1270311), SF (Qufu Hongly Chemical Industry Co. Ltd., catalog no. 24276-84-4), or SF tablets (Chengdu Hengda Pharmaceutical Co. Ltd., guoyaozhunzi: H51023583). Cell concentrations and viability of all cell lines were profiled with a Countess II Automated cell counter (Life Technologies, USA).
Cytotoxicity assay
To test the impact of various types of inducers (FA, SF, and SF tablets) on human cells, 1 × 104 HEK-293 cells per well were seeded in a 96-well plate, transfected with the reporter plasmid pSEAP2-control (PSV40-SEAP-pA; 50 ng), and incubated with different concentrations of chemicals (0 to 2000 μM) for 48 hours. The cell culture supernatant was collected for the quantification of SEAP expression (48).
To test the effects of inducers on cell viability, 1 × 104 HEK-293 cells per well were seeded in a 96-well plate and incubated with different concentrations of inducers (0 to 2000 μM) for 48 hours. Then, the cell culture medium was replaced by 100 μl of fresh medium containing 10 μl of an [3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide] (MTT) solution (5 mg/ml; Sangon Biotech, catalog no. A600799) and incubated for 4 hours at 37°C. After incubation, the medium was replaced with 100 μl of dimethyl sulfoxide (Sangon Biotech, catalog no. A600163) and incubated for 10 min at 37°C. The light absorbance was measured at 540 nm using a Synergy H1 hybrid multimode microplate reader (BioTek Instruments Inc.) using Gen5 software (version 2.04).
SEAP assay
Human placental SEAP in cell culture medium was quantified using a p-nitrophenylphosphate–based light absorbance time course assay, as described previously (18). The SEAP production in mice was quantified with a chemiluminescence-based assay kit (Roche Diagnostics, catalog no. 11779842001) according to the manufacturer’s protocol.
Construction of stable cell lines
The Sleeping Beauty (SB) transposon system was used for selecting stable cell lines as described before (36). The HEKFAR-ON-SEAP cell line, transgenic for SF-inducible SEAP expression, was constructed by cotransfecting HEK-293 cells with pLS180 [ITR-PiPadR5-SEAP-pA::PhCMV-KRAB-Li6-PadR-P2A-puromycin-pA-ITR; 200 ng] and the Sleeping Beauty transposase expression vector PhCMV-T7-SB100 (PhCMV-SB100X-pA; 20 ng) and selected with puromycin (1 μg/ml) for 2 weeks. The living cells were picked for further cultivation and induced with 1 mM SF for 48 hours. The monoclonal HEKFAR-ON-SEAP cell line showing the highest fold induction in gene expression in response to SF was used for subsequent studies.
Mismatch-sensitive T7E1 assay
The genomic DNA of treated HEK-293 cells was extracted using a TIANamp Genomic DNA Extraction Kit (TIANGEN Biotech Inc., catalog no. DP304) according to the manufacturer’s protocol. Then, sgRNA-targeted CCR5 genes were polymerase chain reaction (PCR)–amplified from genomic DNA using the primers listed in table S2. The PCR amplicons were purified from agarose gel with a Universal DNA Purification Kit (TIANGEN Biotech Inc., catalog no. DP214). Twenty microliters of the reaction mixture containing 500 ng of purified PCR products and 2 μl of 10× M buffer (TIANGEN Biotech Inc., catalog no. 1060S) were reannealed (95°C for 5 min, and then slowly cooled to room temperature) to form heteroduplex DNA. Then, 0.5 μl of T7 endonuclease I (New England Biolabs, catalog no. M0302) was added and incubated at 37°C for 2 hours. The digested products were analyzed by 2% agarose gel electrophoresis. The percentage of deletion by Cas9 was calculated with the following formula: 100% × (b + c)/(a + b + c), where a represented the intensity of undigested PCR amplicons and b and c represented the intensities of the T7E1-digested products.
qPCR analysis
Total RNA of treated HEK-293 cells was isolated using the RNAiso Plus Kit (Takara Bio, catalog no. 9108). One microgram of purified RNA was reverse-transcribed into complementary DNA (cDNA) using a PrimeScript Reverse Transcription Kit with the gDNA Eraser (Takara Bio, catalog no. RR047). Quantitative PCR (qPCR) was performed by SYBR Premix Ex Taq (Takara Bio, catalog no. RR420) with primers listed in table S3 and profiled by QuantStudio (Thermo Fisher Scientific Inc.) with the following amplification parameters: 95°C for 10 min, 40 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s, and a final extension at 72°C for 10 min. All samples were normalized against the endogenous housekeeping gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH), and the results were evaluated as the relative mRNA level using the standard ΔΔCt method (49).
Fluorescence imaging
The expression of d2EYFP was visualized using an Olympus microscope (Olympus IX71, TH4-200) equipped with an Olympus digital camera, a pE-100-LED (CoolLED) as the transmission light source, a Spectra X (Lumencor) as the fluorescent light source, a 10× objective, a 488-nm/509-nm (Blue/Green/Red) excitation/emission filter set, and the Image-Pro Express C software (version ipp6.0). Identical settings including exposure times for d2EYFP were used for all fluorescence micrographs.
Flow cytometric analysis
Flow cytometric analysis was performed as described previously (18). In brief, cells were analyzed with a Becton Dickinson FACSCalibur Flow Cytometer (BD Biosciences) equipped for d2EYFP [488-nm laser, 505-nm long pass filter, 530/30 emission filter (passband centered on 530 nm; passband width: 30 nm)] detection and set to exclude dead cells and cell doublets. About 10,000 cells were recorded in each dataset and analyzed with the BD CellQuest Pro software (version no. 6.0). To quantify the expression profiles of the FA-controlled gene deletion and SF/BA-controlled programmable biocomputers in human cells, transfected HEK-293 populations were gated for cells with high d2EYFP fluorescence beyond a threshold of 103 arbitrary fluorescence units. The percentage of gated cells was multiplied by their median fluorescence, resulting in a weighted d2EYFP expression profile that correlated fluorescence intensity with cell number.
Mouse experiments
The HEKFAR-ON-SEAP stable cell line engineered for SF-controlled SEAP expression was encapsulated into coherent alginate-poly-(l-lysine)-alginate beads (400-μm diameter; 200 cells per capsule) using a B-395 Pro Encapsulator (BUCHI Labortechnik AG) according to the manufacturers’ instructions. Microcapsules were generated with the following parameters: 200-μm nozzle with a vibration frequency of 1300 Hz, 20-ml syringe operated at a flow rate of 450 U, and 1.10 kV for bead dispersion. Eight-week-old male wild-type C57BL/6 J mice [East China Normal University (ECNU) Laboratory Animal Centre] were intraperitoneally injected with 2 × 106 microencapsulated HEKFAR-ON-SEAP cells. The mice were then intraperitoneally injected daily with SF or orally administrated with SF tablets at doses ranging from 0 to 1000 mg/kg. Blood samples were collected, and SEAP levels were quantified by a chemiluminescence-based assay kit. The serum was isolated at the indicated points in time after implantation clotting of the blood (37°C for 1 hour and then 4°C for 1.5 hours) and subsequent centrifugation (2700g, 10 min).
Ethics statement
All experiments involving mice were performed according to the directives approved by the ECNU Animal Care and Use Committee (protocol ID: m20140301). All the procedures for samples or data collection used were carried out in compliance with the Ministry of Science and Technology of the People’s Republic of China on Animal Care Guidelines. All mice were euthanized after the experiments.
Statistical analysis
All in vitro data represent means ± SD of three independent experiments (n = 3). For mouse experiments, each treatment group was composed of five to six mice (n = 5 to 6). The blood sample analysis was blinded. Comparisons between groups were analyzed using Student’s t tests, and the values are expressed as means ± SD. Differences were considered statistically significant at P < 0.05. Prism 6 software (version 6.01, GraphPad Software Inc.) was used for statistical analysis.
Supplementary Material
Acknowledgments
We thank the ECNU Multifunctional Platform for Innovation (011) for supporting the mouse experiments and the Instruments Sharing Platform of School of Life Sciences at ECNU for supporting research. We also thank C. Huang for help with the artwork. Funding: This work was financially supported by grants from the National Key R&D Program of China, Synthetic Biology Research (no. 2019YFA0904500), the National Natural Science Foundation of China (NSFC: nos. 31971346 and 31861143016), the Science and Technology Commission of Shanghai Municipality (no. 18JC1411000), the Thousand Youth Talents Plan of China, and the Fundamental Research Funds for the Central Universities to H.Y. This work was also partially supported by NSFC: no. 31901023 to N.G. W.W. was supported by the Deutsche Forschungsgemeinschaft (DFG; German Research Foundation) under Germany’s Excellence Strategy–EXC-2189–Project ID: 390939984. Author contributions: H.Y. conceived this study. H.Y., Y.W., and S.L. designed the project. Y.W., S.L., Y.L., and K.D. performed the experimental work. Y.W., S.L., and K.D. performed the mouse experiments. H.Y., N.G., W.W., and Y.W. analyzed the results and wrote the manuscript. All authors read and approved the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. All genetic components related to this paper are available with a material transfer agreement and may be requested from H.Y. (hfye@bio.ecnu.edu.cn).
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/32/eabb9484/DC1
REFERENCES AND NOTES
- 1.Kohn D. B., Booth C., Kang E. M., Pai S.-Y., Shaw K. L., Santilli G., Armant M., Buckland K. F., Choi U., De Ravin S. S., Dorsey M. J., Kuo C. Y., Leon-Rico D., Rivat C., Izotova N., Gilmour K., Snell K., Dip J. X.-B., Darwish J., Morris E. C., Terrazas D., Wang L. D., Bauser C. A., Paprotka T., Kuhns D. B., Gregg J., Raymond H. E., Everett J. K., Honnet G., Biasco L., Newburger P. E., Bushman F. D., Grez M., Gaspar H. B., Williams D. A., Malech H. L., Galy A., Thrasher A. J.; Net4CGD consortium , Lentiviral gene therapy for X-linked chronic granulomatous disease. Nat. Med. 26, 200–206 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ye L., Park J. J., Dong M. B., Yang Q., Chow R. D., Peng L., Du Y., Guo J., Dai X., Wang G., Errami Y., Chen S., In vivo CRISPR screening in CD8 T cells with AAV-Sleeping Beauty hybrid vectors identifies membrane targets for improving immunotherapy for glioblastoma. Nat. Biotechnol. 37, 1302–1313 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu M.-R., Jusiak B., Lu T. K., Engineering advanced cancer therapies with synthetic biology. Nat. Rev. Cancer 19, 187–195 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Zhong G., Wang H., He W., Li Y., Mou H., Tickner Z. J., Tran M. H., Ou T., Yin Y., Diao H., Farzan M., A reversible RNA on-switch that controls gene expression of AAV-delivered therapeutics in vivo. Nat. Biotechnol. 38, 169–175 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manghwar H., Li B., Ding X., Hussain A., Lindsey K., Zhang X., Jin S., CRISPR/Cas systems in genome editing: Methodologies and tools for sgRNA design, Off-target evaluation, and strategies to mitigate Off-target effects. Adv. Sci. 7, 1902312 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marino N. D., Pinilla-Redondo R., Csörgő B., Bondy-Denomy J., Anti-CRISPR protein applications: natural brakes for CRISPR-Cas technologies. Nat. Methods 17, 471–479 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elsallab M., Levine B. L., Wayne A. S., Abou-El-Enein M., CAR T-cell product performance in haematological malignancies before and after marketing authorisation. Lancet Oncol. 21, E104–E116 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh A. K., McGuirk J. P., CAR T cells: Continuation in a revolution of immunotherapy. Lancet Oncol. 21, E168–E178 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Gossen M., Bujard H., Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. U.S.A. 89, 5547–5551 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fussenegger M., Morris R. P., Fux C., Rimann M., von Stockar B., Thompson C. J., Bailey J. E., Streptogramin-based gene regulation systems for mammalian cells. Nat. Biotechnol. 18, 1203–1208 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Weber W., Fux C., Daoud-el Baba M., Keller B., Weber C. C., Kramer B. P., Heinzen C., Aubel D., Bailey J. E., Fussenegger M., Macrolide-based transgene control in mammalian cells and mice. Nat. Biotechnol. 20, 901–907 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Gitzinger M., Kemmer C., El-Baba M. D., Weber W., Fussenegger M., Controlling transgene expression in subcutaneous implants using a skin lotion containing the apple metabolite phloretin. Proc. Natl. Acad. Sci. U.S.A. 106, 10638–10643 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang H., Ye H., Xie M., Daoud El-Baba M., Fussenegger M., Cosmetics-triggered percutaneous remote control of transgene expression in mice. Nucleic Acids Res. 43, e91 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gitzinger M., Kemmer C., Fluri D. A., El-Baba M. D., Weber W., Fussenegger M., The food additive vanillic acid controls transgene expression in mammalian cells and mice. Nucleic Acids Res. 40, e37 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie M., Ye H., Hamri G. C., Fussenegger M., Antagonistic control of a dual-input mammalian gene switch by food additives. Nucleic Acids Res. 42, e116 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bojar D., Scheller L., Hamri G. C., Xie M., Fussenegger M., Caffeine-inducible gene switches controlling experimental diabetes. Nat. Commun. 9, 2318 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang H., Xie M., Charpin-El Hamri G., Ye H., Fussenegger M., Treatment of chronic pain by designer cells controlled by spearmint aromatherapy. Nat. Biomed. Eng. 2, 114–123 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Yin J., Yang L., Mou L., Dong K., Jiang J., Xue S., Xu Y., Wang X., Lu Y., Ye H., A green tea-triggered genetic control system for treating diabetes in mice and monkeys. Sci. Transl. Med. 11, eaav8826 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Shao J., Xue S., Yu G., Yu Y., Yang X., Bai Y., Zhu S., Yang L., Yin J., Wang Y., Liao S., Guo S., Xie M., Fussenegger M., Ye H., Smartphone-controlled optogenetically engineered cells enable semiautomatic glucose homeostasis in diabetic mice. Sci. Transl. Med. 9, eaal2298 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Ye H., Daoud-El Baba M., Peng R. W., Fussenegger M., A synthetic optogenetic transcription device enhances blood-glucose homeostasis in mice. Science 332, 1565–1568 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Stanley S. A., Gagner J. E., Damanpour S., Yoshida M., Dordick J. S., Friedman J. M., Radio-wave heating of iron oxide nanoparticles can regulate plasma glucose in mice. Science 336, 604–608 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bai P., Liu Y., Xue S., Hamri G. C., Saxena P., Ye H., Xie M., Fussenegger M., A fully human transgene switch to regulate therapeutic protein production by cooling sensation. Nat. Med. 25, 1266–1273 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Xie M., Fussenegger M., Designing cell function: assembly of synthetic gene circuits for cell biology applications. Nat. Rev. Mol. Cell Biol. 19, 507–525 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Wang Y., Wang M., Dong K., Ye H., Engineering mammalian designer cells for the treatment of metabolic diseases. Biotechnol. J. 13, e1700160 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Mathew S., Abraham T. E., Bioconversions of ferulic acid, an hydroxycinnamic acid. Crit. Rev. Microbiol. 32, 115–125 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Zduńska K., Dana A., Kolodziejczak A., Rotsztejn H., Antioxidant properties of ferulic acid and its possible application. Skin Pharmacol. Physiol. 31, 332–336 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Gao J., Yu H., Guo W., Kong Y., Gu L., Li Q., Yang S., Zhang Y., Wang Y., The anticancer effects of ferulic acid is associated with induction of cell cycle arrest and autophagy in cervical cancer cells. Cancer Cell Int. 18, 102 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng Y.-H., Lin F.-H., Wang C.-Y., Hsiao C.-Y., Chen H.-C., Kuo H.-Y., Tsai T.-F., Chiou S.-H., Recovery of oxidative stress-induced damage in Cisd2-deficient cardiomyocytes by sustained release of ferulic acid from injectable hydrogel. Biomaterials 103, 207–218 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Nankar R., Prabhakar P. K., Doble M., Hybrid drug combination: Combination of ferulic acid and metformin as anti-diabetic therapy. Phytomedicine 37, 10–13 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Manore M., Meeusen R., Roelands B., Moran S., Popple A. D., Naylor M. J., Burke L. M., Stear S. J., Castell L. M., BJSM reviews: A-Z of nutritional supplements: Dietary supplements, sports nutrition foods and ergogenic aids for health and performance--Part 16. Br. J. Sports Med. 45, 73–74 (2011). [DOI] [PubMed] [Google Scholar]
- 31.Wang B.-H., Ou-Yang J.-P., Pharmacological actions of sodium ferulate in cardiovascular system. Cardiovasc. Drug Rev. 23, 161–172 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Xu L. N., Ouyang R., Antithrombotic effect of sodium ferulate in rats (author's transl). Zhongguo Yao Li Xue Bao 2, 35–37 (1981). [PubMed] [Google Scholar]
- 33.Hu B., Song J. T., Ji X. F., Liu Z. Q., Cong M. L., Liu D. X., Sodium ferulate protects against angiotensin II-induced cardiac hypertrophy in mice by regulating the MAPK/ERK and JNK pathways. Biomed. Res. Int. 2017, 3754942 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vatlin A. A., Bekker O. B., Lysenkova L. N., Shchekotikhin A. E., Danilenko V. N., A functional study of the global transcriptional regulator PadR from a strain Streptomyces fradiae-nitR+bld, resistant to nitrone-oligomycin. J. Basic Microbiol. 58, 739–746 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Nguyen T. K. C., Tran N. P., Cavin J.-F., Genetic and biochemical analysis of PadR-padC promoter interactions during the phenolic acid stress response in Bacillus subtilis 168. J. Bacteriol. 193, 4180–4191 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kebriaei P., Izsvák Z., Narayanavari S. A., Singh H., Ivics Z., Gene therapy with the sleeping beauty transposon system. Trends Genet. 33, 852–870 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Gilbert L. A., Larson M. H., Morsut L., Liu Z., Brar G. A., Torres S. E., Stern-Ginossar N., Brandman O., Whitehead E. H., Doudna J. A., Lim W. A., Weissman J. S., Qi L. S., CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shao J., Wang M., Yu G., Zhu S., Yu Y., Heng B. C., Wu J., Ye H., Synthetic far-red light-mediated CRISPR-dCas9 device for inducing functional neuronal differentiation. Proc. Natl. Acad. Sci. U.S.A. 115, E6722–E6730 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ausländer D., Ausländer S., Pierrat X., Hellmann L., Rachid L., Fussenegger M., Programmable full-adder computations in communicating three-dimensional cell cultures. Nat. Methods 15, 57–60 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Ausländer S., Ausländer D., Müller M., Wieland M., Fussenegger M., Programmable single-cell mammalian biocomputers. Nature 487, 123–127 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Tamsir A., Tabor J. J., Voigt C. A., Robust multicellular computing using genetically encoded NOR gates and chemical 'wires'. Nature 469, 212–215 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim H., Bojar D., Fussenegger M., A CRISPR/Cas9-based central processing unit to program complex logic computation in human cells. Proc. Natl. Acad. Sci. U.S.A. 116, 7214–7219 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang L., Yin J., Wang M., Ye H., Research progress of mammalian synthetic biology in biomedical field. Sheng Wu Gong Cheng Xue Bao 33, 436–455 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Zhang J., Lan N., Hirudin variants production by genetic engineered microbial factory. Biotechnol. Genet. Eng. Rev. 34, 261–280 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Qiao M., Tu M., Wang Z., Mao F., Chen H., Qin L., Du M., Identification and antithrombotic activity of peptides from Blue Mussel (Mytilus edulis) protein. Int. J. Mol. Sci. 19, 138 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pazos F., Valencia A., Protein co-evolution, co-adaptation and interactions. EMBO J. 27, 2648–2655 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuhlman B., Bradley P., Advances in protein structure prediction and design. Nat. Rev. Mol. Cell Biol. 20, 681–697 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xue S., Yin J., Shao J., Yu Y., Yang L., Wang Y., Xie M., Fussenegger M., Ye H., A synthetic-biology-inspired therapeutic strategy for targeting and treating hepatogenous diabetes. Mol. Ther. 25, 443–455 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hawkins S. F. C., Guest P. C., Multiplex analyses using real-time quantitative PCR. Methods Mol. Biol. 1546, 125–133 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/32/eabb9484/DC1


