Abstract
A major question in human genetics is how sequence variants of broadly expressed genes produce tissue- and cell type-specific molecular phenotypes. Genetic variation of alternative splicing is a prevalent source of transcriptomic and proteomic diversity in human populations. We investigated splicing quantitative trait loci (sQTLs) in 1,209 samples from 13 human brain regions, using RNA sequencing (RNA-seq) and genotype data from the Genotype-Tissue Expression (GTEx) project. Hundreds of sQTLs were identified in each brain region. Some sQTLs were shared across brain regions, whereas others displayed regional specificity. These “regionally ubiquitous” and “regionally specific” sQTLs showed distinct positional distributions of single-nucleotide polymorphisms (SNPs) within and outside essential splice sites, respectively, suggesting their regulation by distinct molecular mechanisms. Integrating the binding motifs and expression patterns of RNA binding proteins with exon splicing profiles, we uncovered likely causal variants underlying brain region-specific sQTLs. Notably, SNP rs17651213 created a putative binding site for the splicing factor RBFOX2 and was associated with increased splicing of MAPT exon 3 in cerebellar tissues, where RBFOX2 was highly expressed. Overall, our study reveals a more comprehensive spectrum and regional variation of sQTLs in human brain and demonstrates that such regional variation can be used to fine map potential causal variants of sQTLs and their associated neurological diseases.
Keywords: alternative splicing, splicing quantitative trait loc, genetic variation, single-nucleotide polymorphism, RNA-seq, transcriptome
Introduction
Alternative splicing is a crucial post-transcriptional regulatory mechanism that enables enormous RNA-level complexity. Through alternative splicing, specific exons may be included or excluded from the final mature messenger RNA (mRNA), allowing a single gene to generate multiple transcript and protein products with unique biological functions.1 In humans, more than 95% of multi-exon genes undergo alternative splicing,2 with the nervous system being a prominent site.3,4 Abnormalities in alternative splicing have been associated with multiple pathologies,5, 6, 7, 8 including neurodegenerative disorders like Parkinson disease (PD),9 Alzheimer disease (AD),10,11 amyotrophic lateral sclerosis (ALS),12 and frontotemporal dementia (FTD),13 as well as neuropsychiatric disorders like autism spectrum disorders (ASD)14, 15, 16, 17 and schizophrenia.18, 19, 20
Genetic variants can alter exon inclusion or splice site usage, by creating or disrupting splice sites21 or other cis splicing regulatory elements within precursor mRNAs.22, 23, 24 Such genetically regulated alternative splicing events were recently recognized as a primary link between genetic variation and disease.25 Splicing quantitative trait loci (sQTL) analysis is a common method for discovering genotype-splicing associations. In an sQTL analysis, the splicing level of an alternative exon or splice site is treated as a quantitative trait and tested for association with genotype across a population. Using this approach, two recent reports described effects of genetic variants on alternative splicing in human brain as well as their associations with schizophrenia20 and AD.26 However, both studies were restricted to a single brain region, and therefore a more comprehensive spectrum and regional variation of sQTLs in human brain were not revealed.
In this study, we systematically discovered sQTLs in 13 regions of human brain using RNA sequencing (RNA-seq) and genotype data from the Genotype-Tissue Expression (GTEx) project (Figure 1A). We decided to study patterns and regional variation of sQTLs in human brain, based on the observation that brain region is the dominant contributing factor to variations in gene expression and alternative splicing in human brain, as compared to other biological factors such as age and sex. By comparing sQTL signals across different brain regions, we found that some sQTLs were shared across brain regions, whereas others displayed regional specificity. These “regionally ubiquitous” and “regionally specific” sQTLs were enriched for genetic variants within and outside essential splice sites, respectively, suggesting their regulation by distinct molecular mechanisms. Integrating RNA binding protein (RBP) motifs with RBP expression and exon splicing profiles, we demonstrate that regional variation of sQTLs can be used to fine map potential causal variants underlying genetically regulated alternative splicing events and their associated neurological diseases.
Figure 1.
Overview of Study and Data
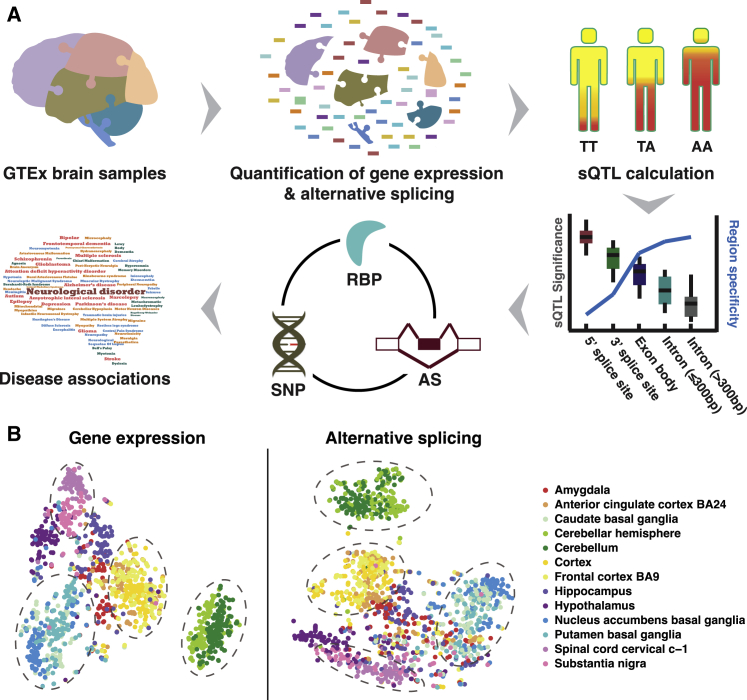
(A) Overview of study. RNA-seq data derived from human brain tissue samples were downloaded from the GTEx Portal. Data were processed to quantify gene expression (in TPM) and alternative splicing (in PSI) across 13 brain regions. Splicing QTLs (sQTLs) were identified for each individual brain region, based on alternative splicing and genotype information. The sQTL data were used to study the relationship among the positional distribution of sQTLs, significance of sQTLs, and brain region specificity. RNA binding protein (RBP) expression was incorporated with alternative splicing (AS) and single-nucleotide polymorphism (SNP) data to identify potential causal cis variants and trans regulators. Data were used to identify sQTLs potentially associated with 14 neurological disorders.
(B) t-SNE clustering of all brain tissue samples based on gene expression (left) and alternative splicing (right). Samples are color coded by brain region. Dashed ovals encircle samples from physically proximate brain regions.
Material and Methods
Datasets and Processing of GTEx Brain Tissue Samples
RNA-seq data (BAM files; v7, June 2017 release) for human brain tissues from postmortem donor specimens were downloaded from the GTEx Portal website (see Web Resources). RNA-seq data were available for 1,409 samples from 13 brain regions of 201 individuals.
An ultra-fast version of rMATS (rMATS-turbo, see Web Resources) was run on BAM files to obtain exon inclusion levels (PSI values). Detailed information regarding the quantification of different types of alternative splicing events can be found in the original rMATS paper.27 Missing PSI values were imputed using k-nearest neighbors. To ensure reliability of the imputation and downstream analyses, we only included an exon in our analyses if it passed the following filters:
-
1)
Average PSI within 0.05 and 0.95,
-
2)
Average total read count (inclusion count + skipping count) ≥10,
-
3)
Percentage of samples with missing PSI value < 5%,
-
4)
max(PSI) – min(PSI) > 0.05
Processed gene expression data (in transcripts per kilobase million, TPM) were downloaded from the GTEx Portal. These data were available for 1,258 of the 1,409 samples with RNA-seq data. Median normalization was performed on each sample for between-sample normalization.
Genotype data of GTEx brain samples based on whole-genome sequencing (WGS) were downloaded from the database of Genotypes and Phenotypes (dbGaP, see Web Resources). Description of the WGS genotype data can be found on the GTEx portal28 (see Web Resources). SNPs with a minor allele frequency < 0.05 as well as SNPs on sex chromosomes were excluded from analysis, leaving 6,317,213 SNPs for downstream analysis. The final dataset for sQTL analysis comprised 1,209 samples for which genotype data and corresponding RNA-seq data, including processed gene expression data, were available.
Age-, Brain Region-, and Sex-Dependent Splicing and Expression
To determine the extent to which splicing (PSI) results were affected by differences in age, brain region, or sex between tissue samples, the following linear mixed model was used:
where is the PSI for exon i of sample j; and Agej, BrainRegionj, and Sexj are the age, brain region, and sex of sample j with regression coefficients , , and , respectively. Random effect term Pij was introduced to account for the fact that multiple GTEx samples may come from the same donor. To control for all other known and unknown confounding factors (e.g., ancestry, BMI, batch effects, etc.), the following surrogate variable(s) estimated by Surrogate Variable Analysis (SVA)29 was used:
where SVkj is the value of surrogate variable (SV) k for sample j with regression coefficient ; N is the total number of SVs that are not correlated with age, brain region or sex; is the error term; and is the regression intercept for exon i. The model was fitted by using the lmer function in the lme4 package in R. For each exon, we used a least-squares approach to estimate regression coefficients and a likelihood ratio test to estimate significance. If , , or significantly deviated from 0, then the exon was considered to be dependent on age, brain region, or sex. False discovery rate (FDR) < 5% was used as the cutoff for significance.
We performed the same analysis on gene expression data by replacing PSI with normalized TPM, but with one difference. For gene expression analysis, SVA estimated 1,232 SVs, which generated more parameters than could be analyzed by the linear mixed model. As t-SNE analysis did not reveal any obvious effect of confounding factors on gene expression, we excluded SVs from the analysis.
Correcting PSI Values for Confounding Factors before sQTL Analysis
Before performing the sQTL analysis, we corrected the PSI value by removing potential confounding factors. First, we performed principal component analysis in PLINK on GTEx genotype data to obtain population structure. This analysis was performed on a genome-wide set of linkage disequilibrium (LD)-pruned variants (R2 > 0.2). The top three principal components were used as covariates to correct for population structure, because they accounted for 5.5% of the genotype variance with diminishing returns (0.25% or smaller) for subsequent PCs. The top three PCs were sufficient to capture the major population structure in GTEx data consisting of African American, American Indian, Asian, and White individuals (Figure S1).
Next, we used SVA to remove potential confounding factors from sources other than brain region. The input of SVA was logit transformed PSI values of all alternative splicing events across all samples. As the aim of the sQTL analysis was to identify sQTLs across different brain regions, we included only one factor (brain region) in the sQTL analysis. Any variation that could not be explained by brain region was estimated by SVA and removed.
We fitted the following linear model:
where M is the total number of SVs uncorrelated with brain region. SVmj is the value of surrogate variable (SV) m for sample j. PCkj is the value of principal component k for sample j. All samples from the same donor have the same values of genotype PCs. The top three principal components were used to account for population structure. We used the residual of the model as the corrected PSI value in sQTL analysis.
Identification of sQTLs
After the corrected PSI value was obtained, each single-nucleotide polymorphism (SNP) within a 200-kb window on each side of an alternative splicing event was fitted with a linear regression separately. We estimated the FDR using a permutation procedure to obtain the null distribution of p values. We used the same permutation approach as in Zhao et al.30 For each alternative splicing event, we permuted the individual label 5 times, recalculated the p values of all SNPs, and recorded the minimum p value for each alternative splicing event for each permutation. This set of minimum p values serves as the empirical null distribution for the p values (denoted as p0). We then compared the true distribution of minimum p values (denoted as p1) to this null distribution to obtain the estimate of FDR at the event level. For example, for FDR = 0.1, we found a p value cutoff z such that P(p0 < z)/ P(p1 < z) = 0.1, where P(p0 < z) is the fraction of minimum p values from permutation less than z and P(p1 < z) is the fraction of minimum p values from observed data less than z. We applied the permutation procedure to estimate FDR values for the three types of alternative splicing events separately. Within each type, the p values from all events and all permutations were used to define a single empirical null distribution for estimating FDR values.
We defined the sQTL SNP of an alternative splicing event as the closest SNP with the most significant association, using a two-tier cutoff for significance: (1) FDR < 10% and (2) uncorrected p < 10−5. The corresponding p value of FDR = 10% is ∼10−6 for each brain region; thus, events meeting this threshold represent a higher confidence set of sQTLs. This calculation was done in all 13 brain regions separately.
LD Calculation
Most GWAS variants have been identified in samples of European ancestry. Therefore, for LD calculations, we used genotype data from the CEU population (Utah residents with ancestry from northern and western Europe). Genotype data were downloaded from the 1000 Genomes Project (HapMap Project Genome Browser version E, data release #28 [Phase II+III], see Web Resources). To calculate LD between SNPs, we used PLINK (with parameters --no-fid --no-parents --r2 --ld-window-kb 1000 --ld-window 99999 --ld-window-r2 0.8). SNPs with r2 > 0.8 were defined as being in high LD.
We calculated the LD of sQTL SNPs for skipped exon (SE), alternative 5′ splice sites (A5SS), and alternative 3′ splice sites (A3SS) events with SNPs from the 1000 Genomes Project. SNPs that were in high LD with sQTL SNPs were annotated by using GWAS variants from the NHGRI-EBI GWAS Catalog v.1.0.1 (see Web Resources). This list includes GWAS SNPs with p values < 1 × 10−5. Therefore, SNPs with p values between 5 × 10−8 and 1 × 10−5 were included in the analysis. We used 14 keywords to classify GWAS traits into disease categories: “Alzheimer,” “amyotrophic lateral sclerosis,” “Parkinson,” “frontotemporal dementia,” “epilepsy,” “autism,” “schizophrenia,” “bipolar,” “depression,” “attention deficit hyperactivity disorder,” “glio,” “multiple sclerosis,” “narcolepsy,” and “stroke.” After identifying GWAS SNPs that were in high LD with sQTL SNPs, we obtained the GWAS SNP disease ontology information (“MAPPED_TRAIT” column in GWAS Catalog table). If the disease ontology information contained any of the 14 keywords, then the sQTL SNP and the corresponding alternative splicing event were considered to be related to that specific disease.
Colocalization Analysis between sQTL and GWAS Signals
We performed a colocalization analysis between sQTL and GWAS signals to test whether a single causal variant underlies the sQTL signal and GWAS signal. For sQTLs associated with GWAS traits, we collected the GWAS SNPs that were in high LD with sQTL SNPs, together with the GWAS studies reporting these GWAS SNPs. Whenever available, we downloaded the harmonized summary statistics of the GWAS studies from the GWAS Catalog (see Web Resources). Colocalization analysis using coloc31 under default parameter settings was performed on the GWASs, using their harmonized GWAS summary statistics along with the associated sQTL’s summary statistics in brain regions with a significant sQTL signal. SNPs within a 200-kb window on each side of an alternative splicing event were used for the colocalization analysis. A posterior probability of ≥75% was considered strong evidence for a single causal variant underlying the sQTL signal and the GWAS signal.32
Selection of the Most Representative sQTL SNP across 13 Brain Regions
For each sQTL event, we collected the sQTL SNPs for each brain region (see Identification of sQTLs). Among all significant (FDR < 10%) sQTL SNPs, we selected the SNP that was the top (i.e., most significant) SNP in the largest number of brain regions as the most representative sQTL SNP. In the event of a tie, we compared competing SNPs and chose the SNP that was significant (FDR < 10%) in the largest number of brain regions. If sQTL SNPs were still tied, then we chose the SNP closest to the sQTL event as the most representative sQTL SNP across all 13 brain regions.
Defining Regionally Ubiquitous and Regionally Specific sQTLs
We considered an sQTL event to be regionally specific if: (1) the uncorrected p value of the most representative SNP across 13 brain regions was < 10−5 in no more than 4 brain regions and (2) the uncorrected p value was less than the FDR cutoff (FDR < 10%) in at least one brain region. We considered an sQTL event to be regionally ubiquitous if the uncorrected p value of the most representative SNP across 13 brain regions was less than the FDR cutoff (FDR < 10%) in at least 10 brain regions. We designed this two-tier cutoff for sQTL significance (FDR < 10%, uncorrected p < 10−5) to ensure a reliable definition of significant and insignificant events.
Predicting the Effect of SNPs on RBP-RNA Binding via DeepBind
We used the DeepBind model33 to quantify the effect of SNPs on RBP-RNA binding. DeepBind takes RNA sequences as input and outputs DeepBind scores, which quantify the binding specificity of different RBPs for the input sequences. The DeepBind scores can be used to generate mutation maps, which visually display the impact of sequence variants on RBP-RNA binding.
First, we collected all significant SNPs (and 20-bp flanking sequences on both sides) within 300 bp of the nearest splice site of each sQTL event across all 13 brain regions. Next, we tested the effect of each significant SNP on the binding of 102 human RBPs for which the DeepBind model had been trained. Following the procedure described in Alipanahi et al.,33 we calculated DeepBind scores and generated mutation maps for 41-bp sliding windows using a 20-bp motif detector. We used a modified approach to generate the mutation map for each SNP-RBP pair. Specifically, whereas Alipanahi et al.33 was primarily concerned with the ability of a sequence variant to decrease the binding score (i.e., base height was unchanged if a sequence variant increased binding), our study was also concerned with the ability of a sequence variant to increase binding. To account for this, we calculated the sum of the scores on each base in the reference genome by using the absolute value of the four DeepBind scores on that base, and then scaled the height of each base by using this sum. In this way, the height of each base was proportional to the ability of sequence variants on the base to change (increase or decrease) binding.
To facilitate the identification of potential causal variants underlying regionally specific sQTLs, we analyzed RBP expression data across the 13 brain regions. If a SNP regulates exon splicing by altering RBP binding, then we would expect to see higher sQTL significance in brain regions where the RBP is highly expressed. To test this possibility, we grouped samples based on whether they were from brain regions where the sQTL was significant or insignificant. For each RBP, we applied a Wilcoxon rank sum test to determine whether the RBP was differentially expressed between the two groups, with FDR < 5% indicating significant differential expression. FDR is calculated using the p values of all exon-SNP-RBP combinations.
Motif Scan of NOVA1
NOVA1 is not included in the DeepBind model but has a well-defined consensus motif (YCAY). Therefore, for NOVA1, instead of running the DeepBind model, we carried out a motif scan on the same set of SNPs as were used in the DeepBind analysis. For each SNP, we checked all possible 4-mer sequences overlapping with the SNP to see if the SNP created or disrupted a NOVA1 binding site.
Results
Gene Expression and Alternative Splicing in Human Brain
We downloaded RNA-seq data from the GTEx project website and collected corresponding gene expression, alternative splicing, and genotype information of 1,209 human brain samples comprising 13 brain regions (Figure S2; see Material and Methods for details). We applied our computational tool rMATS,27 developed for quantifying alternative splicing events from large-scale RNA-seq data, to estimate exon splicing levels (percent spliced in, or PSI) from the GTEx RNA-seq data. After applying filters to select high-confidence alternative splicing events with reliable RNA-seq quantitation, we collected 10,665 skipped exons (SE), 1,443 alternative 5′ splice site (A5SS) events, and 2,434 alternative 3′ splice site (A3SS) events (Figure S3). The t-distributed stochastic neighbor embedding (t-SNE) algorithm was used to inspect relationships among samples. Using this approach, we were able to separate samples from different brain regions and to recapitulate relationships between brain regions using gene expression and alternative splicing information (Figure 1B shows results for SE events; similar patterns were obtained for A5SS and A3SS events). Although we tested additional co-variates such as age and sex, as well as various potential confounding or batch effects, these other factors did not enable clear separation of samples based on gene expression or alternative splicing (Figure S4). Our results indicate that brain region is the dominant contributing factor to variations in gene expression and alternative splicing in human brain.
To quantify contributions of different biological factors to alternative splicing and gene expression, we fit a linear mixed model to each alternative splicing event or gene. We included brain region, age, and sex in the model as main effects and controlled for confounding factors (Material and Methods). Using an FDR of < 5%, we found that gene expression and alternative splicing showed substantial variation depending on brain region, but much less variation depending on age or sex (Figure S5). These findings were consistent with the t-SNE results (full list of brain region-, age-, or sex-dependent genes and alternative splicing events are given in Tables S1 and S2).
Given this strong brain-regional variation, we validated our results using previously reported brain region-specific alternative splicing events and genes; neurexin is one prominent example. Through alternative splicing, neurexins 1, 2, and 3 generate thousands of mRNA and protein products.34, 35, 36 Ehrmann et al. reported that the tissue-dependent RBP KHDRBS3 (referred to as T-STAR) and its paralog KHDRBS1 (referred to as Sam68) regulate brain region-specific alternative splicing of neurexin1-3 exon 4.37 Specifically, exon 4 was included at low levels in brain regions with a high gene expression ratio of KHDRBS3 versus KHDRBS1 (e.g., cortex) and at high levels in regions with a low gene expression ratio (e.g., cerebellum). Plotting the exon inclusion level of neurexin2 exon 4 with the KHDRBS3:KHDRBS1 expression ratio (Figure S6), we found consistent results with those reported previously.37 Exon 4 of either neurexin1 or neurexin3 showed a similar pattern based on our data.
Identification of sQTLs in 13 Brain Regions
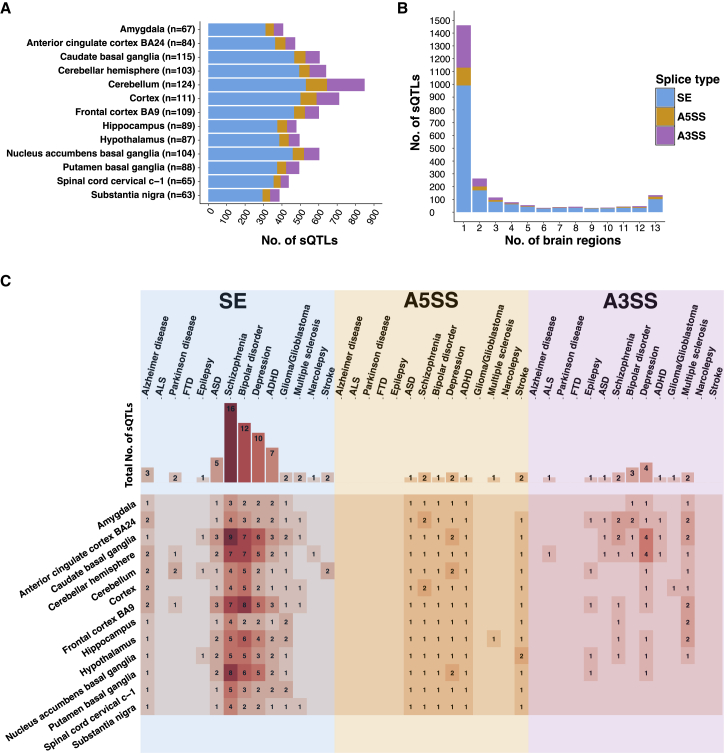
To elucidate genetic regulation of alternative splicing in human brain, we performed sQTL analyses in each of the 13 brain regions separately (see Material and Methods for details). After controlling for confounding factors, we identified sQTLs by using linear regression to calculate the association between exon splicing levels (PSI values) and SNP genotypes in each brain region. The total numbers of tests performed were 111,720,167 for SE, 14,675,960 for A5SS, and 25,059,203 for A3SS events. The closest SNP with the smallest p value within 200 kb of each alternative splicing event (SE, A5SS, or A3SS) was selected as the sQTL SNP in that brain region. The corresponding alternative splicing event was defined as the sQTL event. Between 387 and 849 significant sQTL events (p < 10−5) were found in each brain region (Figure 2A), with most events being specific to only a few regions (Figure 2B). We also identified 133 sQTL events that were significant in all 13 brain regions. These data indicate that sQTLs in human brain can be either “regionally ubiquitous” or “regionally specific.” Of note, in the step of correcting PSI values for confounding factors, we identified 3 surrogate variables based on SVA. Surrogate variable 1 was significantly correlated with RNA integrity number (RIN) (p value = 0.00089) and post mortem interval (PMI) (p value = 0.045). Therefore, although we did not explicitly correct for confounding factors like RIN and PMI, these factors were considered and accounted for by the use of surrogate variables in sQTL discovery.
Figure 2.
Identification of sQTLs in Each Brain Region
(A) Stacked bar plot showing the number of sQTLs (including SE, A5SS, and A3SS events) identified in each brain region, with the number in parentheses referring to the number of tissue samples with available genotype information.
(B) Stacked bar plot showing the histogram for the number of brain regions where sQTLs (including SE, A5SS, and A3SS events) were determined to be significant.
(C) Heatmap showing the number of disease sQTLs (including SE, A5SS, and A3SS events) associated with each neurological disorder in each brain region. Each row represents one brain region. Each column represents one neurological disorder. Bar plot above heatmap shows the total number of unique sQTLs associated with each neurological disorder.
Next, we annotated sQTL events in terms of disease risk by calculating the LD between sQTL SNPs and SNPs identified as disease-associated variants by genome-wide association studies (GWASs) (see Material and Methods for details). We used disease ontology terms in the NHGRI-EBI GWAS Catalog,38 focusing on terms related to 14 neurological disorders: AD, ALS, PD, FTD, epilepsy, ASD, schizophrenia, bipolar, depression, attention deficit hyperactivity disorder (ADHD), glioma/glioblastoma, multiple sclerosis, narcolepsy, and stroke. Any sQTL event with an sQTL SNP in high LD (r2 > 0.8) with a neurological disorder-related GWAS variant was considered to be a disease sQTL event related to that specific disorder. Figure 2C shows the number of disease sQTL events for each disorder in each brain region. Results based on GWAS SNPs reaching genome-wide significance (p value ≤ 5 × 10−8) can be found in Figure S7.
We identified a predominance of disease sQTL events that were related to neuropsychiatric disorders (e.g., schizophrenia). This result was not surprising, given the greater prevalence of reported neuropsychiatric disease-related GWAS variants in the literature.39,40 We also identified sQTL events related to neurodegenerative diseases (e.g., AD and PD). Neurodegenerative disease-related sQTL events tended to be brain region specific, whereas neuropsychiatric disorder-related sQTL events were shared across a larger number of brain regions. Full information on the sQTLs identified in each brain region, together with the disease association, can be found in Table S3. The percentage of GWAS loci associated with brain sQTLs was comparable across the brain disorders analyzed. Likewise, we did not observe significant enrichment for brain sQTLs in brain disorders, as compared to apparently non-brain phenotypes such as body mass index and body height. This is not surprising, given that brain sQTLs could also be sQTLs in non-brain tissues and cell types and that genomic variants can have pleiotropic effects on phenotypes.41
For sQTLs associated with GWAS traits, we also performed a colocalization analysis between sQTL and GWAS signals whenever data were available (see Material and Methods for details). In total, we found 278 brain sQTL-associated GWASs. Among them, 27 had harmonized summary statistics (Table S4). Colocalization analysis using coloc31 was performed on these 27 studies. In total, 124 colocalization tests were performed on 43 sQTL-GWAS pairs, using the GWAS summary statistics and the sQTL summary statistics of every brain region with a significant sQTL signal. Full results of the colocalization tests can be found in Table S5. Results from 77 tests supported the colocalization model of a single causal variant underlying the sQTL signal and the GWAS signal (posterior probability ≥75%). One example is an sQTL involving PGAP3 exon 4 and SNP rs1565922 (Figure S8). SNP rs1565922 was significantly associated with the splicing level of PGAP3 exon 4 in multiple brain regions (cerebellar hemisphere, cerebellum, cortex, and frontal cortex) and was also in high LD with rs2517959, a GWAS variant of bipolar disorder. Figure S8 showed the colocalization at the PGAP3 locus between the sQTL signal (cortex) and the GWAS signal, suggesting a single causal variant affecting PGAP3 exon 4 splicing and bipolar disorder.
Relationship among SNP Position, Significance, and Brain Region Specificity of sQTLs
We examined the relationship among SNP position, significance, and brain region specificity of sQTLs. Here, we describe the result for SE events, but similar results were observed for A5SS and A3SS events.
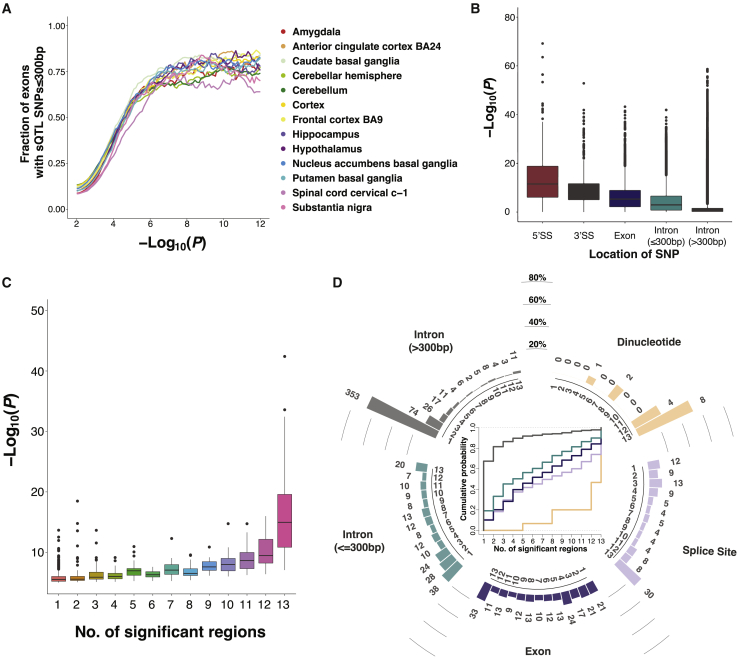
To evaluate the positional distribution of sQTL SNPs, we plotted the –log10 p values of significant sQTL SNPs in all 13 brain regions together, against the distance to the sQTL exons (Figure S9). As expected, and consistent with previous results,20 we found that SNPs closer to the alternative exons tended to be more significant. Next, we examined SNP positions for all exons included in the sQTL analysis for each brain region. For each exon, we obtained the –log10 p values of all SNPs within 200 kb of the exon, together with the distance of the SNPs to the exon. As the p value cutoff for significant sQTLs increased, we observed an increase in the fraction of exons with at least one significant SNP within 300 bp of splice sites (Figure 3A). This result, which was reproducible across all brain regions, indicated enrichment in the local regulation of significant sQTLs by exon-proximal SNPs.
Figure 3.
Relationship among SNP Position, Significance, and Brain Region Specificity of sQTLs (SE Events)
(A) Fraction of sQTL with at least one significant SNP within 300 bp of the splice sites as a function of the significance (−log10(p value)) cutoff for significant sQTLs. Each curve represents the result in one brain region.
(B) Boxplot showing the distribution of significance (−log10(p value)) of all SNPs within 200 kb of all significant (p ≤ 10−5) sQTLs (SE events). SNPs are grouped based on SNP position relative to the splice sites.
(C) Boxplot showing the relationship between overall sQTL significance (−log10(p value)) and the number of brain regions where sQTLs are significant. Each dot represents one sQTL (SE event). An sQTL having at least one significantly associated SNP in a brain region is considered significant in that brain region. Overall sQTL significance is calculated by considering all brain regions where the sQTL is significant, and taking the median of the smallest p values.
(D) Bar plots (outside) and cumulative distribution function (CDF) (inside) showing the relationship between SNP position and brain region specificity of sQTLs (SE events). The sQTLs are grouped based on the position of their significant SNP (e.g., dinucleotide, splice site, etc.). For each group, the bar plot shows the histogram of the percentage of sQTLs that are significant in a given number of brain regions. Each bar is labeled above with the number of significant sQTLs in the given number of brain regions.
To obtain a fine-grained picture of the relationship between SNP position and significance of sQTLs, we classified all SNPs within 200 kb of all sQTL exons into 5 groups, based on the SNP position relative to the corresponding sQTL exon. In the figures that follow, 5′SS represents the 9 bases (3 exonic and 6 intronic) around the 5′ splice site and 3′SS represents the 23 bases (20 intronic and 3 exonic) around the 3′ splice site.42 We found that SNP position was associated with sQTL significance (Figure 3B), with 5′SS SNPs having the highest sQTL significance, followed by 3′SS SNPs, exonic SNPs, and proximal intronic SNPs (≤300 bp of exons). SNPs in distal intronic regions (>300 bp from exons) had the largest overall p values, indicating their relatively minor impact on splicing. These results are consistent with previous sQTL studies on human B-lymphoblastoid cell lines.30,43
Next, for each individual sQTL, we asked whether its number of significant brain regions was associated with its level of sQTL significance. For each sQTL exon, we plotted the sQTL significance against the number of significant regions (Figure 3C). An sQTL exon with at least one significantly associated SNP within the 200 kb window in a brain region was considered significant in that brain region. Overall significance of the sQTL exon was calculated as the median of the smallest p value from all regions in which the sQTL was significant. As expected, we found that sQTL exons that were significant in a greater number of brain regions tended to have higher overall sQTL significance (Figure 3C).
Next, we analyzed the relationship between the SNP position and brain region specificity of sQTLs. For each sQTL exon, we first classified all significant SNPs within 200 kb across all brain regions into 5 categories: splice site dinucleotide (GT and AG for donor and acceptor sites, respectively), splice site (9 nt around 5′SS, 23 nt around 3′SS, excluding the dinucleotide), exon body, proximal intronic region (≤300 bp of the exon), and distal intronic region (>300 bp of the exon). Then, we assigned each sQTL exon to a specific category based on whether it had a significant SNP in that category, prioritized sequentially from splice site dinucleotide to distal intronic region. This information was plotted together with the number of significant brain regions for each sQTL exon (Figure 3D). To restrict this analysis to high-confidence sQTLs, here we used a more stringent cutoff of permutation FDR < 10% instead of the uncorrected p value < 10−5 as the cutoff for defining significant sQTLs. We observed that sQTL exons with significant SNPs closer to their splice sites tended to be significant in a greater number of brain regions. For example, for sQTLs with significant SNPs located at the splice site dinucleotide, 53% were significant in all 13 brain regions and 93% were significant in the majority (≥7 of 13) of brain regions. By contrast, for sQTLs with significant SNPs located only in proximal or distal intronic regions, the majority (50% or 90%, respectively) were significant in no more than 4 brain regions (Figure 3D). We observed the same trend when we used permutation FDR < 5% or permutation FDR < 1% as the cutoff to define significant sQTLs (Figure S10).
Examples of Regionally Ubiquitous and Regionally Specific sQTLs
We used stringent criteria to define high-confidence sets of regionally ubiquitous and regionally specific sQTLs in human brain. Specifically, sQTLs reaching transcriptome-wide significance (permutation FDR < 10%) in at least 10 brain regions were considered regionally ubiquitous (Figure 4A), whereas sQTLs reaching transcriptome-wide significance (permutation FDR < 10%) in at least one brain region but having no more than 4 brain regions with the uncorrected p value < 10−5 were considered regionally specific (Figure 4E) (see Material and Methods for details). The reason for allowing the event to reach this more relaxed cutoff (< 10−5) in up to 4 brain regions is to allow and account for the inherent similarity in alternative splicing profiles and/or RBP expression among certain GTEx brain regions, e.g., cerebellum and cerebellum hemisphere, or cortex and frontal cortex. In total, 148 sQTLs were defined as regionally ubiquitous. 758 sQTLs were defined as regionally specific, among which 653 (86%) were significant in only one or two brain regions.
Figure 4.
Regionally Ubiquitous Versus Regionally Specific sQTLs
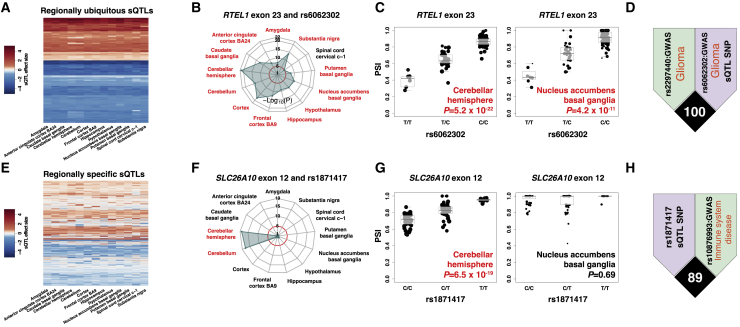
(A) Heatmap showing effect sizes (correlation coefficients between alternative splicing levels and SNP genotypes) of regionally ubiquitous sQTLs. Each row represents one sQTL. Each column represents one brain region.
(B) Radar plot showing significance (−log10(p value)) of a regionally ubiquitous sQTL (RTEL exon 23 and rs6062302) in each brain region. Red circle indicates uncorrected p = 10−5. Significant brain regions are highlighted in red text.
(C) Boxplot showing the significant association (p value) of SNP rs6062302 with PSI value (exon inclusion level) of RTEL exon 23 in two brain regions: cerebellar hemisphere and nucleus accumbens basal ganglia.
(D) LD plot showing a GWAS variant (rs2297440, GWAS p value = 4 × 10−46, glioma; green) in high LD with the sQTL SNP (rs6062302; purple). SNP rs6062302 itself is also a GWAS variant related to glioma (GWAS p value = 1 × 10−13).
(E) Heatmap showing effect sizes (correlation coefficients between alternative splicing levels and SNP genotypes) of regionally specific sQTLs.
(F) Radar plot showing significance (−log10(p value)) of a regionally specific sQTL (SLC26A10 exon 12 and rs1871417) in each brain region. Red circle indicates uncorrected p = 10−5. Significant brain regions are highlighted in red text.
(G) Boxplot showing the association (p value) of SNP rs1871417 with PSI value (exon inclusion level) of SLC26A10 exon 12 in two brain regions: cerebellar hemisphere (significant) and nucleus accumbens basal ganglia (not significant).
(H) LD plot showing a GWAS variant (rs10876993, GWAS p value = 4 × 10−6, immune system disease; green) in high LD with the sQTL SNP (rs1871417; purple).
For each sQTL exon, we obtained and plotted the coefficient from the linear regression analysis of exon splicing levels against genotypes in each brain region, which represented the effect size of the sQTL signal in that region, using the most representative sQTL SNP across the 13 brain regions (Figures 4A and 4E) (see Material and Methods for details). Consistent with Figure 3D, we found that regionally specific sQTLs were enriched for SNPs in exonic and intronic regions, whereas regionally ubiquitous sQTLs were enriched for SNPs in splice sites (Figure S11A, 2 test p < 2.2 × 10−16). We observed the same trend when we defined significant sQTLs using a transcriptome-wide significance cutoff of permutation FDR < 5% (Figure S11B, 2 test p < 2.2 × 10−16) or permutation FDR < 1% (Figure S11C, 2 test p = 1.8 × 10−7).
For regionally ubiquitous sQTLs, we illustrate the example of the sQTL exon in regulator of telomere elongation helicase 1 (RTEL1), which protects telomeres during DNA replication. Several variants in RTEL1 have been reported to be associated with risk of glioma.44,45 We observed significant associations between PSI values of RTEL1 exon 23 and genotypes of rs6062302 in all brain regions except spinal cord (Figure 4B). Figure 4C shows the highly significant correlations in the cerebellar hemisphere and the nucleus accumbens basal ganglia. SNP rs6062302 itself is a glioma-related GWAS variant, and it is in high LD with another glioma-related GWAS variant (Figure 4D).
Beyond the example of RTEL1 (Figures 4B–4D), our analyses identified a total of 148 regionally ubiquitous sQTLs, including those associated with GWAS signals. For example, we identified a significant correlation between rs67573812 and C8orf59 exon 2 (Figure S12A). This SNP, located at the 5′SS (dinucleotide) of C8orf59 exon 2, disrupts a canonical splice site (GT to GA). A second example involved flotillin 1 (FLOT1) exon 5 and rs1059612, located in the proximal intronic region of exon 5 (Figure S12B). FLOT1 is a membrane-raft associated protein that is involved in synaptic transmission and synapse formation.46,47 Expression QTL and GWASs have associated FLOT1 expression with genetic risks for schizophrenia and major depressive disorder.48,49 We found that the SNP was in high LD with GWAS variants related to schizophrenia, bipolar, ADHD, unipolar depression, and ASD. Another example involved SLC39A13 exon 5 and rs2293576 (Figure S12C), located within the exon body of exon 5 and in high LD with the AD-related GWAS variant rs10838725. This finding is consistent with previous reports that SLC39A13 is within the LD block of rs10838725, an AD-risk SNP identified by The International Genomics of Alzheimer’s Project.50,51
For regionally specific sQTLs, we illustrate the correlation between PSI values of SLC26A10 exon 12 and genotypes of rs1871417, which was significant only in cerebellar hemisphere and cerebellum (Figure 4F). Figure 4G shows the correlation in one significant region (cerebellar hemisphere) and one insignificant region (nucleus accumbens basal ganglia). The sQTL SNP of SLC26A10 exon 12 (rs1871417) was in high LD with an immune system disease-related GWAS variant rs10876993 (Figure 4H). More specifically, rs10876993 was reported to be related to celiac disease and rheumatoid arthritis.52 It is somewhat unexpected to observe strong LD between a cerebellum-specific sQTL and GWAS SNP for immune system diseases. In future work, it would be interesting to investigate whether this sQTL is also present in certain types of immune cells.
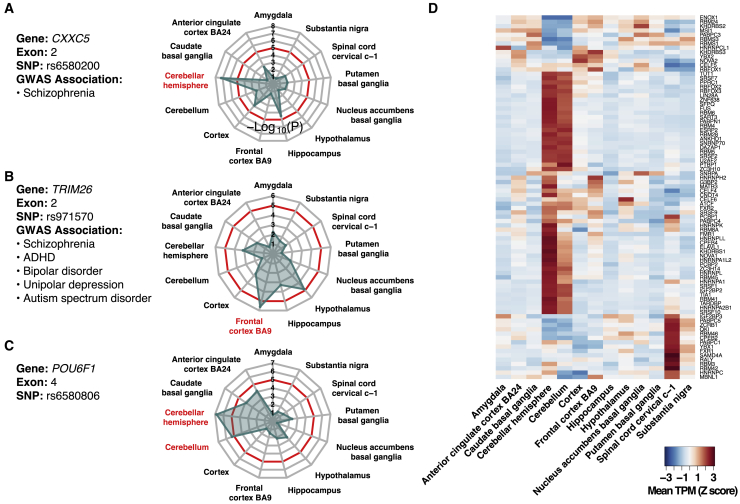
We found many other sQTLs restricted to specific brain regions. For example, the association between rs6580200 and CXXC5 exon 2 was significant only in cerebellar hemisphere (Figure 5A). Previous pathway-based analysis determined CXXC5 to be a schizophrenia-associated gene.53 The SNP that we identified was in high LD with a schizophrenia-associated GWAS variant. In TRIM26, the association between rs971570 and exon 2 was significant only in frontal cortex (Figure 5B). Although the function of TRIM26 is unknown, several studies found that TRIM26 was differentially expressed between individuals with schizophrenia and control subjects.54,55 Here, we found that this SNP was in high LD with GWAS variants related to schizophrenia, as well as ADHD, ASD, bipolar disorder, and unipolar depression. In POU6F1, the association between rs6580806 and exon 4 was significant only in cerebellum and cerebellar hemisphere (Figure 5C).
Figure 5.
Regionally Specific sQTLs and RBP Expression
(A–C) Radar plots showing significance (−log10(p value)) of three regionally specific sQTLs (A: rs6580200 and CXXC5 exon 2; B: rs971570 and TRIM26 exon 2; C: rs6580806 and POU6F1 exon 4) in each brain region.
(D) Heatmap showing gene expression (z-score-transformed TPM) of selected RBPs across all 13 brain regions. Each row represents one RBP. Each column represents one brain region. In addition to all 102 Homo sapiens RBPs with DeepBind RBP-RNA binding models, the heatmap includes RBFOX2, RBFOX3, NOVA1, and NOVA2.
Using Regional Variation of sQTL Signals to Prioritize Causal sQTL cis Variants and trans Regulators
To understand why the genetic regulation of alternative splicing could be restricted to specific brain regions, we investigated the underlying regulatory mechanisms of regionally specific sQTLs. One hypothesis is that the brain region-specific expression of RBPs, functioning as trans-acting regulators of alternative splicing, could contribute to the brain region specificity of sQTLs. To test this hypothesis, we analyzed the gene expression patterns of RBPs across brain regions. Many RBPs showed highly brain region-specific expression patterns (Figure 5D) and, thus, could potentially provide a source of regional regulation.
To prioritize potential causal cis variants and identify potential trans regulators of brain region-specific sQTLs, we used a deep-learning model DeepBind33 to predict the effect of a given SNP on RBP-RNA binding. DeepBind predicts RBP sequence specificities based on sequence features from CLIP-seq (crosslinking and immunoprecipitation followed by high-throughput sequencing) experiments. We assessed the RBP binding effects of all significant SNPs within 300 bp of a given sQTL exon, and then incorporated RBP expression data to further prioritize candidate cis variants and trans regulators (see Material and Methods). Intuitively, if a SNP regulates the splicing of an exon through affecting RBP binding, we would expect to see greater sQTL significance and larger effect size in brain regions where the RBP is highly expressed. To test this, we classified the brain samples into two groups: (1) samples from brain regions where the sQTL is significant and (2) samples from brain regions where the sQTL is insignificant. For each RBP, a Wilcoxon rank sum test was performed to see whether the RBP is differentially expressed between the two groups. Full information of the differential RBP expression analysis can be found in Table S6. RBPs with FDR < 5% and fold change > 1.5 were included.
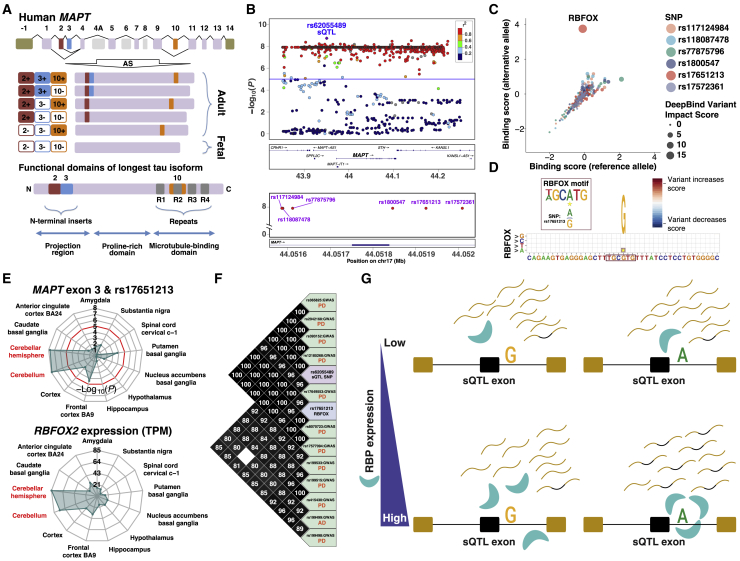
Using this strategy, we fine-mapped possible causal variants for a brain region-specific sQTL for exon 3 of microtubule-associated protein tau (MAPT). MAPT encodes tau, an abundant protein in the nervous system that promotes microtubule assembly and stability. Aggregation of hyperphosphorylated tau protein is a primary marker of AD.10,56,57 Moreover, numerous studies have implicated tau in the pathogenesis of other neurological disorders, including PD.58 Human MAPT contains 16 exons (Figure 6A), with exons 2, 3, and 10 being alternatively spliced to generate six isoforms. The sQTL exon that we identified was exon 3. Figure 6B shows the positional distribution and sQTL p values of SNPs within 200 kb of MAPT exon 3 in cerebellum. In a local window including 300 bp of upstream and downstream intronic regions, there were six significant SNPs that were in high LD with the top sQTL SNP rs62055489 (>100 kb from MAPT exon 3). Testing the effects of these six SNPs against all RBPs in the DeepBind model, we identified a strong effect of rs17651213 (G>A) on RBFOX binding (Figure 6C). We found that the SNP created a consensus binding site of the RBFOX family of splicing factors downstream of MAPT exon 3 (Figure 6D). When we checked the expression levels of RBFOX1, RBFOX2, and RBFOX3, we found that RBFOX2/3 had the highest expression levels in cerebellum and cerebellar hemisphere, two brain regions where the sQTL was also significant (Figures 6E and S13).
Figure 6.
Using Regional Specificity of sQTL Signals to Prioritize Causal sQTL cis Variants and trans Regulators
(A) Top: Gene structure and six isoforms of human MAPT (tau) gene. Exons −1 and 14 are in the untranslated regions (UTRs). Primary transcript of human MAPT contains 13 exons (exons 4A, 6, and 8 are not transcribed in human brain). Among the 13 exons, exons 2, 3, and 10 are alternatively spliced, generating six mRNA isoforms. Bottom: Functional domains of the longest full-length MAPT protein isoform (including exons 2, 3, and 10).
(B) Plots showing distribution of all SNPs within 200 kb (upper) and all significant SNPs within 300 bp (lower) of MAPT exon 3 in cerebellum. Window is extended 30 kb (upper) and 30 bp (lower). Each dot represents one SNP, color-coded according to its LD with the top sQTL SNP (rs62055489). y axis shows the significance of association (−log10(p value)) between each SNP and the sQTL exon. Horizontal line indicates the significance cutoff (p = 10−5). Genes in the UCSC Genome Browser (see Web Resources) are shown in panels below plots.
(C) Bubble plot showing effects of six SNPs on RBP-RNA binding, as predicted by DeepBind. Axes show RBP binding scores of sequences with reference allele (x axis) or alternative allele (y axis) for each RBP. Bubble size is proportional to the difference in DeepBind scores between the two alleles.
(D) DeepBind variant map for SNP rs17651213 with the RBFOX binding site. Star indicates the position of the SNP.
(E) Radar plots showing significance (−log10(p value)) of the sQTL (MAPT exon 3 and rs17651213) and mean gene expression level (TPM) of the RBP (RBFOX2) in each brain region.
(F) LD plot showing SNP rs17651213 (blue) in high LD with the top sQTL SNP (rs62055489; purple), 11 PD GWAS SNPs (GWAS p values ranging from 2 × 10−118 to 2 × 10−6, green), and 1 AD GWAS SNP (GWAS p value = 6 × 10−6, green).
(G) Diagram illustrating a mechanistic model for tissue-/cell-type-specific sQTLs.
RBFOX2 is a well-characterized splicing factor whose binding downstream of alternative exons promotes exon inclusion.59 SNP rs17651213 was located 88 bp downstream of MAPT exon 3. When the alternative allele that creates the RBFOX binding site was present, we observed greater inclusion of the exon (Figures S14 and S15), consistent with the position-dependent effect of RBFOX2 binding on alternative splicing.59 Finally, in accordance with the importance of tau protein in AD and PD pathogenesis, we found that rs17651213 was in high LD with 11 PD GWAS variants and 1 AD GWAS variant (Figure 6F). Overall, by integrating RBP motifs, region-specific RBP gene expression, and sQTL pattern, we identified rs17651213 as the likely causal variant that regulates MAPT exon 3 splicing by creating an RBFOX2 binding site downstream of the exon.
We also identified other brain region-specific sQTLs with potential causal SNPs and RBP regulators. For example, DeepBind prediction suggested that the SNP rs6580200 regulates splicing of CXXC5 exon 2 by disrupting an HNRNPK binding site (Figure S16). This sQTL was significant in cerebellar hemisphere, where HNRNPK had the highest level of expression among 13 brain regions (Figure S17). A second example was rs4077093, which may regulate splicing of POU6F1 exon 4 by disrupting NOVA1 binding (Figure S18). The sQTL was significant in cerebellum and cerebellar hemisphere, where NOVA1 was highly expressed (Figure S19). Using the well-defined consensus motif of NOVA1 (YCAY), we performed a motif scan and identified the effect of rs4077093 on NOVA1 binding. Specifically, rs4077093 (A>C) was predicted to disrupt a NOVA1 motif within the exon and found to be associated with increased exon inclusion, consistent with previous reports that NOVA1 binding within alternative exons promotes exon skipping.60
Discussion
Alternative splicing is known to influence biological functions and disease processes, but much remains unknown about the disease causality and underlying regulatory mechanisms of genetically regulated alternative splicing events. A number of studies have performed sQTL analyses to interrogate genotype-splicing associations in human cell lines or tissues, including brain where alternative splicing is highly prevalent.20,26,61 However, previous studies largely conducted sQTL analyses one tissue at a time and did not compare and contrast sQTL signals across multiple tissues or cell types. In this work, we analyzed sQTLs in 1,209 human brain samples across 13 brain regions. We identified regionally ubiquitous sQTLs with significant signals across a large number of brain regions, as well as regionally specific sQTLs whose significance was restricted to specific brain regions. Many sQTLs were associated with GWAS signals. Together, our study provides a comprehensive catalog of genetically regulated alternative splicing events in human brain and reveals their associations with neurological traits and diseases.
One major challenge in genetic association studies of molecular and phenotypic traits is to identify the causal variants and regulatory mechanisms underlying the observed association. In an sQTL analysis, multiple SNPs in LD with each other can be significantly associated with the levels of exon splicing, and it is difficult to pinpoint the specific variant(s) causal for the observed sQTL signal. Our analysis of SNP significance as a function of distance to sQTL exons suggests that the majority of sQTLs, especially those with strong effects on splicing, likely have their causal variants located within the exonic or proximal intronic (≤300 bp of exon) regions. This observation is consistent with prior findings that sequence information within this window has a high predictive power for alternative splicing patterns.61,62 By examining the SNP positions for regionally ubiquitous and regionally specific sQTLs, we uncover the likely molecular mechanisms responsible for generating these two types of sQTLs. Regionally ubiquitous sQTLs are enriched for SNPs located on the 5′ or 3′ splice site. As splice sites play an essential role in splicing, it is not surprising that SNPs strengthening or weakening splice site signals have ubiquitous effects across different brain regions or tissue types. By contrast, regionally specific sQTLs tend to have SNPs located outside the 5′ or 3′ splice site and are enriched for SNPs located in exonic and intronic regions. Causal SNPs underlying regionally specific sQTLs likely affect splicing by modulating the interactions of tissue- or cell-type-specific splicing factors with the pre-mRNA.
We developed an integrative strategy to fine map the likely causal variants for brain region-specific sQTLs. We assessed SNP effects on RBP-RNA binding and compared brain region-specific RBP gene expression patterns and sQTL signals to identify potential causal cis variants and trans regulators for regionally specific sQTLs. Using this strategy, we identified and highlighted the likely molecular mechanisms for several brain region-specific sQTLs (MAPT, CXXC5, POU6F1), in which SNPs created or disrupted the binding sites of splicing factors with region-specific expression patterns. As the research community continues to accumulate population-scale RNA-seq datasets61 as well as RNA binding profiles and specificities of RBPs,63 we envision that this integrative strategy can be applicable to fine map causal variants of tissue- or cell-type-specific sQTLs in diverse biological systems.
Our study highlights a molecular mechanism of how genetic variants of broadly expressed genes can have tissue- or cell-type-specific effects on splicing. As one example, we found that a G-to-A SNP in MAPT (rs17651213) was associated with increased inclusion of MAPT exon 3 and created the consensus binding site of the splicing factor RBFOX2. This association was specific to cerebellar tissues, where RBFOX2 was highly expressed. This example illustrates a general scenario that a SNP alters the putative binding site of a trans splicing regulator within the RNA, but its molecular impact is dependent on the concentration of the regulator such that the SNP only alters splicing in tissues or cell types where the regulator is highly expressed (Figure 6G).
We should note that in our sQTL analysis, we defined one sQTL SNP for an alternative splicing event in each brain region by selecting the closest SNP with the most significant association. One limitation of this approach is that an alternative splicing event may possibly be influenced by multiple independent (i.e., non-LD) SNPs in each brain region, but this scenario is not currently addressed by our analysis. Another limitation of this study is that it used bulk tissues from the GTEx project, and samples from each brain region represented a mixture of multiple cell types. This may dilute the power of detecting tissue- or cell-type-specific sQTLs, especially if the cell type affected represents a minor population of cells in the bulk tissues. For example, it is well known that RBFOX2 expression is highest in neuronal cells64, 65, 66 and the cerebellum has the highest density of neurons among brain regions.67,68 Therefore, the significant association between rs17651213 and MAPT exon 3 in cerebellar tissues could be related to neuron-specific splicing regulation through RBFOX2, and it is possible that a stronger SNP-splicing association can be observed specifically in neurons. Future studies using population-scale RNA-seq data of purified cell types or single cells may further expand the catalog and our mechanistic understanding of genetically regulated alternative splicing events in human brain.
Declaration of Interests
Y.X. is a scientific co-founder of Panorama Medicine Inc.
Acknowledgments
The authors thank Drs. Douglas Black, Peter Stoilov, and Christopher Ross for helpful discussions. This work was supported by National Institutes of Health grants R01GM088342 (Y.X.), R01GM117624 (Y.X.), R01MH109166 (Y.X.), and R01NS076631 (Y.X. and B.L.D.). The results published here are in part based upon data generated by the Genotype-Tissue Expression (GTEx) Project (https://commonfund.nih.gov/gtex). The GTEx Project was supported by the Common Fund of the Office of the Director of the NIH, and by National Cancer Institute, National Human Genome Research Institute, National Heart, Lung, and Blood Institute, National Institute on Drug Abuse, National Institute of Mental Health, and National Institute of Neurological Disorders and Stroke. Y.Z. was supported by a UCLA Dissertation Year Fellowship.
Published: June 25, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.06.002.
Web Resources
Custom scripts for data analysis, https://github.com/Xinglab/GTEx-brain-sQTL
GTEx WGS genotype data, https://storage.googleapis.com/gtex-public-data/Portal_Analysis_Methods_v7_09052017.pdf
HapMap Project Genome Browser version E, https://www.internationalgenome.org/
NHGRI-EBI GWAS Catalog v1.0.1, ftp://ftp.ebi.ac.uk/pub/databases/gwas/releases/2017/08/01
NHGRI-EBI GWAS Catalog Summary Statistics, https://www.ebi.ac.uk/gwas/downloads/summary-statistics
rMATS-turbo, http://rnaseq-mats.sourceforge.net/rmats4.0.2/
UCSC Genome Browser, https://genome.ucsc.edu/
Supplemental Data
References
- 1.Nilsen T.W., Graveley B.R. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang E.T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S.F., Schroth G.P., Burge C.B. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbosa-Morais N.L., Irimia M., Pan Q., Xiong H.Y., Gueroussov S., Lee L.J., Slobodeniuc V., Kutter C., Watt S., Çolak R. The evolutionary landscape of alternative splicing in vertebrate species. Science. 2012;338:1587–1593. doi: 10.1126/science.1230612. [DOI] [PubMed] [Google Scholar]
- 4.Yeo G., Holste D., Kreiman G., Burge C.B. Variation in alternative splicing across human tissues. Genome Biol. 2004;5:R74. doi: 10.1186/gb-2004-5-10-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Licatalosi D.D., Darnell R.B. Splicing regulation in neurologic disease. Neuron. 2006;52:93–101. doi: 10.1016/j.neuron.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 6.Manning K.S., Cooper T.A. The roles of RNA processing in translating genotype to phenotype. Nat. Rev. Mol. Cell Biol. 2017;18:102–114. doi: 10.1038/nrm.2016.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raj B., Blencowe B.J. Alternative splicing in the mammalian nervous system: recent insights into mechanisms and functional roles. Neuron. 2015;87:14–27. doi: 10.1016/j.neuron.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 8.Vuong C.K., Black D.L., Zheng S. The neurogenetics of alternative splicing. Nat. Rev. Neurosci. 2016;17:265–281. doi: 10.1038/nrn.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trabzuni D., Wray S., Vandrovcova J., Ramasamy A., Walker R., Smith C., Luk C., Gibbs J.R., Dillman A., Hernandez D.G. MAPT expression and splicing is differentially regulated by brain region: relation to genotype and implication for tauopathies. Hum. Mol. Genet. 2012;21:4094–4103. doi: 10.1093/hmg/dds238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buée L., Bussière T., Buée-Scherrer V., Delacourte A., Hof P.R. Tau protein isoforms, phosphorylation and role in neurodegenerative disorders. Brain Res. Brain Res. Rev. 2000;33:95–130. doi: 10.1016/s0165-0173(00)00019-9. [DOI] [PubMed] [Google Scholar]
- 11.Rockenstein E.M., McConlogue L., Tan H., Power M., Masliah E., Mucke L. Levels and alternative splicing of amyloid β protein precursor (APP) transcripts in brains of APP transgenic mice and humans with Alzheimer’s disease. J. Biol. Chem. 1995;270:28257–28267. doi: 10.1074/jbc.270.47.28257. [DOI] [PubMed] [Google Scholar]
- 12.Da Cruz S., Cleveland D.W. Understanding the role of TDP-43 and FUS/TLS in ALS and beyond. Curr. Opin. Neurobiol. 2011;21:904–919. doi: 10.1016/j.conb.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ling S.C., Polymenidou M., Cleveland D.W. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron. 2013;79:416–438. doi: 10.1016/j.neuron.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Rubeis S., He X., Goldberg A.P., Poultney C.S., Samocha K., Cicek A.E., Kou Y., Liu L., Fromer M., Walker S., DDD Study. Homozygosity Mapping Collaborative for Autism. UK10K Consortium Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iossifov I., O’Roak B.J., Sanders S.J., Ronemus M., Krumm N., Levy D., Stessman H.A., Witherspoon K.T., Vives L., Patterson K.E. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takata A., Ionita-Laza I., Gogos J.A., Xu B., Karayiorgou M. De novo synonymous mutations in regulatory elements contribute to the genetic etiology of autism and schizophrenia. Neuron. 2016;89:940–947. doi: 10.1016/j.neuron.2016.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quesnel-Vallières M., Weatheritt R.J., Cordes S.P., Blencowe B.J. Autism spectrum disorder: insights into convergent mechanisms from transcriptomics. Nat. Rev. Genet. 2019;20:51–63. doi: 10.1038/s41576-018-0066-2. [DOI] [PubMed] [Google Scholar]
- 18.Clinton S.M., Haroutunian V., Davis K.L., Meador-Woodruff J.H. Altered transcript expression of NMDA receptor-associated postsynaptic proteins in the thalamus of subjects with schizophrenia. Am. J. Psychiatry. 2003;160:1100–1109. doi: 10.1176/appi.ajp.160.6.1100. [DOI] [PubMed] [Google Scholar]
- 19.Xu B., Ionita-Laza I., Roos J.L., Boone B., Woodrick S., Sun Y., Levy S., Gogos J.A., Karayiorgou M. De novo gene mutations highlight patterns of genetic and neural complexity in schizophrenia. Nat. Genet. 2012;44:1365–1369. doi: 10.1038/ng.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takata A., Matsumoto N., Kato T. Genome-wide identification of splicing QTLs in the human brain and their enrichment among schizophrenia-associated loci. Nat. Commun. 2017;8:14519. doi: 10.1038/ncomms14519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eom T., Zhang C., Wang H., Lay K., Fak J., Noebels J.L., Darnell R.B. NOVA-dependent regulation of cryptic NMD exons controls synaptic protein levels after seizure. eLife. 2013;2:e00178. doi: 10.7554/eLife.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Faustino N.A., Cooper T.A. Pre-mRNA splicing and human disease. Genes Dev. 2003;17:419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- 23.Scotti M.M., Swanson M.S. RNA mis-splicing in disease. Nat. Rev. Genet. 2016;17:19–32. doi: 10.1038/nrg.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sibley C.R., Blazquez L., Ule J. Lessons from non-canonical splicing. Nat. Rev. Genet. 2016;17:407–421. doi: 10.1038/nrg.2016.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y.I., van de Geijn B., Raj A., Knowles D.A., Petti A.A., Golan D., Gilad Y., Pritchard J.K. RNA splicing is a primary link between genetic variation and disease. Science. 2016;352:600–604. doi: 10.1126/science.aad9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raj T., Li Y.I., Wong G., Humphrey J., Wang M., Ramdhani S., Wang Y.C., Ng B., Gupta I., Haroutunian V. Integrative transcriptome analyses of the aging brain implicate altered splicing in Alzheimer’s disease susceptibility. Nat. Genet. 2018;50:1584–1592. doi: 10.1038/s41588-018-0238-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen S., Park J.W., Lu Z.X., Lin L., Henry M.D., Wu Y.N., Zhou Q., Xing Y. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. USA. 2014;111:E5593–E5601. doi: 10.1073/pnas.1419161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.GTEx Consortium Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leek J.T., Johnson W.E., Parker H.S., Fertig E.J., Jaffe A.E., Storey J.D., Zhang Y., Torres L.C. 2017. sva: Surrogate variable analysis. R package version 3. 10.18129. [Google Scholar]
- 30.Zhao K., Lu Z.X., Park J.W., Zhou Q., Xing Y. GLiMMPS: robust statistical model for regulatory variation of alternative splicing using RNA-seq data. Genome Biol. 2013;14:R74. doi: 10.1186/gb-2013-14-7-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giambartolomei C., Vukcevic D., Schadt E.E., Franke L., Hingorani A.D., Wallace C., Plagnol V. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10:e1004383. doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Franceschini N., Giambartolomei C., de Vries P.S., Finan C., Bis J.C., Huntley R.P., Lovering R.C., Tajuddin S.M., Winkler T.W., Graff M., MEGASTROKE Consortium GWAS and colocalization analyses implicate carotid intima-media thickness and carotid plaque loci in cardiovascular outcomes. Nat. Commun. 2018;9:5141. doi: 10.1038/s41467-018-07340-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alipanahi B., Delong A., Weirauch M.T., Frey B.J. Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat. Biotechnol. 2015;33:831–838. doi: 10.1038/nbt.3300. [DOI] [PubMed] [Google Scholar]
- 34.Craig A.M., Kang Y. Neurexin-neuroligin signaling in synapse development. Curr. Opin. Neurobiol. 2007;17:43–52. doi: 10.1016/j.conb.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loya C.M., Van Vactor D., Fulga T.A. Understanding neuronal connectivity through the post-transcriptional toolkit. Genes Dev. 2010;24:625–635. doi: 10.1101/gad.1907710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Traunmüller L., Gomez A.M., Nguyen T.-M., Scheiffele P. Control of neuronal synapse specification by a highly dedicated alternative splicing program. Science. 2016;352:982–986. doi: 10.1126/science.aaf2397. [DOI] [PubMed] [Google Scholar]
- 37.Ehrmann I., Dalgliesh C., Liu Y., Danilenko M., Crosier M., Overman L., Arthur H.M., Lindsay S., Clowry G.J., Venables J.P. The tissue-specific RNA binding protein T-STAR controls regional splicing patterns of neurexin pre-mRNAs in the brain. PLoS Genet. 2013;9:e1003474. doi: 10.1371/journal.pgen.1003474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buniello A., MacArthur J.A.L., Cerezo M., Harris L.W., Hayhurst J., Malangone C., McMahon A., Morales J., Mountjoy E., Sollis E. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47(D1):D1005–D1012. doi: 10.1093/nar/gky1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bray N.J., O’Donovan M.C. The genetics of neuropsychiatric disorders. Brain Neurosci. Adv. 2019;2:2. doi: 10.1177/2398212818799271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCarroll S.A., Feng G., Hyman S.E. Genome-scale neurogenetics: methodology and meaning. Nat. Neurosci. 2014;17:756–763. doi: 10.1038/nn.3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Solovieff N., Cotsapas C., Lee P.H., Purcell S.M., Smoller J.W. Pleiotropy in complex traits: challenges and strategies. Nat. Rev. Genet. 2013;14:483–495. doi: 10.1038/nrg3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yeo G., Burge C.B. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- 43.Pickrell J.K., Marioni J.C., Pai A.A., Degner J.F., Engelhardt B.E., Nkadori E., Veyrieras J.-B., Stephens M., Gilad Y., Pritchard J.K. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464:768–772. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shete S., Hosking F.J., Robertson L.B., Dobbins S.E., Sanson M., Malmer B., Simon M., Marie Y., Boisselier B., Delattre J.Y. Genome-wide association study identifies five susceptibility loci for glioma. Nat. Genet. 2009;41:899–904. doi: 10.1038/ng.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wrensch M., Jenkins R.B., Chang J.S., Yeh R.F., Xiao Y., Decker P.A., Ballman K.V., Berger M., Buckner J.C., Chang S. Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nat. Genet. 2009;41:905–908. doi: 10.1038/ng.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cremona M.L., Matthies H.J., Pau K., Bowton E., Speed N., Lute B.J., Anderson M., Sen N., Robertson S.D., Vaughan R.A. Flotillin-1 is essential for PKC-triggered endocytosis and membrane microdomain localization of DAT. Nat. Neurosci. 2011;14:469–477. doi: 10.1038/nn.2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pizzo A.B., Karam C.S., Zhang Y., Yano H., Freyberg R.J., Karam D.S., Freyberg Z., Yamamoto A., McCabe B.D., Javitch J.A. The membrane raft protein Flotillin-1 is essential in dopamine neurons for amphetamine-induced behavior in Drosophila. Mol. Psychiatry. 2013;18:824–833. doi: 10.1038/mp.2012.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O’Brien H.E., Hannon E., Hill M.J., Toste C.C., Robertson M.J., Morgan J.E., McLaughlin G., Lewis C.M., Schalkwyk L.C., Hall L.S. Expression quantitative trait loci in the developing human brain and their enrichment in neuropsychiatric disorders. Genome Biol. 2018;19:194. doi: 10.1186/s13059-018-1567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhong J., Li S., Zeng W., Li X., Gu C., Liu J., Luo X.J. Integration of GWAS and brain eQTL identifies FLOT1 as a risk gene for major depressive disorder. Neuropsychopharmacology. 2019;44:1542–1551. doi: 10.1038/s41386-019-0345-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Efthymiou A.G., Goate A.M. Late onset Alzheimer’s disease genetics implicates microglial pathways in disease risk. Mol. Neurodegener. 2017;12:43. doi: 10.1186/s13024-017-0184-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karch C.M., Ezerskiy L.A., Bertelsen S., Goate A.M., Alzheimer’s Disease Genetics Consortium (ADGC) Alzheimer’s disease risk polymorphisms regulate gene expression in the ZCWPW1 and the CELF1 loci. PLoS ONE. 2016;11:e0148717. doi: 10.1371/journal.pone.0148717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhernakova A., Stahl E.A., Trynka G., Raychaudhuri S., Festen E.A., Franke L., Westra H.-J., Fehrmann R.S.N., Kurreeman F.A.S., Thomson B. Meta-analysis of genome-wide association studies in celiac disease and rheumatoid arthritis identifies fourteen non-HLA shared loci. PLoS Genet. 2011;7:e1002004. doi: 10.1371/journal.pgen.1002004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu C., Pan W. Integrating eQTL data with GWAS summary statistics in pathway-based analysis with application to schizophrenia. Genet. Epidemiol. 2018;42:303–316. doi: 10.1002/gepi.22110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de Jong S., van Eijk K.R., Zeegers D.W., Strengman E., Janson E., Veldink J.H., van den Berg L.H., Cahn W., Kahn R.S., Boks M.P., Ophoff R.A., PGC Schizophrenia (GWAS) Consortium Expression QTL analysis of top loci from GWAS meta-analysis highlights additional schizophrenia candidate genes. Eur. J. Hum. Genet. 2012;20:1004–1008. doi: 10.1038/ejhg.2012.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Watanabe M., Hatakeyama S. TRIM proteins and diseases. J. Biochem. 2017;161:135–144. doi: 10.1093/jb/mvw087. [DOI] [PubMed] [Google Scholar]
- 56.Ballatore C., Lee V.M., Trojanowski J.Q. Tau-mediated neurodegeneration in Alzheimer’s disease and related disorders. Nat. Rev. Neurosci. 2007;8:663–672. doi: 10.1038/nrn2194. [DOI] [PubMed] [Google Scholar]
- 57.Soto C., Pritzkow S. Protein misfolding, aggregation, and conformational strains in neurodegenerative diseases. Nat. Neurosci. 2018;21:1332–1340. doi: 10.1038/s41593-018-0235-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Irwin D.J., Lee V.M., Trojanowski J.Q. Parkinson’s disease dementia: convergence of α-synuclein, tau and amyloid-β pathologies. Nat. Rev. Neurosci. 2013;14:626–636. doi: 10.1038/nrn3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yeo G.W., Coufal N.G., Liang T.Y., Peng G.E., Fu X.-D., Gage F.H. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat. Struct. Mol. Biol. 2009;16:130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ule J., Stefani G., Mele A., Ruggiu M., Wang X., Taneri B., Gaasterland T., Blencowe B.J., Darnell R.B. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444:580–586. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- 61.Park E., Pan Z., Zhang Z., Lin L., Xing Y. The expanding landscape of alternative splicing variation in human populations. Am. J. Hum. Genet. 2018;102:11–26. doi: 10.1016/j.ajhg.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xiong H.Y., Alipanahi B., Lee L.J., Bretschneider H., Merico D., Yuen R.K.C., Hua Y., Gueroussov S., Najafabadi H.S., Hughes T.R. RNA splicing. The human splicing code reveals new insights into the genetic determinants of disease. Science. 2015;347:1254806. doi: 10.1126/science.1254806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Van Nostrand E.L., Freese P., Pratt G.A., Wang X., Wei X., Blue S.M., Dominguez D., Cody N.A.L., Olson S., Sundararaman B. A large-scale binding and functional map of human RNA binding proteins. bioRxiv. 2018 doi: 10.1101/179648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gehman L.T., Meera P., Stoilov P., Shiue L., O’Brien J.E., Meisler M.H., Ares M., Jr., Otis T.S., Black D.L. The splicing regulator Rbfox2 is required for both cerebellar development and mature motor function. Genes Dev. 2012;26:445–460. doi: 10.1101/gad.182477.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nakahata S., Kawamoto S. Tissue-dependent isoforms of mammalian Fox-1 homologs are associated with tissue-specific splicing activities. Nucleic Acids Res. 2005;33:2078–2089. doi: 10.1093/nar/gki338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Underwood J.G., Boutz P.L., Dougherty J.D., Stoilov P., Black D.L. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol. Cell. Biol. 2005;25:10005–10016. doi: 10.1128/MCB.25.22.10005-10016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hariri A.R. The emerging importance of the cerebellum in broad risk for psychopathology. Neuron. 2019;102:17–20. doi: 10.1016/j.neuron.2019.02.031. [DOI] [PubMed] [Google Scholar]
- 68.Keller D., Erö C., Markram H. Cell densities in the mouse brain: a systematic review. Front. Neuroanat. 2018;12:83. doi: 10.3389/fnana.2018.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.