Abstract
Male infertility affects ∼7% of men, but its causes remain poorly understood. The most severe form is non-obstructive azoospermia (NOA), which is, in part, caused by an arrest at meiosis. So far, only a few validated disease-associated genes have been reported. To address this gap, we performed whole-exome sequencing in 58 men with unexplained meiotic arrest and identified the same homozygous frameshift variant c.676dup (p.Trp226LeufsTer4) in M1AP, encoding meiosis 1 associated protein, in three unrelated men. This variant most likely results in a truncated protein as shown in vitro by heterologous expression of mutant M1AP. Next, we screened four large cohorts of infertile men and identified three additional individuals carrying homozygous c.676dup and three carrying combinations of this and other likely causal variants in M1AP. Moreover, a homozygous missense variant, c.1166C>T (p.Pro389Leu), segregated with infertility in five men from a consanguineous Turkish family. The common phenotype between all affected men was NOA, but occasionally spermatids and rarely a few spermatozoa in the semen were observed. A similar phenotype has been described for mice with disruption of M1ap. Collectively, these findings demonstrate that mutations in M1AP are a relatively frequent cause of autosomal recessive severe spermatogenic failure and male infertility with strong clinical validity.
Keywords: male infertility, non-obstructive azoospermia, spermatogenesis, meiotic arrest, M1AP, cryptozoospermia, oligozoospermia, spermatogenic failure, meiosis 1 associated protein
Main Text
Around 7% of all men in Western societies experience infertility,1 which is primarily diagnosed by semen analysis comprising sperm concentration and count as the most relevant parameters. More than 10% of all infertile men exhibit azoospermia2—the absence of spermatozoa in the ejaculate. Azoospermia constitutes the most challenging and clinically severe form of male infertility and is further classified into obstructive azoospermia (OA) with normal spermatogenesis and non-obstructive azoospermia (NOA) due to impaired spermatogenesis. In some men, a few spermatozoa can be identified after centrifugation of the semen, which is denoted as cryptozoospermia. From the biological point of view, NOA and cryptozoospermia are closely related, just as they are in their clinical implications, i.e., virtually no chance of natural conception.3
The variable spermatogenic impairment in these men correlates to a diverse spectrum of testicular histological phenotypes. This spectrum includes Sertoli cell-only (SCO), over maturation arrest, and hypospermatogenesis, and all of these can be complete, focal, or mixed. Maturation arrest most frequently presents as meiotic arrest in which spermatocytes are the most advanced germ cell types in the testes. If germ cell arrest is complete, no mature spermatozoa develop; testicular biopsy and sperm extraction (TESE) will not be successful and assisted reproductive technology (ART) will not be possible.
In a large fraction of severely disturbed spermatogenesis, a genetic origin is assumed,4 and affected men are routinely screened for chromosomal aberrations and Y chromosome azoospermia factor (AZF) microdeletions. Yet these diagnostic tests only establish a causal diagnosis in 15%–20% of azoospermia-affected individuals.2 Recently, monogenic alterations associated with germ cell arrest in human males have been described.5,6 However, according to a standardized clinical validity assessment, the X chromosome gene TEX11 (MIM: 300311) currently remains the only one in which variants are associated with male infertility with strong evidence.7 Given the large number of genes in which variants are known to cause meiotic arrest in mice, the vast majority of mutations causing this phenotype in humans are yet to be identified.
To this end, we first screened the exomes of well-characterized men with complete bilateral meiotic arrest and identified bi-allelic loss-of-function (LoF) variants in the gene encoding meiosis 1 associated protein (M1AP) in three unrelated men. In mice, M1ap is primarily expressed in male germ cells throughout spermatogenesis, and its knockout leads to infertility due to meiotic arrest and severe oligozoospermia.8,9 Our subsequent analyses in four independent cohorts and a consanguineous Turkish family, as well as in vitro analyses of a recurring M1AP frameshift variant, corroborated that disruption of M1AP is associated with a variable spectrum of severely impaired spermatogenesis, mostly at meiosis and resulting in azoospermia, but also compatible with sparse postmeiotic germ cell development and retrieval of sperm in some instances.
We originally selected 64 azoospermic but otherwise healthy males who attended the Centre of Reproductive Medicine and Andrology (CeRA), University Hospital Münster (n = 51) or the Clinic for Urology, Pediatric Urology and Andrology, Gießen (n = 13) for couple infertility. All men were diagnosed with complete bilateral germ cell arrest at the spermatocyte stage after the evaluation of at least 100 seminiferous tubules in tissue sections of both testes accompanied by a negative TESE outcome, i.e., no sperm could be recovered. This is a subset of all individuals included in our large-scale Male Reproductive Genomics (MERGE) study, which currently comprises 735 men with lacking or severe quantitatively impaired spermatogenesis and 53 individuals with normal spermatogenesis (OA and controls) (Figure S1). Specifically, we performed whole-exome sequencing (WES; for details, see Supplemental Methods) in 569 men with NOA, 116 with cryptozoospermia, and 50 with severe oligozoospermia (sperm concentration < 5 M/mL). Chromosomal aberrations and AZF deletions were excluded in this and all other cohorts and subjects (detailed below). All participants gave written informed consent and the study protocol was approved by the respective ethics committees and institutional review boards (details in Supplemental Data).
We identified likely causal variants in the three genes TEX11, STAG3, and SYCP2 in six of the men with complete meiotic arrest.5,10,11 The WES data of the remaining 58 men were filtered for rare (minor allele frequency [MAF] < 0.01 according to the Genome Aggregation Database12 [gnomAD]) bi-allelic LoF variants. Affected genes were prioritized with regard to the level of expression in the testes and previous evidence for an association with infertility in either human or model species (Figure S1). The highest-ranked gene was M1AP because three unrelated men (M330, M864, and M1792, Table 1) carried the same homozygous LoF variant (c.676dup, MAF = 0.0021, no homozygotes in gnomAD12), the M1AP mRNA displayed the highest expression in the testis (according to both the Genotype-Tissue Expression [GTEx] project13 and the Human Protein Atlas [HPA]), and it was shown to play a crucial role in spermatogenesis in mice.8,9 The c.676dup variant in M1AP was confirmed by Sanger sequencing (GenBank: NM_138804.4, the longest isoform with highest testis expression; for primer sequences see Table S1) in all affected men from the MERGE cohort. Testis biopsies of all three individuals were collected for TESE and research use. These were fixed in Bouin’s solution and embedded in paraffin, and subsequently, sections were stained with periodic acid-Schiff (PAS) as previously described.14 These were re-analyzed to confirm complete meiotic arrest (Figure 1). Because DNA from the three individuals’ parents was not available, and to exclude a hemizygous deletion on the other allele, quantitative PCR (qPCR) of M1AP’s exon 5 was performed on gDNA (primers and conditions in Table S1 and the Supplemental Methods). This excluded an intragenic deletion in all individuals (Table S2). No regions of homozygosity (ROHs) involving M1AP were detected for any of the affected men, rendering consanguinity of their parents unlikely. We also did not notice evidence for consanguinity between the men (analysis by H3M2 and vcftools algorithms,15 data not shown).
Table 1.
Genetic and Clinical Data of Infertile Men with M1AP Variants
| Individual | Age, Origin | M1AP Variant | Fertility Parameters | Testicular Phenotype, TESE Outcome |
|---|---|---|---|---|
| M330 | 38 years, Germany | c.[676dup];[676dup], p.[Trp226LeufsTer4];[Trp226LeufsTer4] | FSH, 9; LH, 5.3; T, 14.6; TV, 17/23; azoospermia | meiotic arrest (0/0% tubules with ES, 0/0% RS, 91/99% SC, 6/1% SG, 3/0% SCO, 0/0% TS), TESE negative |
| M864 | 41 years, Germany | c.[676dup];[676dup], p.[Trp226LeufsTer4];[Trp226LeufsTer4] | FSH, 4.7; LH, 1.5; T, 9.6; TV, 19/26; azoospermia | meiotic arrest (0/0% tubules with ES, 0/0% RS, 71/91% SC, 10/4% SG, 17/1% SCO, 2/4% TS), TESE negative |
| M1792 | 36 years, Germany | c.[676dup];[676dup], p.[Trp226LeufsTer4];[Trp226LeufsTer4] | FSH, 7.8; LH, 5.1; T, 10.1; TV, 15/15; azoospermia | meiotic arrest (0/0% tubules with ES, 0/0% RS, 96/97% SC, 0/2% SG, 3/1% SCO, 1/0% TS), TESE negative |
| M1943 | 43 years, Croatia | c.[676dup];[797G>A], p.[Trp226LeufsTer4];[Arg266Gln] | FSH, 5.5; LH, 3.2; T, 43.6; TV, 28/20; cryptozoospermia or azoospermiaa | N/A |
| M2062 | 26 years, Poland | c.[676dup];[676dup], p.[Trp226LeufsTer4];[Trp226LeufsTer4] | FSH, 3.5; LH, 3.6; T, 18.6; TV, 26/23; cryptozoospermia or azoospermiaa | N/A |
| Y126 | 34 years, Portugal | c.[676dup];[949G>A], p.[Trp226LeufsTer4];[Gly317Arg] | FSH, 6.7; LH, 3.2; T, N/A; TV, N/A; azoospermia | maturation arrest at round spermatid stage, (quantification N/A), TESE negative |
| P86 | 44 years, Portugal | c.[148T>C];[1289T>C], p.[Ser50Pro];[Leu430Pro] | FSH, N/A; LH, N/A; T, N/A; TV, N/A; azoospermia | dispersed Sertoli cells, some tubules contained only spermatogonia, (quantification N/A), TESE negative |
| RU01691 | 41 years, the Netherlands | c.[676dup];[676dup], p.[Trp226LeufsTer4];[Trp226LeufsTer4] | FSH, 5; LH, 2.0; T, 11.3; TV, N/A; azoospermia | predominant meiotic arrest with occasional spermatids, (unilateral TESE: 4% tubules with ES, 5% RS, 88% SC, 2% SG, 0% SCO, 0% TS), TESE positive |
| MI-0006-P | 33 years, UK | c.[676dup];[676dup], p.[Trp226LeufsTer4];[Trp226LeufsTer4] | FSH, 10.4; LH, N/A; T, 15.4; TV, 20/20; azoospermia | predominant meiotic arrest with occasional postmeiotic germ cells (quantification N/A), TESE negative |
| T1024 | 28 years, Turkey | c.[1166C>T];[1166C>T], p.[Pro389Leu];[Pro389Leu] | FSH, 8.3; LH, 4.4; T, 8.8; TV, 15/15; azoospermia | maturation arrest at round spermatid stage, (quantification N/A), TESE negative |
| F1: II-116 (Tu et al.) | 34 years, China | c.[1435−1G>A];[1435−1G>A], p.? | FSH, 3.65; LH, 3.17; T, 18.24; TV, 12/12; severe oligozoospermia | N/A |
Abbreviations are as follows: FSH, follicle-stimulating hormone (IU/L); LH, luteinizing hormone (IU/L); T, testosterone (nmol/L); TV, testicular volume right/left (mL); ES, elongating spermatids; RS, round spermatids; SC, spermatocytes; SG, spermatogonia; SCO, Sertoli cell-only; TS, tubular shadows; N/A, not available. Reference values: FSH 1–7 IU/L, LH 2–10 IU/L, T > 12 nmol/L, TV > 15 mL per testis.
Semen contained none or below 10 spermatozoa/sample on repeated analyses.
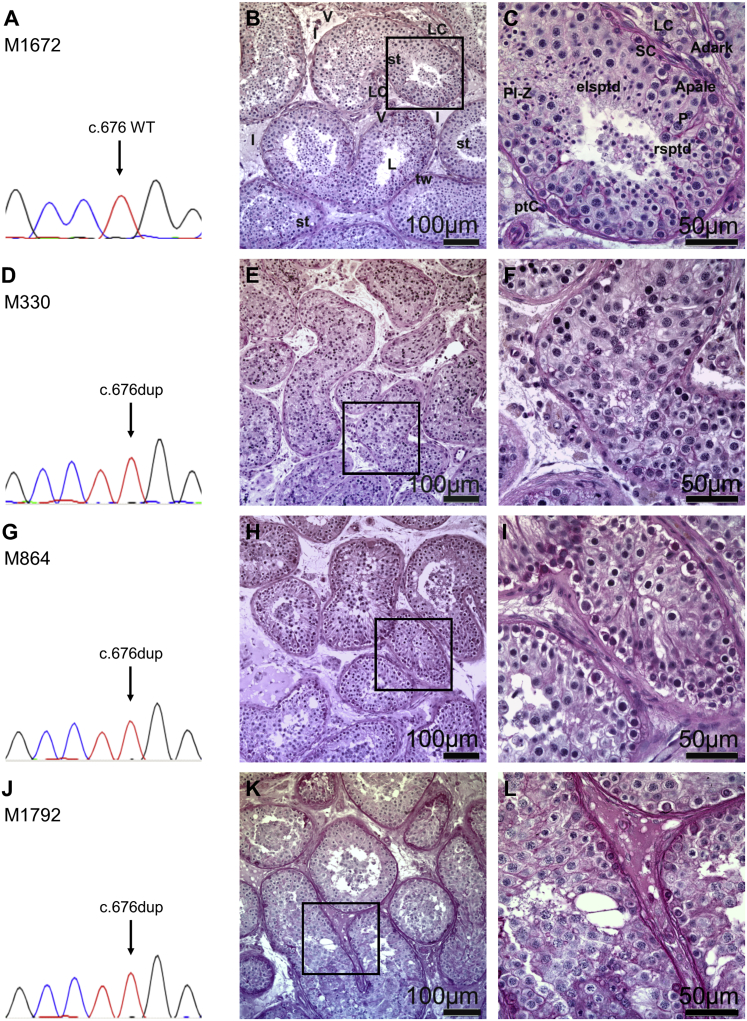
Figure 1.
Recurrent Homozygous Variant c.676dup in M1AP Leading to Complete Bilateral Meiotic Arrest in Three Individuals from the MERGE Study
(A) Electropherogram with the wild-type sequence of M1AP exon 5 (M1672 with obstructive azoospermia).
(B and C) Testicular tissue showing complete spermatogenesis, PAS staining.
(B) Testicular tissues are composed of seminiferous tubules and interstitium. The seminiferous tubules are separated from the interstitial space (I) by tubular walls (tws) formed by myoid peritubular cells and the lamina propria. Inside, the seminiferous epithelium and the lumen (L) are localized. In the interstitium, groups of steroidogenic Leydig cells (LCs) and blood vessels (Vs) are observed. Tubular cross-section showed the regular appearance of a functioning testis exhibiting complete germ cell differentiation.
(C) Detail of B; the tubules are surrounded by the lamina propria and the peritubular cells (ptCs), forming the wall. Within the seminiferous epithelium, somatic Sertoli cells (SCs) are supporting the germ cells differentiating from A spermatogonia (Apale/Adark) via premeiotic spermatocytes (preleptotene to zygotene stage; Pl-Z) into the meiotic pachytene spermatocytes (Ps). After meiosis is completed, haploid round spermatids (rsptds), which mature further into elongated spermatids (elsptds), are formed.
(D–L) Identification of a recurrent homozygous variant in M1AP c.676dup (p.Trp226LeufsTer4). Sanger sequencing verified the variant in M330 (D), M864 (G), and M1792 (J), leading to complete bilateral meiotic arrest as indicated by histological examination of testis biopsies (M330 [E, magnified in F], M864 [H, magnified in I], M1792 [K, magnified in L]), which show spermatocytes as the most advanced germ cells in all tubules. Magnified areas and scale bars are indicated.
By screening the complete MERGE cohort, we identified an additional man (M2062) carrying the same homozygous LoF variant, c.676dup (Figure S2), and another man (M1943) carrying c.676dup in combination with a second rare missense variant, c.797G>A (p.Arg266Gln). This missense variant is consistently predicted as pathogenic by all in silico prediction programs (PolyPhen-2, SIFT, MutationTaster, and HOPE17). Both men had varying azoospermia and cryptozoospermia in repeated semen analyses and did not undergo a testicular biopsy. In contrast, no individuals with two rare M1AP variants predicted as pathogenic via in silico programs were observed in the remaining MERGE cohort or in the individuals with spermatogenic impairment and other testicular phenotypes, such as SCO (n = 213), or normal spermatogenesis (OA or controls, n = 53).
Next, through collaborations established within the International Male Infertility Genomics Consortium (IMIGC), three cohorts of infertile men in whom WES was performed in independent studies (details are provided in the Supplemental Methods) were screened for bi-allelic variants in M1AP: 930 men with unexplained NOA from the Genetics of Male Infertility Initiative (GEMINI) study, 283 men with unexplained azoospermia (n = 214) or oligozoospermia (n = 69) who presented at Radboud University Medical Center (Radboudumc, Nijmegen), and 48 men with unexplained azoospermia (n = 36) or oligozoospermia (n = 12) recruited at The Newcastle upon Tyne Hospitals NHS Foundation Trust (Newcastle, UK). From these 1,261 individuals in total, we identified four additional infertile men with likely bi-allelic variants in M1AP. Clinical data of all individuals carrying M1AP variants is shown in Table 1. The variants, gnomAD frequencies, in silico predictions, and classification according to the guidelines of the American College of Medical Genetics and Genomics (ACMG) and the Association for Molecular Pathology (AMP)18 are given in Table 2 (for detailed variant description and interpretation, see Supplemental Note). On the basis of RNA analyses from public sources (GTEx and HPA), M1AP might also be expressed in the bone marrow and other tissues, such as the pituitary. However, no blood-system-related or hormonal abnormalities were noted in any of the affected men reported herein.
Table 2.
Assessment of M1AP Variants
| cDNA Change | Protein Change | In Silico Prediction for Missense Variants (PolyPhen-2/SIFT/MutationTaster) | MAFa(gnomAD) | MAF (Local Controls) | Conservation | Classification According to ACMG-AMP Guidelines18 |
|---|---|---|---|---|---|---|
| c.676dup | p.Trp226LeufsTer4 | N/A | 0.0021a | 0.0088 | N/A | pathogenic |
| c.1435−1G>A16 | p.? | N/A | 0 | ND | N/A | pathogenic |
| c.148T>C | p.Ser50Pro | T/P/D | 0 | ND | platypus | uncertain significance |
| c.797G>A | p.Arg266Gln | D/D/D | 0.0002 | ND | zebrafish | uncertain significance |
| c.949G>A | p.Gly317Arg | D/D/D | 0.00007 | ND | platypus | uncertain significance |
| c.1166C>T | p.Pro389Leu | D/D/D | 0.00001 | ND | tetraodon | uncertain significance |
| c.1289T>C | p.Leu430Pro | D/D/D | 0.000008 | ND | platypus | uncertain significance |
Abbreviations are as follows: D, damaging, deleterious, or disease-causing; B, benign; T, tolerated; P, polymorphism; MAF, minor allele frequency; N/A, not applicable; ND, not determined.
Overall MAF is presented. This is, for example, slightly higher for the recurring variant c.676dup in non-Finnish Europeans with an MAF of 0.0038. By contrast, this variant has not been reported in East and South Asian populations.
Two individuals of Portuguese origin (Y126 and P86) analyzed within the GEMINI study each carried two different variants in M1AP (Figure S2). Individual Y126 carried the missense variant c. 949G>A (p.Gly317Arg) and the recurrent frameshift variant c.676dup and had germ cell arrest at the round spermatid stage. Individual P86 had the two missense variants c.148T>C (p.Ser50Pro) and c.1289T>C (p.Leu430Pro), suggesting compound heterozygosity. Testicular histology showed severely disturbed spermatogenesis. Individuals RU01691 from the Netherlands and MI-0006-P from the UK were also homozygous for the frameshift variant c.676dup. RU01691’s parents were both heterozygous carriers. His testicular biopsy showed bilateral severe hypospermatogenesis with predominantly meiotic arrest; sporadically, spermatids were present, and material was cryopreserved for intracytoplasmic sperm injection (ICSI) but hitherto not used. Individual MI-0006-P had azoospermia and predominant meiotic arrest with rare postmeiotic germ cells (Figure S2).
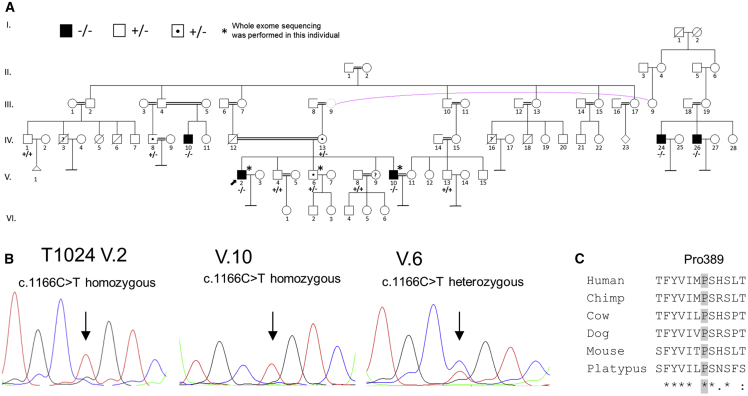
In parallel, WES was performed in two infertile, azoospermic brothers from a consanguineous Turkish family as well as in their fertile brother. The index T1024 (V.2; Figure 2) presented at Istanbul Memorial Hospital, Assisted Reproductive Technologies and Reproductive Genetics Centre and Bursa Uludag University Faculty of Medicine Hospital, Turkey because of couple infertility. Testicular histology demonstrated maturation arrest at round spermatid stage, and no sperm could be recovered by TESE. The WES data were analyzed focusing on rare homozygous variants shared between both infertile brothers but not found in the fertile brother. The two affected men carried rare homozygous missense variants in the autosomal genes AMPD2, CELSR2, CEP164, and M1AP as well as rare hemizygous variants in the X chromosome genes ATG4A and ENOX2. Of these genes, only M1AP has been described in the context of infertility. Both infertile men carried the homozygous missense variant c.1166C>T (p.Pro389Leu) (MAF = 0.00001), which was also found in a heterozygous state in the fertile brother, in M1AP. No homozygous individuals with this variant have been described in any public databases, whereas it was found in a homozygous state in three additional infertile males from this family: two third cousins once removed from the maternal side and one second cousin once removed from the paternal side. We did identify both a fertile man and a fertile woman (IV.13 and V.6, respectively, in Figure 2A) as heterozygous carriers of the same variant (example result of Sanger sequencing for subject V.6 shown in Figure 2B).
Figure 2.
Turkish Consanguineous Family with Infertile, Azoospermic Men Homozygous for M1AP Missense Variant and Fertile Heterozygous Carriers
(A) Pedigree of the Turkish family with five infertile azoospermic men carrying the homozygous M1AP variant c.1166C>T (p.Pro389Leu) indicated with black boxes and −/−. The index individual T1024, who presented at Uludag University Faculty of Medicine Hospital, is marked with an arrow (V.2). Heterozygous carriers of the M1AP variant are marked with a point and +/-. Examined family members with the homozygous M1AP wild-type allele are marked with +/+. Homozygous men are infertile, whereas heterozygous carriers are fertile.
(B) Representative electropherograms of the index affected individual (V.2), his infertile brother (V.10), and his fertile brother (V.6), who is a heterozygous carrier.
(C) The missense variant affects a highly conserved amino acid.
In a complementary approach, an updated version of the population sampling probability (PSAP) pipeline19 was used to prioritize potentially causative variants. PSAP models the significance of observing a single person’s genotype in comparison to genotype frequencies in unaffected populations. This enabled us to rank all variants per individual by following the prioritization criteria MAF ≤ 0.01, CADD ≥ 20, and PopScore ≤ 0.005.20 The bi-allelic M1AP LoF variants were ranked in the first position for M1792 and in the third position for M330, M864, and M2062 in the discovery cohort (Table S3). The missense variant c.797G>A and the duplication c.676dup of M1943 were ranked in the ninth position under a compound heterozygous recessive disease model. The five men identified in the follow-up analyses exhibited highly ranked M1AP variants as well: position seven in RU01691, position four in Y126, position seven in P86, position two in MI-0006-P, and position three in T1024.
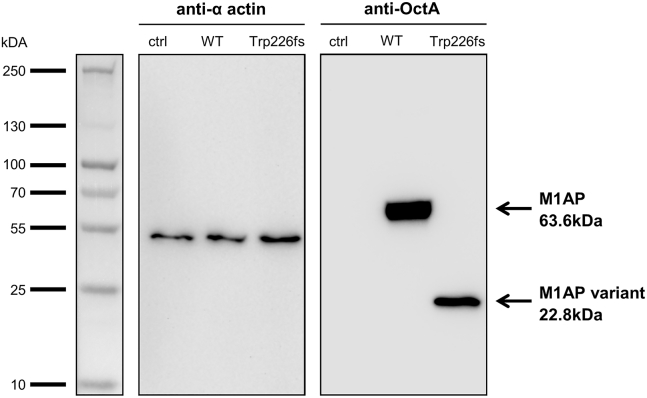
The rare but recurring M1AP variant c.676dup warranted further analyses. It is located in exon 5 of 11 and causes a frameshift and premature stop codon (p.Trp226LeufsTer4) as confirmed by testicular cDNA sequencing of exon 5 of individual M864 (Figure S3). This results in a truncated protein as shown by heterologous expression of mutated M1AP in HEK293T and subsequent immunoblot analysis displaying a protein band of almost 23 kD (Supplemental Methods and Figure 3), indicating a protein lacking 57% of its normal length. This is in line with the analysis of individual M864’s testicular RNA that resulted in an equal band compared to control testis RNA, excluding elimination of the mRNA through nonsense-mediated decay (Figure S3). Still, because of the non-native, ectopic expression of the protein in HEK293T cells, it remains possible that no product is translated by the mutant mRNA in vivo. The relevance of the homozygous frameshift variant c.676dup in M1AP is further supported by the exceptionally low PSAP-PopScore (9.7 × 10−7) and the high prioritization (Table S3). Moreover, the expected mode of inheritance for M1AP is autosomal recessive according to a general prediction,21 fitting our observations of bi-allelic variants in the affected men. According to the ACMG-AMP guidelines,18 this variant is categorized as pathogenic (Table S4).
Figure 3.
Heterologous Expression of M1AP in HEK293T Cells
Non-transfected control (ctrl), wild-type (WT), and mutated c.676dup, p.[Trp226LeufsTer4];[Trp226fs] M1AP cDNA (N-terminal located DYK-tag, in pcDNA3.1) plasmids were transfected into HEK293T cells. 24 h post-transfection, whole-cell lysates were prepared and separated on an SDS-PAGE. A PAGE Ruler Plus Prestained Protein Ladder (Thermo Scientific) was used for validation of protein sizes, and Biorad ImageLab Software was used to calculate protein sizes. Anti-alpha-actin antibody (1:5000, ab5694, abcam) served as a loading control, and M1AP was detected by a monoclonal anti-OctA antibody (1:1000, sc-166355, Santa Cruz) against the integrated DYK-tag (1:1000) (n = 3).
The fact that the same frameshift variant, c.676dup, was also found in individuals from Croatia, Poland, the Netherlands, UK, and Portugal, suggests that it is relatively prevalent in European populations most likely originating from a founder mutation. In total, we screened 1,996 infertile males across the four cohorts (735, 930, 283, and 48 from the MERGE, GEMINI, Nijmegen, and Newcastle studies, respectively), six of whom were homozygous for c.676dup. In contrast, c.676dup is rarely described in global large databases: gnomAD (v2.1.1: 141,421 individuals corresponding to 282,842 alleles) does not contain any homozygous individuals. Hence, subjects homozygous for c.676dup in M1AP are highly significantly overrepresented in our cohort (Fisher’s exact p = 7.2 × 10−12; only for the 76,685 males, p = 2.6 × 10−10). To establish the allele frequency of M1AP c.676dup in an ancestry-matched control group, we performed Sanger sequencing of exon 5 of M1AP in an additional 285 normozoospermic men recruited at the CeRA, Münster. Indeed, five heterozygous but no homozygous subjects were detected. Subsequent sequencing of the complete coding region of M1AP (exon 2 to 11, primers in Table S1) in all five heterozygous carriers ruled out the presence of a second relevant variant in all individuals. Additionally, we queried a previously established database of 5,784 Dutch fertile men and 5,803 fertile women who had conceived at least one child. WES had been performed as part of clinical diagnostic workup of a child with severe development delay (trio-WES, Nijmegen, the Netherlands). Again, 27 heterozygous male and 21 heterozygous female carriers but no homozygous individuals were detected. Also, there were no homozygous subjects of other LoF variants in the whole coding region of M1AP among either fathers or mothers. In summary, statistical evidence strongly supports that homozygous c.676dup in M1AP is associated with severe male infertility, while the different allele frequencies can most likely be explained by population stratification.
To gain insight into the function of M1AP and better assess the relevance of identified variants, we pursued two strategies. First, we attempted to model M1AP’s 3D structure. However, because of the lack of information on M1AP and comparable 3D structures, it was not possible to achieve a reliable prediction (BLAST results for sequence of UniProt: Q8TC57 are below 30% sequence identity to known protein structures, details in Supplemental Methods). Second, we tried to establish immunohistochemistry as well as immunoblot analyses with the two most promising (based on available data from the manufacturers and the HPA) commercially available M1AP antibodies (#PA5-31627, ThermoFisher Scientific and #HPA045420, Sigma-Aldrich) (see Supplemental Methods for details). After optimization, both antibodies did result in a specific signal in immunohistochemistry (Figures S4 and S5) of testicular control sections. However, one of them (#PA5-31627) showed poor results (high amounts of background and antibody precipitates, Figure S4), whereas #HPA045420 seems not to bind M1AP but detect a different target instead (Figure S5). Most importantly, the presumed epitope resides downstream of the variant p.Trp226LeufsTer4 and, therefore, is disrupted in homozygous subjects (further details in Figures S5 and S6). Unfortunately, we were not successful in contacting the colleagues who published the M1ap knockout mice and immunoblot staining with a self-raised antibody.8 In conclusion, the structure of M1AP is currently unknown and it is impossible to predict functional domains of M1AP with sufficient reliability. Moreover, the specific molecular function of M1AP remains to be elucidated in subsequent studies, which would also open the possibility of functional assessment of the missense variants.
The assessment of the other detected M1AP variants has, for the time being, to rely on established in silico tools, and we followed the strict clinical ACMG-AMP criteria (Table S4).18 In addition to the recurrent frameshift variant c.676dup, individuals M1943 and Y126 each carry a missense variant (p.Arg266Gln and p.Gly317Arg, respectively), while individual P86 carries two assumed compound-heterozygous missense variants (p.Ser50Pro and p.Leu430Pro). Although in silico assessment supports the relevance of identified missense variants, all of these are categorized as being of uncertain significance. However, the co-segregation in the Turkish family especially is highly suggestive of the pathogenicity of (at least some) missense variants in M1AP (LOD score = 3.28). Of note, the group of Şehime G. Temel identified M1AP as a candidate independently from the initial identification and the clinical and testicular phenotype of the index individual T1024 fits the spectrum of the other affected men very well. In contrast, all investigated fertile family members had at least one wild-type allele, which further supports the impact of the bi-allelic variant.
Overall, we identified ten unrelated men with likely causal bi-allelic variants in M1AP, nine stemming from four independent study cohorts and one from a Turkish family (Table 1), as well as four additional infertile men from this family. Out of these, eight men underwent testicular biopsy, and histology showed arrested spermatogenesis in all of them. Arrest occurred at meiosis, i.e., the spermatocyte stage, in the majority and was either complete (n = 3, M330, M864, and M1792) or predominant (n = 2, RU1691 and MI-0006-P). Fewer men had a later arrest at the round spermatid stage (n = 2, Y126 and T1024) or earlier with only spermatogonia present (n = 1, P86). Thus, the common phenotype was NOA, but sporadically spermatids and rarely spermatozoa in the semen (below 10 per sample) were observed in two individuals constituting a continuum at the very severe end of spermatogenic failure. A similar phenotype was described for mice with disruption of M1ap that sporadically had some spermatozoa in their semen.8
Very recently, a homozygous splice-site variant (c.1435−1G>A) in M1AP has been published as cause for severe oligozoospermia in a single male from a consanguineous Han Chinese family.16 Of note, this variant resides in the last exon and, on the basis of our in-depth characterization of the same antibody, we put into question Tu et al.’s analyses using the same antibody. Likewise, the proposed localization of M1AP in the sperm midpiece should be critically assessed in light of the clear evidence we provide that the antibody does not seem to stain M1AP specifically (see above and Figures S5 and S6). Still, this variant likely impairs M1APs function, and the finding in this infertile male from yet another population (1) further broadens the phenotypical spectrum of M1AP-associated spermatogenic disturbances and (2) provides more evidence for M1AP’s relevance for male infertility. We therefore included this individual and the variant in Tables 1 and 2 to provide a comprehensive overview of all presumably relevant M1AP variants, affected individuals, and clinical characteristics.
So far, only very few other genes, such as TEX11 and STAG3, with mutations leading to germ cell arrest in both men and mice and validated in independent cohorts have been published.5,7,10,22 From our initial cohort of 64 men with complete bilateral meiotic arrest, we also identified likely causal variants in three other genes, namely TEX11, STAG3, and SYCP2.5,10,11 Among the remaining group of 58 men analyzed herein, we identified three unrelated men with the same homozygous frameshift variant, c.676dup, in M1AP. Thus, disruptive M1AP variants are highly enriched in men affected by male infertility, NOA, and meiotic arrest (5%, 3 out of 64), a frequency comparable to the currently best-characterized TEX11 mutations with 6% (4 out of 64) in this selected group. Concerning the full spectrum of phenotypes associated with M1AP, variants in this gene most likely contribute to less than 1% of the highly heterogeneous individuals with severe spermatogenic defects, i.e., NOA, cryptozoospermia, and oligozoospermia (0.5%, 9 out of 1,996 individuals). Still, M1AP is one of the most commonly mutated genes associated with severe male infertility to date and ranges directly behind the well-established genetic causes Klinefelter syndrome (47,XXY, almost exclusively found in NOA) and Y chromosome AZF deletions (found in azoospermia and severe oligozoospermia).2
Assessing the clinical validity of gene-disease associations is now recognized as being of the utmost importance. Collectively, the presented data, accumulated from several independent groups and populations as well as a consanguineous family, together with previously described evidence from murine studies and the in parallel published second family from China, result in M1AP immediately reaching a strong clinical validity based on a structured assessment according to Smith et al.23 (Table S5). This is an unprecedented example of any gene in which variants are associated with male infertility and clearly shows the power of large-scale collaborative efforts.
The process of meiosis is in part orchestrated similarly in both sexes but initiated at different times in life. Fittingly, a few genes, such as STAG3, have been reported in which variants impair male as well as female meiosis and result in infertility in both sexes.10,22,24 M1AP is predominantly expressed in the adult testis in both men and mice, but it is also reported to be expressed in the fetal mouse ovary.9 However, the structure of ovaries in female M1ap knockout mice appeared normal and fertility was preserved.8 To shed some light onto a potential role of M1AP in human female meiosis, we screened 101 women diagnosed with unexplained premature ovarian insufficiency (POI) (62 with isolated POI and 39 with ovarian dysgenesis) by Sanger sequencing of M1AP’s full coding region. Details of a part of this cohort (n = 25) have been published previously.25 The additional 76 individuals followed the same inclusion and exclusion criteria. No women carrying two rare variants in M1AP were identified. Furthermore, we did not identify a fertile woman carrying a homozygous LoF variant in the large Dutch trio cohort or a woman homozygous for the missense variant in the Turkish family. Thus, we cannot exclude that variants in M1AP might also be a rare cause for POI, but current evidence suggests that M1AP is only required for male meiosis.
In conclusion, the presented data from four independent cohorts strongly supports that both homozygous LoF and deleterious missense variants in M1AP as well as compound heterozygosity for either variant type results in severe spermatogenic failure and male infertility. Co-segregation in multiple affected men from a Turkish family provides further strong and independent evidence. M1AP disruption is associated primarily with germ cell arrest but might also be compatible with the production of a few spermatozoa, similar to the phenotypic spectrum observed in men with Klinefelter’s syndrome or AZFc deletions. On the basis of our data, we cannot reliably predict the probability of successful sperm retrieval by testicular biopsy and TESE, but if we extrapolate the findings from the men reported here, TESE success is quite low (1 of 8, <15%). Our findings provide further evidence that germ cell arrest is often of monogenic origin. According to the commonly applied structured gene assessment,23 M1AP has strong clinical validity for causing NOA, cryptozoospermia, or severe oligozoospermia and should be used as a screening marker before testicular biopsy to estimate the chances of successful TESE. Finally, identifying mutations in M1AP in unexplained infertile men with severe spermatogenic failure provides them with a causal diagnosis for their infertility.
Data and Code Availability
All variants have been submitted to ClinVar (VCV000805830–VCV000805834) and can also be accessed in the Male Fertility Gene Atlas (MFGA), a public platform for collecting and integrating datasets about genetic causes of male infertility produced in a subproject of the Clinical Research Unit “Male Germ Cells: from Genes to Function.” Overall, the published article includes all datasets generated or analyzed during this study. The exceptions are the primary WES data that have not been deposited in a public repository because the individual’s consent did not include this. However, additional information is available from the corresponding author on request.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We are indebted to all individuals consenting to research evaluation of their data and donating their DNA as well as the physicians who took care of them. We thank Christian Ruckert for his bioinformatic support, Martin Bergmann for many years of skillfully evaluating testicular histologies, Nurten Akarsu for encouraging us to start a project with this Turkish Family, Zeliha Görmez for bioinformatics analyses of WES, Şeref Gül and Laurens van de Wiel for their attempts to model M1AP, Joachim Kremerskothen and Verena Höffken for attempting to establish a immunoblot analysis from testicular samples, as well as Christina Burhöi, Nicole Terwort, and Katja Hagen for their excellent technical assistance. We thank Celeste Brennecka for language editing of the manuscript. This work was carried out within the frame of the German Research Foundation Clinical Research Unit “Male Germ Cells: from Genes to Function” (DFG CRU326). Funding for sequencing of the GEMINI cohort was provided by the National Institutes of Health, United States (R01HD078641). The analyses in the Turkish family were supported by a grant from the Bursa University of Uludag Project Unit [KUAP(T)-2014/36].
Published: July 15, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.06.010.
Web Resources
Clinical Research Unit “Male Germ Cells,” https://www.male-germ-cells.de
ClinVar, https://www.ncbi.nlm.nih.gov/clinvar
GEMINI, https://gemini.conradlab.org
GTEx, https://gtexportal.org
Human Protein Atlas, https://www.proteinatlas.org
International Male Infertility Genomics Consortium, http://www.imigc.org
Male Fertility Gene Atlas, https://mfga.uni-muenster.de
MutationTaster, http://www.mutationtaster.org
OMIM, https://www.omim.org
PolyPhen 2, http://genetics.bwh.harvard.edu/pph2
Supplemental Data
References
- 1.Juul S., Karmaus W., Olsen J. Regional differences in waiting time to pregnancy: pregnancy-based surveys from Denmark, France, Germany, Italy and Sweden. The European Infertility and Subfecundity Study Group. Hum. Reprod. 1999;14:1250–1254. doi: 10.1093/humrep/14.5.1250. [DOI] [PubMed] [Google Scholar]
- 2.Tüttelmann F., Ruckert C., Röpke A. Disorders of spermatogenesis: Perspectives for novel genetic diagnostics after 20 years of unchanged routine. Med. Genetik. 2018;30:12–20. doi: 10.1007/s11825-018-0181-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization . World Health Organization; 2010. WHO laboratory manual for the examination and processing of human semen. [Google Scholar]
- 4.Lee J.Y., Dada R., Sabanegh E., Carpi A., Agarwal A. Role of genetics in azoospermia. Urology. 2011;77:598–601. doi: 10.1016/j.urology.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Yatsenko A.N., Georgiadis A.P., Röpke A., Berman A.J., Jaffe T., Olszewska M., Westernströer B., Sanfilippo J., Kurpisz M., Rajkovic A. X-linked TEX11 mutations, meiotic arrest, and azoospermia in infertile men. N. Engl. J. Med. 2015;372:2097–2107. doi: 10.1056/NEJMoa1406192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gershoni M., Hauser R., Yogev L., Lehavi O., Azem F., Yavetz H., Pietrokovski S., Kleiman S.E. A familial study of azoospermic men identifies three novel causative mutations in three new human azoospermia genes. Genet. Med. 2017;19:998–1006. doi: 10.1038/gim.2016.225. [DOI] [PubMed] [Google Scholar]
- 7.Oud M.S., Volozonoka L., Smits R.M., Visser L.E., Ramos L., Veltman J.A. A systematic review and standardized clinical validity assessment of male infertility genes. bioRxiv. 2018 doi: 10.1101/425553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arango N.A., Li L., Dabir D., Nicolau F., Pieretti-Vanmarcke R., Koehler C., McCarrey J.R., Lu N., Donahoe P.K. Meiosis I arrest abnormalities lead to severe oligozoospermia in meiosis 1 arresting protein (M1ap)-deficient mice. Biol. Reprod. 2013;88:76. doi: 10.1095/biolreprod.111.098673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arango N.A., Huang T.T., Fujino A., Pieretti-Vanmarcke R., Donahoe P.K. Expression analysis and evolutionary conservation of the mouse germ cell-specific D6Mm5e gene. Dev. Dyn. 2006;235:2613–2619. doi: 10.1002/dvdy.20907. [DOI] [PubMed] [Google Scholar]
- 10.van der Bijl N., Röpke A., Biswas U., Wöste M., Jessberger R., Kliesch S., Friedrich C., Tüttelmann F. Mutations in the stromal antigen 3 (STAG3) gene cause male infertility due to meiotic arrest. Hum. Reprod. 2019;34:2112–2119. doi: 10.1093/humrep/dez204. [DOI] [PubMed] [Google Scholar]
- 11.Schilit S.L.P., Menon S., Friedrich C., Kammin T., Wilch E., Hanscom C., Jiang S., Kliesch S., Talkowski M.E., Tüttelmann F. SYCP2 Translocation-Mediated Dysregulation and Frameshift Variants Cause Human Male Infertility. Am. J. Hum. Genet. 2020;106:41–57. doi: 10.1016/j.ajhg.2019.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P. The mutational constraint spectrum quantified from variation in 141,456 humans. bioRxiv. 2020 doi: 10.1101/531210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lonsdale J., Thomas J., Salvatore M., Phillips R., Lo E., Shad S., Hasz R., Walters G., Garcia F., Young N., GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nieschlag E., Behre H.M., Nieschlag S. Third Edition. Springer-Verlag Berlin Heidelberg; 2010. Andrology: Male reproductive health and dysfunction. [Google Scholar]
- 15.Magi A., Tattini L., Palombo F., Benelli M., Gialluisi A., Giusti B., Abbate R., Seri M., Gensini G.F., Romeo G., Pippucci T. H3M2: detection of runs of homozygosity from whole-exome sequencing data. Bioinformatics. 2014;30:2852–2859. doi: 10.1093/bioinformatics/btu401. [DOI] [PubMed] [Google Scholar]
- 16.Tu C., Wang Y., Nie H., Meng L., Wang W., Li Y., Li D., Zhang H., Lu G., Lin G. An M1AP homozygous splice-site mutation associated with severe oligozoospermia in a consanguineous family. Clin. Genet. 2020;97:741–746. doi: 10.1111/cge.13712. [DOI] [PubMed] [Google Scholar]
- 17.Venselaar H., Te Beek T.A.H., Kuipers R.K.P., Hekkelman M.L., Vriend G. Protein structure analysis of mutations causing inheritable diseases. An e-Science approach with life scientist friendly interfaces. BMC Bioinformatics. 2010;11:548. doi: 10.1186/1471-2105-11-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilfert A.B., Chao K.R., Kaushal M., Jain S., Zöllner S., Adams D.R., Conrad D.F. Genome-wide significance testing of variation from single case exomes. Nat. Genet. 2016;48:1455–1461. doi: 10.1038/ng.3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kasak L., Punab M., Nagirnaja L., Grigorova M., Minajeva A., Lopes A.M., Punab A.M., Aston K.I., Carvalho F., Laasik E., GEMINI Consortium Bi-allelic Recessive Loss-of-Function Variants in FANCM Cause Non-obstructive Azoospermia. Am. J. Hum. Genet. 2018;103:200–212. doi: 10.1016/j.ajhg.2018.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cassa C.A., Weghorn D., Balick D.J., Jordan D.M., Nusinow D., Samocha K.E., O’Donnell-Luria A., MacArthur D.G., Daly M.J., Beier D.R., Sunyaev S.R. Estimating the selective effects of heterozygous protein-truncating variants from human exome data. Nat. Genet. 2017;49:806–810. doi: 10.1038/ng.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riera-Escamilla A., Enguita-Marruedo A., Moreno-Mendoza D., Chianese C., Sleddens-Linkels E., Contini E., Benelli M., Natali A., Colpi G.M., Ruiz-Castañé E. Sequencing of a ‘mouse azoospermia’ gene panel in azoospermic men: identification of RNF212 and STAG3 mutations as novel genetic causes of meiotic arrest. Hum. Reprod. 2019;34:978–988. doi: 10.1093/humrep/dez042. [DOI] [PubMed] [Google Scholar]
- 23.Smith E.D., Radtke K., Rossi M., Shinde D.N., Darabi S., El-Khechen D., Powis Z., Helbig K., Waller K., Grange D.K. Classification of Genes: Standardized Clinical Validity Assessment of Gene-Disease Associations Aids Diagnostic Exome Analysis and Reclassifications. Hum. Mutat. 2017;38:600–608. doi: 10.1002/humu.23183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caburet S., Vilain É. [STAG3 in premature ovarian failure] Med. Sci. (Paris) 2015;31:129–131. doi: 10.1051/medsci/20153102005. [DOI] [PubMed] [Google Scholar]
- 25.Ledig S., Röpke A., Wieacker P. Copy number variants in premature ovarian failure and ovarian dysgenesis. Sex Dev. 2010;4:225–232. doi: 10.1159/000314958. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All variants have been submitted to ClinVar (VCV000805830–VCV000805834) and can also be accessed in the Male Fertility Gene Atlas (MFGA), a public platform for collecting and integrating datasets about genetic causes of male infertility produced in a subproject of the Clinical Research Unit “Male Germ Cells: from Genes to Function.” Overall, the published article includes all datasets generated or analyzed during this study. The exceptions are the primary WES data that have not been deposited in a public repository because the individual’s consent did not include this. However, additional information is available from the corresponding author on request.