Abstract
Phenylketonuria (PKU), caused by variants in the phenylalanine hydroxylase (PAH) gene, is the most common autosomal-recessive Mendelian phenotype of amino acid metabolism. We estimated that globally 0.45 million individuals have PKU, with global prevalence 1:23,930 live births (range 1:4,500 [Italy]–1:125,000 [Japan]). Comparing genotypes and metabolic phenotypes from 16,092 affected subjects revealed differences in disease severity in 51 countries from 17 world regions, with the global phenotype distribution of 62% classic PKU, 22% mild PKU, and 16% mild hyperphenylalaninemia. A gradient in genotype and phenotype distribution exists across Europe, from classic PKU in the east to mild PKU in the southwest and mild hyperphenylalaninemia in the south. The c.1241A>G (p.Tyr414Cys)-associated genotype can be traced from Northern to Western Europe, from Sweden via Norway, to Denmark, to the Netherlands. The frequency of classic PKU increases from Europe (56%) via Middle East (71%) to Australia (80%). Of 758 PAH variants, c.1222C>T (p.Arg408Trp) (22.2%), c.1066−11G>A (IVS10−11G>A) (6.4%), and c.782G>A (p.Arg261Gln) (5.5%) were most common and responsible for two prevalent genotypes: p.[Arg408Trp];[Arg408Trp] (11.4%) and c.[1066−11G>A];[1066−11G>A] (2.6%). Most genotypes (73%) were compound heterozygous, 27% were homozygous, and 55% of 3,659 different genotypes occurred in only a single individual. PAH variants were scored using an allelic phenotype value and correlated with pre-treatment blood phenylalanine concentrations (n = 6,115) and tetrahydrobiopterin loading test results (n = 4,381), enabling prediction of both a genotype-based phenotype (88%) and tetrahydrobiopterin responsiveness (83%). This study shows that large genotype databases enable accurate phenotype prediction, allowing appropriate targeting of therapies to optimize clinical outcome.
Keywords: PKU, hyperphenylalaninemia, PAH deficiency, tetrahydrobiopterin, BH4, phenylalanine
Introduction
Phenylketonuria (PKU [MIM: 261600]) is the most frequent inborn error of the amino acid metabolism. More than 1,180 bi-allelic variants in the phenylalanine hydroxylase (PAH) gene located on chromosome 12q22–24.1 have been identified.1 These autosomal-recessive inherited variants lead to deficiency in the PAH enzyme, which hydroxylates phenylalanine to tyrosine, with the help of a cofactor (tetrahydrobiopterin; BH4), molecular oxygen, and non-heme iron.2
The metabolic picture is highly heterogenous as it depends on the degree of residual PAH activity and blood phenylalanine (Phe) concentrations. Lower residual enzyme activity usually results in higher blood Phe concentrations and a more severe clinical phenotype if left untreated.3
The severity of PKU is defined by daily Phe tolerance.4 Standard classification is according to the pre-treatment blood Phe concentration and daily dietary Phe tolerance, ranging from the severe classical PKU (cPKU) with pre-treatment blood Phe concentrations of >1,200 μmol/L to mild PKU (mPKU) with pre-treatment blood Phe concentrations of 600–1,200 μmol/L and mild hyperphenylalaninemia (MHP) with pre-treatment Phe blood concentrations of 120–600 μmol/L.1,5
Untreated PKU generally results in global developmental delay or severe irreversible intellectual disability, as well as growth failure, hypopigmentation, motor deficits, ataxia, and seizures.1 The population of PKU-affected individuals is heterogeneous in terms of treatment history and diet compliance.5,6 Early diagnosis and treatment with a low-Phe diet has enabled an almost normal life for the majority of PKU subject.7 Pharmacological treatment with BH4 (sapropterin) and enzyme substitution therapy with Phe ammonia lyase (PAL) provide alternative treatment options for some PKU subjects.8
PKU is one of the most frequent inherited disorders in Europeans, with an incidence of roughly 1:10,000 live births in the USA,9 although the prevalence of PKU varies significantly among ethnicities and geographic regions worldwide. In Europe, the incidence of PKU ranges from 1:850 in the Karachay-Cherkess Republic (Russia)10 to only 1:112,000 live births in Finland.11 PKU occurs less often in Japan, with an incidences of 1:125,000.12
A large number of PAH variants give rise to a wide scale of residual PAH enzyme activities that correspond to different PKU phenotypes.13,14 Associations between genotypes and in vitro residual PAH activity have been documented for many PAH variants.3 Therefore, the molecular genetics of PKU and genotype-based phenotype prediction may be clinically useful, particularly where treatment recommendations are unclear (e.g., due to borderline blood phenylalanine concentrations) or for genetic counseling of patients’ families.
The major goal of this study was the analysis of a large database of PKU phenotype and genotype to elucidate the current distribution of PKU worldwide and to create an overview of the severity of PAH variants, genotypes, and the resulting phenotypes, in various geographic regions and respective countries. Furthermore, this work improves the accuracy of genotypic phenotype prediction by the use of the allelic phenotype value (APV) value and the genotypic BH4 responsiveness prediction.
Subjects and Methods
Literature Search
An electronic search using the databases MEDLINE (via Pub Med), the Cochrane library, and Web of Science was carried out to compare articles that were published between January 1980 and October 2019, covering the epidemiology and genetics of PKU in different world regions. Key words included Phenylketonuria[MeSH Terms] OR “phenylalanine hydroxylase/deficiency”[MeSH Terms] OR hyperphenylalaninaemia[Title/Abstract] OR hyperphenylalaninemia[Title/Abstract] OR PAH deficiency[Title/Abstract] OR phenylalanine hydroxylase deficiency[Title/Abstract] OR phenylketonuri∗[Title/Abstract] OR pku[Title/Abstract]) AND (“1980/01/01”[PDat]: “2019/12/31”[PDat]) AND Humans[Mesh] Filters: Publication date from 1980/01/01 to 2019/12/31; Humans.
This literature search yielded 5,459 records without duplicates, of which 1,118 papers with an appropriate title and abstract were assessed. The final number of relevant records was 256 (Figure S1).
Databases
The PAH locus-specific database PAHvdb, ClinVar, HGMD, and LOVD databases were searched for variants. PAHvdb is linked to the genotype-phenotype BIOPKU database and was used for analyses. The accession number for the PAH is RefSeq: ENSG00000171759; GeneBank: NM_000277.1. All variants were tested using Mutalyzer 2.0 and follow the HGVS guidelines.
The BIOPKU database encompasses information about more than 16,900 PKU-affected subjects from 51 countries, providing information on genotypes, corresponding metabolic phenotypes, BH4 responsiveness (where reported), and highest blood Phe concentrations before starting treatment (where reported). Individual information was collected from the published literature or anonymized records submitted online. Phenotype information was unknown for 690 subjects. Data in the database are anonymized and cannot be traced back to the families. Purpose of the database was to provide an online search tool for reported PAH variants or genotypes, with an output summarizing the number of records, phenotype distribution, BH4 responsiveness, and regional and counties distribution. As mentioned above, BIOPKU is linked with the PAHvdb, thus providing additional information including allelic phenotype values (APV). Data submissions and procedures followed were in accordance with the ethical standards and were approved by local institutional review boards where applicable. Table 1 shows the information included in the database.
Table 1.
Information Included in the Database
| Information Provided | n (%) |
|---|---|
| All subjects | 16,974 |
| Complete genotype | 16,196 |
| Compound heterozygotes | 11,810 (73) |
| Homozygotes | 4,386 (27) |
| Phenotype and genotype | 16,092 |
| cPKU | 9,923 (62) |
| mPKU | 3,521 (22) |
| MHP | 2,648 (16) |
| Genotype and country | 15,357 (91) |
| Blood Phe levels | 6,371 (38) |
| Blood Phe levels and phenotype | 6,369 (38) |
| Blood Phe levels and genotype | 6,115 (36) |
| Phenotype, genotype and BH4 responsiveness | 5,597 |
| Non-responder | 3,191 (57) |
| Responder | 2,316 (43) |
| Number of different genotypes | 3,659 |
| Compound heterozygous | 3,446 (94) |
| Homozygous | 213 (6) |
cPKU, classic PKU; mPKU, mild PKU; MHP, mild hyperphenylalaninemia; BH4, tetrahydrobiopterin
Definition of Phenotypes
Since not all countries use the same nomenclature for the severity of PAH deficiency, in this study the following three metabolic phenotype groups were used: classical PKU (cPKU; pre-treatment blood Phe > 1,200 μmol/L); mild PKU (mPKU; pre-treatment blood Phe 600–1,200 μmol/L), and mild hyperphenylalaninemia (MHP; pre-treatment blood Phe 120–600). Subjects with a moderate PKU (pre-treatment blood Phe 900–1,200 μmol/L) were included in the mPKU group and MHP included MHP-no treatment (blood Phe 120–360 μmol/L) and MHP-gray zone (360–600 μmol/L) categories.5, 6, 7 Any classifications not fitting into one of the above groups (due to different country-specific classifications) were reassigned on the basis of reported pre-treatment blood Phe concentrations. However, neither the method used for blood Phe quantification nor the age at measurement were reported.
Definition of the Allelic (APV) and Genotypic Phenotype Value (GPV)
APV is a value defining the association of a variant with the corresponding metabolic phenotype, thus defining its severity. APV was calculated for variants occurring in a functionally hemizygous constellation (i.e., in a combination with an inactive null allele) in at least five subjects.15 APVs range between 0 and 10, with following classifying definitions cPKU (APV = 0–2.7), mPKU (APV = 2.8–6.6), and MHP (APVs 6.7–10).15
The genotypic phenotype value (GPV) was calculated from the APVs of both alleles and was assigned to a higher APV (APVmax). This calculation was based on the fact that the milder variant (with a higher APV) is always dominant over the severe one.15,16 Possible effects of interallelic complementation and epigenetic factors, which may influence the phenotype,17,18 were not considered in this study.
Definition of BH4 Responsiveness
BH4 responsiveness was defined as a ≥30% reduction of blood Phe concentrations within 24–48 h after the administration of BH4 (20 mg/kg body weight).19,20 The protocols for the BH4 challenge were different in different centers (i.e., 24 h, 48 h, 1 day, 1 week, etc.) and for this reason BH4 responsiveness was simply reported as “yes” or “no.” A linear discriminant analysis was applied to predict BH4 responsiveness based on the APV for untested subjects.
Statistical Analysis
Statistical analysis was performed using R, an open source software and flexible programming language used for the statistical data analysis as well as graphic creations (see Web Resources). A total of 16,196 records with a complete genotype information (variant 1 and 2 known) were analyzed. Two linear discriminant analyses (LDA) were computed: the first LDA was computed to predict the clinical phenotype (cPKU, mPKU, and MHPA) from an individual’s GPV, and the second LDA was computed to predicted BH4 responsiveness by GPV value from untested individuals in BIOPKU.
Results
Prevalence of PKU
Based on the literature search and reports from national screening centers, the prevalence of PKU was estimated for 64 countries. For parts of Africa, Asia, South America, and Caribbean there was no information. The estimated total number of PKU subjects (all phenotypes) from those 64 countries in 2018 was 360,466. For the remaining 257 countries we were unable to find credible PKU prevalence sources; we used the average regional prevalence of 64 countries, multiplied by their populations, resulting in an additional 94,114 PKU-affected individuals (total about 0.45 million PKU subjects). The global PKU prevalence was estimated to be 1:23,930 newborns (Figure 1, Table S1).
Figure 1.
Prevalence of PKU (All Phenotypes) in 64 Countries from 6 World Regions
For exact prevalence numbers see Table S1.
The PKU prevalence was highest in European and certain Middle Eastern populations. Italy (1:4,000) and Ireland (1:4,545) had even higher prevalence than Iran and Jordan (both 1:5,000) or Turkey (1:6,667). However, Saudi Arabia (1:14,245), Iraq (1:14,286), or United Arab Emirates (1:14,493) had lower PKU prevalence.
PKU prevalence was also high in Central European countries, e.g., Germany (1:5,360), Czechia (1:5,521), Austria (1:5,764), and Slovakia (1:5,753). Slovenia (1:7,143) and Poland (1:8,309) had similar rates to Eastern Europe, e.g., Estonia (1:7,143), Russia (1:7,714), Belarus (1:7,692), and Croatia (1:8,333).
PKU occurred slightly less frequently in Western Europe, e.g., in France (1:9,091), United Kingdom (1:10,000), Belgium (1:11,000), or the Netherlands (11,546), and in Southern Europe, e.g., Spain (1:10,115) or Portugal (1:12,500).
Northern Europe showed the lowest PKU rates in Europe, e.g., Norway (1:11,457), Sweden (1:12,681), Denmark (1:13,434), or only 1:112,000 in Finland.
PKU occurred more frequently in Canada (1:15,000) than in the United States of America (1:25,000) or in Latin American countries, e.g., Argentina (1:15,715), Chile (1:19,231), Brazil (1:25,000), Mexico (1:27,778), or Peru (1:46,970).
The lowest PKU prevalence was reported in Asian countries, such as Thailand (1:227,273), Japan (1:125,000), Philippines (1:116,006), or Singapore (1:83,333). One exception was China where the PKU prevalence was 1:15,924, which was comparable to Europe.
Descriptive Analysis of the Phenylalanine Hydroxylase Gene Locus-Specific Database
Substitutions were by far the most frequent variant type in the PAHvdb (80.5%), followed by deletions (12.9%) and duplications (2.1%). Of all variants, 691 were missense variants (58.3%), followed by 165 frameshift variants (13.9%) and 155 splice site (13.1%) variants. Nonsense variants (6.9%), synonymous variants (4.9%), and in-frame variants (1.9%) were less frequent. Extension, complex, and unknown variants, accounted for the remainder. Exon 6 contained the largest number of variants (14.1%), followed by exon 7 (12.2%) and exon 3 (9.9%). Most variants (59.2%) were located in the central catalytic domain, 17.5% in the N-terminal regulatory domain, and 5.4% in the C-terminal oligomerization domain of the PAH monomer. The remaining variants (17.9%) were either in the intronic or UTR regions. Only 7.7% of all variants were located in one of the four cofactor binding regions (Figure S2).
The APV was known for 589 of 1,186 variants. Most variants (441) were defined as severe null alleles (APV = 0), 32 variants as cPKU phenotype, 52 as mPKU, and 64 as MHP alleles. Metabolic phenotype was predicted, on the basis of the known genotype and corresponding GPV from the LDA, in 87.9% of PKU subjects.
Descriptive Analysis of the BIOPKU Database
As of October 2019, the BIOPKU database contained anonymized data on more than 16,900 PKU subjects. A total of 16,196 PKU subjects with 3,659 different genotypes were analyzed. Information on the PKU phenotype was available for 16,092 subjects, resulting in some of them (810) having no genotype information. Maximum pretreatment Phe concentrations were reported for only 6,371 subjects and the BH4 test information was available for 5,597 subjects (Table 1).
Global PKU Phenotype and Genotype Distribution
Information on the PKU phenotype and genotype was available for 16,092 out of 16,196 subjects (99%). Of these, most had cPKU (9,923; 61.7%), 3,521 (21.9%) had mPKU, and 2,648 had MHP (16.4%). Information about the phenotype was unavailable for 104 subjects.
The comparison of pre-treatment Phe level with the reported phenotype (n = 6,369) is illustrated in Figure 2A and with the GPVs (n = 6,115) in Figure 2B. The interquartile range (n, median, 25th–75th percentile) was smallest and lowest for the MHP subject group: 1,283, 320 μmol/L, 242–432 μmol/L, and was larger for mPKU and cPKU (1,487, 793 μmol/L, 660–793 μmol/L, and 3,599, 1,550 μmol/L, 1,270–1,936 μmol/L, respectively).
Figure 2.
Relationship between Blood Phenylalanine, Phenotype, and Genotypic Phenotype Value (GPV)
(A) Boxplot (median, 25th–75th percentile, 1.5) of the maximal pretreatment blood Phe concentrations for three metabolic phenotypes in 6,369 PKU subjects. The circles in the cPKU bar represent ordinary high blood Phe concentrations, since cPKU doesn’t have an upper Phe limit for its classification.
(B) Contour plot of two-dimensional densities of pretreatment blood Phe concentrations and corresponding genotypic phenotype values (GPV) for 6,115 PKU subjects.
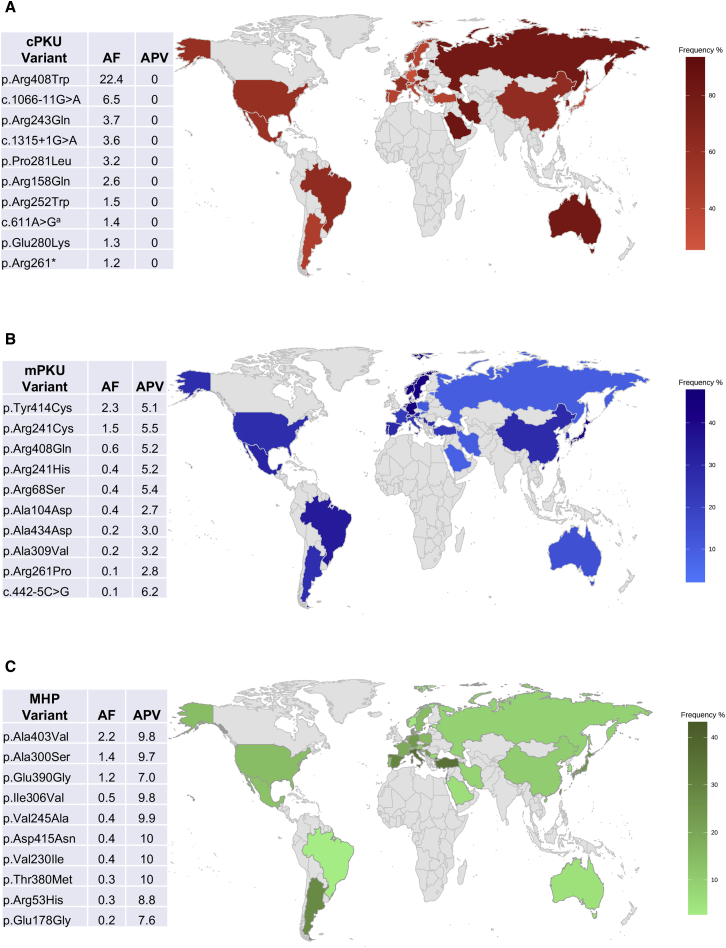
A total of 758 different PAH variants were identified in this study. The three most prevalent variants were c.1222C>T (p.Arg408Trp), with an allele frequency (AF) = 22.2%, c.1066−11G>A (IVS10−11G>A; AF = 6.4%), and c.782G>A (p.Arg261Gln; AF 5.5%). Figure 3 shows the most frequent phenotype-specific variants. A full list of PAH variants is shown in Table S2.
Figure 3.
World Map with Relative Frequency (%) of PKU and the Corresponding Most Common Variants for Classic PKU (cPKU), Mild PKU (mPKU), and Mild Hyperphenylalaninemia (MHP)
Exact frequencies and additional genotypic phenotype values (GPV) for Europe and other world regions are presented in a more granular form in Figure S4. AF, allele frequency; APV, allelic phenotype value (cPKU = 0–2.6; mPKU = 2.7–6.6; MHP = 6.7–10). The accession number for the PAH is RefSeq: ENSG00000171759; GeneBank: NM_000277.1.
ac.611A>G reported as Ex6−96A>G splice variant.
Of all patients, 11,810 (72.9%) were compound heterozygotes and 4,386 (27.1%) were homozygotes. Of 3,659 genotypes, 3,446 (94.2%) were compound heterozygotes and 213 (5.8%) homozygotes. The three most prevalent genotypes were p.[Arg408Trp];[Arg408Trp] with a genotype frequency (GF) = 11.4%, followed by c.[1066−11G>A];[1066−11G>A] (GF = 2.6%) and c.[1222C>T];[1315+1G>A] (p.[Arg408Trp];IVS12+1G>A) (GF = 1.6%). A full list of genotypes is shown in Table S3. Strikingly, 54.5% of all genotypes were specific for only one subject and were not used for the phenotype prediction.
BH4 Responsiveness
Information on BH4 responsiveness was available for 5,597 subjects. About half were classified as BH4 responsive (2,316; 43%). Table S4 shows the distribution of BH4 responsiveness in different phenotype groups. As expected, milder forms of PKU appeared more likely to be BH4 responsive, whereas most patients with cPKU were non-responders. Pretreatment blood Phe concentrations (n, median, 25th–75th percentile) were much lower for BH4-responsive subjects (1,116, 620 μmol/L, 411–853 μmol/L) compared with non-responders (1,180, 1,361 μmol/L, 1,039–1,719 μmol/L.
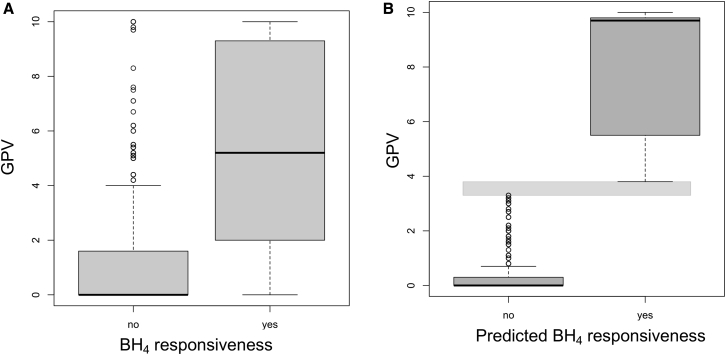
GPV was assigned for 2,246 BH4 non-responder and 1,755 responder PKU subjects (for 2,114 out of 6,115 subjects GVP was not known) (Figure 4A). The 11,584 subjects not tested for BH4 responsiveness were analyzed for potential responsiveness using the GPVs. Patients with a GPV > 3.8 (n = 3,023; 26%) were assigned as potential BH4 responders (p < 0.001 versus patients assigned as non-responders [n = 8,561; 74%]) (Figure 4B).
Figure 4.
Relationship between Genotypic Phenotype Value (GPV) and BH4 Responsiveness
(A) Boxplot (median, 25th–75th percentile, 1.5) of GPV in 2,246 BH4 non-responder and 1,755 responder PKU subjects (for 2,114 out of 6,115 subjects GVP was not known).
(B) Boxplot (median, 25th–75th percentile, 1.5) of GPV (APVmax) for 11,584 PKU subjects with a known genotype, but not tested for BH4 responsiveness. Horizontal gray bar: separation area between GPVs for predicted BH4 responsiveness (3.8–10) and non-responsiveness (0–3.3).
PKU Phenotypes and Genotypes in World Regions
Overview of Global Data
Subject data originated from 51 countries from 17 world regions and a total of 15,357 subjects from 33 countries with at least 35 reported cases were analyzed (Tables S5 and S6, Figure S3). Most cases were reported from Central (18.3%) and Eastern (18.1%) Europe, Eastern Asia (13.8%), Western Europe (9.6%), and the Middle East (9.2%). Very few subjects were reported from the northern and southern parts of Africa (Table S6).
cPKU was the most frequent phenotype in all world regions, with high rates reported in Australia and eastern Europe (81%), and rates of <50% reported only in Serbia, Argentina, Turkey, Netherlands, Sweden, Spain, Italy, Japan, Slovenia, Germany, and Taiwan (Figure 4A). Of 2,835 subjects in Russia, 81% had cPKU, 9.9% had mPKU, and 9.1% had MHP. Estonia, representing the Baltic region, reported 93.5% of subjects with cPKU. The ratio of mPKU:MHP subjects was comparable, except for Eastern Asia, North America, and South America, where mPKU was more common. The phenotype distribution in world regions, as well as Genotypic Phenotype Values (GPV) for Europe and other world regions, is shown in a more granular form in Figure S4.
Reports from individual regions are summarized in Table 2 and below.
Table 2.
Summary of Genotype and Phenotype Information for 51 Countries with at Least 35 Patients Reported
| Country | cPKU (%) | mPKU (%) | MHP (%) | No. 1 Frequent Genotype | GF (%) | No. 2 Frequent Genotype | GF (%) | No. 3 Frequent Genotype | GF (%) | Different Alleles (n) | No. 1 Frequent Allele | AF (%) | No. 2 Frequent Allele | AF (%) | No. 3 Frequent Allele | AF (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Europe | ||||||||||||||||
| Austria | 59.7 | 30.2 | 10.1 | p.[Arg408Trp];[Arg408Trp] | 7.0 | p.[Arg261Gln];[Arg408Trp] | 6.3 | c.[1315+1G>A];[1315+1G>A] | 5.6 | 61 | p.Arg408Trp | 23.4 | c.1315+1G>A | 11.2 | p.Arg261Gln | 6.6 |
| Bulgaria | 64.9 | 27.0 | 8.1 | p.[Arg408Trp];[Arg408Trp] | 16.2 | p.[Leu48Ser];[Arg408Trp] | 8.1 | p.[Leu48Ser]:c.[1066−11G>A] | 5.4 | 20 | p.Arg408Trp | 32.4 | p.Arg261Gln | 17.6 | p.Leu48Ser | 9.5 |
| Croatia | 67.7 | 19.1 | 13.2 | p.[Leu48Ser];[Arg408Trp] | 14.3 | p.[Glu390Gly];[Arg408Trp6] | 8.6 | p.[Arg158Gln];[Arg408Trp] | 5.7 | 26 | p.Arg408Trp | 31.4 | p.Leu48Ser | 14.3 | p.Glu390Gly | 7.1 |
| Czechia | 69.9 | 7.4 | 22.7 | p.[Arg408Trp];[Arg408Trp] | 18.2 | p.[Arg408Trp];[Ala403Val] | 4.0 | p.[Arg158Gln];[Arg408Trp] | 3.8 | 92 | p.Arg408Trp | 41.7 | p.Ala403Val | 5.3 | p.Arg158Gln | 4.6 |
| Denmark | 51.3 | 28.3 | 20.4 | p.[Tyr414Cys]:c.[1315+1G>A] | 8.7 | c.[1315+1G>A];[1315+1G>A] | 7.7 | p.[Arg408Trp]:c.[1315+1G>A] | 7.7 | 83 | c.1315+1G>A | 27.3 | p.Arg408Trp | 17.6 | p.Tyr414Cys | 12.1 |
| Estonia | 93.5 | 2.2 | 4.3 | p.[Arg408Trp];[Arg408Trp] | 66.3 | p.[Arg408Trp];[Arg261Gln] | 5.4 | p.[Arg408Trp];[Leu48Ser] | 4.3 | 16 | p.Arg408Trp∗ | 82.6 | p.Arg261Gln | 2.7 | c.1315+1G>A | 2.2 |
| France | 59.1 | 22.0 | 18.9 | p.[Glu280Lys];[Glu280Lys] | 2.2 | p.[Gly352Valfs∗48];[Gly352Valfs∗48] | 1.7 | p.[Arg261Gln];[Arg261Gln] | 1.5 | 229 | c.1066−11G>A | 7.4 | p.Arg261Gln | 6.5 | p.Arg408Trp | 5.5 |
| Germany | 32.0 | 44.0 | 24.0 | p.[Arg408Trp];[Arg408Trp] | 4.1 | p.[Arg408Trp];[Tyr414Cys] | 2.7 | p.[Arg261Gln];[Arg261Gln] | 2.4 | 102 | p.Arg408Trp | 19.3 | p.Tyr414Cys | 9.7 | c.1315+1G>A | 8.8 |
| Italy | 39.1 | 24.3 | 36.6 | p.[Arg261Gln]:c.[1066−11G>A] | 2.8 | c.[1066−11G>A];[1066−11G>A] | 2.2 | p.[Arg261Gln];[Arg261Gln] | 1.8 | 159 | p.Arg261Gln | 10.7 | c.1066−11G>A | 10.6 | p.Ala403Val | 8.4 |
| Netherlands | 42.0 | 37.2 | 20.8 | p.[Tyr414Cys]:c.[1315+1G>A] | 4.5 | p.[Arg261Gln]:c.[1315+1G>A] | 3.6 | p.[Pro281Leu]:c.[1315+1G>A] | 2.7 | 65 | c.1315+1G>A | 13.7 | p.Arg261Gln | 8.5 | p.Pro281Leu | 7.6 |
| Norway | 52.1 | 43.8 | 4.2 | p.[Gly46Ser]:c.[1315+1G>A] | 12.2 | p.[Gly46Ser]:c.[842+1G>A] | 6.1 | p.[Gly46Ser];[Phe299Cys] | 6.1 | 24 | p.Gly46Ser | 16.3 | p.Tyr414Cys | 13.3 | c.1315+1G>A | 12.2 |
| Poland | 74.0 | 10.9 | 15.1 | p.[Arg408Trp];[Arg408Trp] | 40.0 | p.[Arg408Trp]:c.[1066−11G>A] | 5.6 | p.[Arg408Trp];[Arg158Gln] | 4.4 | 94 | p.Arg408Trp | 64.6 | c.1066−11G>A | 4.3 | p.Arg158Gln | 3.5 |
| Portugal | 52.9 | 34.3 | 12.7 | p.[Arg261Gln];[Arg261Gln] | 7.8 | c.[1066−11G>A];[1066−11G>A] | 5.9 | p.[Arg261Gln];[Val388Met] | 4.9 | 34 | c.1066−11G>A | 16.2 | p.Arg261Gln | 16.2 | p.Val388Met | 11.3 |
| Russia | 81.0 | 9.9 | 9.1 | p.[Arg408Trp];[Arg408Trp] | 31.6 | p.[Arg261Gln];[Arg408Trp] | 5.0 | p.[Pro281Leu];[Arg408Trp] | 3.7 | 148 | p.Arg408Trp | 53.7 | p.Arg261Gln | 5.6 | p.Pro281Leu | 4.1 |
| Serbia | 48.0 | 28.0 | 24.0 | p.[Leu48Ser];[Leu48Ser] | 14.7 | p.[Leu48Ser];[Arg408Trp] | 10.7 | p.[Leu48Ser];[Arg158Gln] | 4.0 | 30 | p.Leu48Ser | 31.3 | p.Arg408Trp | 14.7 | p.Ile306Val | 7.3 |
| Slovakia | 72.6 | 5.9 | 21.5 | p.[Arg408Trp];[Arg408Trp] | 26.5 | p.[Arg408Trp]:c.[1315+1G>A] | 4.6 | p.[Arg408Trp];[Ala403Val] | 4.6 | 51 | p.Arg408Trp | 47.9 | p.Arg158Gln | 5.9 | c.1315+1G>A | 5.0 |
| Slovenia | 34.1 | 22.7 | 43.2 | p.[Arg158Gln];[Glu390Gly] | 9.1 | p.[Arg261Gln];[Glu390Gly] | 9.1 | p.[Pro281Leu];[Ala403Val] | 6.8 | 27 | p.Arg408Trp | 12.5 | p.Glu390Gly | 11.4 | p.Arg158Gln | 11.4 |
| Spain | 41.6 | 28.8 | 29.6 | c.[1066−11G>A];[1066−11G>A] | 5.2 | p.[Ile65Thr]:c.[1066−11G>A] | 2.0 | p.[Arg261Gln]:c.[1066−11G>A] | 2.0 | 155 | c.1066−11G>A | 11.1 | p.Val388Met | 6.8 | p.Ile65Thr | 6.4 |
| Sweden | 41.8 | 43.6 | 14.6 | p.[Arg408Trp]:c.[1315+1G>A] | 16.4 | p.[Arg408Trp];[Arg408Trp] | 10.9 | p.[Tyr414Cys]:c.[1315+1G>A] | 9.1 | 12 | p.Arg408Trp | 27.3 | p.Tyr414Cys | 23.6 | c.1315+1G>A | 21.8 |
| Switzerland | 54.8 | 30.6 | 14.6 | p.[Arg261Gln];[Arg261Gln] | 9.7 | p.[Ile95del];[Tyr414Cys] | 4.8 | p.[Leu48Ser];[Leu48Ser] | 4.8 | 39 | p.Arg261Gln | 15.3 | p.Arg408Trp | 8.1 | c.1066−11G>A | 7.3 |
| Asia | ||||||||||||||||
| China | 62.1 | 27.6 | 10.3 | p.[Arg243Gln];[Arg243Gln] | 6.1 | p.[Arg243Gln]:c.[611A>G]a | 4.6 | p.[Arg241Cys];[Arg243Gln] | 3.7 | 234 | p.Arg243Gln | 23.3 | c.611A>Ga | 10.2 | p.Arg241Cys | 8.0 |
| Japan | 34.5 | 40.0 | 25.5 | p.[Arg413Pro];[Arg241Cys] | 9.1 | p.[Arg413Pro];[Arg413Pro] | 7.3 | p.[Tyr414Cys]:c.[1315+1G>A] | 7.3 | 31 | p.Arg413Pro | 18.2 | p.Arg241Cys | 16.4 | p.Arg111∗ | 7.3 |
| Korea | 71.0 | 24.7 | 4.3 | p.[Arg241Cys];[Ala259Thr] | 4.3 | p.[Arg241Cys];[Arg243Gln] | 4.3 | c.[611A>G];[611A>G]a | 3.2 | 47 | p.Arg243Gln | 13.3 | c.442−1G>A | 10.1 | p.Arg241Cys | 8.5 |
| Taiwan | 25.4 | 47.9 | 26.8 | p.[Arg241Cys];[Arg241Cys] | 8.5 | p.[Arg241Cys]:c.[611A>G]a | 7.0 | p.[Ala434Asp];[Arg408Trp] | 2.8 | 34 | p.Arg241Cys | 24.6 | p.Arg408Gln | 12.0 | c.611A>Ga | 7.7 |
| North America | ||||||||||||||||
| USA | 58.6 | 27.2 | 14.2 | p.[Arg408Trp];[Arg408Trp] | 4.0 | c.[1066−11G>A];[1066−11G>A] | 3.1 | p.[Arg408Gln]:c.[1315+1G>A] | 2.5 | 214 | p.Arg408Trp | 18.5 | c.1066−11G>A | 7.8 | c.1315+1G>A | 6.9 |
| Latin America | ||||||||||||||||
| Argentina | 45.4 | 26.8 | 27.8 | p.[Arg408Trp];[Arg408Trp] | 4.1 | p.[Arg408Trp];[Ala403Val] | 3.1 | p.[Val388Met];[Tyr414Cys] | 3.1 | 47 | p.Arg408Trp | 10.3 | p.Arg261Gln | 9.8 | c.1066−11G>A | 9.3 |
| Brazil | 63.0 | 34.2 | 2.8 | p.[Val388Met];[Val388Met] | 5.0 | p.[Arg261Gln];[Arg261Gln] | 3.7 | c.[1066−11G>A];[1066−11G>A] | 2.3 | 56 | p.Val388Met | 16.2 | p.Arg261Gln | 11.6 | c.1066−11G>A | 9.4 |
| Mexico | 56.5 | 30.4 | 13.1 | c.[60+5G>T];[60+5G>T] | 10.6 | p.[Val388Met]:c.[60+5G>T] | 10.6 | c.[60+5G>A];[441+5G>T] | 4.3 | 33 | c.60+5G>T | 23.4 | p.Val388Met | 9.6 | c.1066−11G>A | 7.4 |
| Middle East | ||||||||||||||||
| Iran | 81.1 | 10.3 | 8.6 | c.[1066−11G>A];[1066−11G>A] | 18.4 | p.[Arg261Gln];[Arg261Gln] | 7.6 | c.[168+5G>C];[168+5G>C] | 6.8 | 106 | c.1066−11G>A | 22.1 | p.Arg261Gln | 9.7 | c.168+5G>C | 8.7 |
| Israel | 50.5 | 21.7 | 27.8 | c.[1066−11G>A];[1066−11G>A] | 9.4 | p.[Leu48Ser];[Leu48Ser] | 5.2 | c.[168+1G>A];[168+1G>A] | 3.8 | 55 | c.1066−11G>A | 15.1 | p.Leu48Ser | 10.8 | p.Ala403Val | 7.3 |
| Saudi Arabia | 85.0 | 9.0 | 6.0 | p.[Arg252Trp];[Arg252Trp] | 25.4 | p.[Val388Met];[Val388Met] | 10.4 | p.[Arg261∗];[Arg261∗] | 9.0 | 21 | p.Arg252Trp | 27.6 | p.Arg261∗ | 11.2 | p.Val388Met | 10.4 |
| Turkey | 42.1 | 22.7 | 35.2 | c.[1066−11G>A];[1066−11G>A] | 13.9 | p.[Arg261Gln];[Arg261Gln] | 8.3 | p.[Pro281Leu];[Pro281Leu] | 4.6 | 64 | c.1066−11G>A | 22.9 | p.Arg261Gln | 11.8 | p.Leu48Ser | 8.8 |
| Australia | ||||||||||||||||
| Australia | 80.4 | 14.1 | 5.4 | p.[Arg261Gln]:c.[1315+1G>A] | 7.4 | p.[Arg408Trp]:c.[1315+1G>A] | 7.4 | c.[1315+1G>A];[1315+1G>A] | 6.3 | 37 | c.1315+1G>A | 23.2 | p.Arg408Trp | 20.5 | p.Ile65Thr | 10.5 |
The accession number for the PAH is RefSeq: ENSG00000171759; GeneBank: NM_000277.1.
c.611A>G reported as Ex6-96A>G splice variant.
Europe
Variant p.Arg408Trp was the most common (AF = 63.7%) throughout Europe, followed by c.1066−11G>A (AF = 11%) and p.Arg261Gln (AF = 11%). Eastern Europe had the highest AF for p.Arg408Trp (54.6%), mostly with homozygous genotype p.[Arg408Trp];[Arg408Trp] (GF = 32.7%); findings were similar for Central Europe (AF = 44.4% and GF = 23.8%). Russia contributed the largest number of records. A single variant, p.Arg408Trp (AF = 53.7%), was dominant in Russia, followed by p.Arg261Gln (AF = 5.6%) and c.842C>T (p.Pro281Leu; AF = 4.1%). The most common genotype was p.[Arg408Trp];[Arg408Trp] (30.6% of 458 different genotypes). In Southeastern Europe, p.Arg408Trp was also the most frequent allele, but c.[143T>C];[1222C>T], p.[Leu48Ser];[Arg408Trp] was the most prevalent genotype (GF = 10.6%)
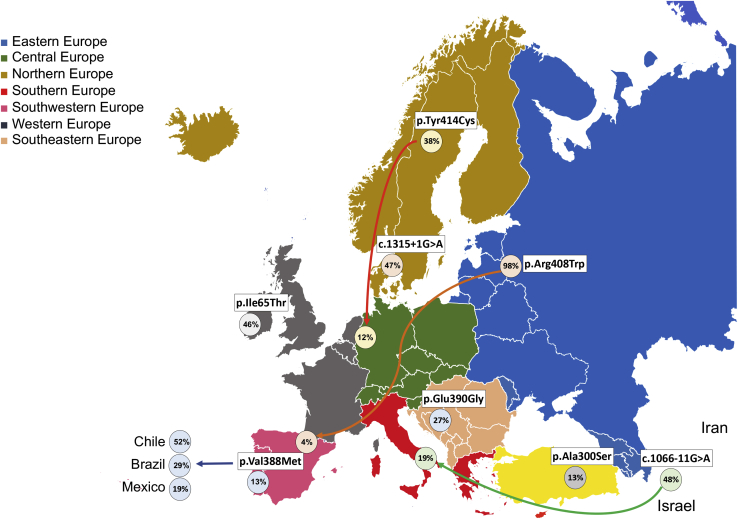
The distribution of patients with p.Arg408Trp on at least one allele (compound heterozygotes and homozygotes) decreased from 98% in Estonia to 89% in Poland, 76% in Russia, 69% in Slovakia, 65% in Czechia, 40% in Austria, 36% in Germany, 10% in France, 6% in Italy, to only 4% in Spain (Table 3, Figure 5). Genotypes with the c.1066−11G>A splice site variant occurred commonly in Armenia (48%), Turkey (32%), Iran (26%), Israel (21%), Spain (20%), and Italy (19%) (Table 4, Figure 5). The prevalence of p.Arg261Gln was 10%–30% in most countries.
Table 3.
Frequency (%) of Subjects with p.Arg408Trp and p.Arg261Gln Variants on at least One Allele (Compound Heterozygotes or Homozygotes) in Different Countries
| p.Arg408Trp | %a | p.Arg261Gln | %a |
|---|---|---|---|
| Estonia | 98.9 | Bulgaria | 29.7 |
| Poland | 89.2 | Portugal | 23.5 |
| Romania | 87.5 | Switzerland | 21.0 |
| Russia | 75.7 | Brazil | 20.5 |
| Slovakia | 69.4 | Italy | 19.7 |
| Czechia | 64.6 | Argentina | 17.5 |
| Croatia | 57.1 | Netherlands | 15.7 |
| Bulgaria | 48.6 | Turkey | 15.3 |
| Sweden | 43.6 | Slovenia | 13.6 |
| Austria | 39.9 | Germany | 13.6 |
| Germany | 35.7 | Croatia | 12.9 |
| Australia | 34.7 | Austria | 12.6 |
| USA | 32.9 | Spain | 11.9 |
| Denmark | 28.8 | Iran | 11.8 |
| Serbia | 28.0 | Australia | 11.6 |
| Slovenia | 22.7 | France | 11.5 |
| Argentina | 16.5 | Russia | 10.3 |
| Switzerland | 14.5 | Serbia | 9.3 |
| France | 9.9 | Israel | 9.0 |
| Turkey | 8.8 | USA | 8.6 |
| Israel | 7.1 | Slovakia | 6.8 |
| Brazil | 6.8 | Estonia | 5.4 |
| Italy | 5.7 | Poland | 3.6 |
| Netherlands | 4.5 | Czechia | 2.3 |
| Spain | 4.0 | Denmark | 2.3 |
| Iran | 1.4 | China | 1.6 |
For a total number of patients in each country, see Table S6. The accession number for the PAH is RefSeq: ENSG00000171759; GeneBank: NM_000277.1. Source BIOPKU database.
Percentage of subjects (of a total number of patients) with a variant occurring in a compound heterozygous or homozygous constellation.
Figure 5.
Patterns of Common PAH Variants Possibly Associated with Migration in Europe
Table 4.
Frequency (%) of Subjects with Common Variants on at least One Allele (Compound Heterozygotes or Homozygotes) in Different Countries
| p.Gly46Ser | %a | p.Leu48Ser | %a | p.Ile65Thr | %a | p.Arg158Gln | %a | p.Arg241Cys | %a |
|---|---|---|---|---|---|---|---|---|---|
| Norway | 32.7 | Serbia | 48.0 | Ireland | 45.5 | Slovenia | 22.7 | Taiwan | 40.8 |
| Denmark | 2.0 | Croatia | 25.7 | Australia | 17.9 | Italy | 10.3 | Japan | 27.3 |
| Spain | 1.8 | Bulgaria | 18.9 | Brazil | 12.8 | Slovakia | 10.0 | China | 15.0 |
| Brazil | 0.9 | Israel | 16.5 | Spain | 12.1 | Netherlands | 9.9 | S. Korea | 14.9 |
| USA | 0.4 | Turkey | 13.4 | Portugal | 10.8 | Czechia | 8.6 | Netherlands | 4.5 |
| p.Arg243Gln | %a | p.Val245Ala | %a | p.Arg252Trp | %a | p.Glu280Lys | %a | p.Pro281Leu | %a |
| China | 40.4 | Italy | 4.9 | Saudi Arabia | 29.9 | France | 5.1 | Netherlands | 14.8 |
| S. Korea | 26.6 | Slovakia | 2.3 | Brazil | 9.6 | USA | 4.3 | Turkey | 11.2 |
| USA | 19.7 | Germany | 2.1 | Portugal | 6.9 | Australia | 4.2 | Portugal | 10.8 |
| Taiwan | 11.3 | Denmark | 1.5 | Slovakia | 6.8 | Spain | 3.5 | Italy | 10.3 |
| Spain | 5.1 | France | 1.2 | Czechia | 5.1 | Russia | 2.5 | Germany | 9.4 |
| p.Phe299Cys | %a | p.Ala300Ser | %a | p.Val388Met | %a | p.Glu390Gly | %a | p.Ala403Val | %a |
| Norway | 14.3 | Turkey | 13.0 | Chile | 52.4 | Slovenia | 22.7 | Italy | 15.6 |
| Australia | 4.2 | Israel | 9.0 | Brazil | 28.8 | Croatia | 14.3 | Slovenia | 13.6 |
| USA | 2.3 | Italy | 7.1 | Portugal | 20.6 | Serbia | 10.7 | Israel | 13.2 |
| France | 1.1 | Iran | 3.8 | Mexico | 19.1 | Turkey | 6.5 | Argentina | 10.3 |
| Russia | 0.2 | Spain | 3.7 | Spain | 13.1 | Austria | 5.6 | Czechia | 10.1 |
| p.Tyr414Cys | %a | c.611A>Gb | %a | c.442−1G>A | %a | c.1066−11G>A | %a | c.1315+1G>A | %a |
| Sweden | 38.2 | China | 18.7 | Korea | 18.1 | Armenia | 47.8 | Denmark | 46.9 |
| Norway | 24.5 | Taiwan | 12.7 | China | 8.0 | Turkey | 31.9 | Sweden | 36.4 |
| Denmark | 22.2 | S. Korea | 11.7 | Japan | 7.3 | Iran | 25.7 | Norway | 26.0 |
| Germany | 18.0 | France | 0.2 | Poland | 0.4 | Israel | 20.8 | Netherlands | 22.4 |
| Netherlands | 11.7 | Russia | 0.1 | Russia | 0.1 | Spain | 20.1 | Germany | 17.4 |
For a total number of patients in each country, see Table S6. The accession number for the PAH is RefSeq: ENSG00000171759; GeneBank: NM_000277.1. Source BIOPKU database.
Percentage of subjects (of a total number of patients) with a variant occurring in a compound heterozygous or homozygous constellation.
c.611G>A reported as Ex6-96A>G splice variant.
PKU phenotypes were more evenly distributed in Southern Europe (39.5% cPKU, 36.6% MHP, 23.9% mPKU), and milder phenotypes were more predominant, e.g., 37% had MHP in Italy. This was consistent with a high frequency of variants with a substantial residual PAH activity, e.g., c.1208C>T (p.Ala403Val; AF = 8.4), c.898G>T (p.Ala300Ser; AF = 3.6), c.734T>C (p.Val245Ala; AF = 2.4), or c.1241A>G (p.Tyr414Cys; AF = 2.4). Variants c.1066−11G>A and p.Arg261Gln, often called the “Mediterranean mutation,” accounted for majority of mutant alleles, and the most frequent genotype was c.[1066−11G>A];[1066−11G>A] (GF = 5.2%). In Portugal and Italy, c.1066−11G>A and p.Arg261Gln occurred at similar rates (AF = 16.2% and AF = 10.7%, respectively). Variant c.1162G>A (p.Val388Met) was also common in Portugal (AF = 11.3%) and Spain (AF = 6.8%).
Low rates of cPKU were found in Southwestern Europe (42.7%) and Northern Europe (50.5%). In Southeastern Europe, Croatia (68%) and Bulgaria (65%) had a higher prevalence of cPKU than Serbia (48%). p.Arg408Trp was the most frequent variant in Croatia (AF = 31.4%) and Bulgaria (AF = 32.4%), but less common in Serbia (AF = 14.7%). The p.Leu48Ser variant, which was initially identified in Turkey,21 has the highest reported AF (31.3%) in Serbia. The most frequent genotypes in other countries locally were p.[Leu48Ser];[Leu48Ser] (14.7%) in Serbia; p.[Leu48Ser];[Arg408Trp] (14.3%) in Croatia; and p.[Arg408Trp];[Arg408Trp] genotype (16.2%) in Bulgaria.
In Northern Europe c.1315+1G>A was the most common splice site variant (AF = 25%). Most patients were compound heterozygous for p.[Arg408Trp];c.[1315+1G>A]. Denmark had the highest number of cases of the mild p.Tyr414Cys variant in the world and Sweden and Norway also a relatively high rate of mPKU (>43%). In Denmark the most frequent variant was c.1315+1G>A with an AF = 27.3%, while c.136G>A (p.Gly46Ser) occurred more commonly in Sweden and Norway. Variants p.Arg408Gln and c.896T>G (p.Phe299Cys) were specific for Norway.
The p.Arg408Trp variant was also the most frequent variant in all Central European countries, except for Switzerland where p.Arg261Gln (AF = 15.3%) was more prevalent. Classical PKU was particularly frequent in Poland and Slovakia (>70%), but not in Czechia, consistent with a higher prevalence of the mild variant, p.Ala403Val. There was a wide spectrum of PKU variants in Germany: of 102 distinct variants, p.Arg408Trp accounted for only 19.3%, and other variants such as p.Tyr414Cys (AF = 9.7%) as well as c.1315+1G>A (AF = 8.8%) were prominent. Almost half (44%) of patients in Germany had the mPKU phenotype, 32% had cPKU, and 24% had MHP.
In France (1,307 patients), 59.1% had cPKU, and there were a total of 229 different variants, e.g., c.1066−11G>A (AF = 7.4%), p.Arg261Gln (AF = 6.5%), p.Arg408Trp (AF = 5.5%), c.1315+1G>A (AF = 4.5%), and c.838G>A (p.Glu280Lys; AF = 3.7%). The p.Arg408Trp variant was also less common in the Netherlands (AF = 2.2%). The predominant genotype in Western Europe was p.[Glu280Lys];[Glu280Lys].
Latin America
The most frequent variants here, p.Val388Met (AF = 13.9%), p.Arg261Gln (AF = 10.7%), and c.1066−11G>A (AF = 9.4%), were also prominent mutations in Southern Europe (see above). Homozygous p.[Val388Met];[Val388Met] (GF = 4.4%) occurred most frequently. Of the three relevant South American countries, Brazil had the highest numbers of cPKU (63%), followed by Mexico (57%) and Argentina (43%). In comparison to Brazil or Argentina, Mexico’s most frequent variant was c.60+5G>T (IVS1+5G>T).
North America
The most prevalent variants in North America also resembled those in European populations: p.Arg408Trp (18.5%), c.1066−11G>A (7.9%), c.1315+1G>A (6.9%), as did the genotype distribution p.[Arg408Trp];[Arg408Trp] (GF = 4.0%)
Middle East
The predominance of c.1066−11G>A variant (AF = 20.1%) and its homozygous genotype (GF = 15.3%) was evident in Iran, Turkey, Israel, and Saudi Arabia. Other frequent variants included p.Arg261Gln, c.168+5G>C (IVS2+5G>C), p.Pro281Leu, and c.727C>T (p.Arg243∗). 81% of the Iranian patients had cPKU. In comparison to Iran, Turkey had a similar AF for c.1066−11G>A (22.9%) and p.Arg261Gln (11.8%), but strongly different phenotypes: 42% had cPKU, 35% MHP, and 23% mPKU. This may be related to the augmented presence of mild variants such as p.Ala300Ser (7.4%) or c.1169A>G (p.Glu390Gly; 4.2%). In contrast to other Middle Eastern countries, the most frequent variant from Saudi Arabia was c.754C>T (p.Arg252Trp; 27.6%), followed by c.781C>T (p.Arg261∗; 11.2%) and p.Val388Met (10.4%); 30% of all patients had a p.Arg252Trp-associated genotype, i.e., compound heterozygous or homozygous (Table 4).
Asia
Most reported cases from Asia came from the east, with the most prevalent being the missense variant c.728G>A (p.Arg243Gln; AF = 21.8%), followed by c.611 > G (9.8%) and c.721C>T (p.Arg241Cys; 8.7%). The most frequent genotype was homozygous p.[Arg243Gln];[Arg243Gln] (GF = 5.6%).
In China, 62% of subjects (n = 2,008) had a cPKU, 28% mPKU and 10% a MHP phenotype. A total of 234 different variants was reported, of which five—p.Arg243Gln, c.611>G, p.Arg241Cys, c.331C>T (p.Arg111∗), and c.1238G>C (p.Arg413Pro)—had a frequency of >5% and accounted for 52.5% of all alleles. Furthermore, 679 different genotypes were reported, the most frequent being p.[Arg243Gln];[Arg243Gln]. Korea was the only country that, like China, had a high rate of cPKU (71%); this was less common in Japan (37%) and Taiwan (25%). p.Arg243Gln, c.611>G, and p.Arg241Cys were detected in Korea, Taiwan, and China, each with allele frequencies >5%. The p.Arg111∗ and p.Arg413Pro variants were common in Japan, China, and Taiwan, but not Korea. The splice variant c.442−1G>A (IVS4−1G>A) was much more prevalent in Korea than in other Eastern Asian countries.
Genotypes with p.Arg243Gln were common in China (40%) and Korea (27%) (Table 4), while those with p.Arg241Cys were prevalent in Taiwan (41%), Japan (27%), and China (15%) (Table 4). Patients with the splice site variant c.442−1G>A were more common in Korea (18%), followed by China (8%) and Japan (7%). The splice variant c.611A>G (Ex6-96A>G), which masquerades as missense variant p.Tyr204Cys,22 was found 19% of patients in China, 13% in Taiwan, and 12% in Korea (Table 4).
Africa
The low number of reports available from Africa were considered not sufficiently representative of this world region. Thus, we did not analyze these data.
Discussion
The aim of this study was to elucidate the prevalence of PKU and distribution of causative PAH variants worldwide and in different countries. We provided a rough estimation of global PKU prevalence by calculating the number of affected patients for countries based on the provided PKU prevalence and the total population in 2018 (see UN World Population Prospects in Web Resources) and an average prevalence for countries not employing newborn screening for PKU. Overall, it appears that there are about 0.45 million PKU-affected individuals worldwide, of whom at least two thirds have PKU that requires treatment (most subjects had a severe, cPKU phenotype). An improvement in early diagnosis via NBS in countries still lacking it should be an urgent priority. With the multiple allelic combinations generated by so many mutations, a full description of all genotype/phenotype correlations is impossible in this report. Nevertheless, it is clear that for many allelic combinations, often the most common in a region or country, such correlations are possible as detailed in the final three sections in the Results along with Table 2 and Figure S4. Recognizing these relationships is important for counseling parents now that newborn screening predominates diagnosis of this disorder, particularly in instances when treatment recommendations are unclear (e.g., due to borderline blood Phe levels).
Although PKU has been found to be most common in European populations,9 its similar prevalence in certain Middle Eastern countries (particularly Turkey and Iran) was a remarkable finding.23,24 A possible contributing factor may be the frequency of consanguinity in Islamic cultures, especially marriages between first cousins, which would favor the autosomal-recessive inheritance of PKU.25 Prevalence and consanguinity was missing for many countries, making it more difficult to assess this hypothesis.26
We identified 758 different (previously reported) PKU variants in this study, emphasizing the strong genetic heterogeneity of PKU. Information about 16,000 genotypes and phenotypes from PKU subjects enabled a more detailed study of their distribution across different regions and countries than previously possible.27,28 Overall, the severe p.Arg408Trp variant was the most common, especially among Eastern European populations, in accordance with previous studies.29,30 Furthermore, p.Arg408Trp dominates most Central European populations (Poland, Slovakia, Czechia, Austria, or Germany), again supporting previous studies.31, 32, 33, 34, 35
Previous research had suggested that some PKU variants appeared to have been carried by migration, e.g., p.Arg408Trp, c.1066−11G>A, c.1315+1G>A, or p.Phe299Cys and p.Arg408Gln.27,36 Our study supports and extends the knowledge on how certain variants appear to have spread within Europe and to different regions and countries worldwide. In particular, the distribution of PKU variants across Europe was consistent with successive waves of historical migration, and Figure 5 summarizes the likely geographical routes of transmission of some variants within Europe. It was previously suggested that the concomitant excess of (unaffected) PKU carriers is at least in part the result of over-dominant selection (“heterozygous advantage”).37
For example, Germany has occupied a “crossroads” location during several migration waves throughout history and displays a broad spectrum of PAH variants, with similarities to some other populations, such as northern European countries (Denmark, Sweden, and Norway).35 This implies a genetic connection of these regions, possibly during the Germanic settlement of Scandinavia. Norway’s most frequent variant, p.Gly46Ser, was not observed in the German population. Furthermore, the occurrence of c.1066−11G>A in Germany was surprisingly high in our analysis, which might be explained by immigration from Turkey. This variant, often described as the “Mediterranean mutation,” may be of Italian origin38 and has been found mainly in Southern European countries or the Middle East.39,40 Our study showed that c.1066−11G>A has the highest AF in Middle Eastern countries. A typical east-west gradient can be seen, originating from Western Asia and the Middle East, i.e., from Armenia (48%) via Turkey, Iran, and Israel to Spain.
The p.Arg408Trp-associated genotype, the most common variant in people with Slavic roots, followed the east-central-southwest European axis, starting from Estonia and Russia, with the highest number of severe PKU-affected individuals, via Poland, Czechia, Slovakia, Germany, France, and Italy, down to Spain. The high prevalence of cPKU in Russia, and the dominance of the severe variant, p.Arg408Trp, were striking, given that Russia has a diverse ethnic composition, with as many as 160 ethnic groups (see Russian Federation Population Data in Web Resources).
Another well-documented axis exists for the p.Tyr414Cys-associated genotype, from Northern to Western Europe, i.e., from Sweden, Norway, and Denmark via Germany to the Netherlands. Some variants, e.g., c.1315+1G>A, seem to have spread within Scandinavian countries at high frequency. The same was true for the p.Glu390Gly-associated genotype concentrating between Slovenia, Croatia, Serbia, and Austria. A trend was evident for very mild p.Ala300Ser-associated genotypes, frequent in Turkey and Israel, Italy to Spain. No trends for the distribution of p.Val245Ala and p.Ala403Val across Europe could be seen.
The influence of migration patterns during history can also be seen in the heterogenous spectrum of PAH variants in Latin America. These countries were strongly marked by immigration of Europeans, especially from Southern Europe, during colonial times.41 An example is p.Val388Met, which was especially prevalent in Brazil and Chile, once colonies of Portugal and Spain. In Argentina variants frequently occurring in Eastern, Central, and Northern Europe, e.g., p.Arg408Trp, p.Arg261Gln, and c.1066−11G>A, were more prevalent. In contrast to Brazil or Argentina, the Mexican population has a high proportion of indigenous individuals, which could be a reason for the exceptional high AF of the c.60+5G>T splice variant there.42
The USA has one of the most multi-racial and ethnic populations in the world. The strongest ancestral influence is European, mainly from Germany, Ireland, England, Italy, and France (see US Census Bureau in Web Resources), which explains a comparable distribution of PKU variants in the USA and Europe.43 African Americans have a much lower PKU incidence than white Americans.44 It would be of interest to follow up this study with regard to the different rates of growth of major ethnic groups (Europeans, African American, Hispanic, Asian).
While the overall PKU prevalence in China was 1:15,924, its distribution across the country varies significantly with higher rates in the north in comparison to the south.45 In accordance with previous reports, p.Arg234Gln accounts for 23% of all variants in China.46 Three additional variants p.Arg243Gln, c.611A>G, and p.Arg241Cys (AF > 5%) were also common in Korea and Taiwan, indicating commonalities of migration movements and evolution among those nationalities. Prevalent PKU variants for Europe, Middle East, Latin America, and the USA were uncommon in the Asian populations.
Despite immense progress in the diagnosis and treatment of PKU in the last decade, there are still too many areas of the world without adequate access to this care starting with newborn screening. In particular, data from Africa and certain Asian and South American as well as Caribbean countries were missing.
Preliminary data indicated the great value of large patient databases with genotype and phenotype information and the introduction of the APV and GPV for the genotypic phenotype prediction.15 This study extends these previous observations with additional information and has confirmed the power of genotyping in prediction of phenotype and BH4 responsiveness in PKU, thus offering a powerful tool of personalized medicine for this inherited metabolic disease.
This study also confirms that functionally mild variants (in a compound heterozygous constellation), with a substantial residual PAH activity are always dominant over inactive severe variants (null alleles) and give rise to a milder phenotype and often potential BH4 responsiveness. Compared with severe classic PKU variants, milder variants were much rarer in number but determined the milder metabolic phenotype. Interestingly, homozygous mild variants have a higher APV and thus a milder phenotype when occurring in a compound heterozygous state with a null allele.15 As a rule, two inactive severe variants are never BH4 responsive, although anecdotal reports sometimes suggest otherwise.47
In conclusion, this study provides an overview of the current distribution of the most important PAH variants and patient genotypes in various world regions. This information, together with the APV value, helps to predict the metabolic phenotype and the possible treatment options for PKU subjects.
Declaration of interests
N.B. has received honoraria and/or consulting fees from BioMarin Pharmaceuticals, Censa, Nestle Pharmaceuticals, and Homology Medicines. A.B. has received advisory board honoraria, speaker fees, and travel support from Biomarin Pharmaceuticals, Nutricia, Cambrooke, PIAM, APR, Sanofi Genzyme, and Takeda. B.K.B. has received honoraria and/or consulting fees from BioMarin, Shire (a Takeda company), Sanofi Genzyme, Ultragenyx, Alexion, Horizon, Denali, JCR Pharma, Regenxbio, Inventiva, Chiesi, Homology Medicines, Aeglea, Agios, and Moderna. G.F.H. received lecture fees from Takeda. J.V. received research funding from Biomarin Pharmaceuticals, Homology Pharmaceuticals, American Gene Technologies, Nestle Pharmaceuticals, Rubius Pharmaceuticals, and Synlogic Pharmaceuticals.
All other authors have no conflicts of interests and nothing to disclose.
Acknowledgments
This article is dedicated in honor of Dr. Charles Scriver, the incomparable physician scientist, whose contributions to PKU and all metabolic disease have taught us the real meaning of medical research.
We are indebted to Suha Daas, who works at the National Newborn screening Program, Ministry of Health, Tel Hashomer Israel for her help and analyses. Farrah Rajabi, Fran Rohr, Ann Wessel, Leslie Martell, Steven Dobrowolski, Per Guldberg, and the late Flemming Guttler were greatly responsible for the data from New England. This work was funded in part by the Fundación Isabel Gemio-Fundación La Caixa (LCF/PR/PR16/11110018), the Regional Government of Madrid (CAM, B2017/BMD3721), and by NIH, United States grants R01DK117916 and R01NR016991. The research conducted at the Murdoch Children’s Research Institute was supported by the Victorian Government’s Operational Infrastructure Support Program. The work of G.F.H. on newborn screening has been generously supported by the Dietmar Hopp Foundation, St. Leon-Rot.
A medical writer (Dr. Mike Gwilt, GT Communications, UK) edited the authors’ manuscript draft for conciseness, at the request of the Corresponding Author and funded by the Corresponding Author’s institution.
Published: July 14, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.06.006.
Contributor Information
Sven F. Garbade, Email: sven.garbade@med.uni-heidelberg.de.
Nenad Blau, Email: nenad.blau@kispi.uzh.ch.
Data and Code Availability
The PAH variants dataset generated during this study is deposited in public PAHvdb repository at http://www.biopku.org/home/pah.asp. The PAH genotypes dataset generated during this study is deposited in public BIOPKU repository at http://www.biopku.org/home/biopku.asp. Specific export data can be obtained on request at the corresponding author.
Web Resources
Human Gene Mutation Database, http://www.hgmd.cf.ac.uk/ac/index.php
HGVS nomenclature recommendations, http://www.hgvs.org/content/guidelines
LOVD, http://www.lovd.nl/
Mutalyzer, https://mutalyzer.nl/index
OMIM, https://www.omim.org/
Russian Federation Embassy to UK, Population Data, https://www.rusemb.org.uk/russianpopulation
United Nations – World Population Prospects – Population Division, https://population.un.org/wpp/
United States Census Bureau, Overview of Race and Hispanic Origin: 2010, https://www.census.gov/library/publications/2011/dec/c2010br-02.html
Supplemental Data
References
- 1.Blau N., van Spronsen F.J., Levy H.L. Phenylketonuria. Lancet. 2010;376:1417–1427. doi: 10.1016/S0140-6736(10)60961-0. [DOI] [PubMed] [Google Scholar]
- 2.Flydal M.I., Martinez A. Phenylalanine hydroxylase: function, structure, and regulation. IUBMB Life. 2013;65:341–349. doi: 10.1002/iub.1150. [DOI] [PubMed] [Google Scholar]
- 3.Himmelreich N., Shen N., Okun J.G., Thiel C., Hoffmann G.F., Blau N. Relationship between genotype, phenylalanine hydroxylase expression and in vitro activity and metabolic phenotype in phenylketonuria. Mol. Genet. Metab. 2018;125:86–95. doi: 10.1016/j.ymgme.2018.06.011. [DOI] [PubMed] [Google Scholar]
- 4.Güttler F. Hyperphenylalaninemia: diagnosis and classification of the various types of phenylalanine hydroxylase deficiency in childhood. Acta Paediatr. Scand. Suppl. 1980;280:1–80. [PubMed] [Google Scholar]
- 5.Camp K.M., Parisi M.A., Acosta P.B., Berry G.T., Bilder D.A., Blau N., Bodamer O.A., Brosco J.P., Brown C.S., Burlina A.B. Phenylketonuria Scientific Review Conference: state of the science and future research needs. Mol. Genet. Metab. 2014;112:87–122. doi: 10.1016/j.ymgme.2014.02.013. [DOI] [PubMed] [Google Scholar]
- 6.van Spronsen F.J., van Wegberg A.M., Ahring K., Bélanger-Quintana A., Blau N., Bosch A.M., Burlina A., Campistol J., Feillet F., Giżewska M. Key European guidelines for the diagnosis and management of patients with phenylketonuria. Lancet Diabetes Endocrinol. 2017;5:743–756. doi: 10.1016/S2213-8587(16)30320-5. [DOI] [PubMed] [Google Scholar]
- 7.Vockley J., Andersson H.C., Antshel K.M., Braverman N.E., Burton B.K., Frazier D.M., Mitchell J., Smith W.E., Thompson B.H., Berry S.A., American College of Medical Genetics and Genomics Therapeutics Committee Phenylalanine hydroxylase deficiency: diagnosis and management guideline. Genet. Med. 2014;16:188–200. doi: 10.1038/gim.2013.157. [DOI] [PubMed] [Google Scholar]
- 8.Blau N., Longo N. Alternative therapies to address the unmet medical needs of patients with phenylketonuria. Expert Opin. Pharmacother. 2015;16:791–800. doi: 10.1517/14656566.2015.1013030. [DOI] [PubMed] [Google Scholar]
- 9.Scriver C.R. The PAH gene, phenylketonuria, and a paradigm shift. Hum. Mutat. 2007;28:831–845. doi: 10.1002/humu.20526. [DOI] [PubMed] [Google Scholar]
- 10.Gundorova P., Zinchenko R.A., Kuznetsova I.A., Bliznetz E.A., Stepanova A.A., Polyakov A.V. Molecular-genetic causes for the high frequency of phenylketonuria in the population from the North Caucasus. PLoS ONE. 2018;13:e0201489. doi: 10.1371/journal.pone.0201489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guldberg P., Henriksen K.F., Sipilä I., Güttler F., de la Chapelle A. Phenylketonuria in a low incidence population: molecular characterisation of mutations in Finland. J. Med. Genet. 1995;32:976–978. doi: 10.1136/jmg.32.12.976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okano Y., Kudo S., Nishi Y., Sakaguchi T., Aso K. Molecular characterization of phenylketonuria and tetrahydrobiopterin-responsive phenylalanine hydroxylase deficiency in Japan. J. Hum. Genet. 2011;56:306–312. doi: 10.1038/jhg.2011.10. [DOI] [PubMed] [Google Scholar]
- 13.Güttler F., Guldberg P., Henriksen K.F., Mikkelsen I., Olsen B., Lou H. Molecular basis for the phenotypical diversity of phenylketonuria and related hyperphenylalaninaemias. J. Inherit. Metab. Dis. 1993;16:602–604. doi: 10.1007/BF00711693. [DOI] [PubMed] [Google Scholar]
- 14.Blau N., Shen N., Carducci C. Molecular genetics and diagnosis of phenylketonuria: state of the art. Expert Rev. Mol. Diagn. 2014;14:655–671. doi: 10.1586/14737159.2014.923760. [DOI] [PubMed] [Google Scholar]
- 15.Garbade S.F., Shen N., Himmelreich N., Haas D., Trefz F.K., Hoffmann G.F., Burgard P., Blau N. Allelic phenotype values: a model for genotype-based phenotype prediction in phenylketonuria. Genet. Med. 2019;21:580–590. doi: 10.1038/s41436-018-0081-x. [DOI] [PubMed] [Google Scholar]
- 16.Zschocke J. Dominant versus recessive: molecular mechanisms in metabolic disease. J. Inherit. Metab. Dis. 2008;31:599–618. doi: 10.1007/s10545-008-1016-5. [DOI] [PubMed] [Google Scholar]
- 17.Scriver C.R., Waters P.J. Monogenic traits are not simple: lessons from phenylketonuria. Trends Genet. 1999;15:267–272. doi: 10.1016/s0168-9525(99)01761-8. [DOI] [PubMed] [Google Scholar]
- 18.Shen N., Heintz C., Thiel C., Okun J.G., Hoffmann G.F., Blau N. Co-expression of phenylalanine hydroxylase variants and effects of interallelic complementation on in vitro enzyme activity and genotype-phenotype correlation. Mol. Genet. Metab. 2016;117:328–335. doi: 10.1016/j.ymgme.2016.01.004. [DOI] [PubMed] [Google Scholar]
- 19.Evers R.A.F., van Wegberg A.M.J., Anjema K., Lubout C.M.A., van Dam E., van Vliet D., Blau N., van Spronsen F.J. The first European guidelines on phenylketonuria: Usefulness and implications for BH4 responsiveness testing. J. Inherit. Metab. Dis. 2020;43:244–250. doi: 10.1002/jimd.12173. [DOI] [PubMed] [Google Scholar]
- 20.Muntau A.C., Adams D.J., Bélanger-Quintana A., Bushueva T.V., Cerone R., Chien Y.H., Chiesa A., Coşkun T., de Las Heras J., Feillet F. International best practice for the evaluation of responsiveness to sapropterin dihydrochloride in patients with phenylketonuria. Mol. Genet. Metab. 2019;127:1–11. doi: 10.1016/j.ymgme.2019.04.004. [DOI] [PubMed] [Google Scholar]
- 21.Konecki D.S., Schlotter M., Trefz F.K., Lichter-Konecki U. The identification of two mis-sense mutations at the PAH gene locus in a Turkish patient with phenylketonuria. Hum. Genet. 1991;87:389–393. doi: 10.1007/BF00197153. [DOI] [PubMed] [Google Scholar]
- 22.Ellingsen S., Knappskog P.M., Eiken H.G. Phenylketonuria splice mutation (EXON6nt-96A-->g) masquerading as missense mutation (Y204C) Hum. Mutat. 1997;9:88–90. doi: 10.1002/(SICI)1098-1004(1997)9:1<88::AID-HUMU21>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 23.Matalon R., Michals K. Phenylketonuria: screening, treatment and maternal PKU. Clin. Biochem. 1991;24:337–342. doi: 10.1016/0009-9120(91)80008-q. [DOI] [PubMed] [Google Scholar]
- 24.El-Metwally A., Yousef Al-Ahaidib L., Ayman Sunqurah A., Al-Surimi K., Househ M., Alshehri A., Da’ar O.B., Abdul Razzak H., AlOdaib A.N. The prevalence of phenylketonuria in Arab countries, Turkey, and Iran: A systematic review. BioMed Res. Int. 2018;2018:7697210. doi: 10.1155/2018/7697210. 10.1155/2018/7697210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shoraka H.R., Haghdoost A.A., Baneshi M.R., Bagherinezhad Z., Zolala F. Global prevalence of classic phenylketonuria based on Neonatal Screening Program Data: systematic review and meta-analysis. Clin Exp Pediatr. 2020;63:34–43. doi: 10.3345/kjp.2019.00465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saadallah A.A., Rashed M.S. Newborn screening: experiences in the Middle East and North Africa. J. Inherit. Metab. Dis. 2007;30:482–489. doi: 10.1007/s10545-007-0660-5. [DOI] [PubMed] [Google Scholar]
- 27.Zschocke J. Phenylketonuria mutations in Europe. Hum. Mutat. 2003;21:345–356. doi: 10.1002/humu.10192. [DOI] [PubMed] [Google Scholar]
- 28.Danecka M.K., Woidy M., Zschocke J., Feillet F., Muntau A.C., Gersting S.W. Mapping the functional landscape of frequent phenylalanine hydroxylase (PAH) genotypes promotes personalised medicine in phenylketonuria. J. Med. Genet. 2015;52:175–185. doi: 10.1136/jmedgenet-2014-102621. [DOI] [PubMed] [Google Scholar]
- 29.Gundorova P., Stepanova A.A., Kuznetsova I.A., Kutsev S.I., Polyakov A.V. Genotypes of 2579 patients with phenylketonuria reveal a high rate of BH4 non-responders in Russia. PLoS ONE. 2019;14:e0211048. doi: 10.1371/journal.pone.0211048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lillevali H., Reinson K., Muru K., Simenson K., Murumets U., Mols T., Ounap K. Hyperphenylalaninaemias in Estonia: Genotype-Phenotype Correlation and Comparative Overview of the Patient Cohort Before and After Nation-Wide Neonatal Screening. JIMD Rep. 2018;40:39–45. doi: 10.1007/8904_2017_61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bik-Multanowski M., Kaluzny L., Mozrzymas R., Oltarzewski M., Starostecka E., Lange A., Didycz B., Gizewska M., Ulewicz-Filipowicz J., Chrobot A. Molecular genetics of PKU in Poland and potential impact of mutations on BH4 responsiveness. Acta Biochim. Pol. 2013;60:613–616. [PubMed] [Google Scholar]
- 32.Kádasi L., Poláková H., Feráková E., Hudecová S., Bohusová T., Szomolayová I., Strnová J., Hruskovic I., Moschonas N.K., Ferák V. PKU in Slovakia: mutation screening and haplotype analysis. Hum. Genet. 1995;95:112–114. doi: 10.1007/BF00225087. [DOI] [PubMed] [Google Scholar]
- 33.Réblová K., Hrubá Z., Procházková D., Pazdírková R., Pouchlá S., Zeman J., Fajkusová L. Hyperphenylalaninemia in the Czech Republic: genotype-phenotype correlations and in silico analysis of novel missense mutations. Clin. Chim. Acta. 2013;419:1–10. doi: 10.1016/j.cca.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 34.Sterl E., Paul K., Paschke E., Zschocke J., Brunner-Krainz M., Windisch E., Konstantopoulou V., Möslinger D., Karall D., Scholl-Bürgi S. Prevalence of tetrahydrobiopterine (BH4)-responsive alleles among Austrian patients with PAH deficiency: comprehensive results from molecular analysis in 147 patients. J. Inherit. Metab. Dis. 2013;36:7–13. doi: 10.1007/s10545-012-9485-y. [DOI] [PubMed] [Google Scholar]
- 35.Zschocke J., Hoffmann G.F. Phenylketonuria mutations in Germany. Hum. Genet. 1999;104:390–398. doi: 10.1007/s004390050973. [DOI] [PubMed] [Google Scholar]
- 36.Zschocke J., Mallory J.P., Eiken H.G., Nevin N.C. Phenylketonuria and the peoples of Northern Ireland. Hum. Genet. 1997;100:189–194. doi: 10.1007/s004390050488. [DOI] [PubMed] [Google Scholar]
- 37.Krawczak M., Zschocke J. A role for overdominant selection in phenylketonuria? Evidence from molecular data. Hum. Mutat. 2003;21:394–397. doi: 10.1002/humu.10205. [DOI] [PubMed] [Google Scholar]
- 38.Eisensmith R.C., Okano Y., Dasovich M., Wang T., Güttler F., Lou H., Guldberg P., Lichter-Konecki U., Konecki D.S., Svensson E. Multiple origins for phenylketonuria in Europe. Am. J. Hum. Genet. 1992;51:1355–1365. [PMC free article] [PubMed] [Google Scholar]
- 39.Okano Y., Wang T., Eisensmith R.C., Longhi R., Riva E., Giovannini M., Cerone R., Romano C., Woo S.L. Phenylketonuria missense mutations in the Mediterranean. Genomics. 1991;9:96–103. doi: 10.1016/0888-7543(91)90225-4. [DOI] [PubMed] [Google Scholar]
- 40.Kleiman S., Avigad S., Vanagaite L., Shmuelevitz A., David M., Eisensmith R.C., Brand N., Schwartz G., Rey F., Munnich A. Origins of hyperphenylalaninemia in Israel. Eur. J. Hum. Genet. 1994;2:24–34. doi: 10.1159/000472338. [DOI] [PubMed] [Google Scholar]
- 41.Desviat L.R., Pérez B., De Lucca M., Cornejo V., Schmidt B., Ugarte M. Evidence in Latin America of recurrence of V388M, a phenylketonuria mutation with high in vitro residual activity. Am. J. Hum. Genet. 1995;57:337–342. [PMC free article] [PubMed] [Google Scholar]
- 42.Vela-Amieva M., Abreu-González M., González-del Angel A., Ibarra-González I., Fernández-Lainez C., Barrientos-Ríos R., Monroy-Santoyo S., Guillén-López S., Alcántara-Ortigoza M.A. Phenylalanine hydroxylase deficiency in Mexico: genotype-phenotype correlations, BH4 responsiveness and evidence of a founder effect. Clin. Genet. 2015;88:62–67. doi: 10.1111/cge.12444. [DOI] [PubMed] [Google Scholar]
- 43.Rajabi F., Rohr F., Wessel A., Martell L., Dobrowolski S.F., Guldberg P., Güttler F., Levy H.L. Phenylalanine hydroxylase genotype-phenotype associations in the United States: A single center study. Mol. Genet. Metab. 2019;128:415–421. doi: 10.1016/j.ymgme.2019.09.004. [DOI] [PubMed] [Google Scholar]
- 44.Hofman K.J., Steel G., Kazazian H.H., Valle D. Phenylketonuria in U.S. blacks: molecular analysis of the phenylalanine hydroxylase gene. Am. J. Hum. Genet. 1991;48:791–798. [PMC free article] [PubMed] [Google Scholar]
- 45.Xiang L., Tao J., Deng K., Li X., Li Q., Yuan X., Liang J., Yu E., Wang M., Wang H. Phenylketonuria incidence in China between 2013 and 2017 based on data from the Chinese newborn screening information system: a descriptive study. BMJ Open. 2019;9:e031474. doi: 10.1136/bmjopen-2019-031474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li N., Jia H., Liu Z., Tao J., Chen S., Li X., Deng Y., Jin X., Song J., Zhang L. Molecular characterisation of phenylketonuria in a Chinese mainland population using next-generation sequencing. Sci. Rep. 2015;5:15769. doi: 10.1038/srep15769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hennermann J.B., Vetter B., Kulozik A.E., Mönch E. Partial und total tetrahydrobiopterin-responsiveness in classical and mild phenylketonuria (PKU) J. Inherit. Metab. Dis. 2002;25:21. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The PAH variants dataset generated during this study is deposited in public PAHvdb repository at http://www.biopku.org/home/pah.asp. The PAH genotypes dataset generated during this study is deposited in public BIOPKU repository at http://www.biopku.org/home/biopku.asp. Specific export data can be obtained on request at the corresponding author.