A high-throughput 3D hPSC differentiation screening strategy reveals basic and translational insight for CNS cell therapies.
Abstract
The emergence of several cell therapy candidates in the clinic is an encouraging sign for human diseases/disorders that currently have no effective treatment; however, scalable production of these cell therapies has become a bottleneck. To overcome this barrier, three-dimensional (3D) cell culture strategies have been considered for enhanced cell production. Here, we demonstrate a high-throughput 3D culture platform used to systematically screen 1200 culture conditions with varying doses, durations, dynamics, and combinations of signaling cues to derive oligodendrocyte progenitor cells and midbrain dopaminergic neurons from human pluripotent stem cells (hPSCs). Statistical models of the robust dataset reveal previously unidentified patterns about cell competence to Wnt, retinoic acid, and sonic hedgehog signals, and their interactions, which may offer insights into the combinatorial roles these signals play in human central nervous system development. These insights can be harnessed to optimize production of hPSC-derived cell replacement therapies for a range of neurological indications.
INTRODUCTION
Stem cells—including adult and pluripotent subtypes—offer tremendous clinical promise for the treatment of a variety of degenerative diseases, as these cells have the capacity to self-renew indefinitely, mature into functional cell types, and thereby serve as a source of cell replacement therapies (CRTs). Human pluripotent stem cells (hPSCs) are of increasing interest for the development of CRTs due to their capacity to differentiate into all cell types in an adult, for which adult tissue–specific stem cells may, in some cases, not exist or may be difficult to isolate or propagate (1). For example, one potential CRT enabled by hPSCs is the treatment of spinal cord injury (SCI) with oligodendrocyte progenitor cells (OPCs). These hPSC-OPCs have recently advanced to a phase 2 clinical trial for the treatment of SCI (2) and are being considered for additional myelin-associated disorders in the central nervous system (CNS), including adrenoleukodystrophy, multiple sclerosis (3, 4), and radiation therapy–induced injury (5). In parallel, hPSC-derived midbrain dopaminergic (mDA) neurons are under consideration for Parkinson’s disease therapy (6, 7).
The promise of hPSC-derived therapeutics such as hPSC-OPCs or mDA neurons motivates the development of manufacturing processes to accommodate the potential associated clinical need. For example, approximately 250,000 patients in the United States suffer from some form of SCI, with an estimated annual incidence of 15,000 new patients (8). Human clinical trials involving hPSC-OPCs have used dosages of 20 million cells per patient (9), such that the hypothetical demand would be over 1 trillion differentiated OPCs. It is therefore imperative to develop systems to enable discovery of efficient and scalable differentiation protocols for these therapies.
Differentiation protocols to direct hPSCs into functional OPCs (10, 11) have been developed to approximate the signaling environment at precise positions within the developing spinal cord. Positional identity of cells is guided patterning cues that form intersecting gradients along the dorsoventral axis, such as Sonic hedgehog (SHH), and rostrocaudal axis, such as retinoic acid (RA). In addition, certain cues are present along both axes, such as Wnts (12–15). These signaling environments vary over time as the embryo develops (16, 17). However, translating this complex developmental biology to an in vitro culture requires optimization of a large combinatorial parameter space of signaling factor identities, doses, durations, dynamics, and combinations over many weeks to achieve efficient yield of the target cell type, and there remains open questions about the impact of cross-talk between patterning cues on the expression of cellular markers present in OPCs such as transcription factors Olig2 and Nkx2.2 (18). Strategies to derive OPCs and other potential CRTs from hPSCs have shown steady progress, especially with application of high-throughput screening technology (19–21); however, current production systems for hPSC-derived CRTs involve two-dimensional (2D) culture formats that are challenging to scale (22–28).
More recently, 3D culture systems have demonstrated strong potential for a larger scale and higher yield (29) of hPSC expansion and differentiation than 2D counterparts, as well as compatibility with good manufacturing practice (GMP) standards (30–33). While high-throughput systems for screening 3D cell culture environments have been applied to basic biological studies of hPSC proliferation (34), we envision that this technology could additionally be applied toward systematically optimizing production strategies for CRTs to accelerate the pace of their discovery and development toward the clinic while simultaneously uncovering new interactions among signaling cues that affect cell fate. Here, we harness the powerful capabilities of a uniquely structured microculture platform (35, 36), to screen dosage, duration, dynamics, and combinations of several cellular signaling factors in 3D for hPSC differentiation (Fig. 1). The independent control of gel-encapsulated cells (on pillar chip) and media (in well chip) enables simultaneous media replenishment for more than 500 independent microcultures in a single chip. Furthermore, we use custom hPSC reporter cell lines (37) to enable live imaging of proliferation and differentiation of OPCs for over 80 days on the microculture chip. One thousand two hundred combinatorial culture conditions, amounting to 4800 independent samples, were screened while consuming less than 0.2% of the reagent volumes of a corresponding 96-well plate format. Furthermore, the robust dataset enabled statistical modeling to identify relative differentiation sensitivities to, and interactions between, various cell culture parameters in an unbiased manner. Last, we demonstrate the generalizability of the platform by applying it toward a screen for differentiation of tyrosine hydroxylase–expressing dopaminergic neurons from hPSCs.
Fig. 1. Micropillar and microwell high-throughput culture system.
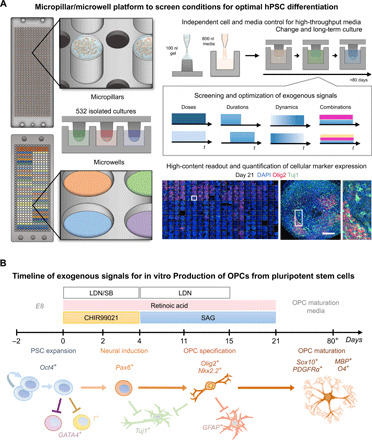
(A) A micropillar chip with cells suspended in a 3D hydrogel is stamped to a complementary microwell chip containing isolated media conditions to generate 532 independent microenvironments. One hundred nanoliters of hPSCs suspended in a hydrogel is automatically dispensed onto the micropillars, and 800 nl of media is automatically dispensed into the microwells by a robotic liquid handling robot programmed to dispense in custom patterns. The independent substrate for cells and media enables screens of combinations of soluble cues at various dosages and timings. Scale bar, 1 mm. (B) Timeline of exogenous signals for in vitro 3D OPC differentiation from hPSCs and anticipated cellular marker expression along various differentiation stages.
RESULTS
Miniaturization and increased throughput of 3D hPSC culture for differentiation screening
Initially, we assessed whether hPSCs could be dispensed in the microculture platform system uniformly and with high viability. Quantification of total, live, and dead cell counts across the microchip indicates uniform culture seeding and cell viability at the initiation of an experiment (fig. S1).
We then used a custom-made Nkx2.2-Cre H9 reporter line, which constitutively expresses DsRed protein but switches to green fluorescent protein (GFP) expression upon exposure to Cre recombinase, to longitudinally monitor proliferation and differentiation of hPSCs to Nkx2.2+ oligodendrocyte progenitors in 3D on the microchip platform. A small range of culture conditions from previously published protocols of OPC differentiation were selected for an initial, pilot differentiation experiment, and the GFP expression was quantified after 21 days of differentiation. Cell morphology changes accompanying neural lineage commitment and maturation were clearly observed at later stages in the 3D differentiation (movie S1 and fig. S2) as cultures were maintained and monitored for up to 80 days on the microchip. We then developed fluorescence image analysis pipelines for quantification of nuclear and cytoplasmic marker expression via immunocytochemistry for endpoint analyses at various times (fig. S3). Together, these results support the robust and long-term culture potential and cellular marker expression readout of this miniaturization methodology for hPSC differentiation screening.
Olig2 expression is substantially modulated by tuning early 3D culture parameters
hPSC seeding density
We first focused on parameters within the first week of 3D differentiation into OPCs (Fig. 2A). The importance of autocrine, paracrine, and juxtacrine signaling mechanisms among cells in many systems led us to anticipate that the density of cells at the start of differentiation could affect the early neural induction efficiency and, consequently, the efficiency of OPC differentiation. We therefore demonstrated the ability of this microculture platform to test a range of initial hPSC seeding densities on day −2 (fig. S1) and assessed the effect of seeding density on Olig2 expression. We observed notable differences in levels of cell-to-cell adhesion in hPSC cultures by day 0, 2 days after initial seeding (Fig. 2Bi). Then, after 15 days of differentiation, we observed a trend that lower hPSC seeding density, between 10 and 50 cells per pillar, increased OPC specification slightly (Fig. 2Bii).
Fig. 2. Early culture parameters play a large role in OPC differentiation efficiency.
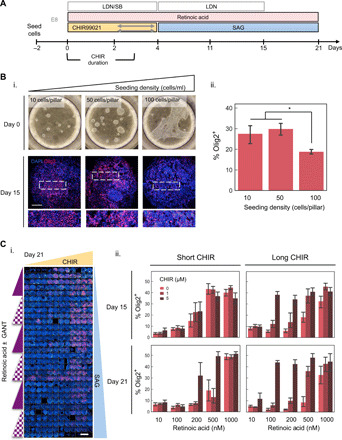
(A) Timeline of key parameters in the early phase of OPC differentiation. (B) i. Bright-field images of 3D H9 microculture sites at day 0 seeded with varying cell densities and the immunocytochemistry images of Olig2 (red) expression at day 15. Scale bar, 100 microns. ii. Quantification of day 15 Olig2 expression with respect to seeding density and SAG dose. *P value < 0.05 using Tukey’s Method for multiple comparisons. (C) i. Montage of 360 fluorescence confocal images representing 90 unique differentiation timelines on a single microchip stained for Hoechst (blue) and Olig2 (red) after 21 days of differentiation. ii. Trends in Olig2 expression at days 15 and 21 in various CHIR and RA concentrations and durations (short CHIR, days 0 to 1; long CHIR, days 0 to 3). Error bars represent 95% confidence intervals from four technical replicates.
Timing of SMAD inhibition relative to RA and Wnt signals
The formation of the neural tube in human development (12) results from cells in the epiblast being exposed to precisely timed developmental signals such as Wnt (38) and RA that then instruct neural subtype specification (39). This led us to hypothesize that the overall differentiation efficiency of hPSCs to OPCs in this 3D context in vitro would be sensitive to the timing at which RA and Wnt signals were introduced during neural induction. Therefore, we induced neuroectodermal differentiation of hPSCs via inhibition of bone morphogenetic protein (BMP) signaling using the dual SMAD inhibition approach (40), with LDN193189 (hereafter referred to as LDN) and SB431542 (hereafter referred to as SB), and tested a range of times (0, 2, and 4 days) at which RA and Wnt signals (by CHIR99021, hereafter referred to as CHIR) were introduced into the culture. We observed a strong correlation between early addition of RA/CHIR and OPC specification such that combined exposure of RA and CHIR signals with SMAD inhibition on day 0 resulted in up to sixfold higher Olig2 expression in some cases (fig. S4), potentially implicating an important role of synchronized exposure of RA and CHIR signals with SMAD inhibition for specifying Olig2+ progenitors. For subsequent experiments, we kept the timing of RA and CHIR addition at day 0 and evaluated how the dose and duration of these signals may affect Olig2+ specification.
Dose and duration of key signaling agonists
We examined the combinatorial and temporal effects of three signaling cues that form gradients across intersecting developmental axes in the neural tube to influence specification of oligodendrocyte progenitors: RA (present along the rostrocaudal axis of the CNS development), SHH (41) (a morphogen that patterns the dorsoventral axis of the developing CNS and is activated by smoothened agonist, hereafter referred to as SAG), and Wnt (present along both the rostrocaudal and dorsoventral axes). Because OPC specification is likely sensitive to the relative concentrations of these cues, for example, given the importance of morphogen gradients in oligodendrocyte differentiation in the developing neural tube (12), we assessed the Olig2 expression resulting from a full factorial combinatorial screen of these cues (fig. S5). Most notably, we observed positive correlations in Olig2 expression in response to increasing RA dose and increasing duration of CHIR exposure from days 0 to 4 of differentiation (Fig. 2C). Without CHIR, an increase in RA from 10 to 1000 nM resulted in a 10-fold increase of Olig2 expression by day 21. A similar 10-fold increase in Olig2 expression was observed at an RA concentration of 100 nM if CHIR was present for the first 3 days of differentiation (Fig. 2C). Analysis of variance (ANOVA) analysis revealed a strong effect size for RA when added early in the differentiation, as well as an interaction between RA dose and longer CHIR duration, in specifying Olig2+ cells in this 3D context (fig. S5), consistent with previous work conducted in 2D in vitro formats (19, 42).
Strategic addition of signaling antagonists to modulate OPC specification
In other developmental systems, the activity of the Wnt signaling pathway was observed to be biphasic (43), whereby activation of the pathway initially enhances cardiac development but later represses it. As this complex signaling profile has been applied to enhance cardiomyocyte differentiation protocols in vitro (44), we analogously investigated whether adding antagonists of key signaling pathways after pathway activation could further enhance the OPC differentiation efficiency by adjusting the dorsoventral and rostrocaudal positioning in vitro. Maintaining the 5 μM CHIR for days 0 to 3 from the previous experiment, we used IWP-2 (an inhibitor of the Wnt pathway), GANTT61 (an antagonist of SHH signaling), and DAPT (a Notch pathway antagonist) (Fig. 3A) to inhibit endogenous autocrine/paracrine and/or basal signaling. We used a full factorial analysis of these cues to additionally probe for combinatorial interactions among the pathway inhibitors.
Fig. 3. Strategic addition of signaling antagonists to modulate OPC specification.
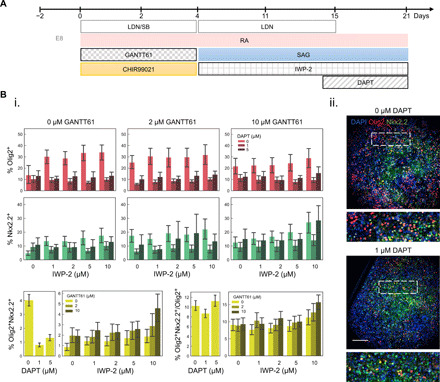
(A) Timing of addition for three inhibitory signaling cues—GANTT61, IWP-2, and DAPT—in the OPC differentiation protocol. (B) i. Olig2+, Nkx2.2+, and the proportion of total Olig2+ that are Nkx2.2+/Olig2+ cells in at day 21 in response to full factorial combinations of selected novel signaling antagonists. ii. Immunocytochemistry images of costained Olig2 (red) and Nkx2.2 (green) cells. Scale bar, 100 μm. Error bars represent 95% confidence intervals from four technical replicates.
To further refine the markers for OPC specification, we measured Nkx2.2 expression in addition to Olig2 and quantified the proportion of cells coexpressing both OPC markers. Most notably, a significant decrease in %Olig2 was observed in response to Notch inhibitor DAPT across all conditions tested (Fig. 3Bi). The same trend was not observed with respect to %Nkx2.2. This result could point to a role for Notch signaling in maintaining or promoting specification of Olig2+ progenitors—a hypothesis not previously examined to our knowledge—and serves as preliminary evidence to test Notch agonists such as DLL-4 in follow-up studies of OPC optimization. This effect may be mediated by an interaction with the SHH pathway (45).
A slight increase in %Olig2+ cells was detected with increasing Wnt inhibitor IWP-2 dose when no SHH inhibitor GANTT61 was present, as was a slight increase in %Nkx2.2+ cells as a function of increasing IWP-2 and GANTT61 dose, pointing to a potential interaction between these two cues in inducing Nkx2.2 expression. The highest proportion of Olig2+Nkx2.2+ cells was observed at the highest IWP-2 and GANTT61 doses and was not influenced by DAPT exposure (Fig. 3Bii). As CHIR was present between days 0 and 3 in the differentiation, it seems that the role of Wnt signaling changes during the 21-day differentiation window of hPSCs to OPCs in that initially (days 0 to 3) it promotes OPC differentiation but shifts to an inhibitory role at later stages (days 4 to 21). To examine the extent of reproducibility of these findings, we tested the effect of temporal modulation of Wnt signals in a human induced pluripotent stem cell (hiPSC) line, TCTF, and found that the general trend of activation followed by inactivation of Wnt signaling would increase the proportion of Olig2+ cells at day 21 (fig. S6).
Temporal sensitivity of OPC specification revealed by varying the signal dose over time
Although the levels of key signaling cues may vary temporally within the natural developmental environment of certain target cell types, such as within the neural tube where a dynamic SHH gradient along the dorsoventral axis patterns pMN development (16, 17), the dosage of signaling cues in the media for in vitro stem cell differentiation protocols is often applied at a constant level throughout the culture period. On the basis of this discrepancy, we applied the micropillar/microwell chip to screen through numerous temporal profiles of SAG, as well as RA due to its analogous role along the rostrocaudal axis during spinal cord development, by dividing the signal window into “early” and “late” stages that were dosed independently to form constant, increasing, and decreasing dose profiles over time (Fig. 4A). To gain additional insights into OPC marker expression, we measured Tuj1 expression and calculated the proportion of Olig2+ cells that coexpressed Tuj1 to potentially identify any modulators of the balance between Olig2+ cells that proceed down a motor neuron fate (which are both Olig2+ and Tuj1+) versus an oligodendrocyte fate (Olig2+/Nkx2.2+).
Fig. 4. Fine-tuning temporal profiles of RA and SAG to influence OPC specification.
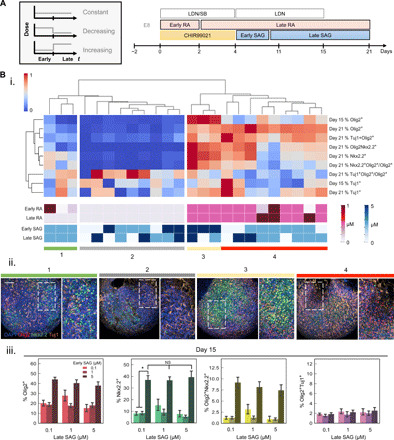
(A) Timeline of early and late windows for RA and SAG exposure. (B) i. Hierarchical cluster analysis of standardized (z score) phenotypic responses to temporal changes in RA and SAG dose during OPC differentiation. ii. Representative immunocytochemistry images of each major category of endpoint population phenotype mix of Olig2 (red), Nkx2.2 (green), and Tuj1 (orange) expression. Scale bar, 100 μm. iii. Olig2, Nkx2.2, and coexpression of Olig2+Nkx2.2+ and Olig2+Tuj1+ at day 15 in response to time-varying doses of SAG. Error bars represent 95% confidence intervals from four technical replicates. *P value < 0.05.
To consider all measured phenotypes simultaneously, we applied a hierarchical cluster analysis from which we were able to identify several patterns. A broad range of endpoint phenotype proportions of Olig2, Nkx2.2, and Tuj1 was found to result from varying the temporal dosing of only two signaling cues, RA and SAG, pointing to a very fine sensitivity to temporal changes in signal exposure in these populations. Four categories of the endpoint marker expression profiles were created to further interpret the cluster analysis. Categories 1 and 2 are composed of phenotypes ranking low on OPC progenitor fate (low Olig2 and/or Nkx2.2 expression), all of which shared the low dosing of RA at 0.1 μM between days 2 and 21 of the differentiation, further emphasizing the strong impact of RA on OPC yield. In contrast, category 3—composed of the highest Olig2 and Nkx2.2 expression as well as Olig2+Nkx2.2+ proportion—correlated with the highest dose of “early SAG” but had negligible differences across doses of “late SAG” (Fig. 4Biii, and fig. S7). Last, category 4 points to a biphasic relationship of Nkx2.2 expression as a function of RA dosage, where a high dose of RA of 1 μM in the late stage of differentiation resulted in lower Nkx2.2 expression (fig. S8) compared with a consistent RA of 0.5 μM throughout the entire differentiation. It appears that Olig2 and Nkx2.2 undergo maxima under different RA dosage profiles (fig. S8), and therefore, the use of coexpressing Olig2+Nkx2.2+ cells as the main metric when optimizing OPC differentiation may be most suitable.
Holistic analysis and prioritization of key parameters to influence OPC specification
We sought a comprehensive, yet concise, analysis to describe individual and combinatorial effects of all 12 culture parameters (e.g., signal agonist and antagonist dosages and timings) on the results of the more than 1000 unique differentiation conditions involved in this study. To this end, we fit generalized linear models to correlate the expression and coexpression of Olig2, Nxk2.2, and Tuj1 to individual input parameters within the 12 culture parameters involved in this study, and the 132 pairwise interactions between them. First, we identified significant parameters of interest for each phenotype measured using a factorial ANOVA (fig. S9). After applying a Benjamini and Hochberg false discovery rate correction for multiple comparisons (46), we fit an ordinary least squares model of the statistically significant terms to the phenotype of interest. The parameter coefficients were analyzed as a measure of relative influence on the expression of a certain endpoint phenotype, such as Olig2+Nkx2.2+ cells, and could be interpreted as a sensitivity analysis of key parameters on the OPC specification process. The most significant parameters were then sorted by their effect magnitude (Fig. 5B).
Fig. 5. Factorial ANOVA model of individual and combinatorial effects of 12 culture parameters on expression of key markers in OPC specification.
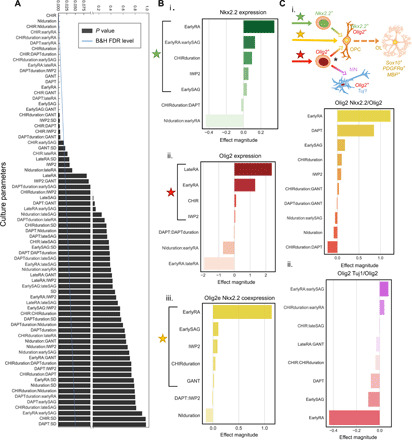
(A) Identification of statistically significant culture parameters using a factorial ANOVA of all single and pairwise effects on Nkx2.2 expression subject to the Benjamini and Hochberg false discovery rate (B&H FDR) correction. (B) Effect magnitude of significant culture parameters for i. Nkx2.2 expression, ii. Olig2 expression, iii. and coexpression of Olig2 and Nkx2.2. (C) i. Diagram summarizing results and effect magnitude of significant culture parameters for Olig2 and Nkx2.2 coexpression within the Olig2+ population and ii. effect magnitude of significant culture parameters for Olig2 and Tuj1 coexpression within the Olig2+ population.
RA, a rostrocaudal patterning cue, was among the most impactful parameters in this study for Olig2 and Nkx2.2 expression (Fig. 5Bi and ii). In particular, a high RA dose (1 μM) early in the differentiation (days 0 and 1) emerged as the most influential culture parameter in the acquisition of OPC fate (coexpression of Olig2 and Nkx2.2) (Fig. 5Bi to iii). In addition, the dose of SAG from days 4 to 10 of differentiation exerted a markedly more significant impact on OPC fate induction than from days 10 to 21 of differentiation, in line with the previous analysis (Fig. 4). IWP-2 and GANT were observed to correlate positively with coexpression of Olig2 and Nkx2.2 as well. Furthermore, this analysis identified two cases of culture parameters interacting in a synergistic manner to promote OPC differentiation. First, higher doses of RA during days 0 to 2 followed by SAG during days 4 to 10 were found to promote higher Nkx2.2 expression. In addition, longer CHIR duration (from days 0 to 4) along with higher GANT dose promoted coexpression of Nkx2.2 and Olig2.
We created a new differentiation protocol from the parameters isolated in this screen to have the most influence in specifying Olig2+Nkx2.2+ progenitors (Fig. 5Biii) and carried out the differentiation into the later stages of OPC maturation in a larger-scale format to assess the ability of this optimized protocol to create mature oligodendrocytes. The protocol was able to produce platelet-derived growth factor receptor α (PDGFRα)–expressing cells by day 60 across multiple hPSC lines, as well as O4-expressing cells by day 75 and myelin basic protein (MBP)– expressing cells and myelination ability at day 100 (fig. S10).
Microculture chip can be applied to multiple types of 3D hPSC differentiation screens
The OPC screening identified new conditions that affect cell differentiation, and we then sought to demonstrate the generalizability of this approach by conducting a different study. Specifically, we screened 90 unique hPSC differentiation protocols for tyrosine hydroxylase+ mDA neurons (Fig. 6). Exposure of CHIR was divided into three periods (early, middle, and late), and dosage for each period was varied independently. This screening strategy uncovered a key window of CHIR competence between days 3 and 7 (early), a negligible effect of CHIR between days 8 and 11 (middle), and an inhibitory effect of CHIR between days 12 and 25 (late) of mDA differentiation. These data further illustrate the existence of biphasic signaling activity during the differentiation process and underscore the need to improve the temporal dosing of several signaling agonists across a range of hPSC-derived CRTs.
Fig. 6. A generalizable platform for screening parameters for optimization of upstream CRT production.
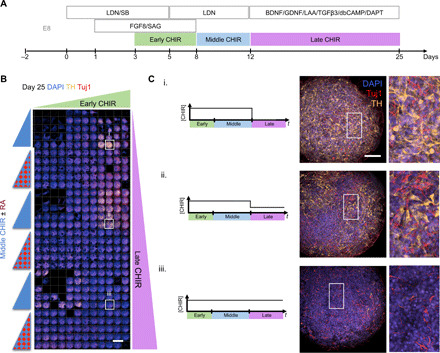
(A) Timeline of small-molecule addition for differentiation of mDA neurons from hPSCs. (B) Montage of 90 unique differentiation timeline to test temporal profiles of CHIR dose stained for tyrosine hydroxylase (TH) and Tuj1. Scale bar, 1 mm. (C) Immunocytochemistry images of i. low, ii. medium, and ii. high proportions of TH+ (yellow) neurons (red) dependent on the temporal profile of CHIR exposure. Scale bar, 100 μm.
DISCUSSION
The clinical emergence of several cell-based therapy candidates (47) is encouraging for human diseases/disorders that currently have no effective small molecule– or biologic-based therapy. As research and development into CRT candidates continues to progress, cell production has emerged as a bottleneck—as delivery vectors recently have in gene therapy—and improved tools will be necessary to enable higher quality and yield in cell manufacturing. Although previous studies have reported ~90% hPSC differentiation efficiency into Olig2+ progenitors using 2D culture formats (19), the 2D culture format constrains the space in which cells can expand to the surface area of the culture plate that limits the overall cell yield that can be produced. The adoption of scalable 3D culture formats, which have demonstrated the ability to produce up to fivefold higher quantities of cells per culture volume, shows promise in surpassing limits of 2D cell expansion (29–33) and could result in a higher overall production quantity of target cells even if differentiation efficiencies were lower than what has been reported in 2D. Therefore, the 3D screening and analysis strategy presented here is relevant for numerous emerging CRT candidates for which conversion of a stem or progenitor cell, such as a hPSCs (48), to a therapeutically relevant cell type requires searching through a large in vitro design space of doses, durations, dynamics, and combinations of signaling cues over several weeks of culture.
Notably, to emulate a ubiquitous and naturally occurring phenomenon in organismal development (16, 49), we dynamically varied key signaling cues in our screening strategy, tuning dosage over time. These analyses revealed new biological insights into the dynamic process by which cell competence to signals and fate are progressively specified (50). For example, by applying this platform to screen through several dynamic signaling levels simultaneously, we observed that the differentiation toward Nkx2.2+ progenitors is very sensitive to the dose of RA between days 0 and 1 and the dose of SAG between days 4 and 10. After these respective time windows, the effect of each respective signal in producing Nkx2.2+ progenitors is decreased, potentially pointing to a decrease in cellular competence to each of these signals over the course of OPC development. These cases of stage-specific responses to signaling cues, revealed by our screening platform, create a new dimension for future optimization of cell production.
To effectively navigate this enormous parameter space across doses, durations, dynamics, and combinations of signaling cues and resulting differentiation outcomes, we developed a robust sensitivity analysis strategy that can rank effect sizes to reveal which parameters should be the focus of optimization to modulate expression of target markers of interest (49) and, by contrast, which parameters exert minimal impact and can thus be neglected. For example, titration of RA dose will exert a significantly higher impact on differentiation efficiency than several other culture parameters combined. Furthermore, insights from this study could reduce the necessary quantity of SHH agonist by more than 50% to achieve similar levels of OPC differentiation. As these cell production processes translate from bench scale to industrial scale, awareness of key parameters that influence critical quality attributes (18) of the cell therapy product (such as expression of specific cellular markers) will be a necessary step in reliably producing these therapeutic cell types at scale for the clinic (51).
The wealth of combinatorial and temporal signaling patterns identified in this study can be analyzed in the context of CNS development as well. We observed a potential case of biphasic activity for the Wnt signaling pathway as both activation and inhibition appeared to increase expression of OPC markers Nkx2.2 and Olig2. In particular, this effect was seen with initial Wnt activation by CHIR during days 0 to 3 of OPC differentiation followed by inhibition by IWP-2 during days 4 to 21 of OPC differentiation. The Wnt pathway has shown stage-specific activity in cardiac and hematopoietic development (43, 44), which may thus be a conserved feature across several developmental systems. Wnt signals play an important role in the gastrulation of the embryo to form the primitive streak (38), yet in the subsequent stages of spinal cord development, Wnt signals induce a dorsalizing effect (52), whereas oligodendrocytes originate from the motor neuron domain on the ventral side. Therefore, suppressing endogenous Wnt signals in vitro after initial activation of Wnt may better recapitulate the natural developmental signaling environment of developing oligodendrocytes. Alternatively, as Wnt signals also play a role in rostrocaudal patterning of the CNS, these insights may further point toward a rostrocaudal region of the CNS during this developmental window that is optimal to recapitulate in vitro for OPC production. The oligodendrocytes created through this protocol, which expressed OTX2 at day 10 (fig. S2C), may resemble OPCs in the midbrain/hindbrain region. It is conceivable that exposure to the Wnt antagonist, IWP-2, induced a position rostral to the spinal cord during the differentiation window. This biphasic Wnt trend was seen again in our analysis of differentiation of mDA neurons, underscoring that stage-specific responses may be a conserved feature across several differentiation processes aiming to recapitulate a precise cellular position across several axes of patterning signals during natural development.
Furthermore, the statistical model identified an interaction between RA and SAG (an SHH agonist) in the early differentiation windows for specifying Nkx2.2+ progenitors (Fig. 5B), which has not been previously reported to our knowledge. In the developing CNS, RA signaling influences rostrocaudal positional identity, whereas SHH signaling specifies dorsoventral positional identity. Therefore, this statistical interaction found in the screen may represent intracellular cross-talk between the RA and SHH signaling pathways to integrate both patterning dimensions into Nkx2.2+ progenitor identity. This finding builds on what is known about RA and SHH signals for Olig2+ progenitor development in the spinal cord (53, 54).
Additionally, the 3D context of this screening platform enables high-throughput investigation into neurodevelopmental model systems that can offer unique perspectives beyond what is capable in 2D screening platforms, for example, by recapitulating cell-to-cell interactions, cytoskeletal arrangement, and multicellular patterning in 3D. The lumen structures that were observed during the neural induction period (fig. S2B and movie S1) in response to caudalizing conditions (high Wnt and RA) could be the basis of future organoid screening strategies to probe early multicellular arrangement and the effect of lumen size and shape on cell fate determination at various positions along the rostrocaudal and dorsoventral axes.
In conclusion, we demonstrate the versatile capabilities of a unique microculture platform for 3D differentiation screening and optimization of hPSC-derived cell therapies, whereby 1200 unique OPC differentiation timelines, and a total of over 4800 independent samples, were investigated using 0.2% of the reagent volumes required in a standard 96-well plate format. The dense dataset enabled subsequent statistical modeling for empirical optimization of the differentiation process and identified differential sensitivities to various culture parameters across time. These insights are important in developing strong process knowledge for manufacturing stem cell therapeutics as they continue to emerge in the clinic, and therefore, such screening strategies may accelerate the pace of discovery and development. Simultaneously, this combinatorial 3D hPSC differentiation screens may provide new insights on the basic biology of human development.
MATERIALS AND METHODS
hPSC culture
Human embryonic stem cells (H9s: National Institutes of Health Stem Cell Registry no. 0062) and hiPSCs (TCTFs: 8FLVY6C2, a gift from S. Li) were subcultured in monolayer format on a layer of 1% Matrigel and maintained in Essential 8 medium during expansion. At 80% confluency, H9s were passaged using Versene solution and replated at a 1:8 split.
3D pluripotent stem cell microculture on micropillar/microwell chip system
H9s were dissociated into single cells using Accutase solution and resuspended in Essential 8 medium containing 10 μM Y-27632 (ROCK Inhibitor). H9s were counted and resuspended at defined densities in 50% Matrigel solution on ice. While chilled, 100 nl of H9s in 50% Matrigel solution was deposited onto the micropillars at a density of 100 cells per pillar, unless otherwise noted, using a custom robotic liquid handling program and then incubated at 37°C for 20 min to promote gelation of 3D cultures. The micropillar chip was then inverted and placed into a fresh microwell chip containing cell culture media (table S1). All liquid dispensing into the microculture platform was performed with a DIGILAB OmniGrid Micro liquid handler with customized programs for deposition patterns. Between days −2 and 0, cells were kept in E8 media supplemented with 10 μM ROCK Inhibitor. Between days 0 and 10, cells were kept in differentiation media made of a base of 50% Dulbecco’s Modified Eagle’s Medium–F12, 50% Neurobasal, 0.5% penicillin/streptomycin (pen/strep), 1:100 GlutaMAX supplement, 1:50 B27 supplement, and 1:50 N2 supplement. Between days 10 and 21, cells were kept in differentiation media made of a base of 100% Neurobasal, 0.5% pen/strep, 1:100 GlutaMAX supplement, 1:50 B27 supplement, and 1:50 N2 supplement. After day 21, OPCs were transitioned to maturation media consisting of 100% Neurobasal, 0.5% pen/strep, 1:100 GlutaMAX supplement, 1:50 B27 supplement, 1:50 N2 supplement, insulin-like growth factor 1 (10 ng/ml), platelet-derived growth factor (PDGF)–AA (10 ng/ml), NT-3 (10 ng/ml), and insulin (25 μg/ml). Media were changed daily by transferring the micropillar chip into a microwell chip containing fresh media every other day using a custom-made mechanical “Chip Swapper” for consistent transfer. Technical replicates included two different dispensing patterns to average out positional effects across the microchip.
On-chip viability assay
At the endpoint of the experiment, the micropillar chip was carefully removed from the microwell chip and placed in new microwell chip containing calcein AM, ethidium homodimer, and Hoechst diluted in sterile phosphate-buffered saline (PBS) (dilution details in table S1). The micropillar chip was incubated for 20 min and then transferred to a new microwell chip containing PBS, and individual microenvironments were imaged using fluorescence microscopy.
On-chip immunofluorescence assays
At the endpoint of the experiment, the micropillar chip was carefully removed from the wellchip and placed into a bath of 4% paraformaldehyde for 15 min to fix cell cultures. Then, the micropillar chip was washed twice in PBS for 5 min each and placed into a bath of 0.25% Triton X-100 + 5% donkey serum in PBS for 10 min to permeabilize cells. After permeabilization, the micropillar chip was washed five times in 5% donkey serum for 5 min each, transferred to a wellchip containing primary antibodies of interest diluted in PBS + donkey serum (dilution details in table S1), and stored overnight at 4°C. After primary staining, the micropillar chip was washed twice in PBS for 5 min each, placed into a microwell chip containing the corresponding secondary antibodies (dilution details in table S1), and incubated at 37°C for 2 hours. After secondary staining, the micropillar chip was washed twice in PBS for 5 min each and placed into a wellchip containing PBS; individual microenvironments were imaged using fluorescence confocal microscopy.
High-throughput fluorescence microscopy
Stained micropillar chips were sealed with a polypropylene film (GeneMate T-2452-1) and imaged with a 20× objective using a Perkin Elmer Opera Phenix automated confocal fluorescence microscope available in the High-Throughput Screening Facility at University of California, Berkeley. Laser exposure time and power were kept constant for a fluorescence channel within an imaging set. Images were scored for marker expression depending on nuclear or cytoplasmic localization (fig. S3).
Two-photon microscopy
Fixed cultures on micropillars at day 15 were stained with 4′,6-diamidino-2-phenylindole (DAPI) and imaged using an upright Olympus BX51WI microscope (Olympus Corporation) equipped with swept field confocal technology (Bruker) and a Ti:sapphire two-photon Chameleon Ultra II laser (Coherent) was used. The two-photon laser was set to 405 nm, and images were captured using an electron multiplying charge-coupled device camera (Photometrics). Prairie View Software (v. 5.3 U3, Bruker) was used to acquire images, and ImageJ software was used to create a video of the z-series.
Statistical methods
Quantified image data were then imported into Python for statistical data analysis (55) and visualization. For comparisons between datasets acquired across different experimental sessions, raw data were scaled and centered by z score, and descriptive statistics were calculated from four technical replicates. Error bars represent 95% confidence intervals, unless otherwise specified. For the hierarchical cluster model, the Euclidean distance was used to measure pairwise distance between each observation, and the unweighted pair group method with arithmetic mean (UPGMA) algorithm was used to calculate the linkage pattern. A Benjamini and Hochberg false discovery rate correction was applied as needed to correct for multiple comparisons. Code is available upon request.
Supplementary Material
Acknowledgments
We thank M. West of the High-Throughput Screening Facility (HTSF) at UC Berkeley and E. Granlund of the College of Chemistry machine shop for machining custom parts. In addition, we are grateful to G. Rodrigues, M. Adil, and J. Zimmermann for participating in the discussions on the work. Funding: This research was supported by the California Institute for Regenerative Medicine (DISC-08982) and the NIH (R01-ES020903) and Instrumentation Grant (S10OD021828) that provided the Perkin Elmer Opera Phenix microscope. R.M. was supported in part by an NSF Graduate Research Fellowship. Author contributions: R.M., D.S.C., and D.V.S. conceived various parts of the project and supervised the study. R.M. designed the experiments and managed the project workflows. X.B. created Nkx2.2-Cre H9 reporter lines. R.M., E.T., and E.C. performed the experiments. R.M. conducted statistical modeling, and A.M. aided in statistical testing. R.M., D.S.C., and D.V.S. analyzed and interpreted the data. R.M. wrote the manuscript with revisions from J.S.D., D.S.C., and D.V.S. Competing interests: R.M., D.S.C., and D.V.S. are inventors on a U.S. patent pending related to this work filed by the University of California, Berkeley (PCT/US2020/029553, filed on 23 April 2020). D.V.S. is the inventor on two U.S. patent pendings related to this work filed by the University of California, Berkeley (PCT/US2016/055362, filed on 4 October 2016; no. PCT/US2016/055361, filed on 5 October 2015). All other authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/32/eaaz1457/DC1
REFERENCES AND NOTES
- 1.Goldman S. A., Stem and progenitor cell-based therapy of the central nervous system: Hopes, hype, and wishful thinking. Cell Stem Cell 18, 174–188 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manley N. C., Priest C. A., Denham J., Wirth E. D. III, Lebkowski J. S., Human embryonic stem cell-derived oligodendrocyte progenitor cells: Preclinical efficacy and safety in cervical spinal cord injury. Stem Cells Transl. Med. 6, 1917–1929 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Douvaras P., Wang J., Zimmer M., Hanchuk S., O’Bara M. A., Sadiq S., Sim F. J., Goldman J., Fossati V., Efficient generation of myelinating oligodendrocytes from primary progressive multiple sclerosis patients by induced pluripotent stem cells. Stem Cell Reports 3, 250–259 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang S., Bates J., Li X., Schanz S., Chandler-Militello D., Levine C., Maherali N., Studer L., Hochedlinger K., Windrem M., Goldman S. A., Human iPSC-derived oligodendrocyte progenitor cells can myelinate and rescue a mouse model of congenital hypomyelination. Cell Stem Cell 12, 252–264 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Piao J., Major T., Auyeung G., Policarpio E., Menon J., Droms L., Gutin P., Uryu K., Tchieu J., Soulet D., Tabar V., Human embryonic stem cell-derived oligodendrocyte progenitors remyelinate the brain and rescue behavioral deficits following radiation. Cell Stem Cell 16, 198–210 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hallett P. J., Deleidi M., Astradsson A., Smith G. A., Cooper O., Osborn T. M., Sundberg M., Moore M. A., Perez-Torres E., Brownell A.-L., Schumacher J. M., Spealman R. D., Isacson O., Successful function of autologous iPSC-derived dopamine neurons following transplantation in a non-human primate model of Parkinson’s disease. Cell Stem Cell 16, 269–274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parmar M., Grealish S., Henchcliffe C., The future of stem cell therapies for Parkinson disease. Nat. Rev. Neurosci. 21, 103–115 (2020). [DOI] [PubMed] [Google Scholar]
- 8.French D. D., Campbell R. R., Sabharwal S., Nelson A. L., Palacios P. A., Gavin-Dreschnack D., Health care costs for patients with chronic spinal cord injury in the Veterans Health Administration. J. Spinal Cord Med. 30, 477–481 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Priest C. A., Manley N. C., Denham J., Wirth E. D. III, Lebkowski J. S., Preclinical safety of human embryonic stem cell-derived oligodendrocyte progenitors supporting clinical trials in spinal cord injury. Regen. Med. 10, 939–958 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Douvaras P., Fossati V., Generation and isolation of oligodendrocyte progenitor cells from human pluripotent stem cells. Nat. Protoc. 10, 1143–1154 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Izrael M., Zhang P., Kaufman R., Shinder V., Ella R., Amit M., Itskovitz-Eldor J., Chebath J., Revel M., Human oligodendrocytes derived from embryonic stem cells: Effect of noggin on phenotypic differentiation in vitro and on myelination in vivo. Mol. Cell. Neurosci. 34, 310–323 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Tao Y., Zhang S.-C., Neural Subtype Specification from Human Pluripotent Stem Cells. Cell Stem Cell 19, 573–586 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edlund T., Jessell T. M., Progression from extrinsic to intrinsic signaling in cell fate specification: A view from the nervous system. Cell 96, 211–224 (1999). [DOI] [PubMed] [Google Scholar]
- 14.Jessell T. M., Neuronal specification in the spinal cord: Inductive signals and transcriptional codes. Nat. Rev. Genet. 1, 20–29 (2000). [DOI] [PubMed] [Google Scholar]
- 15.Nordström U., Jessell T. M., Edlund T., Progressive induction of caudal neural character by graded Wnt signaling. Nat. Neurosci. 5, 525–532 (2002). [DOI] [PubMed] [Google Scholar]
- 16.Sagner A., Briscoe J., Morphogen interpretation: Concentration, time, competence, and signaling dynamics. Wiley Interdiscip. Rev. Dev. Biol. 6, e271 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saha K., Schaffer D. V., Signal dynamics in Sonic hedgehog tissue patterning. Development 133, 889–900 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Zhou Q., Choi G., Anderson D. J., The bHLH transcription factor Olig2 promotes oligodendrocyte differentiation in collaboration with Nkx2.2. Neuron 31, 791–807 (2001). [DOI] [PubMed] [Google Scholar]
- 19.Maury Y., Côme J., Piskorowski R. A., Salah-Mohellibi N., Chevaleyre V., Peschanski M., Martinat C., Nedelec S., Combinatorial analysis of developmental cues efficiently converts human pluripotent stem cells into multiple neuronal subtypes. Nat. Biotechnol. 33, 89–96 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Desbordes S. C., Studer L., Adapting human pluripotent stem cells to high-throughput and high-content screening. Nat. Protoc. 8, 111–130 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Titmarsh D. M., Glass N. R., Mills R. J., Hidalgo A., Wolvetang E. J., Porrello E. R., Hudson J. E., Cooper-White J. J., Induction of human iPSC-derived cardiomyocyte proliferation revealed by combinatorial screening in high density microbioreactor arrays. Sci. Rep. 6, 24637 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grealish S., Diguet E., Kirkeby A., Mattsson B., Heuer A., Bramoulle Y., Van Camp N., Perrier A. L., Hantraye P., Björklund A., Parmar M., Human ESC-derived dopamine neurons show similar preclinical efficacy and potency to fetal neurons when grafted in a rat model of Parkinson’s disease. Cell Stem Cell 15, 653–665 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krohne T. U., Westenskow P. D., Kurihara T., Friedlander D. F., Lehmann M., Dorsey A. L., Li W., Zhu S., Schultz A., Wang J., Siuzdak G., Ding S., Friedlander M., Generation of retinal pigment epithelial cells from small molecules and OCT4 reprogrammed human induced pluripotent stem cells. Stem Cells Transl. Med. 1, 96–109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pennington B. O., Clegg D. O., Melkoumian Z. K., Hikita S. T., Defined culture of human embryonic stem cells and xeno-free derivation of retinal pigmented epithelial cells on a novel, synthetic substrate. Stem Cells Transl. Med. 4, 165–177 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang L., Soonpaa M. H., Adler E. D., Roepke T. K., Kattman S. J., Kennedy M., Henckaerts E., Bonham K., Abbott G. W., Linden R. M., Field L. J., Keller G. M., Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature 453, 524–528 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Butts J. C., McCreedy D. A., Martinez-Vargas J. A., Mendoza-Camacho F. N., Hookway T. A., Gifford C. A., Taneja P., Noble-Haeusslein L., McDevitt T. C., Differentiation of V2a interneurons from human pluripotent stem cells. Proc. Natl. Acad. Sci. U.S.A. 114, 4969–4974 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pagliuca F. W., Millman J. R., Gürtler M., Segel M., Van Dervort A., Ryu J. H., Peterson Q. P., Greiner D., Melton D. A., Generation of functional human pancreatic β cells in vitro. Cell 159, 428–439 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lian X., Zhang J., Azarin S. M., Zhu K., Hazeltine L. B., Bao X., Hsiao C., Kamp T. J., Palecek S. P., Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions. Nat. Protoc. 8, 162–175 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Serra M., Brito C., Correia C., Alves P. M., Process engineering of human pluripotent stem cells for clinical application. Trends Biotechnol. 30, 350–359 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Lei Y., Schaffer D. V., A fully defined and scalable 3D culture system for human pluripotent stem cell expansion and differentiation. Proc. Natl. Acad. Sci. U.S.A. 110, E5039–E5048 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodrigues G. M. C., Gaj T., Adil M. M., Wahba J., Rao A. T., Lorbeer F. K., Kulkarni R. U., Diogo M. M., Cabral J. M. S., Miller E. W., Hockemeyer D., Schaffer D. V., Defined and scalable differentiation of human oligodendrocyte precursors from pluripotent stem cells in a 3d culture system. Stem Cell Reports 8, 1770–1783 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adil M. M., Rodrigues G. M. C., Kulkarni R. U., Rao A. T., Chernavsky N. E., Miller E. W., Schaffer D. V., Efficient generation of hPSC-derived midbrain dopaminergic neurons in a fully defined, scalable, 3D biomaterial platform. Sci. Rep. 7, 40573 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adil M. M., Gaj T., Rao A. T., Kulkarni R. U., Fuentes C. M., Ramadoss G. N., Ekman F. K., Miller E. W., Schaffer D. V., hPSC-derived striatal cells generated using a scalable 3D hydrogel promote recovery in a huntington disease mouse model. Stem Cell Reports 10, 1481–1491 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ranga A., Lutolf M. P., High-throughput approaches for the analysis of extrinsic regulators of stem cell fate. Curr. Opin. Cell Biol. 24, 236–244 (2012). [DOI] [PubMed] [Google Scholar]
- 35.Muckom R., Farland S. M., Yang C., Perea B., Gentes M., Murugappan A., Tran E., Dordick J. S., Clark D. S., Schaffer D. V., High-throughput combinatorial screening reveals interactions between signaling molecules that regulate adult neural stem cell fate. Biotechnol. Bioeng. 116, 193–205 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nierode G. J., Gopal S., Kwon P., Clark D. S., Schaffer D. V., Dordick J. S., High-throughput identification of factors promoting neuronal differentiation of human neural progenitor cells in microscale 3D cell culture. Biotechnol. Bioeng. 116, 168–180 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bao X., Adil M. M., Muckom R., Zimmermann J. A., Tran A., Suhy N., Xu Y., Sampayo R. G., Clark D. S., Schaffer D. V., Gene editing to generate versatile human pluripotent stem cell reporter lines for analysis of differentiation and lineage tracing. Stem Cells 37, 1556–1566 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Liu P., Wakamiya M., Shea M. J., Albrecht U., Behringer R. R., Bradley A., Requirement for Wnt3 in vertebrate axis formation. Nat. Genet. 22, 361–365 (1999). [DOI] [PubMed] [Google Scholar]
- 39.Maden M., Retinoids and spinal cord development. J. Neurobiol. 66, 726–738 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Chambers S. M., Fasano C. A., Papapetrou E. P., Tomishima M., Sadelain M., Studer L., Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 27, 275–280 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roelink H., Porter J. A., Chiang C., Tanabe Y., Chang D. T., Beachy P. A., Jessell T. M., Floor plate and motor neuron induction by different concentrations of the amino-terminal cleavage product of sonic hedgehog autoproteolysis. Cell 81, 445–455 (1995). [DOI] [PubMed] [Google Scholar]
- 42.Amoroso M. W., Croft G. F., Williams D. J., O’Keeffe S., Carrasco M. A., Davis A. R., Roybon L., Oakley D. H., Maniatis T., Henderson C. E., Wichterle H., Accelerated high-yield generation of limb-innervating motor neurons from human stem cells. J. Neurosci. 33, 574–586 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Naito A. T., Naito A. T., Shiojima I., Akazawa H., Hidaka K., Morisaki T., Kikuchi A., Komuro I., Developmental stage-specific biphasic roles of Wnt/β-catenin signaling in cardiomyogenesis and hematopoiesis. Proc. Natl. Acad. Sci. U.S.A. 103, 19812–19817 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lian X., Hsiao C., Wilson G., Zhu K., Hazeltine L. B., Azarin S. M., Raval K. K., Zhang J., Kamp T. J., Palecek S. P., Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc. Natl. Acad. Sci. U.S.A. 109, E1848–E1857 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kong J. H., Yang L., Dessaud E., Chuang K., Moore D. M., Rohatgi R., Briscoe J., Novitch B. G., Notch activity modulates the responsiveness of neural progenitors to sonic hedgehog signaling. Dev. Cell 33, 373–387 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Benjamini Y., Hochberg Y., Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B. Methodol. 57, 289–300 (1995). [Google Scholar]
- 47.Alliance for Regenerative Medicine, Annual Regenerative Medicine Data Report, (2018).
- 48.Trounson A., DeWitt N. D., Pluripotent stem cells progressing to the clinic. Nat. Rev. Mol. Cell Biol. 17, 194–200 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Kutejova E., Briscoe J., Kicheva A., Temporal dynamics of patterning by morphogen gradients. Curr. Opin. Genet. Dev. 19, 315–322 (2009). [DOI] [PubMed] [Google Scholar]
- 50.Spitzer S. O., Sitnikov S., Kamen Y., Evans K. A., Kronenberg-Versteeg D., Dietmann S., de Faria O. Jr., Agathou S., Káradóttir R. T., Oligodendrocyte progenitor cells become regionally diverse and heterogeneous with age. Neuron 101, 459–471.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lipsitz Y. Y., Timmins N. E., Zandstra P. W., Quality cell therapy manufacturing by design. Nat. Biotechnol. 34, 393–400 (2016). [DOI] [PubMed] [Google Scholar]
- 52.Le Dréau G., Martí E., Dorsal-ventral patterning of the neural tube: A tale of three signals. Dev. Neurobiol. 72, 1471–1481 (2012). [DOI] [PubMed] [Google Scholar]
- 53.Novitch B. G., Wichterle H., Jessell T. M., Sockanathan S., A requirement for retinoic acid-mediated transcriptional activation in ventral neural patterning and motor neuron specification. Neuron 40, 81–95 (2003). [DOI] [PubMed] [Google Scholar]
- 54.Ribes V., Le Roux I., Rhinn M., Schuhbaur B., Dollé P., Early mouse caudal development relies on crosstalk between retinoic acid, Shh and Fgf signalling pathways. Development 136, 665–676 (2009). [DOI] [PubMed] [Google Scholar]
- 55.Pal R., Mamidi M. K., Das A. K., Bhonde R., Diverse effects of dimethyl sulfoxide (DMSO) on the differentiation potential of human embryonic stem cells. Arch. Toxicol. 86, 651–661 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/32/eaaz1457/DC1


