Abstract
According to historical records of transatlantic slavery, traders forcibly deported an estimated 12.5 million people from ports along the Atlantic coastline of Africa between the 16th and 19th centuries, with global impacts reaching to the present day, more than a century and a half after slavery’s abolition. Such records have fueled a broad understanding of the forced migration from Africa to the Americas yet remain underexplored in concert with genetic data. Here, we analyzed genotype array data from 50,281 research participants, which—combined with historical shipping documents—illustrate that the current genetic landscape of the Americas is largely concordant with expectations derived from documentation of slave voyages. For instance, genetic connections between people in slave trading regions of Africa and disembarkation regions of the Americas generally mirror the proportion of individuals forcibly moved between those regions. While some discordances can be explained by additional records of deportations within the Americas, other discordances yield insights into variable survival rates and timing of arrival of enslaved people from specific regions of Africa. Furthermore, the greater contribution of African women to the gene pool compared to African men varies across the Americas, consistent with literature documenting regional differences in slavery practices. This investigation of the transatlantic slave trade, which is broad in scope in terms of both datasets and analyses, establishes genetic links between individuals in the Americas and populations across Atlantic Africa, yielding a more comprehensive understanding of the African roots of peoples of the Americas.
Keywords: genetics, population genetics, slave trade, Africa, Americas, identity by descent, ancestry, admixture, migration, history
Introduction
The forced displacement of more than 12.5 million men, women, and children from Africa to the Americas between 1515 and 1865 has had significant social, cultural, health, and genetic impacts across the Americas.1, 2, 3, 4, 5, 6 The consequences of this displacement are myriad and complex, reflecting the varied economic goals and practices of the nations outfitting the ships that transported captive Africans.7,8 The current narrative of this era of forced deportation has been compiled from thousands of historical shipping documents, as well as diaries, personal letters, sketches, and records of slave sales.9 These data suggest that the number of enslaved people disembarking at each port in the Americas, the nation that shipped enslaved people to a given port, and the populations and geographic locations in Africa from which captives were taken differs from region to region within the Americas. For example, shipping manifests indicate that the number of enslaved people disembarking at Brazilian and Caribbean ports was far greater than the number disembarking at other ports in the Americas.5 An estimated 10.1 million enslaved people, primarily male, disembarked in Central America, South America, and the Caribbean (accounting for more than 90% of all captives who were brought to the Americas), with fewer than half a million disembarking in mainland North America5 (3% to 5% of the total). Concurrent with transatlantic voyages, slave traders also transferred nearly 500,000 enslaved people throughout the Americas with most documented intra-American voyages originating in the Caribbean.5,10 Despite a majority of enslaved Africans disembarking into Latin America, estimates of the proportion of African ancestry for Latin Americans with African roots are lower on average than estimates for African Americans in the United States.11, 12, 13, 14, 15 Further, patterns of embarkation of enslaved populations varied over time, whereby people primarily from 48 distinct ethnolinguistic groups were targeted across Atlantic Africa.16 Prior to 1650, slave voyages most often originated in Senegambia and West Central Africa (in locations now known as the Democratic Republic of Congo and Angola). Although West Central Africa remained a primary embarkation region throughout the slave trading period (from 1650 to 1850), these voyages originated with increasing frequency at ports in the Bight of Benin and the Bight of Biafra, in locations now known as Benin, Nigeria, Cameroon, and Gabon.9
While records have shed light on the main trends of the transatlantic slave trade, the effects of under-documented practices, such as illegal slave trading and details of events after disembarkation in the Americas, remain less understood.17, 18, 19, 20 If under-documented events did not shift the overall paradigm of the slave trade and reproduction rates were equivalent among enslaved populations in the Americas, the degree of genetic similarity between the Americas and each slave-trading region of Africa would be expected to be correlated with the recorded number of enslaved people that disembarked into each region. Also, little is known about the extent to which enslaved people and their descendants continued to associate exclusively with those of similar ethnolinguistic origins after generations in the Americas.21 If such association was common, Americans with African roots would be expected to have African ancestry from a single region of Africa in a pattern that is correlated with geography. Finally, previous studies indicate that individuals with mixed ancestry have higher levels of African ancestry in genomic regions inherited through the female line.22, 23, 24 However, the extent of this sex bias in gene pool contributions may vary across the Americas due to regional differences in mortality of enslaved men,25 rape of enslaved African women,26 forced segregation,21 and practices of reducing African representation by promoting reproduction with Europeans (racial whitening).27,28
Previous genetic studies focusing on people of African descent across the Americas found that most African Americans in the United States have more African ancestry from populations that lived near present-day Nigeria than from populations that lived elsewhere in Atlantic Africa.11,15 However, multiple previous studies have lacked representation of West Central Africa, the largest source of enslaved people during the transatlantic slave trade, leading to an incomplete understanding of the dynamics of population interactions during and after the slave trade era. Here, we combined genetic data from individuals representing the major slave trading regions of Atlantic Africa, Europe, and the Americas and explored concordance with centuries of historical documents. We determined ancestral connections using identity by descent, local ancestry inference, and Bayesian dating techniques. In addition, we used ancestral admixture models to estimate the bias in gene pool contributions of African, European, and indigenous American men and women to current populations of the Americas. We used data from this large, diverse cohort of individuals to obtain unprecedented insights into the genetic impact of the transatlantic slave trade.
Subjects and Methods
Human Subjects
We included data from participants drawn from the millions of research participants of 23andMe, Inc., a consumer personal genetics company,29, 30, 31 the 1000 Genomes Project,32 the Human Genome Diversity Project,33 Angola, and previous studies of the Democratic Republic of the Congo,34 Sierra Leone,35 and Khoe-San speaking people.36,37 Angolan participant saliva samples were collected with research consent, which was approved by both the ethics committee of the University 11th of November (Universidade 11 de Novembro), Cabinda, Angola (REf: GD-FM/UoN/2016) and the University of Leicester ethics committee (REf: 11334-sdsb1-genetics). 23andMe research participants provided informed consent and participated in the research online, under a protocol approved by the external AAHRPP-accredited IRB, Ethical & Independent Review Services (E&I Review). 23andMe participants were given the option to fill out web-based questionnaires, including questions on ancestry, ethnicity, and grandparent birth locations. We included in our study data only of customers who signed IRB-approved consent documents, which are explicit about the use of customer data for ancestry research.
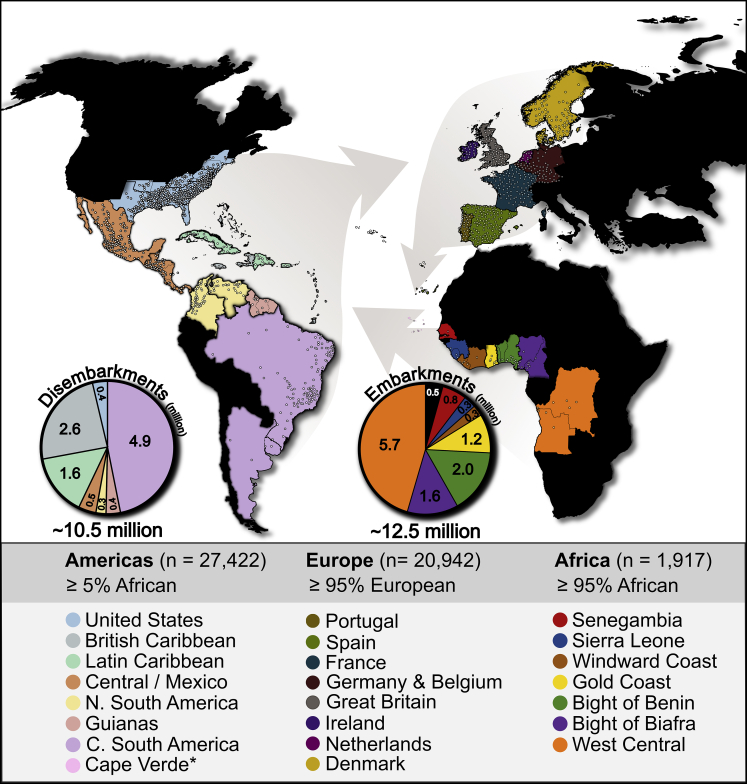
We grouped individuals across the Americas, Atlantic Africa, and Western Europe into cohorts designed to reflect historical regions (Tables S1–S3). Within the United States, we focused solely on coastal and surrounding states known to have been disembarkation locations for enslaved people.5,9 We included the Cape Verde archipelago as control region since it is an Atlantic African country that was colonized by the Portuguese in the 15th century and has been inhabited primarily by the descendants of enslaved people from Senegambia.38 In Africa, we merged contemporary administrative borders to match all major slave trading regions during the transatlantic slave trade except South East Africa, which constituted about 3.8% of all African embarkments, due to limited sample representation in Mozambique and Madagascar5,9 (Figure 1; Table S1). Because records constitute number of captives deported by region of purchase, as opposed to ethnolinguistic affiliation, merging geographic boundaries in this manner allowed us to connect people of African descent directly to slave trading regions from which their African ancestors may have embarked.5 Borders of Atlantic African countries generally aligned with those historical slave trading regions; however, we split Nigeria into western and eastern states along the Niger River because enslaved people from east of the Niger were deported from ports in the Bight of Biafra, whereas people west of the Niger were deported from ports in the Bight of Benin.9 Categorizing Nigeria in this way also corresponds to the historical geographic occupancy of groups with genetic, ethnic, and linguistic differences: the Yoruba in the Bight of Benin and Esan and Igbo in the Bight of Biafra.4 We considered current Atlantic African populations to be suitable proxies for past populations in Africa given limited migration and current population structure following a latitudinal gradient13,39,40 (Figures S1–S6). We combined regions of Europe participating in the transatlantic slave trade by broad historical descriptors7 and population structure (Figures S7–S9; Table S3), which are largely described by geographic proximity.
Figure 1.
Location of Individuals and Cohorts
Arrows highlight the general direction of the triangular trade routes between continents during the transatlantic slave trade. Pie charts indicate the documented number of enslaved people embarking out of regions of Africa (∼12.5 million total) and disembarking in regions of the Americas (∼10.5 million total) between 1515 and 1865. Representatives of regions of the Americas and Europe indicated that they each have four grandparents born within the same country or US state. Representatives of Atlantic Africa either indicated four grandparents born within or historical ties to a country. Points indicate the ∼16,000 unique grandparental geo-coordinates provided by participants. ∗Cape Verde is an Atlantic African island country that, in the 15th century, was colonized by the Portuguese and inhabited primarily by enslaved people from Senegambia.
Study participants from the 23andMe customer base indicated their four grandparents were born within the same country (Latin America and the Caribbean) or state (United States), whereas study participants from public databases and Africa-focused studies were included based on self-described historical ties to an African country relevant to the transatlantic slave trade.32, 33, 34, 35 Using 23andMe’s ancestry inference algorithm,41 we excluded data from individuals in the Americas with less than 5% African ancestry, Africans with less than 95% African ancestry, and Europeans with less than 95% European ancestry. We additionally pruned pairs of related individuals using refinedIBD42 so that no two individuals in the study share more than 500 cM of DNA that is identical by descent. These filtering steps reduced a sample size of 183,915 individuals representing the Americas and Africa to 29,339, and 36,866 individuals representing Europe to 20,942. In remaining Atlantic African representatives, we compiled ethnic affiliations from self-reports in 23andMe customers and sample descriptions in other datasets, finding 37 unique ethnic affiliations (Table S1).
Genotyping
DNA extraction and genotyping of 23andMe customers and Human Genome Diversity Project were performed on saliva samples by the National Genetics Institute (NGI), a CLIA-licensed clinical laboratory and subsidiary of Laboratory Corporation of America. Samples were genotyped on one of five genotyping platforms. The V1 and V2 platforms were variants of the Illumina HumanHap550+ BeadChip, with 560,000 SNPs of which ∼25,000 custom SNPs were selected by 23andMe. The V3 platform was based on the Illumina OmniExpress+ BeadChip, with additional SNPs, totaling about 950,000 SNPs. The V4 and V5 platforms are custom arrays, including a lower-redundancy subset of V2 and V3 SNPs with additional coverage of lower-frequency coding variation and about 570,000 SNPs. Whole-genome genotypes from 1000 Genomes Project32 were subset to match those of V1-V5 platforms and Human Genome Diversity Panel individuals (HGDP)33 were genotyped on both V1 and V5 platforms. Remaining African datasets were genotyped on V3 and V5 platforms. Within each platform, we removed any sample with >5% genotype missingness, producing an average missingness rate of 0.48% across all samples.
Population Structure
We used both ordination techniques and ADMIXTURE43 to assess population structure within Atlantic Africa as well as in Atlantic Africa plus the Americas. For these techniques, we reduced linkage by pruning SNPs overlapping across all genotype platforms with r2 < 0.5 via PLINK,44 resulting in 43,780 SNPs. We included additional Karitiana, Maya, Pima, and Surui individuals from HGDP23 as Native American references. Ordination techniques consisted of principal component analysis (PCA) and uniform manifold approximation and projection43 (UMAP) using 0.8 minimum distance, 20 nearest neighbors, and number of pcs based on the plateau of eigenvalues (8–15). We ran ADMIXTURE,45 with K values from 2 to 20 in Atlantic African, European, and across all samples.
Voyage Records
We accessed transatlantic historical records from the Slave Voyages database.5 The Slave Voyages database contains a collection of records from a wide range of published and archival information on more than 36,000 voyages across the Atlantic between 1492 and the early nineteenth century.5 The database contains more than 100 possible variables for each voyage, including voyage dates, disembarkation/embarkation ports, crew information, and voyage outcomes. Some records that lacked pertinent data were imputed using information such as carrying capacity of vessels and characteristics of similar documented voyages.5 Using the Slave Voyages database, we compiled estimates of enslaved African embarkation (“Imputed total num slaves purchased exact”), disembarkation (“Imputed total slaves disembarked exact”), proportions of males (“Imputed percentage male exact”), and proportion of children (“Imputed percentage child exact”) for each Atlantic African (“Imputed principal region of slave purchase lang en exact”) and American region (“Imputed principal port slave dis lang en exact;” “Imputed principal region slave dis lang en exact”), by voyage year (“Imputed arrival at port of dis exact”). For disembarkation estimates of enslaved Africans into the Americas, we mapped the principal port of disembarkation and/or principal region of disembarkation to American region and grouped by Atlantic African region. We estimated shipment sex bias using the “Voyage - percent males on” field and shipment age bias using the “Voyage - percent children on” field. We similarly utilized the intra-American slave trade database, which contains records of voyages within regions of the Americas that occurred in tandem with the transatlantic slave trade.5,46 We aggregated estimates of the number of enslaved people forcefully deported between regions of the Americas using the “Imputed total disembarked,” “Imputed principal place of purchase,” and “Imputed principal place of slave landing” fields.5,46
Ancestry Inference
We used 23andMe’s Ancestry Composition pipeline to identify the ancestral origins of chromosomal segments in individuals using all available genotype data.41,47 Thus, the number of SNPs used to predict ancestry varied between 560,000 and 950,000 based on the platform an individual was genotyped on. Ancestry Composition uses support vector machine classifiers to assign ancestry labels to locally phased 300-SNP windows, whose ∼1 Mb size increases the power for detecting ancestry origins within a window while still accurately predicting fine-scale local ancestry.48 These initial labels are then processed with an autoregressive pair hidden Markov model to smooth and correct phasing errors. The resulting posterior probabilities are recalibrated with an isotonic regression model. Ancestry Composition assigns 33 local populations, four of which correspond to Atlantic African countries directly impacted by the slave trade, and four of which correspond to western European countries that conducted the slave trade.3 The limited number of populations Ancestry Composition predicts reflects the inherent difficulty to distinguish populations with complex histories, and thus is a balance between accurate assignment of genetic segments and the number of unique populations. In Africa, potential local populations are as follows: Nigerian (Nigeria), Senegambian (The Gambia, Guinea, Guinea Bissau, Senegal), Coastal West African (Sierra Leone, Ghana, Côte d’Ivoire, Liberia), and Congolese (Angola, the Democratic Republic of the Congo). Local European populations are as follows: British and Irish (the United Kingdom and the Republic of Ireland), French and German (Austria, Belgium, France, Germany, the Netherlands, Luxembourg, Switzerland), Iberian (Spain and Portugal), and Scandinavian (Denmark, Iceland, Norway, and Sweden).
Identity by Descent
We estimated identity by descent (IBD) within and between people representing regions in the Americas, Atlantic Africa, and western Europe using the software package refinedIBD.42 We first removed all missing positions and subset markers to the intersection between all platforms, resulting in 87,847 unique positions used in all IBD analyses. While lowering SNP density may reduce the ability to detect smaller segments, removing missing positions minimizes the detection of false-positive segments.49 With the preference of reducing false-positive calls, we additionally specified a minimum LOD score of 3.3 and a minimum reported length of 3 cM. All other parameters were set to default. We used mean pairwise IBD (msIBD) to assess the overall IBD being shared between (or within) regions whereby msIBD is the sum of IBD shared between populations divided by the total pairwise comparisons of individuals in populations. We used this metric because it accounts for imbalances in sample sizes—although other metrics such as the proportion of individuals with IBD match, skewness of IBD segments, and number of IBD segments shared yield identical patterns (Table S4). We also determined the mean number of regions within Europe or Africa with which individuals from the Americas share a ≥5 cM segment (mcIBD region). That is, an individual from a region in the Americas must share at least 5 cM with a single individual from a region in Europe or Africa to be considered to have an IBD connection to that region.
Time to Most Recent Common Ancestor
To compare the timing of genetic connections between African populations and individuals from the Americas with historical records of the transatlantic shipping of enslaved Africans, we estimated the age of IBD segments using a Bayesian method.50 The length of an IBD segment shared between two individuals given the time to their most recent common ancestor (MRCA) is Erlang-2 distributed with , where is the number of generations back in time to the MRCA.50 If we let represent an IBD segment length observed in cM, and we allow the discrete number of generations to be continuous , this distribution is:
However, in practice, we only observe IBD segments larger than some length predetermined by our method to estimate IBD. Thus, we must condition the distribution of IBD segment lengths observed given the TMRCA on the minimum possible segment size
where is the predetermined minimum IBD segment length in cM.
From the set of all IBD segments observed between sampled individuals in two populations we are interested in estimating the time of a MRCA that may have occurred during the transatlantic slave trade. However, not all of the MRCAs between each pair of individuals in the two populations lived during this time period. Some of the IBD segments may come from recent, post-slave trade immigration, and some of the IBD segments may be from more distant shared ancestors that predate the slave trade. We modeled the probability that an observed IBD segment comes from each of the three time periods post-slave trade, slave trade, and pre-slave trade as , , and . Given the possibility of ancestors from each of those time periods, the distribution of IBD segment lengths becomes:
where , , and are the TMRCA for post-slave trade, slave trade, and pre-slave trade, respectively, and is the set of all IBD segment lengths observed in cM.
We placed uniform prior distributions on , , and . These distributions assume we know the time (in generations) of the start and end of the transatlantic slave trade. Assuming a generation time of 20 to 27 years, the transatlantic slave trade began roughly 14 to 20 generations ago.9,22 We set our priors accordingly:
, , , and . We found that the relative timings estimated by our results were robust to a range of different priors, for example assigning the slave trade start and end times as 25 and 4 generations ago. In order to estimate posterior distributions of t_1, t_2, t_3, w_1, w_2, and w_3, we ran an MCMC analysis for 200,000 iterations, sampling parameter values every 40 iterations, and discarding the first 25% of samples as burnin. We assessed convergence of the MCMC by checking that trace plots exhibited apparent stationarity and ensuring that the effective sample size for each estimated parameter was more than 600.
Ancestral Sex Bias
QA We determined unequal sex contributions to the gene pool using an admixture model with X chromosome ancestry estimates, autosome ancestry estimates,22,51 and haplogroup imbalance between Y and mitochondrial haplogroups.52 We estimated sex-specific contributions, whereby multiple source populations contribute to an admixed population, assuming a single admixture event (for generations 1 to 15).51 We used this mechanistic admixture model to explicitly compare autosome to X chromosome ancestry to determine female (sf) and male contributions (sm) to the gene pools of populations with African ancestry. This discrete-time model specifically derives the moment of the distribution of the autosomal admixture fraction from a specific source population and allows uneven sex-specific contribution from each source population at each generation.51 We determined mitochondrial haplogroups using HaploGrep2 with Phylotree 1753 and Y-haplogroups using yHaplo,54 which uses phylogenetically informative SNPs to associate individuals with specific branches of the Y chromosome tree.55 We assigned the origin of haplogroups based on previous studies associating haplogroups with either African, European, or indigenous American ancestry.24,52,56
Results
Genetic Connections Compared to Historical Documents
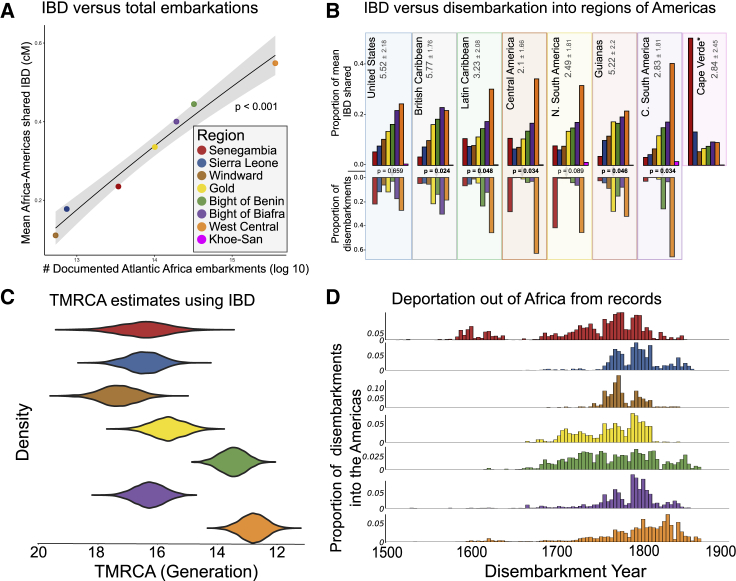
The overall correlation between embarkation counts from Africa and mean proportion of identity by descent shared42 (msIBD) between regions of Atlantic Africa and all of the Americas (Figure 2A) is highly significant (Spearman’s coefficient permutation; p < 0.001). However, when we subset shipping records to specific disembarkation regions of the Americas, msIBD and enslaved disembarkation were not significantly correlated across all regions of the Americas (Figures 2B and S10). For example, the study cohorts representing the United States (p = 0.66) and northern South America (p = 0.09) have very little msIBD with Senegambia, compared to the proportion of captives from Senegambia disembarking in those two regions (Figure 2B). Many regions also demonstrate over-representation of msIBD with the Bights of Biafra and Benin compared to historical documents.
Figure 2.
IBD Sharing between the Americas and Atlantic Africa versus Documented Counts of Deportation
(A) Comparison between the number of enslaved people embarking out of each region of Atlantic Africa to the mean shared IBD (msIBD; cM) between each Atlantic Africa region and all individuals in the Americas.
(B) msIBD between regions of Africa and of the Americas (top) and the estimated proportions of enslaved disembarkation into each region of the Americas (bottom). p values indicate significant correlations between msIBD and disembarkation based on a permutation test using Spearman’s coefficient of correlation. Numbers right of the region labels indicate the mean number (±STD) of mcIBD connections (≥5 cM) an individual representing that region of the Americas has to the seven primary slave trading regions.
(C) Posterior distributions of the estimated TMRCA of IBD segments during the transatlantic slave trade between each Africa region and all of the Americas.
(D) The proportion of disembarkation of each Atlantic Africa region into the Americas over time, according to historical documents. All Atlantic Africa regions correspond to the colors in key in (A). msIBD is the total, cumulative IBD shared between two populations (cM), divided by the total pairwise comparisons between two populations.
The mean number of IBD connections (mcIBDreg1-reg2; the mean number of regions an individual shares a ≥5 cM IBD segment with) an individual in each region of the Americas has to the seven primary slave trading regions of Africa shows representative individuals from the United States match more slave trading regions (mcIBDUS-Africa = 5.5 ± 2.2) than representative individuals from Central America and Mexico who tend to have IBD connections with fewer regions (mcIBDCentral America - Africa = 2.1 ± 1.7, Figure 2B; Table S5). Of note, there is a particularly strong discordance between disembarkations and mcIBD in Central America and northern South America. There is also a stark difference between mcIBD in the British Caribbean (mcIBD British Caribbean - Africa = 5.8 ± 1.8) versus Latin Caribbean (mcIBD Latin Caribbean - Africa = 3.2 ± 2.1), even though these regions are geographically proximate. These results correspond with comparisons between eight regions of western Europe and regions of the Americas, which indicated that Latin Americans tend to have higher mcIBDLatin America -Europe (x̄ = 4.9 ± 0.76; primarily from Spanish Portuguese) than British occupied regions (x̄ = 3.52 ± 1.1; Figures S11,S12; Table S6).
Our Bayesian MCMC approach to estimate time to most recent common ancestor living during the slave trade (TMRCA) from IBD segment lengths predicted that West Central Africa shares more recent common ancestors with individuals in the Americas, whereas Senegambia, Sierra Leone, the Windward Coast, and the Bight of Biafra share earlier common ancestors (Figure 2C). This trend mirrors the majority of embarkation counts out of Africa from temporal historical documents (Figure 2D).
Variation in African Ancestry across the Americas
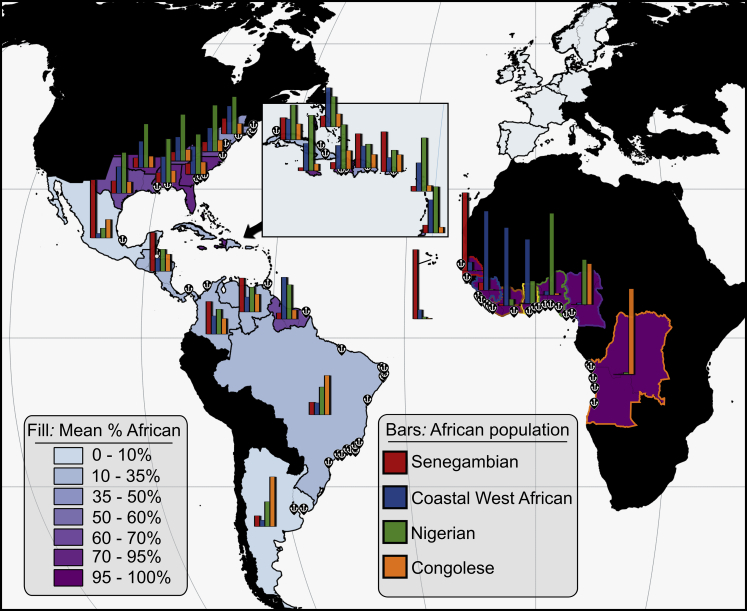
Our estimations of African ancestry with 23andMe’s ancestry inference algorithm indicated that mean sub-Saharan African ancestry in Americans with ≥ 5% African ancestry was highest in the British Caribbean (76% ± 20.3%), the United States (71.3% ± 22%), the Guianas (59.7% ± 32.6%), and Cape Verde (45.5% ± 12.6%; Figure 3). Mean sub-Saharan African ancestry was lowest in Latin America: Latin Caribbean (19.9% ± 12.9%), central South America (13.7% ± 10.5%), northern South America (11.9% ± 8.1%), and Central America plus Mexico (8.9% ± 8.7%). These proportions also correspond with the proportion of African haplogroups in each region (Tables S7 and S8).
Figure 3.
Estimated Proportions of African Ancestry across Atlantic Africa, the Americas, and Europe
Fill: Mean proportion of continent-level African Ancestry in each region. Bars: Mean local ancestry proportions of the four Atlantic African populations 23andMe identifies. Proportions are derived from local ancestry inference, which employs support vector machines (SVM) to assign one of 33 ancestries to each window along the genome and an autoregressive pair hidden Markov model to combine information across window-level SVM calls. Anchor symbols indicate major shipping port locations used during the slave trade.
Within 23andMe’s Ancestry Composition hierarchy, there are four local ancestries corresponding to regions in Atlantic Africa: Nigerian (Nigeria), Senegambian (Gambia, Guinea, Guinea-Bissau, Senegal), Coastal West African (Sierra Leone, Ghana, Côte d’Ivoire, Liberia), and Congolese (Angola, Democratic Republic of the Congo). Ancestry inference in the Americas reveals that the majority of individuals within the United States (93%) as well as from the British and French Caribbean (82%) tend to have ancestry from all four of these Atlantic African populations, with Nigerian being the most common of the four. A high proportion of individuals from the Guianas (64%) and the Latin Caribbean (44%) also have ancestry from all four populations, with Coastal West African ancestry in the Guianas being most common relative to other Atlantic African ancestries, and Senegambian being most common in the Latin Caribbean. The plurality (47%) of Mexican and Central American individuals have only Senegambian ancestry, whereas 32% of individuals from northern South America have both Senegambian and Nigerian ancestry. In central South America, the plurality (31%) of individuals have only Congolese ancestry (Figure 3; Table S7).
Biases in Contributions of Males and Females to Current Gene Pools
We identified a bias toward European male and African female genetic contributions across regions of the Americas using the sex-specific model for admixture,22 which assumes admixture events during the transatlantic slave trade. We observed an African female bias (African sf/sm > 1) in every region of the Americas with highest levels in northern South America (17.17; Table 1; Tables S9 and S10). We also observed a European male sex bias (European sm/sf > 1) in all explored regions except the Latin Caribbean and Central America. Additionally, haplogroup assignments in males indicated that Y chromosome haplogroups tend to be of European origin whereas mitochondrial haplogroups tend to be of African origin across all regions of the Americas (Table S8). Historical documents indicate the overall proportion of enslaved people on voyages were male (Mp > 0.60) across all regions5 (Table 1).
Table 1.
Inferred Ancestry Sex Bias versus Documented Sex Bias
| America Region |
African |
European |
Native American |
Mp | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| sf/sm | X | A | sm/sf | X | A | sf/sm | X | A | ||
| Guianas | 1.73 | 0.65 | 0.59 | 4.60 | 0.12 | 0.15 | – | 0.01 | 0.01 | 0.64 |
| United States | 1.47 | 0.76 | 0.71 | 3.03 | 0.21 | 0.25 | – | 0.02 | 0.01 | 0.69 |
| British Caribbean | 1.88 | 0.82 | 0.75 | 25.53 | 0.12 | 0.18 | – | <0.01 | <0.01 | 0.62 |
| Latin Caribbean | 13.31 | 0.24 | 0.19 | 0.90 | 0.53 | 0.52 | 3.78 | 0.12 | 0.06 | 0.66 |
| C. South America | 4.62 | 0.18 | 0.12 | 1.09 | 0.63 | 0.64 | 2.26 | 0.09 | 0.04 | 0.67 |
| N. South America | 17.17 | 0.14 | 0.11 | 2.27 | 0.43 | 0.49 | 3.38 | 0.30 | 0.17 | 0.63 |
| Central America | 15.60 | 0.11 | 0.08 | 0.89 | 0.32 | 0.31 | 28.04 | 0.44 | 0.27 | 0.68 |
| Cape Verde | 22.56 | 0.59 | 0.44 | 71.00 | 0.35 | 0.50 | – | <0.01 | <0.01 | – |
Female to male (sf/sm) and male to female (sm/sf) genetic contribution bias based on ancestry inference of the X chromosome and autosomes, assuming a single admixture event during the transatlantic slave trade (15 generations ago). For example, sf/sm = 2 for African ancestry means ∼2 African women for every African man. Mean Ancestry Composition based solely on the X chromosome (X) and solely on the autosomes (A) for each ancestry category. Mp is the proportion of enslaved males on voyages to each region of the Americas based on historical documents. sf/sm is not calculated for regions with less than 2% mean ancestry in either the X chromosome or autosomes. Assuming more recent admixture generations yielded near-identical results (Table S9).
Discussion
Overall Concordance with Historical Documents
We found that, overall, genetic evidence of Atlantic African ancestry across the Americas is consistent with historical documents of the transatlantic shipping of enslaved Africans.3,5,9 The expectation that more embarkation out of Africa corresponds with more ancestral connections to the Americas is borne out by the comparison between msIBDAfrica-Americas and the number of captives deported from each African region (Figure 2A). For example, the highest proportion of msIBD is found between West Central Africa and the Americas, consistent with the highest proportion of enslaved embarkation from this slave-trading region (5.7 million people5). West Central Africa also experienced the largest proportion of deportations in the mid-1800s5 (Figure 2D), consistent with our finding that people of African descent in the Americas share the most recent common ancestors with people from West Central Africa (Figure 2C). The mean number of IBD connections (mcIBD) between regions of the Americas and Africa also support embarkation estimates from voyage records. The majority of enslaved people arriving in Latin America (Latin Caribbean, Central America, N. South America, and C. South America) were generally deported from one or two slave trading regions, whereas enslaved people arriving in the United States and British Caribbean were taken from all regions of Atlantic Africa (Figure 2B).5,9,10 Consequently, individuals from Latin America tend to have mcIBD with two or three Atlantic African regions, whereas individuals from the United States and the Caribbean tend to have genetic connections to five or six regions (Figure 2B).
Potential Inconsistencies with Historical Documents
While genetic connections between African populations and individuals from the Americas are strongly correlated with historical documents (Figure 2A), a granular breakdown of disembarkation in discrete regions of the Americas identifies discordance. For example, msIBD with the Bights of Benin and Biafra, as well as Nigerian ancestry, is overrepresented in the United States and Latin America considering the recorded number of enslaved people that embarked from ports along the coast of present-day Nigeria into these regions of the Americas (Figures 2B and 3). However, this discordance is explained by records from the intra-American slave trade, an intercolonial trade that occurred primarily between 1619 and 1807 whose impact has recently been understood through collections of port records, newspapers, and merchant accounts from disparate parts of the Americas.5,46 Documented intra-American voyages indicate that the vast majority of enslaved people were transported from the British Caribbean to other parts of the Americas, presumably to maintain the slave economy as transatlantic slave trading was increasingly prohibited (Figure S13).46 These voyages would spread African ancestry common in the British Caribbean to other regions of the Americas that were not in direct trade with specific regions of Africa. In Latin America, transatlantic slave trading voyages virtually ceased between 1660 and 1780, yet a large proportion of all documented captives continued to be transported throughout the early 1800s to Latin America primarily from the British Caribbean.10 In the United States, nearly 10% of the total enslaved population arrived via intra-America trading with the British Caribbean and consequently msIBD between the United States and the British Caribbean (primarily the Bahamas, British Windward, and Leeward Islands) is high (Figure S14). Therefore, embarkation out of Africa and msIBD between Africa and all of the Americas is correlated (Figure 2A), whereas disembarkation in specific regions of the Americas and msIBD with Africa are not consistently correlated (Figure 2B) because of the asymmetric transportation of enslaved people within the Americas.
Another apparent discordance between historical documents and genetic data is seen in msIBD between most regions of the Americas and Senegambia. Senegambia is underrepresented given the overall proportion of enslaved people who were deported from this region to the Americas (Figure 2B). This is likely because Senegambia was one of the first African regions from which large numbers of people were enslaved.5,9,57 Historical documents and TMRCA estimates from IBD segments support this explanation as well, indicating that Senegambia’s connection to the Americas is older (and less well documented) than that of other regions (Figures 2C and 2D). Having distant ancestors from Senegambia would increase the likelihood of people in the Americas sharing very short IBD segments with Senegambians, which, in turn, may be undetected by estimating IBD segments with reduced SNP densities.49 This pattern of reduced msIBD and older TMRCA estimates is most evident in the disembarkation regions of northern South America and Central America where, according to records, nearly half of the enslaved people who disembarked at ports in these regions came from Senegambia, but less than 10% of msIBD between these regions and Africa is with Senegambia (Figure 2B). However, almost no captives from Senegambia disembarked directly in either northern South America or Central America after 1650,5,9,10 consistent with our finding of an older TMRCA between Senegambia and these two regions, and the likelihood that earlier African disembarkation resulted in smaller IBD segments between Africans and Americans of the present day.
The underrepresentation of ancestral connections to Senegambia also occurs in regions of the Americas with more recent (1700–1865) disembarkation from Senegambia, such as the United States and Latin Caribbean. This pattern, in part, may result from the increasing rates of deportation of children from Senegambia over time (Figure S15). Enslavement of children and unsanitary conditions in the holds of ships led to high rates of malnourishment and illness, which in turn led to lower survival rates in later decades of the slave trade.58,59 It is also possible that Senegambians experienced higher mortality due to dangerous plantation conditions. A large proportion of enslaved Senegambians were rice cultivators who worked on rice plantations in the Americas, which were generally rampant with malaria.57,60,61 Clearing of swamplands was also generally performed by men, which could explain higher male mortality.59 While detailed records exist for very few plantations, reduced genetic representation from Senegambia in parts of the Americas is likely correlated with Senegambians being forced to work under life-threatening conditions.
Ancestry Proportions Discordance
Continental African ancestry estimates are unexpected given the number of enslaved people disembarking in the Americas. Nearly 70% of the 10.5 million enslaved people who survived the Middle Passage disembarked in Latin America, yet regions of Latin America have the lowest overall proportion of African ancestry (Figure 3). On the contrary, individuals in this study from the United States and British Caribbean have the highest African ancestry proportion on average. Together, these results indicate that practices and treatment of enslaved people likely varied across the Americas. For instance, literature suggests higher overall mortality and smaller effective population sizes in enslaved populations brought to Latin America,62,63 which would have reduced the contribution of African ancestry to the gene pool. In addition, enslaved indigenous people often admixed with enslaved Africans in Latin America,59 which is evident in the Native American ancestry and haplogroups common in individuals from Latin America (Tables S7 and S8). Other investigations of genetic ancestry have identified comparable estimates of African ancestry across the United States;11, 12, 13, 14, 15 however, other studies have found that African ancestry is higher in admixed populations in Latin America compared to our study.13,64 This is likely due to the non-random representation of global populations in our study cohort, which consists primarily of US customers and does not target specific admixed populations in Latin America. Although our estimates are lower (about 5%–10% on average), the trend of lower African ancestry in Latin America is still supported by other research.64
Sex-Bias Discordance
Despite more than 60% of enslaved people brought to each region of the Americas being men,5 comparisons of ancestry estimates for the X chromosome and autosomes, as well as the comparison of mitochondrial (maternal) and Y (paternal) haplogroups, revealed a bias toward African female contributions to gene pools across all of the Americas.64,65 However, this African female sex bias is more extreme in Latin America (between 4 and 17 African women for every African man contributing to the gene pool) than in British-colonized Americas (between 1.5 and 2 African women for every African man contributing to the gene pool; Table 1). An Americas-wide African female sex-bias can be attributed to known accounts of rape of enslaved African women by slave owners and other sexual exploitation.19,26,62,66 Regional differences may be due to higher mortality in enslaved males in Latin America as well as a common practice called branqueamento, or racial whitening, which involved women marrying lighter-skinned men with the intention of producing lighter-skinned children.1,25,27,62,67 National branqueamento policies were implemented in multiple Latin American countries, funding and subsidizing European immigrant travels with the intention to dilute African Ancestry through reproduction with light-skinned Europeans.68 Conversely, the smaller African female sex-bias seen in former British colonies could be due to the practice of coercing enslaved people to have children as a means of maintaining enslaved workforces nearing the abolition of the transatlantic slave trade.69 In some areas, such as the United States, enslaved women were incentivized to reproduce with the promise of freedom following the birth of many children.20 Furthermore, racist ideologies in the United States led to the segregation of people of African descent as opposed to promotion of European admixture.70 Overall, the inhumane practices associated with institutionalized slavery, though differing across the Americas, all resulted in an African-female sex bias despite the preponderance of males among those enslaved.
Discordance in Timing
Our Bayesian method of dating IBD segments estimates the number of generations to a shared common ancestor between individuals in the Americas and Africa and is generally consistent with counts of enslaved disembarkation over time, with some exceptions (Figures 2C and 2D). One clear discordance is early genetic connections between the Americas and the Windward Coast relative to historical documents, which indicate a more recent connection. This discordance may be due to ethnolinguistic representation of the Windward Coast in this study. Participants representing the Windward Coast indicated primary ethnolinguistic associations with the Grebo, Kpelle, and Mano peoples, none of whom were known to be significantly impacted by the transatlantic slave trade.4 Therefore, the detected IBD connections with this region may be with more distant common ancestors from the Windward Coast. Other African regions are represented in this study by ethnolinguistic groups more directly impacted by the slave trade (approximately half of the significantly impacted tribes;4 Table S1). In general, our African cohort appears to be a suitable proxy of historical populations impacted by the slave trade given population structure (Figures S1–S3). IBD shared within regions of Africa also highlights general stationarity of African populations (Figure S17) yet signifies some impact of intercontinental migration. For instance, some mixing is apparent between the Bight of Biafra, the Bight of Benin, and West Central Africa. High shared IBD between certain regions may be due to ancient movement, such as the Bantu Migration,71 or the more recent transportation of enslaved people within Africa (8 million people were enslaved and retained in Africa).72
Other discordances highlight differences in mortality following the transatlantic voyage among enslaved people who originated from different regions of Africa. For instance, the earlier-than-expected TMRCA for the Bight of Biafra may be due to the high suicide rates documented in enslaved Igbo people.73,74 Most documented suicides, such as the Igbo Landing mass suicide of 1803 in the United States, were recorded later in the slave trade. If suicide by means of revolt became more common in enslaved people from the Bight of Biafra as the slave trade continued, that TMRCA estimate would appear older than expected given recorded shipping of enslaved people from the Bight of Biafra.
Regional Discordance
Our results indicate that African ancestry and msIBD do not vary much within broad regions of the Americas despite enslaved people disembarking at different rates in different ports. This observation is expected, given that slavers transported enslaved people regionally, and extensive local migration followed the transatlantic slave trade.75 For instance, the domestic slave trade in the United States between 1790 and 1860 would see the transportation of one million enslaved people across the southeastern United States.20 Similarly, “The Trail of Tears” would force Native Americans and the enslaved people they owned west toward Oklahoma.76 Also, the “Great Migrations” that occurred between 1916 and 1970 to escape segregation and poor economic conditions would bring former slaves and their descendants to all parts of the Northeast and Midwest United States18 (Figure S16). Similar movements occurred elsewhere such as migrations from northeastern Brazil (Bahia and Pernambuco) to southern and central Brazil (Rio de Janeiro and Minas Gerais-Goiás).1,2 These large migrations could easily obscure pre-existing local population structure in African-descendant communities as a result of admixture. Because the majority of individuals in this study have connections to multiple populations in Atlantic Africa, it is unlikely that members of distinct African communities remained isolated from one another after they reached the Americas. However, based on limited marriage records of enslaved people,21,77,78 it is likely that communities from distinct Atlantic African regions initially remained isolated, but interbred later, which would lead to genetic evidence of post-transport mixing of African populations over generations.
Despite extensive genetic evidence of internal or local migrations, we found no evidence for long-distance migrations between broad regions in the Americas after disembarkation except between the United States and the British Caribbean (Figures S5, S6, and S15). Representative individuals in the United States and British Caribbean share little IBD with those in Latin America, and very little IBD sharing is found between regions within Latin America. This is striking when comparing the British Caribbean to the Latin Caribbean: although some countries are geographically proximate (e.g., Haiti and the Dominican Republic), ancestry estimates and IBD connections within the Americas and to Atlantic Africa differ substantially.
Discordance between IBD and Ancestry Composition
Local ancestry estimates across the Americas (except for central South America) indicate a lower mean proportion of Congolese (West Central African) ancestry than expected (Figure 3), considering West Central Africa shares more msIBD with the Americas than does any other region in Atlantic Africa (Figure 2A). This pattern likely reflects the fact that 23andMe’s local ancestry analysis differentiates only four regions along the Atlantic African coast and can detect genetic connections at a deeper timescale than IBD analysis. Specifically, local ancestry inference identifies the reference population with haplotypes most similar to those of a test sample for small genomic regions, rather than identifying identical-by-descent segments, so the ancestry it detects is likely more distant in time than that of IBD-based methods.47
Conclusions
We present a broad-scale investigation into the transatlantic slave trade, advancing the understanding of its consequences by comparing genetic connections with historical records. While the current genetic landscape of African ancestry in the Americas largely agrees with transatlantic voyage records, additional historical accounts shed light on discordances. For example, overrepresentation of Nigerian ancestry in parts of the Americas is explained by the intra-American trade of enslaved people from the British Caribbean, and underrepresentation of Senegambian ancestry across the Americas is supported by accounts of early trading and high mortality from this region. Patterns of variation across the Americas, such as lower African ancestry and a higher African female sex bias can be attributed to socioeconomic factors, non-African male admixture, and inhumane treatment of enslaved people. While transatlantic records have told us a large part of the story, insights from this study in combination with other historical accounts shed light on details of the genetic impact of the transatlantic slave trade on present-day populations in the Americas.
Data and Code Availability
There are restrictions to the availability of Angolan and 23andMe genotype data due to 23andMe consent and privacy guidelines. These data will not be made available.
Consortia
The 23andMe Research Team: M. Agee, S. Aslibekyan, A. Auton, R. Bell, S. Clark, S. Das, S. Elson, K. Fletez-Brant, P. Fontanillas, P. Gandhi, K. Heilbron, B. Hicks, D. Hinds, K. Huber, E. Jewett, Y. Jiang, A. Kleinman, K. Lin, N. Litterman, J. McCreight, M. McIntyre, K. McManus, S. Mozaffari, P. Nandakumar, L. Noblin, C. Northover, J. O’Connell, A. Petrakovitz, S. Pitts, J. Shelton, S. Shringarpure, C. Tian, J. Tung, R. Tunney, V. Vacic, X. Wang, A. Zare.
Acknowledgments
We thank the customers and others who consented to participate in research for enabling this study. We acknowledge other employees of 23andMe who contributed to the development of the infrastructure that made this research possible, and the suggestions of Scott Hadly, Altovise Ewing, Vence Bonham, Alvin Tillery, John Thornton, Linda Heywood, Beatriz Marcheco-Teruel, Richard Roberts, David Robinson, David Eltis, the slavevoyages.org Scholars Group, and two anonymous reviewers. We also thank those who made additional samples available and provided guidance and discussion around those samples: Nathan Nunn (DRC), Bert Ely (Sierra Leone), and Brenna Henn (Khoe-San speakers). Finally, we would like to acknowledge the many people who were taken from Africa but never made it to the Americas, whose DNA is not directly part of this paper. Funding for this research was provided by 23andMe.
Declaration of Interests
S.J.M., K.B., S.G.A.E., W.A.F., M.E.M., G.D.P., A.J.S., J.L.M., and members of the 23andMe Research Team are employees of and have stock, stock options, or both, in 23andMe. The remaining authors declare no competing interests.
Published: July 23, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.06.012.
Contributor Information
Steven J. Micheletti, Email: smicheletti@ucdavis.edu.
23andMe Research Team:
M. Agee, S. Aslibekyan, A. Auton, R. Bell, S. Clark, S. Das, S. Elson, K. Fletez-Brant, P. Fontanillas, P. Gandhi, K. Heilbron, B. Hicks, D. Hinds, K. Huber, E. Jewett, Y. Jiang, A. Kleinman, K. Lin, N. Litterman, J. McCreight, M. McIntyre, K. McManus, S. Mozaffari, P. Nandakumar, L. Noblin, C. Northover, J. O’Connell, A. Petrakovitz, S. Pitts, J. Shelton, S. Shringarpure, C. Tian, J. Tung, R. Tunney, V. Vacic, X. Wang, and A. Zare
Web Resources
23andMe’s Ancestry Composition, https://www.23andme.com/ancestry-composition-guide/
1000 Genomes Project, https://www.internationalgenome.org
Ethical and Independent Review Services, http://www.eandireview.com
Human Genome Diversity Project, https://www.hagsc.org/hgdp/
Intra-American Slave Trade Database, https://slavevoyages.org/american/database/
Transatlantic Slave Trade Database, https://slavevoyages.org/voyage/database/
Supplemental Data
Comparisons (A) within Atlantic Africa, (B) within the Americas, (C) within Europe, and (D) between Atlantic African and the Americas. msIBD is the total amount of IBD (cM) shared between individuals in two regions, divided by the total possible number of pairwise comparisons.
(A) African leaf populations (out of 4) that individuals are assigned (> 0.5%); this includes the median number, mean, and proportion of individuals that are assigned 1-4 of the African populations. (B) Mean Ancestry Composition for populations of interest. Populations are organized into continental (broad) and population (granular) estimates for ancestries of interest. (C) Standard deviations for mean estimates in (B).
Research participants were subset into Males to compare the origin continent of their Y and mitochondrial haplogroups. (A) Proportion of male participants representing regions in the Americas with the Y and mitochondrial haplogroup assignment above. (B) The mean continental Ancestry Composition of individuals with the combination of Y and mitochondrial haplogroups above.
Estimates represent all (A) regions of the Americas and (B) broad regions of the Americas using ancestry inference on the X chromosome versus the autosomes. Estimates were withheld when either autosomal or X ancestry estimates are below 5% on average. sf is the estimated contribution of females to the gene pool, sm is the estimated contribution of males, ga is the mean ancestry estimate for autosomes, and gx is the estimated mean ancestry estimate for the X chromosome (either African, Native American, or European).
pf refers to the proportion of females and pm refers to the proportion of males representing each region. ∗ Ancestry categories within Atlantic Africa.
References
- 1.Martins R.B. Growing in silence: the slave economy of nineteenth-century Minas Gerais, Brazil. J. Econ. Hist. 1982;42:222–223. [Google Scholar]
- 2.Bethell L. Cambridge University Press; 2009. The Abolition of the Brazilian Slave Trade: Britain, Brazil and the Slave Trade Question. [Google Scholar]
- 3.Eltis D. The volume and structure of the transatlantic slave trade: a reassessment. William Mary Q. 2001;58:17–46. [PubMed] [Google Scholar]
- 4.Heywood L.M., Thornton J.K. Cambridge University Press; 2007. Central Africans, Atlantic creoles, and the foundation of the Americas, 1585-1660. [Google Scholar]
- 5.Eltis D. 2007. A brief overview of the Trans-Atlantic Slave Trade. Voyages: The Trans-Atlantic Slave Trade Database 1700–1810.http://www.slavevoyages.org/assessment/essay [Google Scholar]
- 6.Rotimi C.N., Tekola-Ayele F., Baker J.L., Shriner D. The African diaspora: history, adaptation and health. Curr. Opin. Genet. Dev. 2016;41:77–84. doi: 10.1016/j.gde.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Acemoglu D., Johnson S., Robinson J. The rise of Europe: Atlantic trade, institutional change, and economic growth. Am. Econ. Rev. 2005;95:546–579. [Google Scholar]
- 8.Keller A.G. Ginn; 1908. Colonization: a study of the founding of new societies. [Google Scholar]
- 9.Eltis D., Richardson D. Yale University Press; 2010. Atlas of the Transatlantic Slave Trade. [Google Scholar]
- 10.Borucki A., Eltis D., Wheat D. Atlantic history and the slave trade to Spanish America. Am. Hist. Rev. 2015;120:433–461. [Google Scholar]
- 11.Gouveia M.H., Borda V., Leal T.P., Moreira R.G., Bergen A.W., Kehdy F.S.G., Alvim I., Aquino M.M., Araujo G.S., Araujo N.M. Origins, admixture dynamics and homogenization of the African gene pool in the Americas. Mol. Biol. Evol. 2020;37:1647–1656. doi: 10.1093/molbev/msaa033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santos H.C., Horimoto A.V., Tarazona-Santos E., Rodrigues-Soares F., Barreto M.L., Horta B.L., Lima-Costa M.F., Gouveia M.H., Machado M., Silva T.M., Brazilian EPIGEN Project Consortium A minimum set of ancestry informative markers for determining admixture proportions in a mixed American population: the Brazilian set. Eur. J. Hum. Genet. 2016;24:725–731. doi: 10.1038/ejhg.2015.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fortes-Lima C., Bybjerg-Grauholm J., Marin-Padrón L.C., Gomez-Cabezas E.J., Bækvad-Hansen M., Hansen C.S., Le P., Hougaard D.M., Verdu P., Mors O. Exploring Cuba’s population structure and demographic history using genome-wide data. Sci. Rep. 2018;8:11422. doi: 10.1038/s41598-018-29851-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang S., Ray N., Rojas W., Parra M.V., Bedoya G., Gallo C., Poletti G., Mazzotti G., Hill K., Hurtado A.M. Geographic patterns of genome admixture in Latin American Mestizos. PLoS Genet. 2008;4:e1000037. doi: 10.1371/journal.pgen.1000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ongaro L., Scliar M.O., Flores R., Raveane A., Marnetto D., Sarno S., Gnecchi-Ruscone G.A., Alarcón-Riquelme M.E., Patin E., Wangkumhang P. The Genomic Impact of European Colonization of the Americas. Curr. Biol. 2019;29:3974–3986.e4. doi: 10.1016/j.cub.2019.09.076. [DOI] [PubMed] [Google Scholar]
- 16.Hall G.M. Univ of North Carolina Press; 2005. Slavery and African ethnicities in the Americas: restoring the links. [Google Scholar]
- 17.Head D. Slave Smuggling by Foreign Privateers: The Illegal Slave Trade and the Geopolitics of the Early Republic. J. Early Repub. 2013;33:433–462. [Google Scholar]
- 18.Baharian S., Barakatt M., Gignoux C.R., Shringarpure S., Errington J., Blot W.J., Bustamante C.D., Kenny E.E., Williams S.M., Aldrich M.C., Gravel S. The great migration and African-American genomic diversity. PLoS Genet. 2016;12:e1006059. doi: 10.1371/journal.pgen.1006059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baptist E.E. “Cuffy,”“fancy maids,” and “one-eyed men”: rape, commodification, and the domestic slave trade in the United States. Am. Hist. Rev. 2001;106:1619–1650. [Google Scholar]
- 20.Deyle S. Oxford University Press on Demand; 2005. Carry me back: the domestic slave trade in American life. [Google Scholar]
- 21.Edwards L.F. “The Marriage Covenant is at the Foundation of all Our Rights”: The Politics of Slave Marriages in North Carolina after Emancipation. Law Hist. Rev. 1996;14:81–124. [Google Scholar]
- 22.Goldberg A., Rosenberg N.A. Beyond 2/3 and 1/3: the complex signatures of sex-biased admixture on the X chromosome. Genetics. 2015;201:263–279. doi: 10.1534/genetics.115.178509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lind J.M., Hutcheson-Dilks H.B., Williams S.M., Moore J.H., Essex M., Ruiz-Pesini E., Wallace D.C., Tishkoff S.A., O’Brien S.J., Smith M.W. Elevated male European and female African contributions to the genomes of African American individuals. Hum. Genet. 2007;120:713–722. doi: 10.1007/s00439-006-0261-7. [DOI] [PubMed] [Google Scholar]
- 24.Bryc K., Durand E.Y., Macpherson J.M., Reich D., Mountain J.L. The genetic ancestry of African Americans, Latinos, and European Americans across the United States. Am. J. Hum. Genet. 2015;96:37–53. doi: 10.1016/j.ajhg.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meade T.A. John Wiley & Sons; 2016. History of modern Latin America: 1800 to the present. [Google Scholar]
- 26.Malarek V. Skyhorse Publishing Inc.; 2011. The natashas: The horrific inside story of slavery, rape, and murder in the global sex trade. [Google Scholar]
- 27.Montalvo F.F., Codina G.E. Skin color and Latinos in the United States. Ethnicities. 2001;1:321–341. [Google Scholar]
- 28.Skidmore T.E. Duke University Press; 1993. Black into white: race and nationality in Brazilian thought. [Google Scholar]
- 29.Bryc K., Auton A., Nelson M.R., Oksenberg J.R., Hauser S.L., Williams S., Froment A., Bodo J.-M., Wambebe C., Tishkoff S.A., Bustamante C.D. Genome-wide patterns of population structure and admixture in West Africans and African Americans. Proc. Natl. Acad. Sci. USA. 2010;107:786–791. doi: 10.1073/pnas.0909559107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DeFrancesco L. 23andMe protects consumers’ data. Nat. Biotechnol. 2015;33:1220. doi: 10.1038/nbt1215-1220. [DOI] [PubMed] [Google Scholar]
- 31.Eriksson N., Macpherson J.M., Tung J.Y., Hon L.S., Naughton B., Saxonov S., Avey L., Wojcicki A., Pe’er I., Mountain J. Web-based, participant-driven studies yield novel genetic associations for common traits. PLoS Genet. 2010;6:e1000993. doi: 10.1371/journal.pgen.1000993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cann H.M., de Toma C., Cazes L., Legrand M.-F., Morel V., Piouffre L., Bodmer J., Bodmer W.F., Bonne-Tamir B., Cambon-Thomsen A. A human genome diversity cell line panel. Science. 2002;296:261–262. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- 34.van Dorp L., Lowes S., Weigel J.L., Ansari-Pour N., López S., Mendoza-Revilla J., Robinson J.A., Henrich J., Thomas M.G., Nunn N., Hellenthal G. Genetic legacy of state centralization in the Kuba Kingdom of the Democratic Republic of the Congo. Proc. Natl. Acad. Sci. USA. 2019;116:593–598. doi: 10.1073/pnas.1811211115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jackson B.A., Wilson J.L., Kirbah S., Sidney S.S., Rosenberger J., Bassie L., Alie J.A., McLean D.C., Garvey W.T., Ely B. Mitochondrial DNA genetic diversity among four ethnic groups in Sierra Leone. Am. J. Phys. Anthropol. 2005;128:156–163. doi: 10.1002/ajpa.20040. [DOI] [PubMed] [Google Scholar]
- 36.Henn B.M., Gignoux C.R., Jobin M., Granka J.M., Macpherson J.M., Kidd J.M., Rodríguez-Botigué L., Ramachandran S., Hon L., Brisbin A. Hunter-gatherer genomic diversity suggests a southern African origin for modern humans. Proc. Natl. Acad. Sci. USA. 2011;108:5154–5162. doi: 10.1073/pnas.1017511108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knight A., Underhill P.A., Mortensen H.M., Zhivotovsky L.A., Lin A.A., Henn B.M., Louis D., Ruhlen M., Mountain J.L. African Y chromosome and mtDNA divergence provides insight into the history of click languages. Curr. Biol. 2003;13:464–473. doi: 10.1016/s0960-9822(03)00130-1. [DOI] [PubMed] [Google Scholar]
- 38.Beleza S., Campos J., Lopes J., Araújo I.I., Hoppfer Almada A., Correia e Silva A., Parra E.J., Rocha J. The admixture structure and genetic variation of the archipelago of Cape Verde and its implications for admixture mapping studies. PLoS ONE. 2012;7:e51103. doi: 10.1371/journal.pone.0051103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Busby G.B., Band G., Si Le Q., Jallow M., Bougama E., Mangano V.D., Amenga-Etego L.N., Enimil A., Apinjoh T., Ndila C.M., Malaria Genomic Epidemiology Network Admixture into and within sub-Saharan Africa. eLife. 2016;5:e15266. doi: 10.7554/eLife.15266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murdock G.P. McGraw Hill Text; 1959. Africa its peoples and their culture history. [Google Scholar]
- 41.Durand E.Y., Do C.B., Mountain J.L., Macpherson J.M. Ancestry Composition: A Novel, Efficient Pipeline for Ancestry Deconvolution. bioRxiv. 2014 doi: 10.1101/010512. [DOI] [Google Scholar]
- 42.Browning B.L., Browning S.R. Improving the accuracy and efficiency of identity-by-descent detection in population data. Genetics. 2013;194:459–471. doi: 10.1534/genetics.113.150029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McInnes L., Healy J., Melville J. 2018. Umap: Uniform manifold approximation and projection for dimension reduction. ArXiv Preprint ArXiv:1802.03426. [Google Scholar]
- 44.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alexander D.H., Novembre J., Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O’Malley G.E. UNC Press Books; 2014. Final passages: the intercolonial slave trade of British America, 1619-1807. [Google Scholar]
- 47.Do C., Durand E.Y.J.-M., Macpherson J.M. Google Patents; 2016. Scalable pipeline for local ancestry inference. [Google Scholar]
- 48.Loh P.-R., Palamara P.F., Price A.L. Fast and accurate long-range phasing in a UK Biobank cohort. Nat. Genet. 2016;48:811–816. doi: 10.1038/ng.3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Powell J.E., Visscher P.M., Goddard M.E. Reconciling the analysis of IBD and IBS in complex trait studies. Nat. Rev. Genet. 2010;11:800–805. doi: 10.1038/nrg2865. [DOI] [PubMed] [Google Scholar]
- 50.Palamara P.F., Lencz T., Darvasi A., Pe’er I. Length distributions of identity by descent reveal fine-scale demographic history. Am. J. Hum. Genet. 2012;91:809–822. doi: 10.1016/j.ajhg.2012.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goldberg A., Verdu P., Rosenberg N.A. Autosomal admixture levels are informative about sex bias in admixed populations. Genetics. 2014;198:1209–1229. doi: 10.1534/genetics.114.166793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haber M., Jones A.L., Connell B.A., Asan, Arciero E., Yang H., Thomas M.G., Xue Y., Tyler-Smith C. A Rare Deep-Rooting D0 African Y-Chromosomal Haplogroup and Its Implications for the Expansion of Modern Humans Out of Africa. Genetics. 2019;212:1421–1428. doi: 10.1534/genetics.119.302368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weissensteiner H., Pacher D., Kloss-Brandstätter A., Forer L., Specht G., Bandelt H.-J., Kronenberg F., Salas A., Schönherr S. HaploGrep 2: mitochondrial haplogroup classification in the era of high-throughput sequencing. Nucleic Acids Res. 2016;44(W1):W58-63. doi: 10.1093/nar/gkw233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Poznik G.D. Identifying Y-chromosome haplogroups in arbitrarily large samples of sequenced or genotyped men. bioRxiv. 2016 doi: 10.1101/088716. [DOI] [Google Scholar]
- 55.Consortium Y.C., Y Chromosome Consortium A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res. 2002;12:339–348. doi: 10.1101/gr.217602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kivisild T. Maternal ancestry and population history from whole mitochondrial genomes. Investig. Genet. 2015;6:3. doi: 10.1186/s13323-015-0022-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baum R.M. Oxford University Press; 1999. Shrines of the slave trade: Diola religion and society in precolonial Senegambia. [Google Scholar]
- 58.King W. Indiana University Press; 2011. Stolen childhood: Slave youth in nineteenth-century America. [Google Scholar]
- 59.Schneider D., Schneider C.J. Infobase Publishing; 2014. Slavery in America. [Google Scholar]
- 60.National Research Council . National Academies Press; 1996. Lost crops of Africa: volume I: grains. [Google Scholar]
- 61.Carney J. The African origins of Carolina rice culture. Ecumene. 2000;7:125–149. [Google Scholar]
- 62.Bergad L. Cambridge University Press; 2007. The comparative histories of slavery in Brazil, Cuba, and the United States. [Google Scholar]
- 63.Browning S.R., Browning B.L., Daviglus M.L., Durazo-Arvizu R.A., Schneiderman N., Kaplan R.C., Laurie C.C. Ancestry-specific recent effective population size in the Americas. PLoS Genet. 2018;14:e1007385. doi: 10.1371/journal.pgen.1007385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marcheco-Teruel B., Parra E.J., Fuentes-Smith E., Salas A., Buttenschøn H.N., Demontis D., Torres-Español M., Marín-Padrón L.C., Gómez-Cabezas E.J., Álvarez-Iglesias V. Cuba: exploring the history of admixture and the genetic basis of pigmentation using autosomal and uniparental markers. PLoS Genet. 2014;10:e1004488. doi: 10.1371/journal.pgen.1004488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Conomos M.P., Laurie C.A., Stilp A.M., Gogarten S.M., McHugh C.P., Nelson S.C., Sofer T., Fernández-Rhodes L., Justice A.E., Graff M. Genetic Diversity and Association Studies in US Hispanic/Latino Populations: Applications in the Hispanic Community Health Study/Study of Latinos. Am. J. Hum. Genet. 2016;98:165–184. doi: 10.1016/j.ajhg.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cash F.G. Leeds Anti-Slavery Society; 1853. Slavery a System of Inherent Cruelty. [Google Scholar]
- 67.Klimentidis Y.C., Miller G.F., Shriver M.D. Genetic admixture, self-reported ethnicity, self-estimated admixture, and skin pigmentation among Hispanics and Native Americans. Am. J. Phys. Anthropol. 2009;138:375–383. doi: 10.1002/ajpa.20945. [DOI] [PubMed] [Google Scholar]
- 68.Agier M. Racism, culture and black identity in Brazil. Bull. Lat. Am. Res. 1995;14:245–264. [Google Scholar]
- 69.Marable M. Haymarket Books; 2015. How capitalism underdeveloped Black America: Problems in race, political economy, and society. [Google Scholar]
- 70.Laveist T.A. Segregation, poverty, and empowerment: health consequences for African Americans. Milbank Q. 1993;71:41–64. [PubMed] [Google Scholar]
- 71.Vansina J. New linguistic evidence and ‘the Bantu expansion.’. J. Afr. Hist. 1995;36:173–195. [Google Scholar]
- 72.Manning P. The enslavement of Africans: a demographic model. Canadian Journal of African Studies/La Revue Canadienne Des Études Africaines. 1981;15:499–526. [Google Scholar]
- 73.Rucker W.C. LSU Press; 2008. The river flows on: Black resistance, culture, and identity formation in early America. [Google Scholar]
- 74.Snyder T.L. Suicide, slavery, and memory in North America. J. Am. Hist. 2010;97:39–62. [Google Scholar]
- 75.Gates H.L. Alfred a Knopf Incorporated; 2011. Life upon these shores: Looking at African American history, 1513-2008. [Google Scholar]
- 76.Henderson G., Olasiji T.D. University Press of America; 1995. Migrants, immigrants, and slaves: Racial and ethnic groups in America. [Google Scholar]
- 77.Graham R. Slave families on a rural estate in colonial Brazil. J. Soc. Hist. 1976;9:382–402. [Google Scholar]
- 78.Holt K. Marriage choices in a plantation society: Bahia, Brazil. Int. Rev. Soc. Hist. 2005;50:25–41. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparisons (A) within Atlantic Africa, (B) within the Americas, (C) within Europe, and (D) between Atlantic African and the Americas. msIBD is the total amount of IBD (cM) shared between individuals in two regions, divided by the total possible number of pairwise comparisons.
(A) African leaf populations (out of 4) that individuals are assigned (> 0.5%); this includes the median number, mean, and proportion of individuals that are assigned 1-4 of the African populations. (B) Mean Ancestry Composition for populations of interest. Populations are organized into continental (broad) and population (granular) estimates for ancestries of interest. (C) Standard deviations for mean estimates in (B).
Research participants were subset into Males to compare the origin continent of their Y and mitochondrial haplogroups. (A) Proportion of male participants representing regions in the Americas with the Y and mitochondrial haplogroup assignment above. (B) The mean continental Ancestry Composition of individuals with the combination of Y and mitochondrial haplogroups above.
Estimates represent all (A) regions of the Americas and (B) broad regions of the Americas using ancestry inference on the X chromosome versus the autosomes. Estimates were withheld when either autosomal or X ancestry estimates are below 5% on average. sf is the estimated contribution of females to the gene pool, sm is the estimated contribution of males, ga is the mean ancestry estimate for autosomes, and gx is the estimated mean ancestry estimate for the X chromosome (either African, Native American, or European).
pf refers to the proportion of females and pm refers to the proportion of males representing each region. ∗ Ancestry categories within Atlantic Africa.
Data Availability Statement
There are restrictions to the availability of Angolan and 23andMe genotype data due to 23andMe consent and privacy guidelines. These data will not be made available.