Abstract
Aminoacyl-tRNA synthetases (ARSs) are ubiquitous, ancient enzymes that charge amino acids to cognate tRNA molecules, the essential first step of protein translation. Here, we describe 32 individuals from 21 families, presenting with microcephaly, neurodevelopmental delay, seizures, peripheral neuropathy, and ataxia, with de novo heterozygous and bi-allelic mutations in asparaginyl-tRNA synthetase (NARS1). We demonstrate a reduction in NARS1 mRNA expression as well as in NARS1 enzyme levels and activity in both individual fibroblasts and induced neural progenitor cells (iNPCs). Molecular modeling of the recessive c.1633C>T (p.Arg545Cys) variant shows weaker spatial positioning and tRNA selectivity. We conclude that de novo and bi-allelic mutations in NARS1 are a significant cause of neurodevelopmental disease, where the mechanism for de novo variants could be toxic gain-of-function and for recessive variants, partial loss-of-function.
Keywords: aminoacyl-tRNA synthetase, neurodevelopment, next generation sequencing, developmental delay, epilepsy, neuropathy
Introduction
The attachment of tRNA to cognate amino acids is essential for protein translation. Aminoacyl-tRNA synthetases (ARSs) are a group of enzymes encoded by ancient genes which are ubiquitously expressed and highly conserved.1, 2, 3 These enzymes play a fundamental role in the esterification of proteinogenic amino acids to cognate tRNA. In total, 37 genes encoding ARS enzymes have been described. Of these, 20 encode enzymes that function in the cytoplasm, and the remainder relate exclusively to mitochondrial enzymes. Despite the essential canonical function and ubiquitous expression of ARS enzymes, mutations in these genes have been implicated in a variety of human diseases with both recessive and dominant inheritance patterns.2,4, 5, 6 These mutations result in neurological disorders, ranging from mild late-onset peripheral neuropathy to severe multi-systemic neurodevelopmental disorders4,5,7, 8, 9 (Table S1).
Mutations in cytoplasmic ARS-encoding genes cause peripheral nervous system degeneration resulting in Charcot-Marie-Tooth neuropathies (GARS1 and AARS1 [MIM: 601065]) and brain stem and spinal cord hypomyelination (DARS1 [MIM: 603084]). ARSs, and ARSs interacting genes, including DARS1, RARS1 (MIM: 107820), AIMP1 (MIM: 603605), and AARS1, have been implicated in neurodevelopmental disorders and epilepsies. Furthermore, mitochondrial ARS2 mutations are often associated with leukoencephalopathy (AARS2 [MIM: 615889] and DARS2 [MIM: 611105]) or pontocerebellar hypoplasia (RARS2 [MIM: 611524]). More recently, recessive mutations in FARSA (MIM: 602918), VARS1 (MIM: 192150), CARS1 (MIM: 123859), and TARS1 (MIM: 187790), with subsequent partial loss of the ARS protein, have been linked to neurodevelopmental phenotypes.10, 11, 12, 13, 14 Modes of inheritance can be dominant or recessive; in cases such as AARS1, YARS1 (MIM: 603623), MARS1 (MIM: 156560), HARS1 (MIM: 142810), and GARS1, both patterns can occur.6
The loss of function associated with mutations in ARSs is attributed to decreased aminoacylation efficiency or misfolding, causing protein instability with lower steady-state levels.13 However, in some cases (GARS1, YARS1, and AARS1), it has not been possible to ascribe the phenotype to a loss of primary aminoacylation.15, 16, 17 Overall, the physiological functions of ARS genes and previously identified disease associations indicate an essential biological role for these proteins, implying that defects in all ARSs incur disease.6
Asparaginyl-tRNAAsn is generated by asparaginyl-tRNA synthetase (NARS1 (MIM: 108410; RefSeq accession number NM_004539.4] in a reaction involving two steps. NARS1 first catalyzes the ATP-dependent activation of asparagine (Asn) into Asn∼AMP with the release of pyrophosphate, and then transfers the activated Asn onto tRNAAsn with the release of AMP (Figure 1A). Here, we report the clinical phenotypes associated with de novo dominant and bi-allelic, autosomal recessive mutations in NARS1 in 32 affected individuals from 21 families. We provide genetic proof for these mutations and analyze their impact through the use of individual cell lines, neural progenitor cells, and molecular modeling.
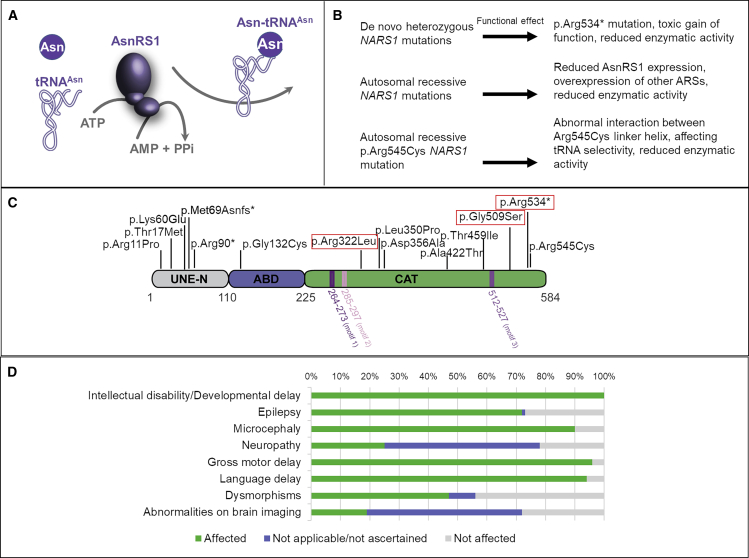
Figure 1.
AsnRS1 Protein Structure and Function
(A) AA asparagine (Asn) is ligated to tRNAAsn and catalyzed by AsnRS1 and ATP to produce Asn-tRNA (Asn), AMP, and pyrophosphate.
(B) NARS1 mutations and their predicted functional effect.
(C) Schematic representation of human ARS1 primary structure. Three main domains are depicted: the unique domain (UNE-N), the anticodon binding domain (ABD), and the catalytic domain (CAT). The nature and position of the mutants are shown above the primary structure, de novo boxed in red, and the positions of the domains are indicated below, including motif 1 (involved in AsnRS1 dimerization) and motifs 2 and 3 (which form the active site).
(D) Bar graph summarizing proportions of various clinical findings affecting individuals with NARS1 mutations.
Subjects and Methods
Study Participants
Individuals were recruited via an international collaborative network of research and diagnostic sequencing laboratories. Samples and clinical information were obtained, with informed consent, from each institution using local institutional review board (IRB) ethics for functional analysis of human DNA and biomaterial. Clinical data collection involved a detailed review of medical records, photographs, videos, and phone interviews, as well as a clinical re-evaluation by a neurologist. Tables S2–S4 summarize the clinical and demographic details of the included cases.
Sequencing
Exome sequencing was carried out using a number of methods in different centers with different analysis platforms and pipelines used (see Supplemental Methods, Section 2).
Bioinformatic Analysis
cDNA and protein sequence variants are described in accordance with the recommendations of the Human Genome Variation Society using Ensembl ENSG00000134440 and ENST00000256854.10 as the reference sequences. Evolutionary conservation of nucleotides was assessed using PhyloP (46 vertebrate species) and genomic evolutionary rate profiling (GERP) scores.18 These were accessed through the University of California—San Francisco (UCSC) Genome Browser19 using genomic coordinates from GRCh37/hg19. Grantham scores were used to assess the physicochemical nature of the amino acid (AA) substitutions. In silico analyses of sequence variants were performed using the pathogenicity prediction tools SIFT, PolyPhen-2, and Mutation Taster version 2.
Our bioinformatics filtering strategy screened for exonic and donor/acceptor splicing variants. In accordance with the pedigree and phenotype, priority was given to rare variants (<0.01% in public databases, including 1000 Genomes Project; National Heart, Lung, and Blood Institute [NHLBI] Exome Variant Server; Complete Genomics 69; and Exome Aggregation Consortium [ExAC v0.2]) fitting a recessive (homozygous or compound heterozygous) or a de novo model and/or variants in genes previously linked to epilepsy, developmental delay, intellectual disability, and other neurological disorders. Upon whole-exome sequencing (WES) analysis of the index family (F9), the NARS1 variant c.1633C>T (p.Arg545Cys) was picked up according to its frequency and prediction tool scores (SIFT—damaging [score = 1], PolyPhen—damaging [score = 1], GERP—5.5, Mutation Taster—0.999992). All the candidate variants were further verified through the use of Sanger sequencing.
Generation of the nrs1 Vector
The pJR1-41XU-nrs1 expression vector was used to express SchizoSaccharomyces pombe nrs1 by amplifying the coding sequence of nrs1 from S. pombe DNA with the Nrs1-PJR-F and Nrs1-PJR-R primers (all primers are provided in Table S5) through the use of Phusion HF polymerase from New England Biolabs (NEB). The PCR product was cloned into XhoI digested pJR1-41XU20 through the use of CloneEZ from Genscript. Plasmids were sequenced to confirm the correct insertion of the fragment.
Deletion of nrs1 Gene in S. pombe Cells
JB775 (h- ade6-M216 ura4-D18 leu1-32) cells were synchronized, made competent, and transformed as previously described.21 Cells were transformed using the plasmid containing the nrs1 gene, pJR1-41XU-nrs1, and transformants were selected according to growth in Edinburgh minimal medium (EMM) + ade + leu, generating the strain MR397. The nrs1 gene was deleted in MR397 cells through the use of the standard method via homologous recombination with the NatMx6 cassette22,23 using the primers Nrs1DelFw and Nrs1DelRv (Table S5). Transformants were selected in EMM + Nat with no thiamine to promote the expression of the nrs1 gene from the plasmid. Deletions were checked via PCR using primers Nrs1ck-L and kanR and Nrs1ck-R and kanF (Table S5). The strain generated was named MR409. MR409 cells were synchronized, made competent, and transformed as previously described.21 The plasmids of the pJR-41XL series contained either the empty vector, wild-type NARS1, or the NARS1 variants described. Transformants were selected in EMM + ade strains.
Cell Culture
Fibroblasts of affected individuals carrying the homozygous c.50C>T (p.Thr17Met), c.32G>C (p.Arg11Pro), and c.1633C>T (p.Arg545Cys) and compound heterozygous c.1067A>C (p.Asp356Ala) and c.203dupA (p.Met69Aspfs∗4), as well as of corresponding controls, were grown in high-glucose Dulbecco’s modified Eagle’s medium (Sigma) supplemented with 10% fetal bovine serum and 1% penicillin and streptomycin.
Semiquantitative RT-PCR for Individual Lymphoblasts
Using TRIzol (Zymo research), as per manufacturer’s instructions, total RNA was extracted from immortalized lymphoblasts available from P2 and parents. The concentration and purity of RNA was determined spectrophotometrically. 1 μg of RNA was reverse transcribed to first strand cDNA through the use of random primers and Moloney murine leukemia virus reverse transcriptase (Promega). GoTaq® Green Master Mix (Promega) was used and PCR reactions were performed with the following protocol: 95°C—2 min (95°C—30 s, 60°C—30 s, 73°C—1 min) for 35 cycles, 73°C—5 min, and 4°C hold. Two exponential curves representing the product formation were determined for both primer pairs. Cycles 28 and 29 were chosen for NARS1 and GAPDH, respectively,we so that amplification rates were in the linear range for semiquantitative comparisons. Reactions were repeated in triplicate.
Western Blotting
For western blotting analysis, protein lysates were obtained from cultured fibroblasts and total protein concentration was measured by means of a Bradford assay. Aliquots of total protein (15 μg) were loaded on 4%–12% sodium dodecyl sulfate (SDS)-polyacrylamide gels (NuPAGE 4%–12% Bis-Tris Protein Gels, ThermoFisher Scientific), transferred to polyvinylidene fluoride membranes, and blocked and incubated overnight with a polyclonal antibody recognizing AsnRS1 (anti-rabbit 1:1000; Proteintech). Secondary antibody was added for 1 h, and signal was detected using enhanced chemiluminescence (ECL) reagents (Amersham Biosciences). Anti-beta-actin antibody (Sigma Aldrich, A3853; 1 in 5,000) was used as a loading control. Blots were repeated in triplicate and statistics were performed using Prism 6. Data are presented as mean ± standard error of the mean (SEM). The significance between the variables was shown based on the p value obtained (ns indicates p > 0.05, ∗p < 0.05, ∗∗p < 0.005, ∗∗∗p < 0.0005, ∗∗∗∗p < 0.00005).
Blue Native Polyacrylamide Gel Electrophoresis (BN-PAGE)
Fibroblast pellets were lysed using 10mM Tris (pH 8), 150mM NaCl, 0.1% NP40 with physical agitation for 30 min, centrifuged at 8,000xg to remove debris. The supernatant was removed, and total protein was quantified with the Bradford assay. Protein concentrations were equalized and prepared to 20μl at 1μg/μl using NativePAGE sample buffer (Thermo) and 1ul of NuPAGE 5% G-250 Sample Additive (Thermo), and then loaded to a NativePAGE 3%–12% Bis-Tris Protein Gel (Thermo). Proteins were transferred to polyvinylidene fluoride (PVDF) membrane through the use of an iBlot2 PVDF Mini transfer stack (ThermoFisher Scientific) and probed with anti-NARS1 monoclonal antibody (Abcam ab129162, 1:5000) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH; Santa Cruz). Blots were repeated in triplicate, and differences were analyzed using Welch-corrected t test.
Induced Neuronal Progenitor Cell (iNPC) Conversion
Based on the protocol published by Meyer et al.,24 iNPCs were generated from primary fibroblasts by transduction with Oct4-, Klf4, and c-Myc-Sendai virus, followed by culturing in neuronal progenitor cell (NPC) induction media (1:1 DMEM/F-12: Neurobasal, 2× N2, 2× B27, 1% GlutaMAX, 10ng/mL hLIF, 3μM CHIR99021, and 2 μM SB431542). Neuroepithelial colonies were formed after 3–4 weeks of culturing. These were then isolated and expanded before we extracted total RNA from individual fibroblasts, age and sex matched healthy control fibroblasts, and iNPCs cells using the mirVana miRNA Isolation Kit (Ambion) for gene expression analysis by qPCR to confirm iNPC lineage and RNAseq in control and individual iNPCs in order to identify differentially expressed genes.
qPCR
Cell pellets from individual fibroblasts and iNPCs were lysed using a Trizol reagent. Following the addition of chloroform, the aqueous phase was transferred to RNeasy spin column (QIAGEN) for RNA isolation and resuspension. cDNA was generated using the reverse transcriptase kit (Applied Biosystems) and qPCR (Applied Biosystems 7900HT) was performed in triplicates using SYBR Green PCR Master Mix (Invitrogen, 4309155). Samples were normalized to expression of GAPDH and β-actin and repeated in triplicate.
RNaseq
Libraries were prepared using Illumina TruSeq Stranded Total RNA with Ribo-Zero Human kit and were sequenced on an Illumina HiSeq 2500 using a paired-end protocol. Quality of sequencing reads were ensured using FastQC. Reads were aligned using STAR aligner, and variants were called using the two-pass protocol outlined in the GATK documentation (see Web Resources). The numbers of reads were counted using HTSeq-count.25 Differentially expressed genes were identified using the DESeq2 Bioconductor package.26 Differentially expressed genes with a false discovery rate of ≤0.1 and a log2 (fold change) ≥1 were considered significant. Gene set enrichment analysis was performed using the CPDB web tool.
NARS1 Enzyme Assay
Aminoacylation was assessed by measuring NARS1 activity in cultured fibroblasts and lymphoblasts. Cell lysates (cytosolic fraction) were incubated in triplicate at 37°C for 10 min in a reaction buffer containing 50mmol/L Tris buffer pH 7.5; 12mmol/L MgCl2; 25mmol/L KCl; 1 mg/mL bovine serum albumin; 0.5mmol/L spermine; 1mmol/L ATP; 0.2mmol/L yeast total tRNA; 1 mmol/L dithiothreitol; and 0.3mmol/L [15N2]-asparagine, [13C4,15N]-threonine, [D2]-glycine, [15N2]-arginine, and [D4]-lysine. The reaction was terminated using trichloroacetic acid. Ammonia was then added to release the labeled AAs from the tRNAs. [13C2,15N]-glycine and [13C6]-arginine were added as internal standards, and the labeled AAs were quantified via LC-MS/MS. Intra-assay variation was determined as <15% of TARS1, GARS1, RARS1 KARS1 activity which were simultaneously detected as control enzymes. AsnRS1 activities were measured blind, and testing was repeated in triplicate. Data are presented as mean ± SEM. The statistical significance of the difference of AsnRS1 activity between controls and affected individuals and/or carriers was determined using a Student’s t test with a 95% confidence interval through the use of SPSS 26.
Molecular Modeling Analysis
The crystal structure of Brugia malayi AsnRS1 with a 65% identity to human AsnRS1, stored under the 2XGT code in the Protein Data Bank, was used for the molecular modeling analysis. The homology model of the dimeric human AsnRS1 overlapped with the S. cerevisiae DARS-tRNAAsp ligase, co-crystallized with tRNA molecule PDB 4WJ4, thus having a similar domain organization and sharing 27.5% of sequence identity with human AsnRS1. The protein was visualized with the University of California–San Francisco Chimera software.27
Results
Genetic Analysis
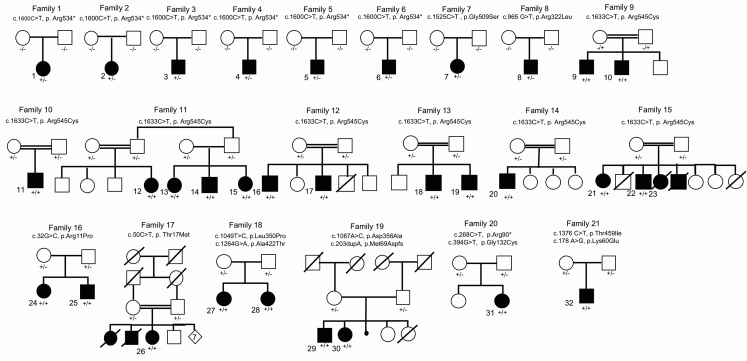
We identified 21 families (F1–F21) and 32 affected individuals (P1–P32) with mutations in NARS1 (Figure 1B shows NARS1 variant schematic and Figure 2 illustrates pedigrees). Eight families had de novo heterozygous variants; six had c.1600C>T (p.Arg534∗) (F1–F6, P1–P6); one had c.1525G>A (p.Gly509Ser) (F7, P7); and one had c.965G>T (p.Arg322Leu) (F8, P8). These variants were not present in our 652 normal brain series or in the gnomAD database.
Figure 2.
Pedigrees of the 21 Families and 32 Affected Individuals Identified in This Study with de novo and Bi-allelic Mutations in NARS1
Filled symbols represent affected individuals and double bars represent consanguinity in the family. −/−, +/−, and +/+ represent wild-type, heterozygous, and homozygous variants, respectively.
Bi-allelic variants were found in thirteen families. Seven have homozygous c.1633C>T (p.Arg545Cys) variants (F9–F15, P9–P23); one has homozygous c.50C>T (p.Thr17Met) (F17, P26); and one has two siblings with homozygous c.32G>C (p.Arg11Pro) (F16, P24 and 25). For compound variants, one family has two siblings with compound heterozygous c.1067A>C (p.Asp356Ala) and c.203dupA (p.Met69Aspfs∗4) (F19, P29 and P30). Two siblings had compound heterozygous c.1049T>C (p.Leu350Pro) and c.1264G>A (p.Ala422Thr) variants (F18, P27 and P28). There was one case with the compound heterozygous variants c.268C>T (p.Arg90∗) and c.394G>T (p.Gly132Cys) (F20, P31) and a final individual with compound heterozygous c.1376C>T (p.Thr459Ile) and c.178A>G (p.Lys60Glu) variants (F21, P32). In gnomAD, c.1264G>A (p.Ala422Thr) is present in six heterozygote individuals, whereas c.1633C>T (p.Arg545Cys) and c.50C>T (p.Thr17Met) were present in five and four heterozygotes, respectively. The c.100 A>T (p.Met34Leu), c.203dupA (p.Met69Aspfs∗4), and c.1049T>C (p.Leu350Pro) variants were absent, while c.32G>C (p.Arg11Pro) was present in one individual. The c.1067A>C (p.Asp356Ala) variant in family 19 was present in 264 heterozygotes, suggesting that this variant may modify the phenotype and be pathogenic only when in trans with a severe variant such as c.203dupA (p.Met69Aspfs∗4).
Clinical Characteristics
Table 1 summarizes the core clinical features of affected individuals with NARS1 defects (see Tables S2–S4 for additional details). All individuals had global developmental delay (GDD) and intellectual disability, which varied in severity from moderate to profound. They had marked delays in language development. Motor development was also severely impaired, and one individual never acquired autonomous ambulation. Microcephaly was observed in the majority of cases (90%). These cases predominantly presented with primary microcephaly; however, secondary microcephaly was also noted. Epilepsy was highly associated with the phenotype, affecting 23 cases (74.2%), with six individuals experiencing seizures below the age of one. The semiology of these attacks varied, with a mixture of partial, myoclonic, and generalized tonic-clonic seizures described. An ataxic gait, poor balance, and dysarthria were frequently detected on examination; this suggests an additional neurodegenerative process; however, no structural abnormality of the cerebellum was observed on imaging. A demyelinating peripheral neuropathy occurred in eight individuals (25%) who had distal leg muscle atrophy. Dysmorphic features described included abnormal hands (e.g., clinodactyly, fetal finger pad, two-to-three-toe syndactyly, slender fingers) and/or feet (e.g., small feet, toe syndactyly, slender feet). Upslanting palpebral fissures was the most common facial dysmorphism reported. A broad forehead, wide mouth, wide-set teeth, and low-set ears with overfolded helices were also described. Skeletal abnormalities including scoliosis, pronounced thoracic kyphosis, and pes-cavus were also noted. Behavioral traits associated with the phenotype included impulsivity, stereotypies with repetitive speech and/or hand movements, and selective feeding rituals.
Table 1.
Summary of NARS1 Variants and Clinical Features of Affected Individuals
| Variant: Nucleotide, Protein | c.1600C>T, p.Arg534∗ | c.1525G>A, p.Gly509Ser | c.965G>T, p.Arg322Leu | c.1633 >T, p.Arg545Cys | c.32G>C, p.Arg11Pro | c.50C>T, p.Thr17Met | c.1049T>C c.1264G>A, p.Leu350Pro p.Ala422Thr | c.1067A>C c.203dupA, p.Asp356Ala p.Met69Aspfs∗4 | c.268 C>T c.394G>T, p.Arg90∗ p.Gly132Cys | c.1376 C>T, c.178 A>G, p.Thr459Ile, p.Lys60Glu |
|---|---|---|---|---|---|---|---|---|---|---|
| Variant type | de novo heterozygous |
de novo heterozygous |
de novo heterozygous |
homozygous | homozygous | homozygous | compound heterozygous |
compound heterozygous |
compound heterozygous |
compound heterozygous |
| Inheritance | AD de novo | AD de novo | AD de novo | AR | AR | AR | AR | AR | AR | AR |
| Family | 1–6 | 7 | 8 | 9–15 | 16 | 17 | 18 | 19 | 20 | 21 |
| Affected Individual(s) | 1–6 | 7 | 8 | 9–23 | 24–25 | 26 | 27–28 | 29–30 | 31 | 32 |
| Ethnicity/country of origin | European | UK | European | Pakistan/North India | Kosovo | Libya | German | Turkey | Canada | USA |
| Age at onset | birth | birth | birth | childhood | childhood | birth | birth | birth | birth | childhood |
| Consanguinity | no | no | no | yes | no | yes | No | no | no | no |
| Presentation | severe GDD | severe GDD | severe GDD | severe GDD | seizures | seizures | mod GDD | mod GDD | severe GDD | severe GDD |
| ID | yes | yes | yes | yes | yes | yes | yes | yes | yes | yes |
| Microcephaly | yes | no | NA | yes | yes | yes | yes | yes | yes | yes |
| Dysmorphic | yes | yes | yes | yes | no | NA | no | no | yes | no |
| Seizures Affected Individuals |
yes 1, 2, 4, 5, 6 |
yes | yes | yes 9, 14, 15, 18, 19, 21, 22, 23 |
yes all individuals |
yes | yes 27 |
yes all individuals |
yes | yes |
| Spasticity Affected Individuals |
yes 3, 4, 6 |
no | yes | no hypotonia in 9, 10, 16, 17 |
yes 24 |
na | no hypotonia |
na | no hypotonia |
yes |
| Neuropathy Affected Individuals |
Yes 1, 2, 5 |
NA | NA | yes 9, 10, 20 |
NA | NA | yes | NA | NA | NA |
| Ataxia Affected Individuals |
yes all individuals |
NA | Yes | yes 9–12, 21 |
NA | NA | yes | NA | yes | yes |
AD = autosomal dominant, AR = autosomal recessive, GDD = global developmental delay, ID = intellectual disability Mod = moderate, NA = not available.
Genotype-Phenotype Correlations
Family 16, with the homozygous variant c.32G>C (p.Arg11Pro), had a particularly severe clinical picture comprised of severe developmental delay, progressive microcephaly, refractory seizures from infancy, and arrested myelination with pronounced cerebral atrophy on MRI (see Supplemental Note, Table S3, and Figure 3).
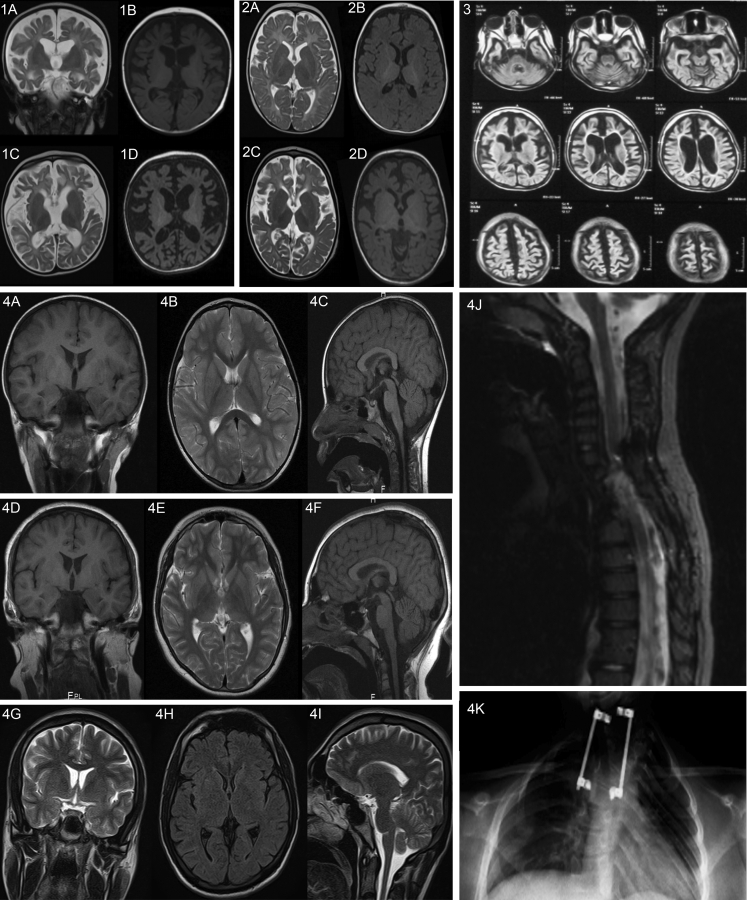
Figure 3.
Radiological Findings of Individuals in Our Cohort
Set 1: Individual homozygous for c.32G>C (p.Arg11Pro). Upper row images (coronal T2-WI [1A] and axial T1-WI [1B]) at the age of 10 months show severely delayed myelination and fronto-temporal atrophy. Lower row images (axial T2-WI [1C] and axial T1-WI [1D]) repeated at the age of 18 months show progressive and global brain atrophy with an emerging pattern of severe hypomyelination.
Set 2: An additional homozygous c.32G>C (p.Arg11Pro) individual. Upper row images (axial T2-WI [2A] and axial T1-WI [2B]) at the age of 8 months show mild fronto-temporal underdevelopment and severely delayed myelination. Lower row images (axial T2-WI [2C] and axial T1-W1 [2D]) repeated at the age of 2 years shows progressive and global brain atrophy along with severe hypomyelination.
Set 3: Individual homozygous for c.50C>T (p.Thr17Met). Axial fluid-attenuated inversion recovery (FLAIR) images at the age of 9 months show global atrophy involving the cerebral and cerebellar hemispheres along with severe hypomyelination.
Set 4: MRI images of an individual with the homozygous c.1633C>T (p.Arg545Cyc) variant. Coronal T1-WI (4A), axial T2-WI (4B), and sagittal T1-WI (4C) at the age of 4 years; coronal T1-WI (4D), axial T2-WI (4E), and sagittal T1-WI (4F) at the age of 11 years; and coronal T2-WI (4G), axial FLAIR (4H), and sagittal T2-WI (4I) at the age of 20 years. These demonstrate normal intracranial appearances across the three different ages. This individual had an upper thoracic scoliosis, which was operatively corrected at the age of 4, demonstrated on the sagittal T2-WI of the spine (4J) and frontal projection radiograph of the chest/thoracic spine (4K).
Otherwise, imaging was normal apart from microcephaly. There was no common structural change across all cases. Individuals with the de novo c.1600C>T (p.Arg534∗) variant showed severe microcephaly. In one family with this variant (F6), mild atrophy was observed (see Supplemental Information, Tables S2–S4, and Figure 3).
Individuals homozygous for c.1633C>T (p.Arg545Cys) demonstrated hypotonia and predominantly distal weakness. Spasticity was observed in individuals with the c.32G>C (p.Arg11Pro) or de novo variants.
A demyelinating polyneuropathy was documented in individuals homozygous for the c.1633C>T (p.Arg545Cys) variant (P9, P10, and P20), and in one case, this was confirmed with a sural nerve biopsy (F9, P9). It was also described in individuals with the de novo c.1600C>T (p.Arg534∗) variant (P1, P2, and P5) and in the family with the compound heterozygous c.1049T>C (p.Leu350Pro) and c.1264G>A (p.Ala422Thr) variants (F18, P27 and P28).
Pathogenicity of NARS1 Variants
NARS1 is intolerant to loss of function (missense variants constraint is Z = 0.87). We identified de novo NARS1 mutations in eight families (F1–F8, P1–P8) with similar phenotypes. A variant at codon 534 recurred in six families (F1–F6, P1–P6). The two other de novo variants altered codons 322 and 509. The c.1600C>T (p.Arg534∗) variant is located 15 AAs from the end of the 548-AA protein, representing a potential hotspot for pathogenic mutations. Arginine at codons 534 and 545 IS universally conserved in AsnRS1 from all three major taxonomic groupings, implying a significant structural or functional role.
The homozygous c.1633C>T (p.Arg545Cys) variant was observed in seven families with recessive disease. This variant affects the same C-terminal catalytic stretch as does c.1600C>T (p.Arg534∗), and therefore it might have a comparable mechanistic effect to c.1600C>T (p.Arg534∗).
The c.1067A>C (p.Asp356Ala) variant was found in trans with the only recessive truncating allele observed thus far at c.203dupA (p.Met69Aspfs∗4) (P29 and P30). Two missense variants (c.965G>T [p.Arg322Leu] and c.653T>C, p.Asn218Ser) were found in P8; however, because c.965G>T (p.Arg322Leu) occurred de novo, it could not be determined whether these variants were in cis or in trans. Moreover, the frequency of c.653T>C (p.Asn218Ser) in the gnomAD database (78 heterozygotes) suggests it is unlikely to be associated with a severe phenotype, leaving c.965G>T (p.Arg322Leu) as the most likely disease-causing variant. The Arg322 residue is essential for enzymatic activity and therefore is predicted to cause impaired enzyme activity. Both the c.50C>T (p.Thr17Met) and c.32G>C (p.Arg11Pro) variants are in the N-terminal UNE-N appended domain of AsnRS1, which is specific to eukaryotes, and has recently been shown to have chemokine activity.28
Functional Characterization
Western Blotting and RT-PCR
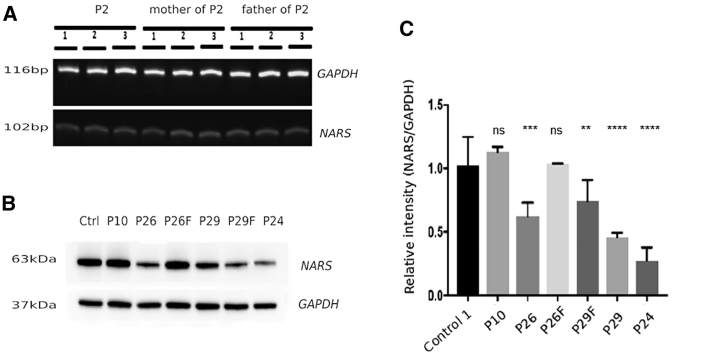
Given the potential loss of function in homozygous NARS1 individuals, we investigated gene expression levels through the use of semiquantitative PCR and protein levels through western blotting of AsnRS1 from lymphoblasts and fibroblasts from families harboring the p.Arg545Cys, p.Thr17Met, p.Asp356Ala, p.Met69Aspfs∗4, and p.Arg11Pro variants. In all instances, both gene expression and protein levels are reduced (Figure 4A–4E).
Figure 4.
Protein Levels of AsnRS1 Are Reduced in Individual-Derived Cells
(A) RT-PCR of the de novo c.1600C>T (p.Arg534∗) variant in P2 and parents (B) western blotting and (C) quantification graph of individuals with NARS1 mutations compared with controls. Ctrl = control, P10 = homozygous c.1633C>T (p.Arg545Cys), P26 = homozygous c.50C>T (p.Thr17Met), P29 = compound heterozygous (c.1067A>C (p.Asp356Ala) and c.203dupA (p.Met69Aspfs∗4) (F denotes father of individuals), P24 = homozygous c.32G>C (p.Arg11Pro).
iNPCs
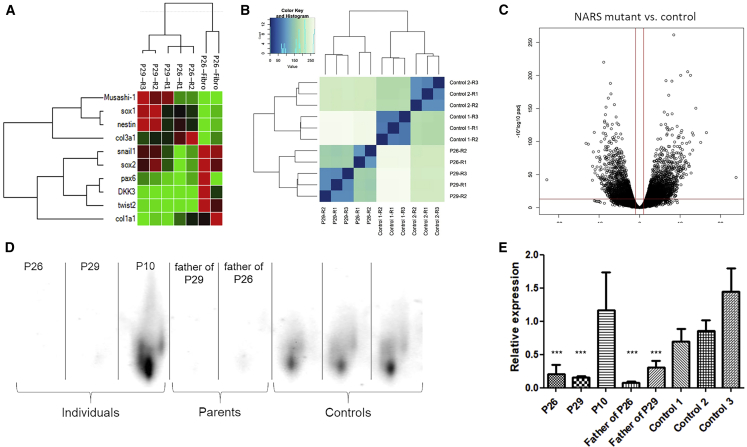
iNPC colonies were produced and isolated from fibroblasts from P26 (c.50C>T), P29 (c.1067A>C), and P30 (c.203dupA). From the isolated colonies, gene expression was determined by using qPCR to select iNPC populations which presented decreased expression of fibroblast markers COL1A1, COL3A1, TWIST2, and DKK3, as well as increased numbers of NPC markers NES, SOX1, and MSI1, and the iNPC population was expanded to be subsequently used for RNA sequencing (RNaseq). RNaseq showed normal NARS1 expression in iNPCs from affected individuals carrying the c.50C>T and c.1067A>C mutations, and decreased expression of the c.203dupA NARS1 allele in the P29 cells. Interestingly, iNPCs from affected individuals show increased expression of several other ARSs (DARS1, GARS1, RARS1, SARS1, TARS1, WARS1, and YARS1) (Figure 5). This could be explained by the fact that the NARS1 mutant(s) are inducing the integrated stress response (ISR), which activated a number of ARS genes as a result of the loss-of-function homozygous recessive variants. Impaired synthetase function may reduce the amount of charged tRNA available for translation elongation, with a possible increase in the levels of uncharged tRNA. Uncharged tRNAs produced as a result of AA deprivation have been reported to bind GCN2, leading to the activation of the ISR.29Analysis of the cellular pathways (Reactome, Gene Ontology) associated with genes with significantly altered mRNA levels showed that upregulated genes were enriched (adjusted p value < 0.01) for pathways heavily associated with protein translation and processing such as endoplasmic reticulum (ER) and Golgi protein processing and ribosomal homeostasis. In addition, increased action of VEGFR1/2 (upregulated by ATF4, which is one of the key transcription factors in the ISR) was suggested.
Figure 5.
BN-PAGE and iNPC RNA-Sequencing
(A) iNPCs from P26 (c.50C>T [p.Thr17Met]) and P29 (c.203dupA [p.Met69Aspfs∗4] and c.1067A>C [p.Asp356Ala]) exhibit increased expression of most iNPC markers (sox1, sox2, nestin, snail1, pax6, DKK3, twist2, and Musashi-1) compared to fibroblast (fbb) as measured by qPCR, shown with hierarchal clustering.
(B) Heatmap with hierarchal clustering generated using all gene counts from RNaseq distinction of control (Ctrl1 a–c, Ctrl2 a–c) and individual-derived (P26 a–b, P29 a–c) iNPCs.
(C) Volcano plot showing log2 of fold change in NARS mutant iNPCs compared to controls and −log10 (adjusted p value).
(D) BN-PAGE western blot showing reduced levels of the AsnRS1 dimer in individuals P26 and P29 and fathers compared to control, but not for individual P10.
(E) Quantification of BN-PAGE western blot AsnRS1 dimer formation, showing significantly (∗∗∗p < 0.001) reduced levels of the AsnRS1 in individuals P26 and P29 and fathers compared to control but not change for P10.
P26 = homozygous c.50C>T (p.Thr17Met), P29 = c.203dupA (p.Met69Aspfs∗4), c.1067A>C (p.Asp356Ala), P10 = c.1633C>T (p.Arh545Cys), father of P26 = heterozygous c.50C>T (p.Thr17Met), father of P29 = c.1067A>C (p.Asp356Ala).
Blue-Native Polyacrylamide Gel Electrophoresis (BN-PAGE)
Similar to most other disease-associated ARSs, AsnRS1 functions as a class II homodimer.30 We showed severely reduced dimer formation in P26 (c.50C>T [p.Thr17Met]) and P29 (c.1067A>C [p.Asn356Ala] and c.203dupA [p.Met69Aspfs∗4]) compared to healthy controls (Figure 5E). The unaffected parents carrying one heterozygous mutation each also appeared to show a decreased level of the AsnRS1 dimer. P10 (c.1633C>T [p.Arg545Cys]) showed an AsnRS1 dimer amount comparable to that healthy controls. The decreased AsnRS1 dimer formation observed in fibroblasts from P26 and P29 shown by BN-PAGE accounts for the apparent deficit in aminoacylation capacity, despite showing no consistent decrease in the levels of AsnRS1 monomers. This idea is further supported by the molecular model simulation (Figure 6) that predicts an unstable dimer for the p.Asn356Ala mutant because this substitution is located at the interface between the two AsnRS1 monomers.
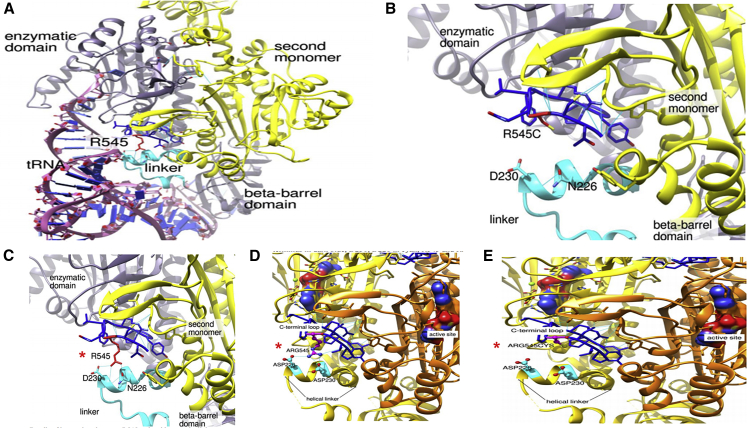
Figure 6.
Molecular Modeling of the NARS1 p.Arg545Cys Homozygous Variant
The crystal structure is based on B.malayi AsnRS1. AsnRS1 is a homodimer; one AsnRS1 monomer is given in yellow and one in orange. Analog of the transition state presented in the surface representation, C terminus in dark blue, Asp230 and Asp226 in cyan, Arg545 and Arg545Cys in magenta.
(A) Interaction between AsnRS1 and tRNA with residues on the helical linker.
(B–E) Zoom in on the C terminus helical linker region, (B) and (E) show loss of molecular interaction and folding of the p.Arg545Cys variant (∗).
ARS Enzymatic Assays
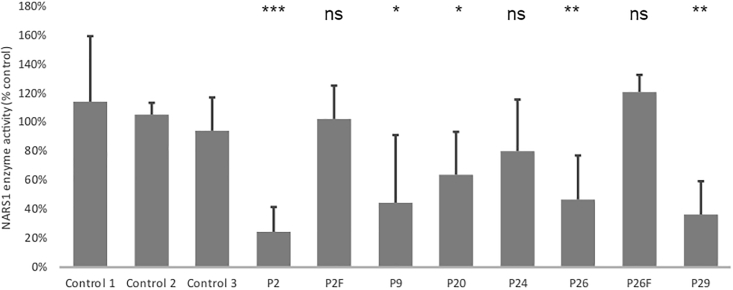
In comparison with controls, AsnRS1 enzymatic activity was reduced in proband-derived fibroblasts and lymphoblasts. The most dramatic decrease was observed for P2 (de novo c.1600C>T [p.Arg534∗]), and the mildest decrease was observed for P24 (c.32G>C [p.Arg11Pro], 80% of the controls). AA residue Arg11 is located in the 5′ end of the non-canonical UNE-N domain (Figure 7 and Figure S11), which has recently been shown to elicit cell migration of human immune cells via migration of CC chemokine receptor 3 (CCR3) in an autoimmune disease associated with ARS genes.28
Figure 7.
Reduced Asparaginyl-tRNA Synthetase Activity in Individuals with Homozygous NARS1 Variants
c.1600C>T (p.Arg534∗) (P2), c.1633C>T (p.Arg545Cys) (P9 and P20), c.32G>C (p.Arg11Pro) (P24), c.50C>T (p.Thr17Met) (P26), and c.1067A>C (p.Asp356Ala)/c.203dupA (p.M69Aspfs∗4) (P29) NARS1 variants in comparison to the average of three unrelated fibroblast cell lines. (All cell lines are fibroblast except P2, which is a lymphoblast cell line. Control values for lymphoblast are similar to fibroblasts.) n = 9, p value FDR < 0.01.
Discussion
We identified de novo heterozygous and bi-allelic mutations in NARS1 in 32 individuals with a neurodevelopmental phenotype. Mutations included recessive mutation hotspots affecting AA residues Arg534 and Arg545, respectively, both located in the last 40 AAs of the protein. Two homozygous variants identified at the 5′ end, c.32G>C (p.Arg11Pro) and c.50C>T (p.Thr17Met), were associated with a severe clinical phenotype. Other mutations in NARS1 were spread throughout and did not cluster in any particular region of the gene.
The clinical phenotypes associated with homozygous variants c.32G>C (p.Arg11Pro) and c.50C>T (p.Thr17Met) correlate with reduced protein levels and could reflect impaired protein stability as suggested by the structural modeling of c.1633C>T (p.Arg545Cys) (Figure 6). Interestingly, MRI imaging of individuals harboring the c.32G>C (p.Arg11Pro) and c.50C>T (p.Thr17Met) variants showed atrophy and white matter abnormalities. In contrast, no such changes were identified in individuals with the p.Arg545Cys variant. The clustering of variants and associated phenotypes at the N and C termini suggests these regions are functionally important and disrupt the protein homodimer and ATP-binding and/or catalytic domain in NARS1. These two variants produced elevated AsnRS1 enzyme activity, which can be attributed to their location in the N-terminal extension domain. This domain has additional non-translational functions, enabling enzymatic activity of the modified protein. When we examine protein expression, protein synthesis, and the aminoacetylation activity, it is clear that the non-translational functions of such ARS proteins, regulated by the newly evolved appended domains such as UNE-N, don’t seem necessary for ARS activity (Figure 7 and Figure S11).
Our functional data, including fibroblasts and iNPCs transcriptomics, suggest that the majority of NARS1 mutations cause a loss of the enzymatic protein by reduced expression and disruption of dimer formation. This results in abnormal protein synthesis and processing with a compensatory increase in expression of other ARSs (Figures 4, 5, and 7 and Figure S11). The increased activity of VEGFR1 and VEGFR 2 was of interest considering the reported actions of other ARSs, as in GARS and EPRS via GAIT complex, SARS1, TARS1, and mini WARS1 on VEGF-related signaling.31 Pathways associated with downregulated genes were typically associated with cell cycle progression, DNA repair and replication such as G2/M checkpoint, homology directed repair, and telomere maintenance pathways; this suggests that this alteration of cellular proliferation could be a result of decreased protein synthesis (Figures S5–S10). In general, the mutations in NARS1 resulted in loss of function in both studied iNPC cell lines (P26 and P29), leading to a transcriptomic signature of induced ISR, upregulation of protein translation and processing in the ER and Golgi, and altered ribosomal homeostasis. This is similar to the results of other studies in cells with reduced aminoacylation activity in disease-associated mutations in other cytosolic ARSs.5,31,32
The recurrent homozygous c.1633C>T (p.Arg545Cys) variant in the western blot (Figure 4) and yeast model (Figure S12) showed near normal protein levels and an increased yeast growth suggestive of a gain-of-function mechanism. However, protein modeling of this variant demonstrated loss of the helix linker, and this indicates reduced tRNA interaction and catalytic activity. This loss-of-function effect was evidenced by the reduced aminoacylation activity to 40% compared to controls (Figure 7 and Figure S11). This effect could potentially be more harmful for cells of the nervous system than for a unicellular organism. One of the possibilities is that the NARS1 mutant mischarges a tRNA in the human cells that might be less conserved in fungi. Thus, the mischarging would be reduced in yeast, hence the better growth without side effects. Molecular modeling has shown that the p.Arg545Cys variant lies within a region that probably interacts with the sugar-phosphate backbone of the tRNA (at positions 68–69), close to the active site of the enzyme.33 Replacing the bulky arginine with a cysteine does not seems to perturb the enzyme’s overall structure (Figure 6). However, by disrupting the tRNA-enzyme contact, this variant may alter the enzyme selectivity toward tRNA, decreasing the overall affinity for tRNA. AsnRS1 enzyme activity for individuals homozygous for this variant (P9 and P20) showed decreased activity (Figure 7 and Figure S11). Similarly, the de novo c.1600C>T (p.Arg534∗) variant, located adjacent to the end of the protein, has a gain-of-function effect that interferes with normal protein function. It is likely a protein that lacks the 15 AAs containing the ATP-binding domain is produced. This region is crucial for enzymatic function, and it escapes mRNA decay, as shown by semiquantitative RT-PCR from family 2 (Figure 4A). We further developed a zebrafish model of this variant (Figure S13) which elicits a dominant negative effect on the wild-type allele, causing a dose-dependent phenotype, specifically cyclopia and gastrulation defects at 200–500pg. Similar cyclopia defects in zebrafish were reported for microcephaly gene ORC1.34
Given the essential function and constraint metrics of NARS1, in conjunction with the clinical phenotypes of included individuals, we propose that genotypes with dominant heterozygous variants produce a toxic gain of function. This is compared with the homozygous recessive variants that probably experience a loss of function, though this can perhaps be least partially compensated for by other ARS genes. Taking into consideration the aminoacylation assay and yeast model for de novo mutations, and the western blot, aminoacylation assay, and modeling for homozygous recessive mutations, we have confirmed the pathogenicity of all NARS1 mutations mentioned. Similar effects are seen in Aicardi-Goutières syndrome, which is caused by pathogenic variants in ADAR. There are relatively frequent alleles that are pathogenic when in trans to a null, but are never found in individuals with the homozygous state.35 For distal C terminus mutations, such as the homozygous c.1633C>T (p.Arg545Cys) variant, the mechanism is likely due to abnormal protein structure and catalytic activity (Figures 1C and 6 and Figures S1 and S2).
Affected individuals had both central and peripheral nervous system involvement and a broad neurodevelopmental phenotype characterized by GDD, microcephaly, ataxia, neuropathy, and seizures. This is reflective of high NARS1 expression in the cortex, cerebellum, and brainstem as demonstrated in mouse brains36,37 (Figure S3). Mutations have been reported for the majority of ARSs. AsnRS2, a mitochondrial ARS protein coded by NARS2, has recently been linked with an overlapping phenotype consisting of multisystem mitochondrial disorder (MID). Intellectual disability, epilepsy in childhood, hearing loss, and myopathy have also been seen in NARS1 individuals.38, 39, 40, 41 In addition, ARS interacting multifunctional proteins 1–3 (AIMP1–3) participate together with nine cytosolic ARSs to constitute the so-called multi-synthetase complex, and have also been associated with a variety of human diseases.42 In considering their critical cellular functions, we expect that all ARSs will have a disease association.7, 8, 9 The NARS1 data bring the number of characterized ARSs to 35 out of 37. On a modified Taylor’s Venn diagram of AA properties, NARS1 is placed in close proximity to other AAs with similar properties (IARS1, LARS1, DARS1, EPRS1, NARS1, RARS1, and QARS1) which also have more severe phenotypes43 (Figure S4).
Our functional work supports the likelihood that there is a loss-of-function mechanism in homozygotes and has helped to further understand the role of NARS1 mutations in disease. The development of CRISPR/Cas9 heterozygous knockin and homozygous knockout animal models is the next important step in understanding the molecular rationale of these NARS1 variants. Considering the high number of individuals and variants identified here, the addition of NARS1 to genetic testing panels for children and young adults presenting with NDD, epilepsy, and/or a demyelinating neuropathy may be of clinical benefit.
Data and Code Availability
The variants reported in this paper have been submitted to the Leiden Open Variation Database, and the accession numbers are: LOVD: 668185, LOVD: 668186, LOVD: 668187, LOVD: 668188, LOVD: 668189, LOVD: 668190, LOVD: 668191, LOVD: 668192, LOVD: 668193, LOVD: 668194, LOVD: 668195, LOVD: 668196, LOVD: 668197, and LOVD: 668198.
Declaration of Interests
Maria J. Guillen Sacoto, Lindsay B. Henderson, Yue Si, Aida Telegrafi, and Ingrid M. Wentzensen are employees of GeneDx. The other authors declare no competing interests.
Acknowledgments
We are grateful to individuals and families for taking part in our research project. We heartfully thank James Burns for reading and correcting our manuscript. We thank the Gene Expression Nervous System Atlas (GENSAT) Project, National Institute of Neurological Disorders and Stroke (NINDS) contracts N01NS02331 and HHSN271200723701C to The Rockefeller University (New York, NY). H.H. is grateful to the Medical Research Council (MRC), The Wellcome Trust Synaptopathies Award, MRC Centre grant G0601943, Ataxia UK, the Rosetrees Trust, Brain Research UK, the University College London (UCL) Official Development Assistance (ODA) and Low and Middle Income Country (LMIC) award, the Multiple System Atrophy (MSA) Trust, Muscular Dystrophy (MDUK). and the Muscular Dystrophy Association (MDA). This research was also supported by the UCL/UCL Hospital (UCLH) National Institute for Health Research University College London Hospitals Biomedical Research Centre.
Published: July 30, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.06.016.
Web Resources
1000 Genomes Project, https://www.genome.gov/27528684/1000-genomes-project
Complete Genomics 69, https://www.completegenomics.com/public-data/69-genomes/
CPDB web tool, http://cpdb.molgen.mpg.de/
Ensembl, http://www.ensembl.org/i
FastQC, http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
GATK documentation, https://software.broadinstitute.org/gatk/
Human Genome Variation Society, http://www.hgvs.org
Mutation Taster version 2, http://www.mutationtaster.org/
National Heart, Lung, and Blood Institute (NHLBI) Exome Variant Server, https://evs.gs.washington.edu/EVS/
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
SIFT, http://sift.jcvi.org/
University of California—San Francisco (UCSC) Genome Browser, https://genome.ucsc.edu/
Supplemental Data
References
- 1.Lee E.Y., Kim S., Kim M.H. Aminoacyl-tRNA synthetases, therapeutic targets for infectious diseases. Biochem. Pharmacol. 2018;154:424–434. doi: 10.1016/j.bcp.2018.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ognjenović J., Simonović M. Human aminoacyl-tRNA synthetases in diseases of the nervous system. RNA Biol. 2018;15:623–634. doi: 10.1080/15476286.2017.1330245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rajendran V., Kalita P., Shukla H., Kumar A., Tripathi T. Aminoacyl-tRNA synthetases: Structure, function, and drug discovery. Int. J. Biol. Macromol. 2018;111:400–414. doi: 10.1016/j.ijbiomac.2017.12.157. [DOI] [PubMed] [Google Scholar]
- 4.Antonellis A., Green E.D. The role of aminoacyl-tRNA synthetases in genetic diseases. Annu. Rev. Genomics Hum. Genet. 2008;9:87–107. doi: 10.1146/annurev.genom.9.081307.164204. [DOI] [PubMed] [Google Scholar]
- 5.Meyer-Schuman R., Antonellis A. Emerging mechanisms of aminoacyl-tRNA synthetase mutations in recessive and dominant human disease. Hum. Mol. Genet. 2017;26(R2):R114–R127. doi: 10.1093/hmg/ddx231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oprescu S.N., Griffin L.B., Beg A.A., Antonellis A. Predicting the pathogenicity of aminoacyl-tRNA synthetase mutations. Methods. 2017;113:139–151. doi: 10.1016/j.ymeth.2016.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Francklyn C.S., Mullen P. Progress and challenges in aminoacyl-tRNA synthetase-based therapeutics. J. Biol. Chem. 2019;294:5365–5385. doi: 10.1074/jbc.REV118.002956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rogers S.O. Evolution of the genetic code based on conservative changes of codons, amino acids, and aminoacyl tRNA synthetases. J. Theor. Biol. 2019;466:1–10. doi: 10.1016/j.jtbi.2019.01.022. [DOI] [PubMed] [Google Scholar]
- 9.González-Serrano L.E., Chihade J.W., Sissler M. When a common biological role does not imply common disease outcomes: Disparate pathology linked to human mitochondrial aminoacyl-tRNA synthetases. J. Biol. Chem. 2019;294:5309–5320. doi: 10.1074/jbc.REV118.002953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okur V., Ganapathi M., Wilson A., Chung W.K. Biallelic variants in VARS in a family with two siblings with intellectual disability and microcephaly: case report and review of the literature. Cold Spring Harb. Mol. Case Stud. 2018;4:a003301. doi: 10.1101/mcs.a003301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stephen J., Nampoothiri S., Banerjee A., Tolman N.J., Penninger J.M., Elling U., Agu C.A., Burke J.D., Devadathan K., Kannan R. Loss of function mutations in VARS encoding cytoplasmic valyl-tRNA synthetase cause microcephaly, seizures, and progressive cerebral atrophy. Hum. Genet. 2018;137:293–303. doi: 10.1007/s00439-018-1882-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siekierska A., Stamberger H., Deconinck T., Oprescu S.N., Partoens M., Zhang Y., Sourbron J., Adriaenssens E., Mullen P., Wiencek P., C4RCD Research Group. AR working group of the EuroEPINOMICS RES Consortium Biallelic VARS variants cause developmental encephalopathy with microcephaly that is recapitulated in vars knockout zebrafish. Nat. Commun. 2019;10:708. doi: 10.1038/s41467-018-07953-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedman J., Smith D.E., Issa M.Y., Stanley V., Wang R., Mendes M.I., Wright M.S., Wigby K., Hildreth A., Crawford J.R. Biallelic mutations in valyl-tRNA synthetase gene VARS are associated with a progressive neurodevelopmental epileptic encephalopathy. Nat. Commun. 2019;10:707. doi: 10.1038/s41467-018-07067-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krenke K., Szczałuba K., Bielecka T., Rydzanicz M., Lange J., Koppolu A., Płoski R. FARSA mutations mimic phenylalanyl-tRNA synthetase deficiency caused by FARSB defects. Clin. Genet. 2019;96:468–472. doi: 10.1111/cge.13614. [DOI] [PubMed] [Google Scholar]
- 15.Forrester N., Rattihalli R., Horvath R., Maggi L., Manzur A., Fuller G., Gutowski N., Rankin J., Dick D., Buxton C. Clinical and Genetic Features in a Series of Eight Unrelated Patients with Neuropathy Due to Glycyl-tRNA Synthetase (GARS) Variants. J. Neuromuscul. Dis. 2020;7:137–143. doi: 10.3233/JND-200472. [DOI] [PubMed] [Google Scholar]
- 16.Lee A.J., Nam D.E., Choi Y.J., Nam S.H., Choi B.O., Chung K.W. Alanyl-tRNA synthetase 1 (AARS1) gene mutation in a family with intermediate Charcot-Marie-Tooth neuropathy. Genes Genomics. 2020;42:663–672. doi: 10.1007/s13258-020-00933-9. [DOI] [PubMed] [Google Scholar]
- 17.Williams K.B., Brigatti K.W., Puffenberger E.G., Gonzaga-Jauregui C., Griffin L.B., Martinez E.D., Wenger O.K., Yoder M.A., Kandula V.V.R., Fox M.D. Homozygosity for a mutation affecting the catalytic domain of tyrosyl-tRNA synthetase (YARS) causes multisystem disease. Hum. Mol. Genet. 2019;28:525–538. doi: 10.1093/hmg/ddy344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cooper G.M., Stone E.A., Asimenos G., Green E.D., Batzoglou S., Sidow A., NISC Comparative Sequencing Program Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuhn R.M., Karolchik D., Zweig A.S., Wang T., Smith K.E., Rosenbloom K.R., Rhead B., Raney B.J., Pohl A., Pheasant M. The UCSC Genome Browser Database: update 2009. Nucleic Acids Res. 2009;37:D755–D761. doi: 10.1093/nar/gkn875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moreno M.B., Durán A., Ribas J.C. A family of multifunctional thiamine-repressible expression vectors for fission yeast. Yeast. 2000;16:861–872. doi: 10.1002/1097-0061(20000630)16:9<861::AID-YEA577>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 21.Rodríguez-López M., Cotobal C., Fernández-Sánchez O., Borbarán Bravo N., Oktriani R., Abendroth H., Uka D., Hoti M., Wang J., Zaratiegui M., Bähler J. A CRISPR/Cas9-based method and primer design tool for seamless genome editing in fission yeast. Wellcome Open Res. 2017;1:19. doi: 10.12688/wellcomeopenres.10038.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bähler J., Wu J.Q., Longtine M.S., Shah N.G., McKenzie A., 3rd, Steever A.B., Wach A., Philippsen P., Pringle J.R. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 23.Sato M., Dhut S., Toda T. New drug-resistant cassettes for gene disruption and epitope tagging in Schizosaccharomyces pombe. Yeast. 2005;22:583–591. doi: 10.1002/yea.1233. [DOI] [PubMed] [Google Scholar]
- 24.Meyer J., Novak M., Hamel A., Rosenberg K. Extraction and analysis of cortisol from human and monkey hair. J. Vis. Exp. 2014;83:e50882. doi: 10.3791/50882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anders S., Pyl P.T., Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 28.Park J.S., Park M.C., Lee K.Y., Goughnour P.C., Jeong S.J., Kim H.S., Kim H.J., Lee B.J., Kim S., Han B.W. Unique N-terminal extension domain of human asparaginyl-tRNA synthetase elicits CCR3-mediated chemokine activity. Int. J. Biol. Macromol. 2018;120(Pt A):835–845. doi: 10.1016/j.ijbiomac.2018.08.171. [DOI] [PubMed] [Google Scholar]
- 29.Dong J., Qiu H., Garcia-Barrio M., Anderson J., Hinnebusch A.G. Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA-binding domain. Mol. Cell. 2000;6:269–279. doi: 10.1016/s1097-2765(00)00028-9. [DOI] [PubMed] [Google Scholar]
- 30.Vijayakumar R., Tripathi T. Soluble expression and purification of a full-length asparaginyl tRNA synthetase from Fasciola gigantica. Protein Expr. Purif. 2018;143:9–13. doi: 10.1016/j.pep.2017.10.009. [DOI] [PubMed] [Google Scholar]
- 31.He W., Bai G., Zhou H., Wei N., White N.M., Lauer J., Liu H., Shi Y., Dumitru C.D., Lettieri K. CMT2D neuropathy is linked to the neomorphic binding activity of glycyl-tRNA synthetase. Nature. 2015;526:710–714. doi: 10.1038/nature15510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boczonadi V., Meyer K., Gonczarowska-Jorge H., Griffin H., Roos A., Bartsakoulia M., Bansagi B., Ricci G., Palinkas F., Zahedi R.P. Mutations in glycyl-tRNA synthetase impair mitochondrial metabolism in neurons. Hum. Mol. Genet. 2018;27:2187–2204. doi: 10.1093/hmg/ddy127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McClain W.H., Schneider J., Bhattacharya S., Gabriel K. The importance of tRNA backbone-mediated interactions with synthetase for aminoacylation. Proc. Natl. Acad. Sci. USA. 1998;95:460–465. doi: 10.1073/pnas.95.2.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bicknell L.S., Bongers E.M., Leitch A., Brown S., Schoots J., Harley M.E., Aftimos S., Al-Aama J.Y., Bober M., Brown P.A. Mutations in the pre-replication complex cause Meier-Gorlin syndrome. Nat. Genet. 2011;43:356–359. doi: 10.1038/ng.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmelzer L., Smitka M., Wolf C., Lucas N., Tüngler V., Hahn G., Tzschach A., Di Donato N., Lee-Kirsch M.A., von der Hagen M. Variable clinical phenotype in two siblings with Aicardi-Goutières syndrome type 6 and a novel mutation in the ADAR gene. Eur. J. Paediatr. Neurol. 2018;22:186–189. doi: 10.1016/j.ejpn.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 36.Melé M., Ferreira P.G., Reverter F., DeLuca D.S., Monlong J., Sammeth M., Young T.R., Goldmann J.M., Pervouchine D.D., Sullivan T.J., GTEx Consortium Human genomics. The human transcriptome across tissues and individuals. Science. 2015;348:660–665. doi: 10.1126/science.aaa0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Consortium G.T., GTEx Consortium Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seaver L.H., DeRoos S., Betz B., Rajasekaran S. Reply to Finsterer Regarding Lethal NARS2-Related Disorder Associated With Rapidly Progressive Intractable Epilepsy and Global Brain Atrophy. Pediatr. Neurol. 2019;93:65. doi: 10.1016/j.pediatrneurol.2018.11.013. [DOI] [PubMed] [Google Scholar]
- 39.Simon M., Richard E.M., Wang X., Shahzad M., Huang V.H., Qaiser T.A., Potluri P., Mahl S.E., Davila A., Nazli S. Mutations of human NARS2, encoding the mitochondrial asparaginyl-tRNA synthetase, cause nonsyndromic deafness and Leigh syndrome. PLoS Genet. 2015;11:e1005097. doi: 10.1371/journal.pgen.1005097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sofou K., Kollberg G., Holmström M., Dávila M., Darin N., Gustafsson C.M., Holme E., Oldfors A., Tulinius M., Asin-Cayuela J. Whole exome sequencing reveals mutations in NARS2 and PARS2, encoding the mitochondrial asparaginyl-tRNA synthetase and prolyl-tRNA synthetase, in patients with Alpers syndrome. Mol. Genet. Genomic Med. 2015;3:59–68. doi: 10.1002/mgg3.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vanlander A.V., Menten B., Smet J., De Meirleir L., Sante T., De Paepe B., Seneca S., Pearce S.F., Powell C.A., Vergult S. Two siblings with homozygous pathogenic splice-site variant in mitochondrial asparaginyl-tRNA synthetase (NARS2) Hum. Mutat. 2015;36:222–231. doi: 10.1002/humu.22728. [DOI] [PubMed] [Google Scholar]
- 42.Boczonadi V., Jennings M.J., Horvath R. The role of tRNA synthetases in neurological and neuromuscular disorders. FEBS Lett. 2018;592:703–717. doi: 10.1002/1873-3468.12962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taylor W.R. The classification of amino acid conservation. J. Theor. Biol. 1986;119:205–218. doi: 10.1016/s0022-5193(86)80075-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The variants reported in this paper have been submitted to the Leiden Open Variation Database, and the accession numbers are: LOVD: 668185, LOVD: 668186, LOVD: 668187, LOVD: 668188, LOVD: 668189, LOVD: 668190, LOVD: 668191, LOVD: 668192, LOVD: 668193, LOVD: 668194, LOVD: 668195, LOVD: 668196, LOVD: 668197, and LOVD: 668198.