Abstract
Dominantly inherited disorders are not typically considered to be therapeutic candidates for gene augmentation. Here, we utilized induced pluripotent stem cell-derived retinal pigment epithelium (iPSC-RPE) to test the potential of gene augmentation to treat Best disease, a dominant macular dystrophy caused by over 200 missense mutations in BEST1. Gene augmentation in iPSC-RPE fully restored BEST1 calcium-activated chloride channel activity and improved rhodopsin degradation in an iPSC-RPE model of recessive bestrophinopathy as well as in two models of dominant Best disease caused by different mutations in regions encoding ion-binding domains. A third dominant Best disease iPSC-RPE model did not respond to gene augmentation, but showed normalization of BEST1 channel activity following CRISPR-Cas9 editing of the mutant allele. We then subjected all three dominant Best disease iPSC-RPE models to gene editing, which produced premature stop codons specifically within the mutant BEST1 alleles. Single-cell profiling demonstrated no adverse perturbation of retinal pigment epithelium (RPE) transcriptional programs in any model, although off-target analysis detected a silent genomic alteration in one model. These results suggest that gene augmentation is a viable first-line approach for some individuals with dominant Best disease and that non-responders are candidates for alternate approaches such as gene editing. However, testing gene editing strategies for on-target efficiency and off-target events using personalized iPSC-RPE model systems is warranted. In summary, personalized iPSC-RPE models can be used to select among a growing list of gene therapy options to maximize safety and efficacy while minimizing time and cost. Similar scenarios likely exist for other genotypically diverse channelopathies, expanding the therapeutic landscape for affected individuals.
Keywords: human pluripotent stem cells, gene therapy, gene augmentation, autosomal dominant disease, macular degeneration, orphan disease, channelopathy, somatic cell gene editing, CRISPR-Cas9
Introduction
Genotypically heterogeneous dominant diseases pose significant challenges and opportunities for precision medicine.1 Among gene therapies, gene augmentation for recessive disorders is the most developed, having spurred multiple clinical trials2, 3, 4 and gained FDA approval for one ocular disease.5 However, gene augmentation is generally ruled out as a stand-alone therapy for dominant disorders due to a perceived need to eliminate the deleterious effects of the mutant allele. Gene editing approaches to silence or repair mutant alleles hold promise in this regard,6, 7, 8 but testing safety and efficacy for every mutant-allele-specific genome editor presents practical and economic challenges in diseases with high mutational diversity. Further, gene editing may not be able to target all mutations6,9,10 and could lead to off-target mutagenesis—particularly within a wild-type (WT) allele—or other adverse events.11 Another consideration for gene therapy development is the need for preclinical model systems with phenotypes and/or genotypes that are relevant to the human disease. This requirement is particularly challenging for genome editing strategies, which utilize sequence-specific tools and thus require human model systems to test safety and efficacy.12 Humanized animal models have also been employed for this purpose,13 although they provide limited information on genome-wide off-target analysis.
One disorder that faces a full array of these therapeutic obstacles is Best disease (MIM: 153700), a major cause of inherited macular degeneration that currently has no treatment options. Best disease exclusively targets the retinal pigment epithelium (RPE), a monolayer of cells essential for the survival and function of photoreceptors. Although Best disease is often diagnosed in early childhood based on its distinctive ophthalmological findings,14 its effects on central vision are generally mild at first. Vision loss occurs progressively and irreversibly over several decades, thus providing a wide time window for therapeutic intervention.
Best disease is a genotypically diverse disorder transmitted primarily in an autosomal dominant fashion, although rare cases of autosomal recessive bestrophinopathy (ARB; MIM: 611809) are known.15 Together, autosomal dominant Best disease (adBD) and ARB are linked to over 200 mutations in BEST1 (MIM: 607854), which encodes a putative homo-pentameric calcium-activated chloride channel (CaCC) found in the RPE. Recent elucidation of the high-resolution crystal structure of chicken Best1 reinforced its role as a CaCC and revealed that disease-associated mutations cluster within regions encoding calcium or chloride ion-binding sites or within structural regions of the channel.16
A significant impediment to the development of therapies for adBD is the lack of model systems that adequately mimic the genotypic and phenotypic characteristics of the disorder. While canine models of ARB mirror the human ARB phenotype,14 no suitable animal models of adBD exist. To provide a therapeutic testing platform for adBD, we previously developed the first human pluripotent stem cell-derived retinal pigment epithelium (iPSC-RPE) models of the disease, which demonstrated relevant cellular dysfunction—most notably, delayed degradation of phagocytosed photoreceptor outer segment (POS) proteins.17,18 These adBD iPSC-RPE models were then used to test the potential for selected pharmacological interventions to ameliorate the cellular phenotype of this disorder.18
In the present study, we examined whether gene therapy could definitively correct the functional defects present in adBD iPSC-RPE. Given that BEST1 forms a homo-pentameric CaCC, we hypothesized that gene augmentation could potentially mitigate the cellular disease phenotype in adBD by increasing the ratio of WT to mutant BEST1 monomers available for channel assembly. This theory presumes that the deleterious effects of the mutant allele can be diluted sufficiently to restore CaCC function, preferably in a controlled manner without the risks associated with unregulated transgene expression.
To test our hypothesis, we employed three iPSC-RPE models of adBD, along with one iPSC-RPE model of ARB as a control. Importantly, the iPSC lines were generated from individuals with BEST1 mutations affecting different functional regions of the channel (i.e., calcium-binding, chloride-binding, and structural).16 We then ectopically expressed WT BEST1 in iPSC-RPE using a viral vector that incorporated the native BEST1 promoter, VMD2, in order to maintain RPE specificity and to keep transgene expression levels in check. Using this strategy, we obtained a >3-fold increase in WT BEST1 levels across all adBD iPSC-RPE models. Single-cell electrophysiology and cell-population-based assays revealed that two of the adBD mutations were exceedingly responsive to gene augmentation alone. Indeed, the correction of the cellular disease phenotype observed in these adBD iPSC-RPE models following gene augmentation was on par with that seen in the ARB iPSC-RPE model.
To address the adBD mutation that failed to respond to gene augmentation, as well as others that may also be refractory to this broad therapeutic strategy, we examined whether CRISPR-Cas9 gene editing could specifically target the mutant BEST1 allele, leaving the normal WT allele intact. We found that gene editing was highly efficient at modifying the mutant allele and restoring iPSC-RPE CaCC activity in all three adBD models. These results bode well for the use of CRISPR-Cas9 to treat adBD mutations that are not candidates for gene augmentation, contingent on the availability of suitable guide RNAs. We then investigated whether gene editing caused untoward effects on the RPE transcriptome or induced off-target genome alterations in any of the adBD models. While no transcriptomic perturbations were detected, a single significant—albeit functionally silent—off-target site contained genomic insertions and deletion mutations (indels) in one adBD model.
Based on our findings, we propose a two-tiered approach to adBD gene therapy that uses iPSC-RPE testing to first determine which mutations are likely to respond to frontline treatment with gene augmentation. BEST1 mutant iPSC-RPE models that do not demonstrate phenotypic correction with gene augmentation would then undergo next-level safety and efficacy testing to assess candidacy for customized gene editing.
Material and Methods
Ethical Guidance and Human Subjects
All work with iPSC lines was carried out in accordance with institutional, national, and international guidelines, and were approved by the Institutional Review Board and Stem Cell Research Oversight Committee at the University of Wisconsin—Madison. Blood samples from affected individuals were collected with informed written consent in accordance with the Declaration of Helsinki and approval from the Institutional Review Board at the University of Wisconsin—Madison or the University of Iowa. This manuscript does not contain data from animal studies or from clinical studies using human subjects.
iPSC Lines
A total of six iPSC lines, two control and four harboring mutations in BEST1, were used in this study. In addition to one control iPSC line (normal) and two iPSC lines from individuals with adBD mutations (RefSeq: NM_004183.4, c.436_437delinsAA [p.Ala146Lys] and RefSeq: NM_004183.4, c.886A>C [p.Asn296His]) previously used by our group for Best disease modeling,17 we used three new iPSC lines. Two of the new iPSC lines harbored the following pathogenic mutations in BEST1: c.652C>T (p.Arg218Cys) (adBD) (RefSeq: NM_004183.4) and c.[422G>A];[584C>T] (p.Arg141His; p.Ala195Val) (ARB) (RefSeq: NM_004183.4). The third new line was an isogenic control iPSC line generated by CRISPR/Cas9-based gene correction of the Arg218Cys adBD iPSC line.19 For the Arg218Cys adBD and ARB iPSC lines, peripheral blood mononuclear cells were reprogrammed using non-integrating episomal vectors to deliver reprogramming factors using methods similar to those previously published.20 The Arg218Cys adBD iPSC line was generated by Fujifilm Cellular Dynamics International, and Cytotune 2.0 (ThermoFisher Scientific; Cat# A16518) was used to reprogram the ARB iPSC line. All iPSC lines were cultured either on mouse embryonic fibroblasts (MEFs) or on Matrigel prior to differentiation. Lines cultured on MEFs were maintained using iPSC media (Dulbecco’s Modified Eagle’s Medium [DMEM]/F12 [1:1], 20% Knockout Serum Replacement [KOSR], 1% MEM non-essential amino acids, 1% L-glutamine, 0.2 mM β-mercaptoethanol, 100 ng/mL fibroblast growth factor 2 [FGF-2]), and iPSCs cultured on Matrigel were cultured with either mTeSR1 or StemFlex media. MEFs, FGF-2, and Matrigel were purchased from WiCell. All other cell culture reagents were purchased from ThermoFisher Scientific. Karyotype analysis was performed as a quality control.
Differentiation of iPSC Lines to RPE
Differentiation of iPSCs to RPE was performed as previously described.17,21 In brief, iPSCs were enzymatically lifted (1 mg/mL dispase for cells cultured on MEFs; 2 mg/mL dispase or 1 mL ReLeSR for cells cultured on Matrigel) to form aggregates, also referred to as embryoid bodies (EBs). EBs were maintained in suspension culture either in EB media (iPS media without FGF-2) and then switched to neural induction media (NIM) on day 4, or gradually weaned off mTeSR1/StemFlex and transitioned to NIM by day 4. NIM is composed of 500 mL DMEM/F12 (1:1), 1% N2 supplement, 1% MEM non-essential amino acids, 1% L-glutamine, 2 μg/mL heparin. EBs were plated on laminin (Cat# 23017015) coated 6-well plates (Nunc; Thermo Fisher Scientific) on day 7. On day 16, neural rosettes were mechanically lifted, leaving adherent cells behind that were maintained in retinal differentiation media (RDM; DMEM:F12 (3:1), 2% B27 without retinoic acid, 1% antibiotic-antimycotic solution). For the first four media changes, RDM was supplemented with 10 μM SU5402 and 3 μM CHIR99021.
After 60 days of differentiation, pigmented patches of RPE were micro-dissected, dissociated using Trypsin-EDTA (0.25%), and plated on laminin-coated surfaces in RDM with 10% fetal bovine serum (FBS) and Rho kinase inhibitor (ROCKi; Y-27632). After 2 days, the media was changed to RDM with 2% FBS, and eventually to RDM once the cells were fully confluent. There were no differences observed between RPE differentiated from iPSCs cultured on MEFs and and those cultured on Matrigel. Mutant and WT genotypes of iPSC-RPE were verified via Sanger sequencing periodically. Heparin (Cat# H-3149) and SU5402 (Cat# SML0443-25MG) were from Sigma-Aldrich, CHIR99021 (Cat# 4423) was from Tocris Bioscience, and ReLeSR was purchased from STEMCELL Technologies. All other differentiation reagents were purchased from ThermoFisher Scientific.
Gene Expression Analysis
Reverse transcriptase-PCR was used to assess RPE-specific gene expression in RPE derived from different iPSC lines, as described previously.17 Primers used are listed in Table S1.
Generation of Lentiviral Vectors
Lentiviral plasmid with the human VMD2 promoter driving expression of BEST1-T2A-GFP was provided by Alfred S. Lewin (University of Florida). LentiCRISPR v2 (LCv2) plasmid was purchased from Addgene (Cat# 52961). Lentiviral gene editing plasmids containing specific sgRNA sequences and the human VMD2 promoter driving expression of spCas9-T2A-GFP were then generated as described hereafter (all primers and sgRNA sequences are listed in the Supplemental Tables). To begin, the “T2A-GFP-WPRE” sequence was amplified from the VMD2-BEST1-T2A-GFP plasmid using LCv2-GFP.Gib.F and LCv2-GFP.Gib.R primers and the Q5 High-Fidelity 2× Master Mix (Q5 2× MM, New England Biolabs [NEB], Cat# M0492L). The “2A-Puro-WPRE” sequence was then removed from the LCv2 plasmid via restriction digestion with PmeI (NEB, Cat# R0560S) and BamHI (NEB, Cat# R3136S). The digestion product was resolved on a 0.7% agarose gel and the plasmid backbone was purified using the Monarch gel purification kit (NEB, Cat# T1020S). The “T2A-GFP-WPRE” sequence was inserted into the digested backbone using the Gibson Assembly kit (Codex, Cat# GA1100) per the manufacturer’s instructions. The completed Gibson Assembly reaction was then amplified using chemically competent E. coli (NEB, Cat# C3040H) and Sanger sequenced to confirm insertion of “T2A-GFP-WPRE” using LCv2-GFP.seq.L and LCv2-GFP.seq.R primers. This intermediate plasmid product (pLCv2-GFP) was digested with AfeI (NEB, Cat# R0652S) and EcoRI-HF (NEB, Cat R310S) to remove the constitutive EF-1 alpha core promoter. The desired digestion product was purified as described above. The VMD2 promoter was then PCR amplified from VMD2-BEST1-T2A-GFP using Q5 2× MM and VMD2.LCv2.GFP.Gib.F and VMD2.LCv2.GFP.Gib.R primers, followed by insertion into the digested LCv2-GFP backbone via Gibson Assembly. Next, the completed Gibson reaction was transformed into chemically competent E. coli, and the sequence of the final product VMD2-spCas9-T2A-GFP was confirmed via Sanger sequencing using VMD2.LCv2.GFP.seq.L and VMD2.LCv2.GFP.seq.R primers. Subsequently, specific sgRNAs were cloned into VMD2-spCas9-T2A-GFP through the use of the restriction digest and Gibson Assembly protocol.
Lentivirus Production and Cell Transduction
Lentivirus stocks were generated by the Cell Culture Core of the University of Wisconsin (UW) Department of Dermatology Skin Disease Research Center. In brief, HEK293 cells cultured on 10 cm dishes were transfected with lentiviral plasmids—10 μg of sgRNA encoding lentiviral plasmid (VMD2-BEST1-T2A-GFP or VMD2-spCas9-T2A-GFP); 5 μg of psPax2 (Addgene, Cat# 12260), and 2 μg of pMD2.G (Addgene, Cat# 12259)—using Lipofectamine (ThermoFisher; Cat# 11668019). After 15 h, culture medium (DMEM with 10% FBS) was replaced with fresh media containing 1% Penicillin-Streptomycin. Media containing lentiviruses was collected the next day and viral titers were calculated using QuickTiter Lentivirus Titer Kit (Cell Biolabs, Cat# VPK-107). Titers for lentiviral stock are listed in Table S6.
For iPSC-RPE transduction, monolayers of iPSC-RPE on transwells were treated with Figure S3 150 μl of specified lentivirus preparation for all experiments. Media was changed on day 2 to RDM, and cells were maintained in culture with media changes every 3 days until used for sequencing or other analyses. Transduction efficiency of iPSC-RPE cultures was calculated by GFP+ cell counting of representative fluorescent images from all four mutant iPSC lines.
Transepithelial Electrical Resistance (TER) Measurements
Monolayers of RPE cultured on transwell inserts (Corning, #3470) were used for all TER measurements. To perform the measurements, we employed an epithelial voltohmmeter (EVOM2) with chopstick electrodes (STX2) from World Precision Instruments according to manufacturer’s instructions. Electrodes were sterilized with ethanol and then rinsed in sterile Milli-Q water followed by Hank’s balanced salt solution (HBSS) before measuring electrical resistance of RPE monolayers. Differences between TER values of transwells with cultured RPE monolayers versus background measurements of cell-free transwell inserts were multiplied by the surface area of the transwell membrane to obtain net TER values in Ω · cm2.
Calcium-Activated Chloride Channel Current Density Measurements
All iPSC-RPE cells used for chloride current measurements were cultured as a monolayer on transwells. To singularize cells prior to measurement, transwells were washed twice with 0 Na-CMF solution (135 mM N-Methyl-D-glucamine [NMDG]-Cl, 5 mM KCl, 10 mM HEPES, 10 mM glucose, 2 mM EDTA-KOH, pH adjusted to 7.4) and then incubated with papain enzyme solution (0 Na-CMF solution containing 2.5 μl/mL papain [46 mg/mL, MP Biomedicals LLC, Cat#100921], 0.375 mg/mL adenosine, 0.3mg/mL L-cysteine, 0.25 mg/mL L- glutathione, and 0.05mg/ ml taurine) for 30 min at 37°C/5% CO2. To stop the reaction, 0.01% BSA was added to the enzymatic solution. After washing twice with 0 Na-CMF solution, cells were dispersed in an extracellular solution containing 140 mM NaCl, 10 mM HEPES, 3 mM KCl, 2 mM CaCl2, 2 mM MgCl2, and 5.5 mM glucose adjusted to pH 7.4 with NaOH by gentle pipetting.
Cells with polarized RPE morphology post-dissociation were used to measure chloride currents. To test effects of gene augmentation or gene editing on BEST1 mutant iPSC-RPE via single-cell patch clamp analysis, only cells with GFP fluorescence (from transduction with VMD2-BEST1-T2A-GFP for gene augmentation or VMD2-spCas9-T2A-GFP encoding AAVS1 sgRNA or mutant allele-targeted sgRNAs for gene editing) were used. Current recordings on these cells were performed using the conventional whole-cell patch clamp technique with an Axopatch 200A amplifier controlled by the Clampex software program via the digidata 1550 data acquisition system (Axon Instruments). Fire-polished borosilicate glass pipettes with 3–5 MΩ resistance were filled with pipette solution containing 4.5 μM calcium or no calcium.
Recordings were carried out at room temperature, and current-voltage tracings were established using ramps from −100 to +100 mV for 1000 ms. The pipette solution with calcium was comprised of (in mM) 146 CsCl, 5 (Ca2+)-EGTA-NMDG, 2 MgCl2, 8 HEPES, and 10 sucrose at pH 7.3, adjusted with NMDG. Another pipette solution devoid of calcium was comprised of (in mM) 146 CsCl, 5 EGTA-NMDG, 2 MgCl2, 8 HEPES, and 10 Sucrose at pH 7.3, adjusted with NMDG. Both of these pipette solutions were mixed to make the solution containing 4.5 μM free calcium as described previously,22 which was then used for patch clamping.
Current density values were obtained by dividing current amplitude by cell capacitance measurements. CaCC current densities for iPSC-RPE are represented as differences between mean 4.5 μM calcium response and mean no calcium response from a total of at least five cells for each condition. At least two differentiations were used as replicates to obtain data for each line.
Immunocytochemistry
iPSC-RPE cultured on transwell inserts were washed with PBS and fixed with 4% paraformaldehyde for 10 min at room temperature (RT). After washing fixed cells three times with PBS, we placed transwell membranes in blocking solution (10% normal donkey serum with 5% BSA, 1% fish gelatin and 0.5% Triton X-100 in PBS) for 1 h at RT, and then incubated overnight at 4°C in primary antibody (1:100 mouse anti-Bestrophin [Millipore, Cat# MAB5466]; 1:100 rabbit anti-ZO-1 [ThermoFisher Scientific, Cat# 61-7300]) prepared in blocking solution. Cells were then washed three times in PBS and incubated for 30 min at RT in appropriate secondary antibody (ThermoFisher Scientific; 1:500 Donkey anti-Mouse IgG [Cat# A31571]; 1:500 Donkey anti-Rabbit Immunoglobulin G [IgG] [Cat# A10040]) prepared in blocking solution. Cells were again washed three times in PBS, incubated in 4′,6-diamidino-2-phenylindole (DAPI; 1:500; ThermoFisher; Cat# D1306) for 30 min, mounted using prolong gold with DAPI (ThermoFisher; Cat# P36931), and imaged using a Nikon A1R confocal microscope with NIS Elements AR 5.0 software.
Rhodopsin Degradation Assay
POS feeding of iPSC-RPE was performed as described previously.17 In brief, bovine POSs (InVision BioResources) were gently resuspended in DMEM. 100 μl media was then removed from each transwell insert, 6.25 × 106 POS were added, and cells were incubated at 37°C and 5% CO2 for 2 h. Afterward, POS containing RDM was removed, and each transwell was washed thoroughly three times using Dulbecco's phosphate-buffered saline (DPBS). Following the washes, cells were harvested (0 time point) or further incubated in fresh RDM for prescribed periods of time. At each time point, transwells were washed, 100 μl radioimmunoprecipitation assay (RIPA) buffer (ThermoFisher; Cat# 89900) containing protease inhibitor cocktail (Sigma-Aldrich; Cat# P8340) was added, and cells were incubated on ice for 30 min to extract total cell protein. Protein quantification was performed using the DC Protein assay kit II (Bio-Rad, Cat# 5000112).
Immunoblots were then performed to monitor rhodopsin degradation as described.17,18 In brief, protein lysates were denatured in 1× Laemmli buffer (reducing) and kept on ice for 10 min. Protein samples were then separated on 4%–20% mini-Protean TGX (Tris-Glycine eXtended) gels (Bio-Rad; Cat# 4568095) and electroblotted onto polyvinylidene fluoride (PVDF) membranes (Millipore; IPFL10100). After blotting, membranes were dried at RT for 15 min, re-activated in methanol for 1 min, and then incubated in blocking buffer (1:1 Odyssey blocking buffer [LI-COR Biosciences; Cat# 927-40000]:PBS) for 1 h. Post-blocking, blots were incubated in primary antibodies (1:500 mouse anti-rhodopsin [Millipore, Cat# MABN15]; 0.1 μg/mL rabbit anti-beta actin [Abcam, Cat# ab8227]) in blocking buffer with 0.1% Tween-20 overnight, washed three times for 5 min each in PBS with 0.1% Tween-20, incubated for 1.5 h at RT in appropriate secondary antibody (LI-COR Biosciences; 1:20,000 Donkey anti-Rabbit IgG [Cat# 926-32213]; 1:20,000 Donkey anti-Mouse IgG [Cat# 926-68022) in blocking buffer with 0.1% Tween-20 and 0.01% SDS, and then washed three times for 5 min each in PBS with 0.1% Tween-20. An Odyssey infrared Imager (LI-COR Biosciences) was used to image blots through the use of Image Studio software. ImageJ was used for quantification of relevant protein bands. Samples from rhodopsin degradation assays were also used to assess levels of BEST1 before and after gene augmentation. Immunoblots were performed as described above, using 1:1000 rabbit anti-Bestrophin1 antibody (LAgen Laboratories; Cat# 016-Best1-01) and 1:1000 mouse anti-Actin antibody (Millipore; Cat# MAB1501) as primary antibodies.
Deep Sequencing Analysis of DNA and RNA Read Frequency
Cells were singularized with TrypLE Express (Gibco, Cat# 12605010) per manufacturer’s instructions. Total DNA and/or RNA was extracted using QuickExtract DNA (Epicenter, Cat# QE09050) or QuickExtract RNA (Epicenter, Cat# QER090150), respectively. Both DNA and RNA extractions were performed per manufacturer’s instructions with the following minor modifications: (1) a ratio of 10,000–25,000 cells per 50 μl of QuickExtract solution was routinely used, and (2) an optional DNase 1 treatment was omitted from the RNA extraction protocol. All samples were stored at −80°C until use.
RNA was reverse transcribed to cDNA through the use of the ProtoScript II First Strand synthesis kit (NEB, Cat# E6560S) and synthesis was performed with the “random primer” option included within the kit. 4 μl of crude RNA extract was added to each cDNA reaction.
In preparation for targeted deep sequencing, Illumina adaptor sequences and sample-specific barcodes were appended to genomic or cDNA amplicons via overhang PCR as described.19 Purified amplicon libraries were assembled into 2 nM total DNA in DNase/RNase free H2O and sequenced using 150 nucleotide paired end reads using MiSeq (6M or 15M total reads) at the UW Biotech Center with the following loading condition: 8 pmol total DNA and 15% PhiX DNA. Raw FASTQ files were read and aligned to expected amplicons by using a command line implementation of CRISPResso (v1.0.8).23 Full CRISPResso progam calls for each sample are listed in Data S1. “Percent allele identity” and “percent edited” were determined using the software’s standard output table of individual read identities. Sequencing reads with counts <100 were not included in the analysis. All FASTQ files are available via NCBI BioProject (accession number: PRJNA633668).
Single-Cell RNA Sequencing (scRNA-seq)
iPSC-RPE cultures derived from the Ala146Lys, Asn296His, and Arg218Cys adBD mutant lines and from an isogenic gene-corrected control line derived from the Arg218Cys line (Arg218Cys>WT) were transduced with 150 μl of VMD2-spCas9-T2A-GFP encoding specific sgRNAs as described in the “Lentivirus Production and Cell Transduction” section. For each sample, sgRNAs were targeted either to mutant BEST1 or to the AAVS1 locus (control). On day 14, cells were dissociated from transwells through the use of a papain dissociation kit (Worthington Biochemical, Cat# LK003150) and filtered using a Flowmi cell strainer (Bel-Art SP Scienceware, Cat# H13680-0040) to obtain a single-cell suspension. Cells were then prepared for scRNA-seq using the droplet-based 10× Genomics GemCode platform according to the manufacturer’s instructions (Chromium Single Cell v2 3′ Reagent Kit). In brief, singularized cells were encapsulated in oil beads containing a unique molecular identifier (UMI) barcode. The cells were then lysed, and cDNA libraries were created featuring cell and transcript-specific molecular identifiers. Libraries were sequenced using an Illumina HiSeq2500 Rapid Run, and reads were aligned to a custom reference genome consisting of the human hg19 GRCh38 genome and an added gene for the spCas9-T2A-GFP transcript.
scRNA-seq Data Analysis
Gene edited iPSC-RPE were clustered based on their genome-wide transcriptome through the use of the t-Distributed Stochastic Neighbor Embedding (t-SNE) algorithm with the 10× Genomics Loupe Cell Browser software (v2.0.0). Reads for each pair of samples (BEST1 mutant allele-targeted sgRNA versus AAVS1 sgRNA control) were aligned, analyzed, clustered with Cell Ranger (v2.1.1; 10× Genomics), and compared in order to detect significant differences in gene expression, with p values adjusted using the Benjamini-Hochberg correction for multiple tests. p < 0.01 was used as the significance threshold for all analyses. Cell Ranger using the aggregate feature was run to concatenate each pair of samples with the same genotype, and differential gene expression within each pair (with gene editing at either the AAVS1 or BEST1 locus) was then analyzed. Potential adverse events were probed using gene lists curated from gene ontology terms associated with the cell cycle, apoptosis, DNA damage response, and the innate immune response, as well as a list of 149 validated marker genes associated with human RPE24 (Data S2; gene ontology sets are available on the Molecular Signatures Database). Differentially expressed genes with p < 0.01 were deemed to be significant. All significantly differentially expressed genes per cluster are reported, with the exception of genes identified by Cell Ranger as having low average UMI counts. Volcano plots were generated in RStudio (v.1.1.456) using the ggplot2 package.
Non-negative Matrix Factorization-Based Comparison of scRNA-seq Datasets
To enable comparison of transcriptional signatures from each sample, non-negative matrix factorization (NMF)25 was applied to each scRNA-seq dataset. NMF is a popular dimensionality reduction and clustering approach that is used to project data into low-dimensional non-negative factors, and thus it can be used to derive a clustering of cells and genes. NMF followed by clustering of genes using the NMF factors was used for Figure S4 to project each dataset into a gene group. The input data for this analysis were a set of gene barcode matrices generated using the Cell Ranger algorithm. The matrices were filtered to remove background barcodes in order to include only detected cellular barcodes, and then further filtered to exclude cells expressing fewer than 2,000 total counts, followed by depth normalization.
NMF with k = 10 factors was applied with a total of five NMF runs. Next, the similarity of NMF results was compared between two samples by using the average best Jaccard coefficient between clusters of one versus another sample. 1-average Jaccard coefficient was then used as the distance to apply hierarchical clustering on the samples. This procedure was repeated five times, and the tree that appeared most often was used. The trees learned in different iterations were largely similar and always grouped the individual-specific iPSC lines first before grouping different lines together.
Quantification and Statistical Analysis
Unless otherwise specified, all analyses were performed using GraphPad Prism (v.8.0.1), and error bars represent mean ± SD; ns = p ≥ 0.05, ∗ for p < 0.05, ∗∗ for p < 0.01, ∗∗∗ for p < 0.001, ∗∗∗∗ for p < 0.0001. Further detail for each analysis is provided within this section. Statistical analyses for CaCC conductance measurements were performed using Origin 2018b. Student’s t test was performed to measure the significance between the groups. p values < 0.05 were considered statistically significant. Statistical significance for differential gene expression in scRNAseq datasets was determined using the Cell Ranger 2.1.1 algorithm. Sample pairs with each genotype were analyzed and clustered with individual Cell Ranger runs for each pair and then analyzed using the Loupe Cell Browser (v.2.0.0). Differential expression was calculated using a negative binomial exact test, and p values were adjusted using the Benjamini-Hochberg correction for multiple tests. p < 0.01 was used as the threshold for assigning significant versus non-significant changes in gene expression.
Results
BEST1 Mutations Result in Decreased CaCC Activity in iPSC-RPE
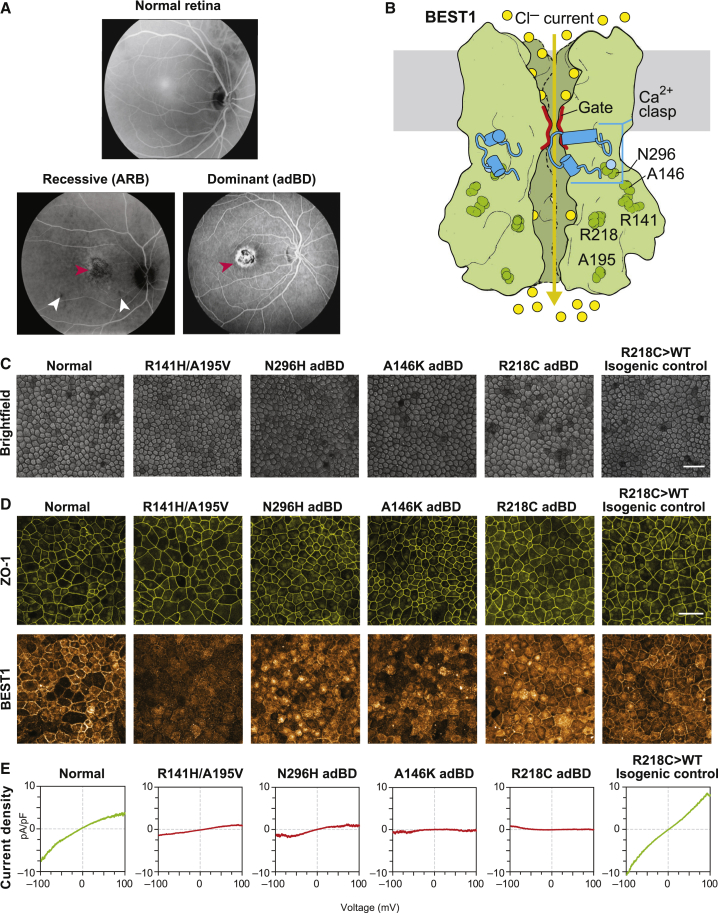
In addition to the Asn296His and Ala146Lys adBD iPSC lines previously reported,17,18 we generated iPSCs from a third individual with adBD with p.Arg218Cys BEST1 and an individual with ARB harboring compound heterozygous mutations in BEST1 (c.422G>A[p.Arg141His]; c.584C>T [p.Ala195Val[) (Figure 1A). Based on the crystallographic studies, each of these mutations alters residues within a different functional region of the BEST1 channel (Figure 1B).16 We also employed two control iPSC lines: a WT iPSC line and an isogenic iPSC line generated via CRISPR-based gene correction of Arg218Cys adBD iPSCs (Arg218Cys>WT).19 All six iPSC lines were tested for pluripotency, differentiated to RPE, and characterized (Figure 1C–1D, and Figure S1A–S1D). iPSC-RPE monolayers for all adBD and control lines, but not for the ARB line, showed robust levels of BEST1 (Figure 1D). The profoundly decreased BEST1 level in our ARB cultures is consistent with reports using heterologous or iPSC-RPE systems that showed low or undetectable levels of p.Arg141His or p.Ala195Val BEST1.26,27 As a measurement of CaCC activity, single-cell patch-clamp recordings of calcium-activated chloride current density were performed and found to be greatly diminished in all BEST1 mutant iPSC-RPE relative to WT control iPSC-RPE (Figure 1E and Figure S1E–S1I). Gene-corrected Arg218Cys>WT isogenic iPSC-RPE control showed CaCC current density at levels similar to those of native WT control lines (Figure 1E and Figure S1J), indicating that the decreased CaCC activity was indeed the result of the BEST1 mutation.
Figure 1.
BEST1 Mutations Reduce CaCC Current in Best Disease iPSC-RPE
(A) Top, image (in grayscale) of a normal fundus; bottom left, fundus image of an individual with ARB harboring compound heterozygous mutations in BEST1 (resulting in p.Arg141His; p.Ala195Val) showing a vitelliform lesion in the macula (red arrowhead) as well as small lesions outside the macula (white arrowheads); bottom right, fundus image showing a vitelliform macular lesion (red arrowhead) in an individual with adBD caused by a heterozygous p.Arg218Cys encoding mutation in BEST1.
(B) A fully functional homo-pentameric BEST1 channel is formed by assembly of WT subunits (green), allowing movement of chloride ions (yellow circles) upon binding of calcium ions (light blue circle) (based on the eukaryotic Best1 crystal structure16).
(C) Light microscopic images of normal, mutant-specific, and isogenic control iPSC-RPE used in this study. Scale bar = 50 μm (applies to all images in C).
(D) Immunocytochemical analyses of ZO-1 and BEST1 localization in iPSC-RPE cells. Scale bar = 50 μm (applies to all images in D).
(E) CaCC current density-voltage plots from WT, ARB, or adBD iPSC-RPE cells, as determined by calculating the difference in average chloride currents in the presence or absence of calcium (Figure S1). For +calcium, n = 6 cells for WT, 12 cells for Arg141His/Ala195Val ARB, 7 cells for Asn296His adBD, 5 cells for Ala146Lys adBD, 5 cells for Arg218Cys adBD, and 10 cells for Arg218Cys>WT isogenic control; for no calcium, n = 8 cells for WT, 12 cells for Arg141His/Ala195Val ARB, 8 cells for Asn296His adBD, 7 cells for Ala146Lys adBD, 8 cells for Arg218Cys adBD, and 9 cells for Arg218Cys>WT isogenic control iPSC-RPE (data combined from at least two replicates).
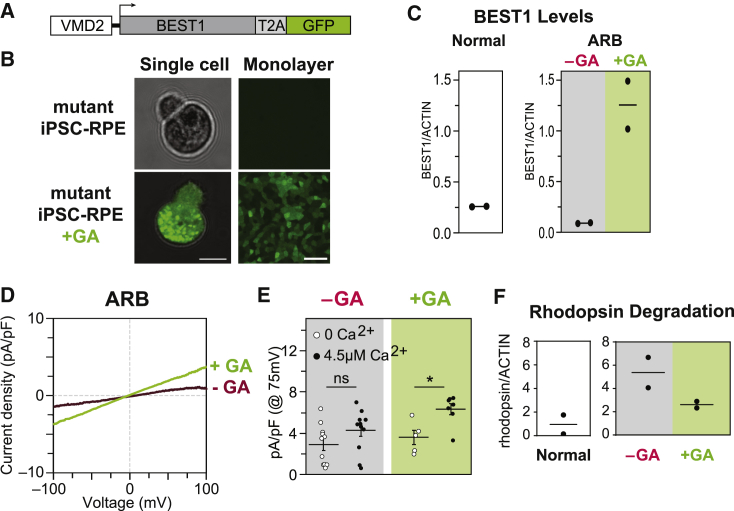
BEST1 Augmentation Restores CaCC Activity and Enhances Rhodopsin Degradation in ARB iPSC-RPE
We next sought to confirm that ectopic expression of WT BEST1 could ameliorate the disease phenotype in our ARB iPSC-RPE model, as has been shown in ARB canines or other iPSC-RPE model systems for ARB.28,29 Single-cell patch clamp recordings of calcium-activated chloride current density were used as a readout of efficacy in iPSC-RPE cells. In addition, we monitored degradation of rhodopsin following POS feeding as an assay of intact RPE monolayer function.
For gene augmentation, we used a lentivirus construct (VMD2-BEST1-T2A-GFP) designed to co-express BEST1 and green fluorescent protein (GFP) transcripts under control of the BEST1 promoter (VMD2), which assures both RPE-specific expression and BEST1-specific gene regulation (Figure 2A and 2B). Lentivirus was chosen for transgene delivery based on its safe use in human retinal gene therapy trials30 (National Institutes of Health [NIH] ClinicalTrials website identifiers: NCT01367444, NCT01736592) and its superior transduction efficiency in cultured human RPE.17,31 Consistent with these prior observations, we noted a transduction efficiency of 83.21(4.62)% across all mutant lines in our study (n = 4). GFP was observed in ARB iPSC-RPE cells post-transduction, and ICC and immunoblot analysis confirmed enhanced levels of BEST1 in treated cultures (Figure 2C and Figure S2A–S2C). By ≥ 4weeks post-transduction, CaCC current density in ARB iPSC-RPE increased significantly, reaching levels comparable to those in WT iPSC-RPE (Figure 2D and 2E and Figure S2E). Furthermore, transduced monolayers of ARB iPSC-RPE demonstrated enhanced degradation of rhodopsin following POS feeding (Figure 2F and Figure S2I). These findings, together with those reported by Guziewicz et al.28 and Li et al.,29 support BEST1 augmentation as a treatment for ARB.
Figure 2.
Gene Augmentation Rescues the ARB iPSC-RPE Cell Phenotype
(A) Construct used for BEST1 augmentation (GA).
(B) Presence or absence of GFP fluorescence in a single dissociated iPSC-RPE cell (left) or iPSC-RPE monolayers (right) before (top) or after (bottom) gene augmentation. Scale bar = 10 μm (left); 50 μm (right).
(C) Immunoblot-based quantification of BEST1 levels (normalized to ACTIN) in WT iPSC-RPE (left), and in ARB iPSC-RPE before or after BEST1 augmentation (right).
(D) CaCC current density-voltage plots before and after gene augmentation in ARB iPSC-RPE. The -GA trace is the same as shown in Figure 1E. For the +GA trace, n = 7 cells for +calcium and 5 cells for no calcium (data combined from two replicates [Figure S2]).
(E) CaCC conductance for individual ARB iPSC-RPE cells at 75 mV before or after gene augmentation. The number of cells is the same as for panels 1E and 2D. Error bars represent mean ± SEM; ns = p ≥ 0.05, ∗ for p < 0.05. Black circles, +calcium condition; white circles, no calcium condition.
(F) Immunoblot-based quantification of rhodopsin levels 120 h after photoreceptor outer segment (POS) feeding in WT iPSC-RPE or in ARB iPSC-RPE before or after WT BEST1 augmentation.
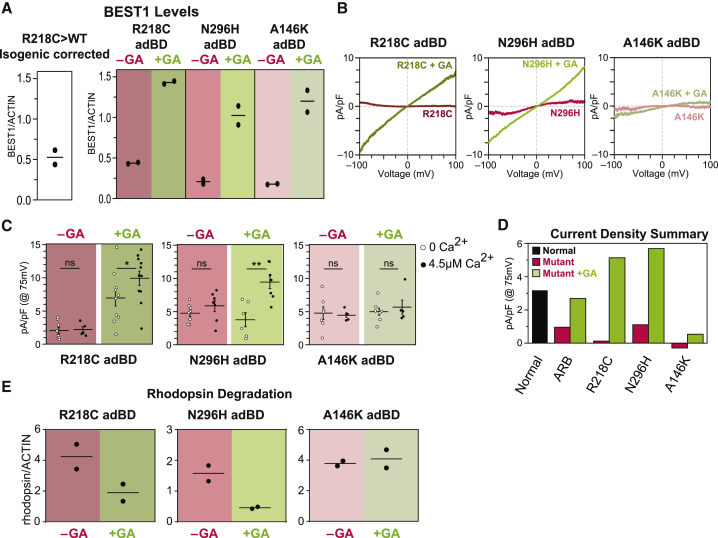
BEST1 Augmentation Restores CaCC Activity and Enhances Rhodopsin Degradation in Arg218Cys and Asn296His adBD iPSC-RPE, but not in Ala146Lys adBD iPSC-RPE
Although it is not as intuitive, we suspected that gene augmentation might also be a viable solo therapeutic strategy for adBD-causing BEST1 mutations. More specifically, we hypothesized that CaCC activity could be restored by increasing the intracellular ratio of WT to mutant BEST1 monomers available to form the homo-pentameric channel.
The same VMD2-BEST1-T2A-GFP lentiviral construct that was tested in ARB iPSC-RPE was used to transduce iPSC-RPE from all three individuals with adBD (Figure S2D). Following gene augmentation, BEST1 levels in each adBD iPSC-RPE model were comparable to those achieved in gene-augmented ARB-iPSC-RPE and >3-fold higher than BEST1 levels present in parallel cultures of untreated adBD iPSC-RPE (Figure 3A and Figure S2C). At ≥4 weeks post-transduction, CaCC activity was fully restored in the Arg218Cys and Asn296His adBD iPSC-RPE models, whereas the Ala146Lys adBD iPSC-RPE model remained unresponsive (Figure 3B-D and Figure S2F–S2H) despite displaying the highest fold increase in BEST1 levels (Figure 3A). Consistent with these single-cell electrophysiological findings, gene augmentation improved rhodopsin degradation in Arg218Cys and Asn296His iPSC-RPE, but not in Ala146Lys iPSC-RPE (Figure 3E and Figure S2J–S2L).
Figure 3.
Gene Augmentation Rescues the Cell Phenotype in Some, but Not All, adBD iPSC-RPE Models
(A) Immunoblot-based quantification of BEST1 levels (normalized to ACTIN) in WT iPSC-RPE and in the adBD iPSC-RPE models before and after BEST1 augmentation (GA).
(B) CaCC current density-voltage plots before and after gene augmentation in adBD iPSC-RPE. The -GA trace is the same as shown in Figure 1E. For the +GA traces, +calcium, n = 11 cells for Arg218Cys C, 7 cells for Asn296His, and 5 cells for Ala146Lys; for no calcium, n = 9 cells for Arg218Cys, 6 cells for Asn296His, and 8 cells for Ala146Lys iPSC-RPE (data combined from two replicates).
(C) CaCC conductance for individual adBD iPSC-RPE cells at 75 mV before and after gene augmentation. The number of cells is the same as for panels 1E and 3B. Error bars represent mean ± SEM; ns = p ≥ 0.05, ∗ for p < 0.05, ∗∗ for p < 0.01.
(D) Mean CaCC conductance at 75 mV before or after gene augmentation for all iPSC-RPE tested.
(E) Rhodopsin levels 48 h after feeding POS to adBD iPSC-RPE before or after WT BEST1 augmentation.
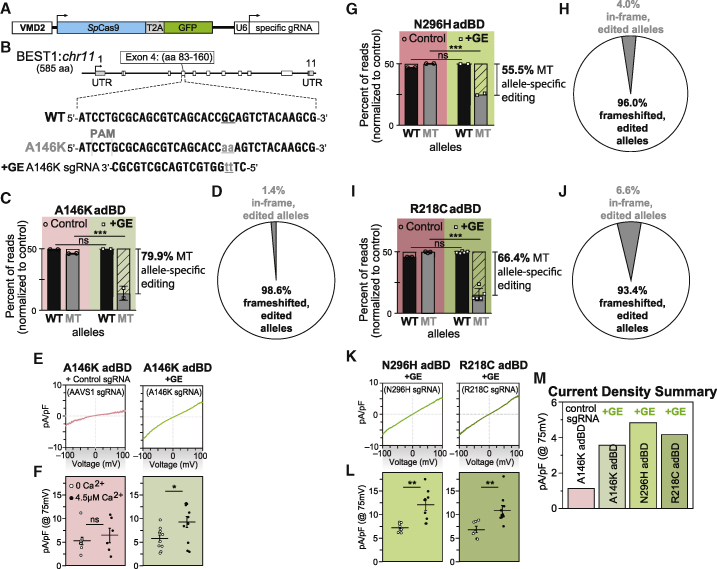
Gene Editing Specifically Targets the Mutant Allele in Ala146Lys adBD iPSC-RPE and Restores CaCC Activity
To determine whether Ala146Lys iPSC-RPE would respond to an alternative therapeutic approach, we tested gene editing as a means to eliminate expression of the mutant BEST1 allele. Gene editing with CRISPR-Cas9 creates targeted double-strand breaks in genomic DNA that are primarily repaired by endogenous non-homologous end joining (NHEJ),32 leading to indels. These indels can cause transcriptional frameshifts that lead to premature termination codons, activation of intrinsic nonsense-mediated decay (NMD) pathways, and degradation of transcription products.33,34
An sgRNA (A146K sgRNA) sequence targeting the locus corresponding to p.Ala146Lys (c.436_437delinsAA) in the mutant BEST1 allele was cloned into a lentiviral plasmid that encoded both the sgRNA (expressed via a U6 promoter) and a human codon optimized Streptococcus pyogenes Cas9 (spCas9)-T2A-GFP transcript (expressed via a VMD2 promoter) (Figure 4A and 4B). We also cloned an sgRNA sequence targeting the AAVS1 safe harbor locus35 into the same lentiviral plasmid backbone to serve as an experimental control.
Figure 4.
Gene Editing Specifically and Efficiently Introduces Frameshifts within the Mutant Allele in adBD iPSC-RPE and Rescues CaCC Activity
(A) Lentiviral genome editing construct used to express spCas9 and mutant allele-targeted sgRNAs.
(B) Diagram showing the heterozygous base pair substitutions in Ala146Lys adBD and the design of the A146K sgRNA. The WT allele is shown above, while the mutant allele encoding p.Ala146Lys (c.436_437delinsAA) is shown below, with the mutated bases indicated in lowercase and underlined.
(C) Percentage of WT and mutant (MT; unedited and edited) allele sequencing reads in Ala146Lys iPSC-RPE treated with A146K sgRNA lentiviral genome editor (“+GE”), respectively, normalized to control (“Control,” genome edited with safe harbor AAVS1-targeting sgRNA).
(D) Indel frameshift and in-frame frequency for mutant allele-edited reads from Ala146Lys adBD iPSC-RPE (corresponds to (C)).
(E and F) (E) CaCC current density-voltage plots and (F) CaCC conductance for individual iPSC-RPE cells from single-cell patch clamp experiments for Ala146Lys iPSC-RPE treated with control (AAVS1) or mutant allele-targeted sgRNA lentiviral genome editor.
(G–J) Percentage of WT and mutant (MT; unedited and edited) allele sequencing reads in Asn296His (G) or Arg218Cys (I) adBD iPSC-RPE treated with N296H or R218C sgRNA lentiviral genome editor, respectively, normalized to control (AAVS1 sgRNA). Indel frameshift and in-frame frequency in Asn296His (H) or Arg218Cys (J) adBD iPSC-RPE treated with N296H or R218C sgRNA lentiviral genome editor, respectively (correspond to (G) and (I), respectively).
(K and L) (K) CaCC current density-voltage plots and (L) CaCC conductance for individual iPSC-RPE cells from single-cell patch clamp experiments for Asn296His or Arg218Cys adBD iPSC-RPE treated with corresponding mutant allele-targeted sgRNA lentiviral genome editor.
(M) Mean CaCC conductance at 75 mV for each adBD iPSC-RPE model. The number of cells is the same as (E) and (K).
For gene editing experiments ((C), (D), and (G–J)), n = 2 (Ala146Lys iPSC-RPE and Asn296His iPSC-RPE) and n = 5 (Arg218Cys iPSC-RPE). For electrophysiology experiments ((E), (F), and (K–M)), for +calcium, n = 6 cells for AAVS1, 11 cells for Ala146Lys, 9 cells for Asn296His, 10 cells for Arg218Cys; for no calcium, n = 9 cells for AAVS1, 10 cells for Ala146Lys, 9 cells for Asn296His, 7 cells for Arg218Cys iPSC-RPE (data combined from two replicates). Error bars in (C), (G), and (I) represent mean ± SD; ns = p ≥ 0.05, ∗∗∗ for p < 0.001. Error bars in (F) and (L) represent mean ± SEM; ns = p ≥ 0.05, ∗ for p < 0.05, ∗∗ for p < 0.01.
Two weeks after transduction of Ala146Lys adBD iPSC-RPE with A146K sgRNA or control (AAVS1 sgRNA) lentiviral genome editor, we quantified the average frequency of deep sequencing reads corresponding to WT, mutant, and edited alleles in genomic DNA. We detected a nearly 80% editing frequency of the mutant allele encoding p.Ala146Lys (c.436_437delinsAA) with no decrease in WT allele frequency post-editing (Figure 4C). Together, these results reflect efficient editing with high specificity for the mutant BEST1 allele encoding p.Ala146Lys versus the WT BEST1 allele.
Using deep sequencing, we next examined specific indels that were introduced into Ala146Lys iPSC-RPE two weeks post-transduction with the A146K sgRNA genome editor (Data S1). An average of 98.6% of the edited alleles resulted in a frameshift mutation (Figure 4D and Data S1), a percentage that is higher than that of out-of-frame indels predicted by a recent machine learning algorithm (Data S1).36 This finding indicates a high likelihood that indels resulting from gene editing at the locus encoding p.Ala146Lys in the mutant BEST1 allele will trigger NMD of the transcribed RNA, effectively knocking out expression of the mutant allele in the vast majority of edited RPE cells.
Lastly, we assessed functional rescue of BEST1 channel activity in AAVS1 control versus mutant allele gene-edited Ala146Lys iPSC-RPE. Single-cell patch-clamp experiments revealed restoration of CaCC activity in Ala146Lys iPSC-RPE following editing with mutant allele-targeted sgRNA, but not after editing with control (AAVS1) sgRNA (Figure 4E and 4F and Figure S3).
Mutant Allele-Specific Gene Editing Restores CaCC Activity in All Tested adBD iPSC-RPE
While the gene editing results obtained in the Ala146Lys adBD iPSC-RPE model were highly encouraging, it is possible that this locus is unique in its potential to be targeted by a mutant allele-specific sgRNA. To extend this investigation, we also evaluated the specificity and efficacy of mutant allele editing in the Asn296His and Arg218Cys adBD iPSC-RPE models. c.886A>C (p.Asn296His) and c.652C>T (p.Arg218Cys) mutant allele-targeted sgRNAs (N296H and R218C sgRNAs) were designed and cloned into separate lentiviral plasmids as described for the A146K sgRNA. Asn296His iPSC-RPE and Arg218Cys iPSC-RPE were transduced with lentiviral genome editors encoding either control (AAVS1) or corresponding mutant allele-targeted sgRNA, and editing outcomes were measured via deep sequencing of genomic DNA (Data S1). Quantification of WT and mutant allele frequency revealed efficient targeting of the mutant alleles encoding p.Asn296His or p.Arg218Cys with their respective sgRNAs (55.5% and 66.4%, respectively) with no demonstrable targeting of the WT alleles (Figure 4G and 4I). A high proportion of editing in these two models resulted in out-of-frame indels (96.0% and 93.4% for Asn296His and Arg218Cys iPSC-RPE, respectively) (Figure 4H and 4J). Subsequent single-cell patch-clamp measurements of CaCC current density confirmed restoration of channel activity post-gene editing in both Arg218Cys and Asn296His iPSC-RPE (Figure 4K–4M and Figure S3). Thus, while some variation in gene editing efficiency was observed using the three different sgRNAs (as expected), more than half of the mutant alleles were edited (with a high percentage of out-of-frame indels) in the three adBD iPSC-RPE models, with no detectable editing of the WT allele.
Mutant Allele-Specific Gene Editing Does not Perturb Global iPSC-RPE Transcriptional Programs, Although Off-Target Editing Can Occur
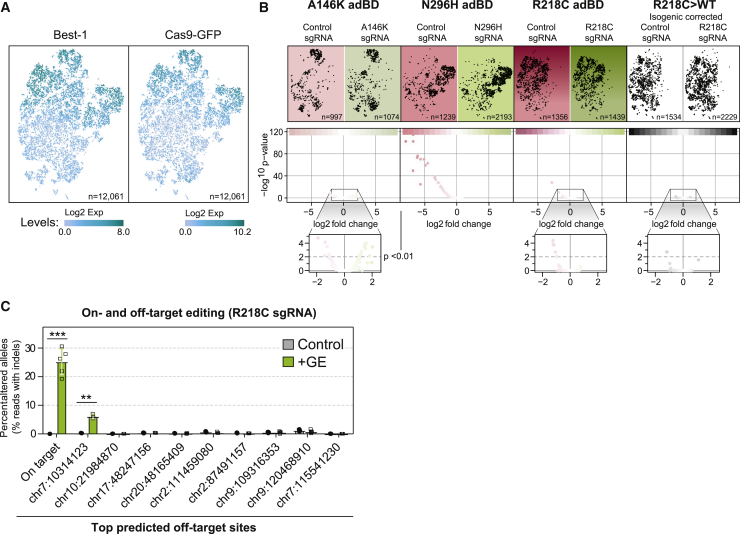
While the mutant allele-specific sgRNAs tested in the three adBD iPSC-RPE models did not target the fellow WT alleles in any of our experiments, the potential still exists for off-target adverse effects elsewhere within the genome. To detect untoward transcriptional effects from gene editing, we performed scRNA-seq for 12,061 individual iPSC-RPE cells treated with genome editors. iPSC-RPE (Arg218Cys, Asn296His, Ala146Lys, or isogenic control Arg218Cys>WT) were edited with genome editors encoding either a mutant allele-targeted sgRNA or a control sgRNA targeting the AAVS1 site, resulting in a total of eight separate samples (Figure S4A).
Clustering of cells across all eight samples indicated that, by virtue of using the VMD2 promoter, spCas9-T2A-GFP transcript levels closely corresponded with BEST1 transcript levels (Figure 5A). Visual comparison of clustering of each individual sample via t-SNE demonstrated that transcriptional signatures are grossly similar between iPSC-RPE lines, whether treated with mutant allele-targeted (+GE) or control (AAVS1) sgRNA (Figure 5B, top). This observation was supported quantitatively by NMF. NMF analysis demonstrated that greater transcriptome variation exists between iPSC-RPE from different lines than between iPSC-RPE from the same line treated with mutant allele-targeted or control sgRNA (Figure S4B).
Figure 5.
Gene Editing Did Not Disrupt iPSC-RPE Transcriptional Programs
(A) t-SNE plot of single iPSC-RPE cells across all eight samples with relative expression of BEST1 (left) and spCas9-T2A-GFP (right) depicted via increasing shades of blue. Total number of cells analyzed (n) is shown.
(B) Top, t-SNE plot of single cells (black dots) from each treated sample. Number of cells analyzed (n) for each sample is shown. Bottom, Volcano plots of transcriptome-wide differences in expression of individual genes (red or green dots) between iPSC-RPE of the same genotype treated with mutant allele-targeted sgRNA (green) versus control (AAVS1, red) sgRNA lentiviral genome editor. p < 0.01 was the threshold for determining significant versus non-significant changes in gene expression.
(C) Frequency of edited alleles at on-target and top nine ranked off-target loci in Arg218Cys adBD iPSC-RPE treated with R218C sgRNA lentiviral genome editor (n = 3 for control and n = 5 for +GE, except n = 3 at first chr 7 off-target locus). Off-target sites are annotated by the location of the first base of the predicted off-target site (further detailed in Data S3). Error bars represent mean ± SD; ∗∗ for p < 0.01, ∗∗∗ for p < 0.001.
Additional analysis of global gene expression (Figure 5B, bottom) and of a focused set of genes related to negative or off-target effects (including cell cycle regulation, apoptosis, DNA damage response, or innate immune response; Figure S4C, Data S2) did not reveal significant upregulation of those gene sets in mutant allele-targeted (+GE) versus control sgRNA-treated samples. However, examination of the top nine potential off-target sites for the R218C sgRNA revealed a low, yet significant percentage of editing at a single site within a non-coding region of chromosome 7 (Figure 5C). Although this finding is not predicted to have a deleterious effect on RPE cell function, it emphasizes the importance of performing comprehensive on- and off-target genome editing analyses using a human model system.
Discussion
The observation that a subset of adBD mutations may be amenable to gene augmentation greatly expands the population of individuals affected by Best disease who might benefit from this therapeutic approach. Based on the crystallographic studies by Dickson et al.,16 the two mutations that responded to gene augmentation encode residues that lie within calcium clasp (p.Asn296His) or chloride-binding (p.Arg218Cys) sites within the BEST1 channel, whereas the mutation that failed to respond (c.436_437delinsAA [p.Ala146Lys]) localizes to a putative structural region. Among the over 200 known BEST1 mutations, many are predicted to encode residues that are directly or indirectly involved in ion binding.16,37 Importantly, a recent study by Ji et al. using baculovirus supports our finding that mutations associated with chloride- and calcium-binding sites in BEST1 can be receptive to gene augmentation.38 However, the fact that not all adBD iPSC-RPE models respond to gene augmentation underscores the need to vet candidacy for gene augmentation carefully.
The mechanism underlying selective responsivity of adBD mutations to gene augmentation cannot be due to traditional allelic haploinsufficiency, in which half the normal amount of WT protein and no mutant protein is produced, resulting in fewer (but fully WT) BEST1 channels. Such a situation exists in parents of individuals with ARB, who have no demonstrable disease phenotype. Rather, adBD mutant monomers must be incorporated alongside WT monomers in all (or nearly all) BEST1 channels.39 We propose that in the case of p.Asn296His and p.Arg218Cys, this commingling of WT and mutant monomers causes ion-binding site insufficiency and channel impermeability, a condition that is surmountable by WT BEST1 augmentation. In contrast, we hypothesize that BEST1 mutations such as the one resulting in p.Ala146Lys, which converts a nonpolar amino acid to a polar amino acid in a compact structural region of the protein, have more pervasive functional consequences, resulting in greater resistance to gene augmentation.
We did consider the possibility that mutation-specific resistance to gene augmentation was due to variability in transgene expression (i.e., there was insufficient WT transgene expression in the non-responsive Ala146Lys adBD iPSC-RPE model). However, we found that BEST1 levels were similar in all models following gene augmentation. In fact, a slightly higher fold-increase in BEST1 levels was achieved in Ala146Lys adBD iPSC-RPE compared with the two adBD models that were rescued by gene augmentation (Arg218Cys and Asn296His). Thus, it is highly unlikely that the differences in functional response observed between Arg218Cys or Asn296His adBD iPSC-RPE and Ala146Lys adBD iPSC-RPE are due to variability in transgene expression. Resistance of the Ala146Lys adBD iPSC-RPE to functional recovery after gene augmentation also cannot be explained by occult artifacts inherent to the iPSC line or its RPE progeny, since gene editing was ultimately successful in restoring CaCC activity in the same differentiated Ala146Lys adBD iPSC-RPE population.
It is also notable that our lentiviral constructs employed the VMD2 promoter, which is ideal from a translational standpoint because it specifies expression in RPE and supports native regulation of BEST1. Use of alternative promoters poses risks of off-target cell effects and/or undesirably low (ineffectual) or high (toxic) levels of protein. Establishing a safe dose range for WT BEST1 overexpression is an important future pursuit, since it is conceivable that higher ratios of WT to mutant BEST1 monomer production could promote CaCC recovery even for refractory adBD mutations. For construct delivery, we selected lentivirus based on its excellent in vitro RPE transduction efficiency17,31 and its current use in RPE gene therapy trials (NIH ClinicalTrials website identifiers: NCT01367444, NCT01736592).30 However, our findings are likely applicable across all in vivo transgene delivery platforms that possess comparable safety and transduction efficiency profiles. Indeed, Ji et al. observed improvement in CaCC activity in isolated Arg218His adBD iPSC-RPE cells following constitutive overexpression of WT BEST1 using an AAV delivery vector.38
There is precedence for using human disease-specific iPSCs as preclinical efficacy models for gene therapy clinical trials.40 Our work extends this utility by providing a framework for preclinical testing of mutation-specific responses in a genotypically heterogenous disease using the affected cell type. It remains to be determined whether separate adBD iPSC-RPE models will be required to assess suitability of gene augmentation versus gene editing for every mutation, or if a few models can sufficiently represent larger categories of mutations (e.g., those that alter residues within ion-binding sites or structural regions).41
For adBD mutations that are not readily amenable to gene augmentation, we showed that targeted gene editing holds great promise as an alternative therapy. Indeed, there is a wide spectrum of BEST1 mutations that could be treated using CRISPR-Cas9 by designing unique mutation-targeted sgRNAs (examples shown in Data S4). While this approach would be costly and time-consuming if separate testing were required for each mutation-specific sgRNA, rapid advances in gene editing technologies and strategies may overcome such limitations. Other gene therapy strategies also exist for dominant ocular diseases; for example, knockdown of both WT and mutant allele transcripts with simultaneous introduction of a modified WT gene.42 Whether such an approach would be safe and effective for adBD mutations that fail to respond to straightforward gene augmentation is not known, but could be tested using the iPSC-RPE model systems employed here.
In our gene editing experiments, we observed higher efficiency out-of-frame editing in iPSC-RPE when compared to a prior study using undifferentiated iPSCs.19 This finding is consistent with recent reports of variable mutation bias across different cell types,36 and it points to the importance of evaluating gene editing using the specific cell type(s) targeted by disease. In addition, editing at BEST1 in iPSC-RPE did not provoke an increase in expression of genes associated with cell cycle regulation, apoptosis, DNA damage response, or innate immune response in comparison to editing at a well-characterized safe-harbor locus35 with a previously described sgRNA.43 Undesirable effects such as these have been reported in other cell types following Cas9-mediated gene editing.11,44 Despite our reassuring findings, the potential remains for off-target genomic alterations, as was observed at a single locus in a small percentage of iPSC-RPE cells in the Arg218Cys adBD model. Although these particular off-target indels are in a non-coding region and are thus predicted to be functionally silent, their presence emphasizes the value of employing human model systems for preclinical genome editing safety studies. Interestingly, no off-target indels were detected in our prior study which used the same sgRNA in undifferentiated Arg218Cys iPSCs19; this further indicates the need to perform off-target analyses in iPSC-RPE and not in surrogate cell types.
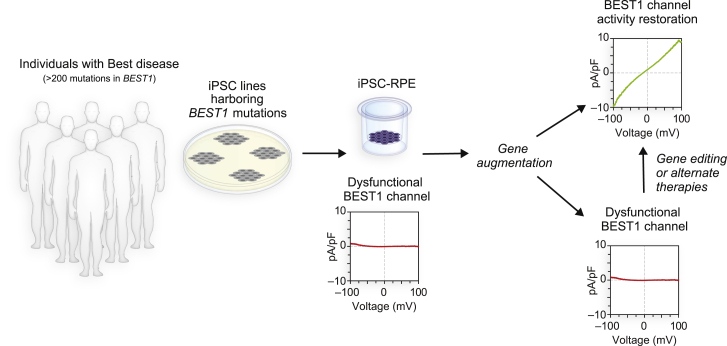
Overall, our results provide a blueprint to guide gene therapy choice in the era of gene augmentation and gene editing (Figure 6). With its inherently larger target populations and established track record in treated individuals, it is practical to utilize gene augmentation when possible, reserving gene editing for mutations that require allele repair or knockout or are otherwise untreatable by gene augmentation. It is noteworthy that the two adBD lines that demonstrated restoration of CaCC activity with gene augmentation or gene editing did so with equal efficacy, underscoring the suitability of either approach. Other desirable characteristics of Best disease as a clinical candidate for gene therapy include (1) a wide time window for gene therapy intervention, (2) accessibility of RPE using standard surgical techniques, (3) a small (∼5.5 mm diameter) treatment area, (4) availability of noninvasive retinal imaging and functional assessment tools, and (5) growing safety data from other RPE-based gene therapy trials.2, 3, 4 As such, Best disease is well positioned to become the first genotypically heterogeneous disorder with dominant and recessive inheritance patterns to have a full menu of therapeutics for all affected individuals. Furthermore, implications of this work likely extend beyond the eye and Best disease to other intractable monogenic conditions caused by mutations in genes that encode multimeric ion channels, including congenital myasthenic syndromes and some forms of epilepsy.45, 46, 47
Figure 6.
In Vitro Gene Therapy Testing Strategy for adBD
The amenability of adBD mutations to correction via gene augmentation can be evaluated for efficacy and safety in a dish using BEST1 mutant iPSC-RPE models. Those individuals with mutations that fail to respond to gene augmentation would then undergo further testing for gene editing (or another alternative strategy) using the same adBD iPSC-RPE model systems.
Data and Code Availability
Raw targeted sequencing files for DNA and RNA sequencing data as well as scRNA-seq data are available via NCBI BioProject: PRJNA633668. Raw patch clamp data are available upon request. Other experimental data are provided in the Supplemental Information, and all source data are available upon request.
Acknowledgments
The authors thank Alfred Lewin (University of Florida) for the VMD2-BEST1-T2A-GFP plasmid construct; the Cellular Imaging and Analysis core at the University of Wisconsin-Madison Waisman Center; and Andrew Thliveris for helpful discussions. This work was supported by National Institutes of Health (NIH) grant R01EY024588 to D.M.G., B.A.T., and E.M.S.; Foundation Fighting Blindness, Research to Prevent Blindness, Retina Research Foundation Emmett Humble Chair, Sarah E. Slack Prevention of Blindness Fund (a component fund of the Muskingum County Community Foundation), and McPherson Eye Research Institute Sandra Lemke Trout Chair in Eye Research to D.M.G.; National Science Foundation (NSF) grant CBET-1350178, NIH grant R35GM119644, Burroughs Wellcome Fund, and Retina Research Foundation Kathryn and Latimer Murfee Chair to K.S.; NIH grant R01EY024995 and Retina Research Foundation M.D. Mathews Professorship to B.R.P.; NIH grants T32HG002760 and F30EY027699, and the VitreoRetinal Surgery Foundation to B.S.; and the NSF’s Division of Graduate Education (DGE) grant DGE-1747503 to K.P.M.. This study was supported in part by a core grant to the Waisman Center (National Institute of Child Health and Human Development [NICHHD] grant U54 HD090256), a University of Wisconsin Data Science Initiative grant to S.R., and the UW2020 Initiative. We thank members of the Gamm and Saha labs for comments on the manuscript, plasmid depositors to Addgene, H. Adam Steinberg and Art for Science for help in figure preparation, the University of Wisconsin-Madison Biotechnology Center for DNA sequencing, and the University of Wisconsin-Madison Skin Disease Research Center for assistance with virus preparation.
Published: July 23, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.06.011.
Contributor Information
Krishanu Saha, Email: ksaha@wisc.edu.
David M. Gamm, Email: dgamm@wisc.edu.
Web Resources
ClinicalTrials.gov, https://clinicaltrials.gov/
Molecular Signatures Database, https://www.gsea-msigdb.org/gsea/msigdb
OMIM, https://www.omim.org
Declaration of Interests
The authors declare no competing interests.
Supplemental Data
References
- 1.Doudna J.A., Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 2.Lam B.L., Davis J.L., Gregori N.Z., MacLaren R.E., Girach A., Verriotto J.D., Rodriguez B., Rosa P.R., Zhang X., Feuer W.J. Choroideremia Gene Therapy Phase 2 Clinical Trial: 24-Month Results. Am. J. Ophthalmol. 2019;197:65–73. doi: 10.1016/j.ajo.2018.09.012. [DOI] [PubMed] [Google Scholar]
- 3.Cukras C., Wiley H.E., Jeffrey B.G., Sen H.N., Turriff A., Zeng Y., Vijayasarathy C., Marangoni D., Ziccardi L., Kjellstrom S. Retinal AAV8-RS1 Gene Therapy for X-Linked Retinoschisis: Initial Findings from a Phase I/IIa Trial by Intravitreal Delivery. Mol. Ther. 2018;26:2282–2294. doi: 10.1016/j.ymthe.2018.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Russell S., Bennett J., Wellman J.A., Chung D.C., Yu Z.F., Tillman A., Wittes J., Pappas J., Elci O., McCague S. Efficacy and safety of voretigene neparvovec (AAV2-hRPE65v2) in patients with RPE65-mediated inherited retinal dystrophy: a randomised, controlled, open-label, phase 3 trial. Lancet. 2017;390:849–860. doi: 10.1016/S0140-6736(17)31868-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ledford H. FDA advisers back gene therapy for rare form of blindness. Nature. 2017;550:314. doi: 10.1038/nature.2017.22819. [DOI] [PubMed] [Google Scholar]
- 6.Bakondi B., Lv W., Lu B., Jones M.K., Tsai Y., Kim K.J., Levy R., Akhtar A.A., Breunig J.J., Svendsen C.N., Wang S. In Vivo CRISPR/Cas9 Gene Editing Corrects Retinal Dystrophy in the S334ter-3 Rat Model of Autosomal Dominant Retinitis Pigmentosa. Mol. Ther. 2016;24:556–563. doi: 10.1038/mt.2015.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsai Y.T., Wu W.H., Lee T.T., Wu W.P., Xu C.L., Park K.S., Cui X., Justus S., Lin C.S., Jauregui R. Clustered Regularly Interspaced Short Palindromic Repeats-Based Genome Surgery for the Treatment of Autosomal Dominant Retinitis Pigmentosa. Ophthalmology. 2018;125:1421–1430. doi: 10.1016/j.ophtha.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li P., Kleinstiver B.P., Leon M.Y., Prew M.S., Navarro-Gomez D., Greenwald S.H., Pierce E.A., Joung J.K., Liu Q. Allele-Specific CRISPR-Cas9 Genome Editing of the Single-Base P23H Mutation for Rhodopsin-Associated Dominant Retinitis Pigmentosa. CRISPR J. 2018;1:55–64. doi: 10.1089/crispr.2017.0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pattanayak V., Lin S., Guilinger J.P., Ma E., Doudna J.A., Liu D.R. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat. Biotechnol. 2013;31:839–843. doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Courtney D.G., Moore J.E., Atkinson S.D., Maurizi E., Allen E.H., Pedrioli D.M., McLean W.H., Nesbit M.A., Moore C.B. CRISPR/Cas9 DNA cleavage at SNP-derived PAM enables both in vitro and in vivo KRT12 mutation-specific targeting. Gene Ther. 2016;23:108–112. doi: 10.1038/gt.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cromer M.K., Vaidyanathan S., Ryan D.E., Curry B., Lucas A.B., Camarena J., Kaushik M., Hay S.R., Martin R.M., Steinfeld I. Global Transcriptional Response to CRISPR/Cas9-AAV6-Based Genome Editing in CD34+ Hematopoietic Stem and Progenitor Cells. Mol. Ther. 2018;26:2431–2442. doi: 10.1016/j.ymthe.2018.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lessard S., Francioli L., Alfoldi J., Tardif J.C., Ellinor P.T., MacArthur D.G., Lettre G., Orkin S.H., Canver M.C. Human genetic variation alters CRISPR-Cas9 on- and off-targeting specificity at therapeutically implicated loci. Proc. Natl. Acad. Sci. USA. 2017;114:E11257–E11266. doi: 10.1073/pnas.1714640114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maeder M.L., Stefanidakis M., Wilson C.J., Baral R., Barrera L.A., Bounoutas G.S., Bumcrot D., Chao H., Ciulla D.M., DaSilva J.A. Development of a gene-editing approach to restore vision loss in Leber congenital amaurosis type 10. Nat. Med. 2019;25:229–233. doi: 10.1038/s41591-018-0327-9. [DOI] [PubMed] [Google Scholar]
- 14.Guziewicz K.E., Sinha D., Gómez N.M., Zorych K., Dutrow E.V., Dhingra A., Mullins R.F., Stone E.M., Gamm D.M., Boesze-Battaglia K., Aguirre G.D. Bestrophinopathy: An RPE-photoreceptor interface disease. Prog. Retin. Eye Res. 2017;58:70–88. doi: 10.1016/j.preteyeres.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson A.A., Guziewicz K.E., Lee C.J., Kalathur R.C., Pulido J.S., Marmorstein L.Y., Marmorstein A.D. Bestrophin 1 and retinal disease. Prog. Retin. Eye Res. 2017;58:45–69. doi: 10.1016/j.preteyeres.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kane Dickson V., Pedi L., Long S.B. Structure and insights into the function of a Ca(2+)-activated Cl(-) channel. Nature. 2014;516:213–218. doi: 10.1038/nature13913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh R., Shen W., Kuai D., Martin J.M., Guo X., Smith M.A., Perez E.T., Phillips M.J., Simonett J.M., Wallace K.A. iPS cell modeling of Best disease: insights into the pathophysiology of an inherited macular degeneration. Hum. Mol. Genet. 2013;22:593–607. doi: 10.1093/hmg/dds469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh R., Kuai D., Guziewicz K.E., Meyer J., Wilson M., Lu J., Smith M., Clark E., Verhoeven A., Aguirre G.D., Gamm D.M. Pharmacological Modulation of Photoreceptor Outer Segment Degradation in a Human iPS Cell Model of Inherited Macular Degeneration. Mol. Ther. 2015;23:1700–1711. doi: 10.1038/mt.2015.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steyer B., Bu Q., Cory E., Jiang K., Duong S., Sinha D., Steltzer S., Gamm D., Chang Q., Saha K. Scarless Genome Editing of Human Pluripotent Stem Cells via Transient Puromycin Selection. Stem Cell Reports. 2018;10:642–654. doi: 10.1016/j.stemcr.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu K., Yu J., Suknuntha K., Tian S., Montgomery K., Choi K.D., Stewart R., Thomson J.A., Slukvin I.I. Efficient generation of transgene-free induced pluripotent stem cells from normal and neoplastic bone marrow and cord blood mononuclear cells. Blood. 2011;117:e109–e119. doi: 10.1182/blood-2010-07-298331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh R., Phillips M.J., Kuai D., Meyer J., Martin J.M., Smith M.A., Perez E.T., Shen W., Wallace K.A., Capowski E.E. Functional analysis of serially expanded human iPS cell-derived RPE cultures. Invest. Ophthalmol. Vis. Sci. 2013;54:6767–6778. doi: 10.1167/iovs.13-11943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuruma A., Hartzell H.C. Bimodal control of a Ca(2+)-activated Cl(-) channel by different Ca(2+) signals. J. Gen. Physiol. 2000;115:59–80. doi: 10.1085/jgp.115.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pinello L., Canver M.C., Hoban M.D., Orkin S.H., Kohn D.B., Bauer D.E., Yuan G.C. Analyzing CRISPR genome-editing experiments with CRISPResso. Nat. Biotechnol. 2016;34:695–697. doi: 10.1038/nbt.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strunnikova N.V., Maminishkis A., Barb J.J., Wang F., Zhi C., Sergeev Y., Chen W., Edwards A.O., Stambolian D., Abecasis G. Transcriptome analysis and molecular signature of human retinal pigment epithelium. Hum. Mol. Genet. 2010;19:2468–2486. doi: 10.1093/hmg/ddq129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee D.D., Seung H.S. Algorithms for Non-negative Matrix Factorization. Adv. Neural Inf. Process. Syst. 2000;13:556–562. [Google Scholar]
- 26.Milenkovic A., Milenkovic V.M., Wetzel C.H., Weber B.H.F. BEST1 protein stability and degradation pathways differ between autosomal dominant Best disease and autosomal recessive bestrophinopathy accounting for the distinct retinal phenotypes. Hum. Mol. Genet. 2018;27:1630–1641. doi: 10.1093/hmg/ddy070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marmorstein A.D., Johnson A.A., Bachman L.A., Andrews-Pfannkoch C., Knudsen T., Gilles B.J., Hill M., Gandhi J.K., Marmorstein L.Y., Pulido J.S. Mutant Best1 Expression and Impaired Phagocytosis in an iPSC Model of Autosomal Recessive Bestrophinopathy. Sci. Rep. 2018;8:4487. doi: 10.1038/s41598-018-21651-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guziewicz K.E., Cideciyan A.V., Beltran W.A., Komáromy A.M., Dufour V.L., Swider M., Iwabe S., Sumaroka A., Kendrick B.T., Ruthel G. BEST1 gene therapy corrects a diffuse retina-wide microdetachment modulated by light exposure. Proc. Natl. Acad. Sci. USA. 2018;115:E2839–E2848. doi: 10.1073/pnas.1720662115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y., Zhang Y., Xu Y., Kittredge A., Ward N., Chen S., Tsang S.H., Yang T. Patient-specific mutations impair BESTROPHIN1's essential role in mediating Ca2+-dependent Cl- currents in human RPE. eLife. 2017;6:e29914. doi: 10.7554/eLife.29914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waugh N., Loveman E., Colquitt J., Royle P., Yeong J.L., Hoad G., Lois N. Treatments for dry age-related macular degeneration and Stargardt disease: a systematic review. Health Technol. Assess. 2018;22:1–168. doi: 10.3310/hta22270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shahi P.K., Hermans D., Sinha D., Brar S., Moulton H., Stulo S., Borys K.D., Capowski E., Pillers D.M., Gamm D.M., Pattnaik B.R. Gene Augmentation and Readthrough Rescue Channelopathy in an iPSC-RPE Model of Congenital Blindness. Am. J. Hum. Genet. 2019;104:310–318. doi: 10.1016/j.ajhg.2018.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cox D.B., Platt R.J., Zhang F. Therapeutic genome editing: prospects and challenges. Nat. Med. 2015;21:121–131. doi: 10.1038/nm.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Popp M.W., Maquat L.E. Leveraging Rules of Nonsense-Mediated mRNA Decay for Genome Engineering and Personalized Medicine. Cell. 2016;165:1319–1322. doi: 10.1016/j.cell.2016.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lindeboom R.G.H., Vermeulen M., Lehner B., Supek F. The impact of nonsense-mediated mRNA decay on genetic disease, gene editing and cancer immunotherapy. Nat. Genet. 2019;51:1645–1651. doi: 10.1038/s41588-019-0517-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sadelain M., Papapetrou E.P., Bushman F.D. Safe harbours for the integration of new DNA in the human genome. Nat. Rev. Cancer. 2011;12:51–58. doi: 10.1038/nrc3179. [DOI] [PubMed] [Google Scholar]
- 36.Shen M.W., Arbab M., Hsu J.Y., Worstell D., Culbertson S.J., Krabbe O., Cassa C.A., Liu D.R., Gifford D.K., Sherwood R.I. Predictable and precise template-free CRISPR editing of pathogenic variants. Nature. 2018;563:646–651. doi: 10.1038/s41586-018-0686-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boon C.J., Klevering B.J., Leroy B.P., Hoyng C.B., Keunen J.E., den Hollander A.I. The spectrum of ocular phenotypes caused by mutations in the BEST1 gene. Prog. Retin. Eye Res. 2009;28:187–205. doi: 10.1016/j.preteyeres.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 38.Ji C., Li Y., Kittredge A., Hopiavuori A., Ward N., Yao P., Fukuda Y., Zhang Y., Tsang S.H., Yang T. Investigation and Restoration of BEST1 Activity in Patient-derived RPEs with Dominant Mutations. Sci. Rep. 2019;9:19026. doi: 10.1038/s41598-019-54892-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson A.A., Lee Y.S., Chadburn A.J., Tammaro P., Manson F.D., Marmorstein L.Y., Marmorstein A.D. Disease-causing mutations associated with four bestrophinopathies exhibit disparate effects on the localization, but not the oligomerization, of Bestrophin-1. Exp. Eye Res. 2014;121:74–85. doi: 10.1016/j.exer.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vasireddy V., Mills J.A., Gaddameedi R., Basner-Tschakarjan E., Kohnke M., Black A.D., Alexandrov K., Zhou S., Maguire A.M., Chung D.C. AAV-mediated gene therapy for choroideremia: preclinical studies in personalized models. PLoS ONE. 2013;8:e61396. doi: 10.1371/journal.pone.0061396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vaisey G., Miller A.N., Long S.B. Distinct regions that control ion selectivity and calcium-dependent activation in the bestrophin ion channel. Proc. Natl. Acad. Sci. USA. 2016;113:E7399–E7408. doi: 10.1073/pnas.1614688113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cideciyan A.V., Sudharsan R., Dufour V.L., Massengill M.T., Iwabe S., Swider M., Lisi B., Sumaroka A., Marinho L.F., Appelbaum T. Mutation-independent rhodopsin gene therapy by knockdown and replacement with a single AAV vector. Proc. Natl. Acad. Sci. USA. 2018;115:E8547–E8556. doi: 10.1073/pnas.1805055115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wienert B., Shin J., Zelin E., Pestal K., Corn J.E. In vitro-transcribed guide RNAs trigger an innate immune response via the RIG-I pathway. PLoS Biol. 2018;16:e2005840. doi: 10.1371/journal.pbio.2005840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.George A.L., Jr. Inherited Channelopathies Associated with Epilepsy. Epilepsy Curr. 2004;4:65–70. doi: 10.1111/j.1535-7597.2004.42010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schaaf C.P. Nicotinic acetylcholine receptors in human genetic disease. Genet. Med. 2014;16:649–656. doi: 10.1038/gim.2014.9. [DOI] [PubMed] [Google Scholar]
- 47.Villa C., Combi R. Potassium Channels and Human Epileptic Phenotypes: An Updated Overview. Front. Cell. Neurosci. 2016;10:81. doi: 10.3389/fncel.2016.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw targeted sequencing files for DNA and RNA sequencing data as well as scRNA-seq data are available via NCBI BioProject: PRJNA633668. Raw patch clamp data are available upon request. Other experimental data are provided in the Supplemental Information, and all source data are available upon request.