Significance
Neutralizing antibodies (NAbs) against HIV-1 are under clinical development. Despite demonstrated antiviral activity when used as treatment in HIV-infected people, the mechanism of action of NAbs remains controversial. Quantification of the relative contribution of Fab-mediated (neutralization of free virus) and Fc-mediated functions such as antibody-dependent cellular cytotoxicity (ADCC), phagocytosis, and complement fixation is essential to develop better therapies. Here, we treated chronically SHIV-infected rhesus macaques with a single dose of NAb VRC07-523LS with or without Fc modifications. Fc function contributes 21% to the antiviral activity of VRC07-523LS with an unmodified Fc region, when compared to NAb with Fc knockout mutations. However, NAbs with improved ADCC function in vitro did not show enhanced antiviral activity in vivo.
Keywords: HIV, neutralizing antibodies, effector function, Fc, mechanism of action
Abstract
Treatment of HIV infection with either antiretroviral (ARV) therapy or neutralizing monoclonal antibodies (NAbs) leads to a reduction in HIV plasma virus. Both ARVs and NAbs prevent new rounds of viral infection, but NAbs may have the additional capacity to accelerate the loss of virus-infected cells through Fc gamma receptor (FcγR)-mediated effector functions, which should affect the kinetics of plasma-virus decline. Here, we formally test the role of effector function in vivo by comparing the rate and timing of plasma-virus clearance in response to a single-dose treatment with either unmodified NAb or those with either reduced or augmented Fc function. When infused into viremic simian HIV (SHIV)-infected rhesus macaques, there was a 21% difference in slope of plasma-virus decline between NAb and NAb with reduced Fc function. NAb engineered to increase FcγRIII binding and improve antibody-dependent cellular cytotoxicity (ADCC) in vitro resulted in arming of effector cells in vivo, yet led to viral-decay kinetics similar to NAbs with reduced Fc function. These studies show that the predominant mechanism of antiviral activity of HIV NAbs is through inhibition of viral entry, but that Fc function can contribute to the overall antiviral activity, making them distinct from standard ARVs.
Neutralizing monoclonal antibodies against HIV-1 are being pursued for prevention, treatment (during acute and chronic phases of infection), sustained antiretroviral (ARV)-free virologic remission during analytic treatment interruption (ATI), and cure of infection. Clinical studies with neutralizing antibodies (NAbs) containing an unmodified Fc region have shown efficacy in both treatment (1–4) and ATI (5–7), and a large clinical study for prevention is ongoing (8). None of these clinical trials address the potential role of Fc-mediated effector function in the therapeutic efficacy of NAbs. While studies addressing the role of Fc function in HIV NAb therapy in animal models have been performed, they have often come to conflicting conclusions (9–13). Quantifying the relative contribution of Fab-mediated neutralization of free-virus versus Fc-mediated effector function against virions and infected cells to the in vivo antiviral effect of NAbs is essential to develop better therapies and potentially cure.
Both ARVs and NAbs lead to a reduction of plasma HIV RNA (1–4). ARVs mainly prevent new rounds of viral infection with subsequent passive loss of both actively and latently infected cells, the latter estimated to take decades to clear. NAbs could be superior to ARVs due to their potential ability to recognize and clear virus-infected cells through Fc-dependent functions such as antibody-dependent cellular cytotoxicity (ADCC) and phagocytosis (5–7, 9, 12). Of note, passive therapy with single or combination NAbs followed by ATI has delayed viral rebound compared to historic controls (5–7). However, the delay in virus rebound from these studies was not associated with a concomitant change in the size of the viral reservoir, highlighting the complex mechanisms operational during NAb therapy.
The ability of ADCC to have an antiviral impact in vivo may be determined by several factors, including the availability and timing of Env expression on the infected cell surface, expression of maturation and exhaustion markers on effector-cell populations, anatomical location and trafficking of target and effector cells, biodistribution of the antibody, and competition between antibody-mediated versus major histocompatibility complex-mediated mechanisms of killing. The contribution of ADCC and other Fc-mediated effector mechanisms to the overall in vivo antiviral activity of an NAb can best be addressed by applying models of plasma-virus dynamics that were initially used to accurately describe plasma-virus decline after treatment with ARVs (14–18). These time-tested models show that when plasma-virus levels are at steady state, the rate of production of plasma virus from infected cells is equal to the rate of clearance of virus from the plasma. The vast majority of the plasma virus is derived from newly infected activated CD4 T cells. ARVs, by blocking new rounds of replication, have no effect on already-infected cells, which continue to produce virus with a half-life of 1.6 d. After the delay inherent in the fact that these already-infected cells will continue to produce virus until they die, plasma virus declines as the rate of natural virus clearance far exceeds the rate of plasma-virus production (which approaches zero on ARVs).
By applying these virus dynamic models, infusion of NAbs into viremic animals can be used to determine if Fc-mediated functions contribute to the antiviral activity of NAbs in vivo. If Fc functions are not involved in the antiviral activity of a NAb, then the plasma-virus dynamics would mimic that of an entry inhibitor. In contrast, if Fc-mediated functions, such as ADCC, are operative for an HIV NAb in vivo, these models predict that plasma-virus decline would begin during the first 1.6 d after NAb infusion, as killing of cells that were infected before NAb infusion would occur. In addition, Fc-mediated mechanisms could accelerate plasma-virus clearance. We therefore used a well-controlled therapeutic intervention model in which simian HIV (SHIV)-infected rhesus macaques were infused with Fc-modified and unmodified NAbs in order to quantify the relative contribution of neutralization versus Fc-mediated effector function to the overall antiviral activity of a NAb in vivo.
Results
Characterization of Fc-Modified Antibodies.
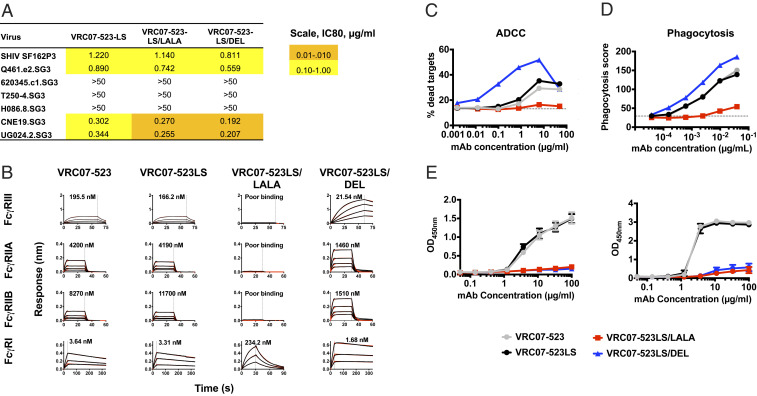
VRC07-523 is a CD4 binding-site NAb with broad neutralizing activity against multiple HIV strains (19). Importantly, it is highly active against SHIVSF162P3, a SHIV that consistently establishes chronic infection with stable plasma viremia and slow loss of CD4 T cells in Indian-origin rhesus macaques (20).
We made mutations in the Fc region of VRC07-523. The LS mutation was introduced to extend in vivo half-life (21). In addition, two other mutations were introduced onto the LS background: LALA (L234A/L235A), which effectively eliminates ADCC and phagocytosis; and DEL (S239D/A330L/I332E), which increases ADCC and phagocytosis (22, 23). The mutations in the Fc region did not affect neutralization against SHIVSF162P3 or a panel of HIVs (Fig. 1A). When introduced onto VRC07-523LS, the LALA mutation greatly reduced binding to rhesus Fc gamma receptor III (FcγRIII) and FcγRII, whereas the DEL mutation increased binding. FcγRI binding by DEL was unaffected, while that of the LALA mutation was decreased 86-fold (Fig. 1B and SI Appendix, Fig. S1). These binding characteristics of the DEL NAb resulted in improved in vitro function with an almost 50-fold improvement in ADCC mediated by natural killer (NK) cells isolated from macaque blood (Fig. 1C). Phagocytosis assays were performed by using the standard THP-1 bead-based assay due to the lack of the nonhuman primate (NHP) cell line. Phagocytosis was improved marginally by the DEL mutation and significantly reduced by the LALA mutation (Fig. 1D). Both LALA and DEL mutations ablated binding to complement from macaque serum (Fig. 1E). There was no effect of the LS mutation on FcγR binding or effector function, but it did have a global impact on binding to rhesus neonatal Fc receptors (FcRns) at pH 6.0 and 7.4 (Fig. 1 B–E and SI Appendix, Fig. S1).
Fig. 1.
In vitro characterization of Fc-modified variants of VRC07-523-LS. The indicated mutations were introduced on the backbone of the VRC07-523LS antibody and tested for the following. (A) Neutralization of SHIVSF162P3 and a six-virus HIV panel (values are IC80, µg/mL). (B) Binding to rhesus macaque Fcγ receptors by biolayer interferometry. KD values are shown within each graph. (C) ADCC against CEM cells chronically infected with SHIVSF162P3. Data are representative of at least three experiments using NK cells isolated from peripheral blood of naïve monkeys. (D) Phagocytosis of beads coated with RSC3 by THP-1 cells. The y axis shows phagocytosis score calculated as percentage of cells bound by beads multiplied with the MFI of beads. Data are representative of at least three experiments. (E) Binding to C1q and C3c from rhesus macaque serum, n = 3. Data are mean ± SD and representative of at least two independent experiments.
Fc Modifications Affect Pharmacokinetics in SHIVSF162P3-Infected Rhesus Macaques.
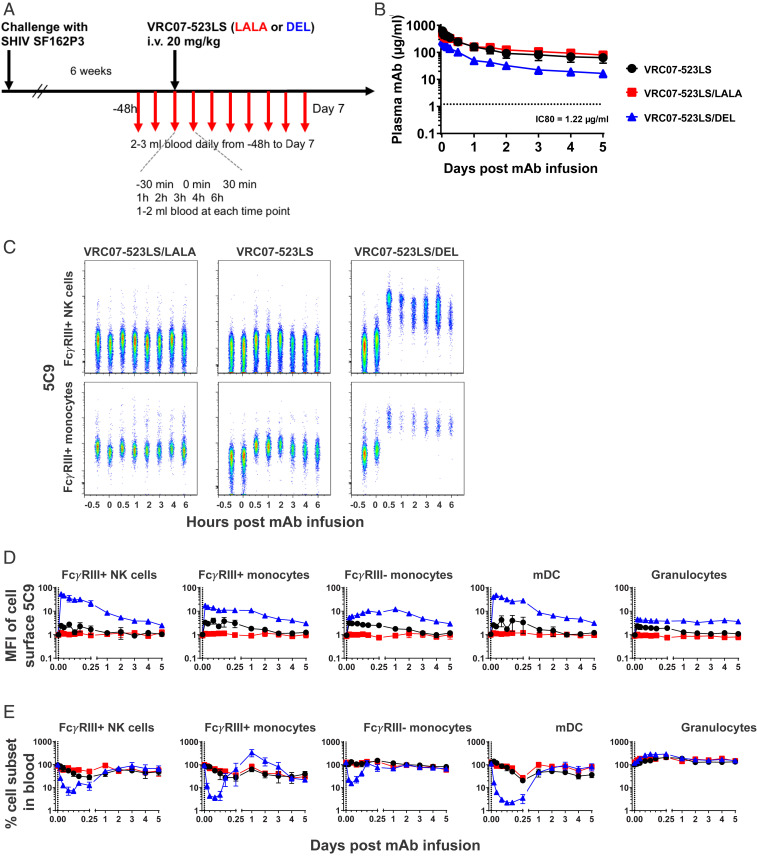
Indian-origin rhesus macaques were infected intravenously (i.v.) with SHIVSF162P3. At 6 wk after challenge, when plasma-virus loads (PVLs) were greater than 1,000 copies per mL, 29 monkeys were assigned to different treatment arms based on pretreatment PVL and given a single i.v. dose of 20 mg/kg antibody, after which they were intensively monitored (Fig. 2A). We chose the 20-mg/kg dose in order to be within the 10- to 40-mg/kg range in which anti-HIV antibodies VRC01, 3BNC117, and 10-1074 have demonstrated antiviral effects in viremic individuals living with HIV.
Fig. 2.
Circulating and cell-bound antibody pharmacokinetics. (A) Experimental scheme of SHIV challenge, antibody treatment, and biosampling in rhesus macaques. (B) Plasma levels of antibodies measured by binding to RSC3. Horizontal dotted lines indicate IC80 of neutralization of SHIVSF162P3 by VRC07-523LS. n = 9, 10, and 10 for LS, LS/LALA, and LS/DEL, respectively. Data are mean ± SD. (C) Representative comparison of anti-Id staining and cell-subset levels on NK cells and CD16+ monocytes up to 6 h after antibody treatment. (D) Levels of cell-surface-bound NAb. The y axis represents the median fluorescent intensity on the detection channel of the anti-Id (5C9) used to detect the infused NAb, normalized to baseline at 30 min preinfusion. Data are mean ± SEM. n = 4, 5, and 4 for LS, LS/LALA, and LS/DEL, respectively. (E) Changes in cell-subset frequency normalized to baseline at 30 min preinfusion. Data are mean ± SEM. n = 5, 5, and 4 for LS, LS/LALA, and LS/DEL, respectively.
Post hoc analysis confirmed that baseline viral loads across the NAb groups were comparable (SI Appendix, Fig. S2; P = 0.8849). Six additional monkeys served as controls and were given either the Ebola-virus-specific NAb mAb114LS (24) (n = 3) or no antibody (n = 3). All VRC07-523 NAbs showed typical pharmacokinetic profiles, but differed in half-lives. The LS/LALA antibody consistently maintained higher circulating concentrations than the LS antibody, whereas the LS/DEL antibody had lower concentrations (Fig. 2B). Nevertheless, all NAbs were present at levels greater than 10-fold the 80% inhibitory concentration (IC80) titer for in vitro SHIVSF162P3 neutralization through 5 d after infusion.
Fc Modifications Affect the Binding to Cell-Surface FcγR In Vivo.
We hypothesized that the difference in circulating antibody concentrations was due to the difference in their ability to bind cell-surface-expressed FcγRs. We therefore measured whether the infused NAbs were detectable on the surface of FcγR-expressing cells (Fig. 2C and SI Appendix, Fig. S3). Consistent with the lack of in vitro binding to FcγRs, we did not detect the LS/LALA antibody on any of the immune cells in the blood (Fig. 2C). At 30 min postinfusion, the LS antibody was detected exclusively on FcγR-expressing cells in the blood. Importantly, we detected binding on NK cells and monocytes that express FcγRIII and are ADCC-proficient. Cell-surface binding of the LS antibody became undetectable 24 to 48 h after NAb infusion. As expected from in vitro binding characteristics, LS/DEL antibody showed stronger cell-surface binding and, at 30 min, appeared to have saturated all cells expressing FcγRs, except B cells (Fig. 2C and SI Appendix, Fig. S4). The kinetics of the cell-bound LS/DEL antibody varied among the different cell types, but cell-bound antibody was readily detectable through day 5. Thus, the LS/DEL antibody effectively competed with endogenous macaque immunoglobulin Gs (IgGs) to engage FcγR-expressing cells. In addition, this engagement of FcγRs by the LS/DEL NAb was associated with a specific loss of FcγR-expressing cells as early as 30 min after infusion (Fig. 2D). However, this decrease was transient and normalized to baseline levels by 24 h (with the exception that FcγRIII+ monocytes took longer) (Fig. 2D).
Effector Functions Contribute to Plasma-Virus Clearance.
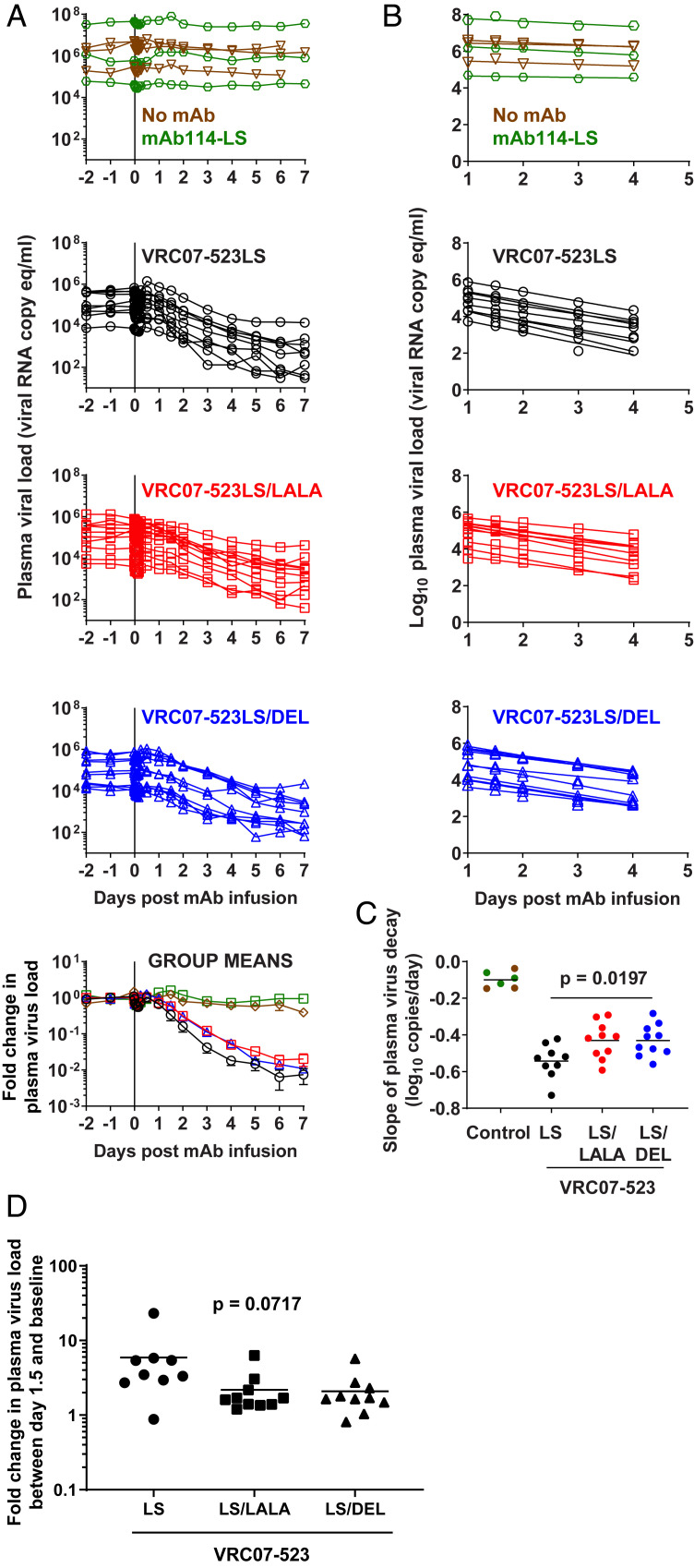
PVL decline was evaluated by using models derived from studies of HIV plasma-virus dynamics after ART (14–18). PVLs were stable for the 2 d before NAb infusion in all monkeys (Fig. 3A). In all groups, there was a small, but consistent, decrease in plasma virus until 4 to 6 h after antibody infusion, followed by an increase at 6 to 12 h and return to baseline by 24 h (SI Appendix, Fig. S5). All monkeys treated with variants of VRC07-523LS, but not with MAb114LS or no antibody, showed an exponential decline in plasma virus beginning ∼24 h after antibody infusion. Importantly, PVL was quantifiable in all monkeys at all time points, thereby allowing for an accurate calculation of the slope of plasma-virus decline (Fig. 3A). PVL values between 1 and 4 d were used to calculate the initial slope of plasma-virus decline for each monkey (Fig. 3B). Later time points were excluded, as plasma-virus decline slowed after day 4 in some monkeys. The LS/LALA- and LS/DEL-treated monkeys showed a very similar pattern of plasma-virus decline, which was slower than what was seen in the LS-treated monkeys (P = 0.0197 by one-way ANOVA; Fig. 3C). Specifically, the slopes of plasma-virus decline for the LS/LALA- and LS/DEL-treated monkeys were 21% less than for the LS-treated monkeys (−0.43 and −0.43 versus −0.54 per day). In addition to this faster virus clearance, we asked if the LS-treated monkeys showed earlier virus clearance. Because of the early fluctuations in plasma virus (Fig. 3A), we used 24 h as the first time point for this analysis. We found that PVL began decreasing in LS-treated monkeys at a mean of 1.5 d after treatment, as compared to 2 and 1.9 d for the LS/LALA and LS/DEL groups, respectively. At 1.5 d, 89% of animals in the LS group had a decrease in PVL, as compared to only 30% of animals in the LS/LALA and LS/DEL groups (P = 0.0717; Fig. 3D). Together, these data show that altering the FcγR binding affinity of a CD4bs antibody can affect the dynamics of in vivo plasma-virus decline. Consistent with the minor difference in PVL, we did not see any significant changes in the slopes of decay of cell-associated viral DNA (SI Appendix, Fig. S6).
Fig. 3.
Decay kinetics of plasma virus following treatment with Fc-modified VRC07-523LS. Rhesus macaques were infected i.v. with SHIVSF162P3 for 6 wk and treated with 20 mg/kg indicated antibodies (n = 3 each for mAb114-LS and no mAb groups; n = 9 for VRC07-523LS; and n = 10 each for VRC07-523LS/LALA and VRC07-523LS/DEL). (A) PVL shown for individual animals and then as normalized to baseline (average of three time points before NAb infusion, day 2, day 1, and 30 min) and expressed as fold change. After normalization, data are shown as group means, and error bars represent ± SEM. (B) Viral loads were log-transformed and fit by using linear regression. (C) Slopes were compared by one-way ANOVA between the VRC07-523LS variant groups. Average slopes in each group are shown by horizontal lines. (D) Fold change in PVL between baseline and day 1.5. P values were calculated by using one-way ANOVA.
Increasing FcγRIII Affinity Led to Necroptosis of Effector Cells and Prozone Effect.
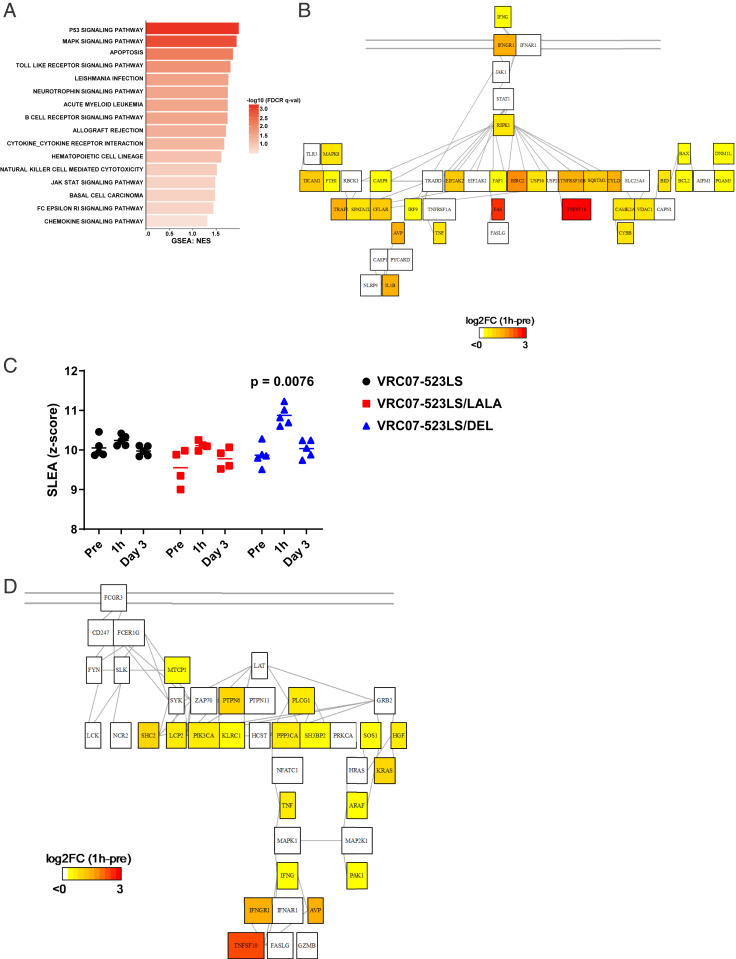
Our initial premise was that the LS/LALA mutation would eliminate ADCC and thereby slow the decline in PVL (which was observed), whereas the LS/DEL mutation would increase ADCC and lead to a faster decline in PVL (the opposite was observed). This paradoxical result was seen, despite the fact that the LS/DEL antibody had saturated FcγRs on ADCC effector cells for at least 5 d after NAb infusion (Fig. 2C), effectively prearming these cells for killing of Env-expressing targets. To investigate the paradoxical effect of the LS/DEL antibodies on PVL in vivo, we first evaluated the transcriptomic profile of NK cells before and 1 h and 3 d after NAb infusion (Fig. 4A). A transcriptomic profile consistent with activation of necroptosis pathways in NK cells was observed 1 h after VRC07-523LS/DEL infusion, but not in other NAb-treated monkeys (Fig. 4A). Pathway analysis indicated the up-regulation of interferon gamma (IFNγ) (likely through FcγRIII cross-linking) that induced the tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-mediated apoptotic pathway (TNFS10, TNFRSF10B, and CASP8) (Fig. 4B) that likely led to loss of NK cells (Fig. 2C). In addition to apoptosis, activation of NK-cell cytotoxicity pathways (MTCP1, PI3KCA, and KRAS) was observed 1 h after VRC07-523LS/DEL infusion (Fig. 4 C and D and SI Appendix, Fig. S7). Apoptosis pathways were also up-regulated in monocytes (P = 0.0221; SI Appendix, Fig. S7). However, these transcriptomic profile changes were transient and returned to baseline by 3 d after LS/DEL NAb infusion.
Fig. 4.
Transcriptomic analysis following antibody treatment. (A) Bar plot presenting pathways induced 1 h after infusion of the LS/DEL antibody in NK cells compared to the preinfusion timepoint (−48 h). (B) Network showing the induction of TRAIL-mediated apoptosis by IFNγ in the NK cells at 1 h after infusion of VRC07-523LS/DEL. (C) Jitter plot of ADCC pathway score of individual animals. (D) Network showing the FcγRIII signaling genes and induction of necroptosis in the NK cells at 1 h after infusion of VRC07-523LS/DEL.
We further explored other potential reasons for the lack of in vivo FcγR-mediated function of the LS/DEL antibody by performing in vitro ADCC assays using targets and effectors precoated with antibody and cultured in the presence or absence of additional antibody to mimic in vivo conditions (SI Appendix, Fig. S9). These assays show that ADCC is inhibited when both targets and effectors are independently preengaged by the antibody. Of particular relevance, effector cells were coated with NAb at 100-fold lower concentrations of the LS/DEL than the LS antibody, leading to a severe loss of ADCC activity for the LS/DEL NAb at concentrations far below what may be required for effective in vivo virus neutralization.
Effector Functions Did Not Contribute to Long-Term Effects.
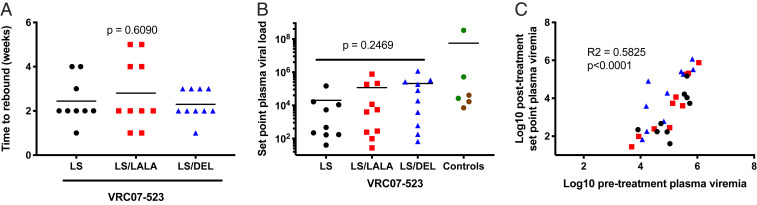
We asked if the altered Fc function could contribute to other measures of reduction in infected cell numbers. Neither the time to rebound nor the postrebound set-point viremia was significantly different across the NAb groups (Fig. 5 and SI Appendix, Fig. S10). Rebound was also associated with changes in the Env sequence that potentially corresponded to NAb escape (SI Appendix, Fig. S11). Of note, some NAb-treated animals showed low to undetectable viremia after rebound. However, postrebound set-point viremia was significantly associated with pretreatment PVL, and not with NAb-related effects (Fig. 5C).
Fig. 5.
Long-term follow-up following antibody treatment. Rhesus macaques were followed for up to 20 wk following antibody treatment. (A) Time to rebound was calculated as time at which viral load was greater than at the previously sampled time point. P values were calculated by one-way ANOVA. (B) Set-point viral load calculated as mean viremia between weeks 10 and 20 following antibody administration. P values were calculated by one-way ANOVA. (C) Pearson correlation analysis between pretreatment baseline viremia (average of three time points before NAb infusion, day 2, day 1, and 30 min) and posttreatment baseline viremia (average of two or three time points between weeks 10 and 20 following antibody administration).
Discussion
Our results indicate that Fc-mediated effector functions of NAbs contribute a measurable and significant, albeit minor, effect on the decline in PVL in SHIV-infected macaques, but do not indicate the exact mechanism involved. Specifically, elimination of effector activity through the introduction of the LALA mutation in the Fc region of an NAb decreased the rate of virus clearance by 21%. By inference, we conclude that 79% of the in vivo anti-SHIV SF162P3 activity of VRC07-523LS in viremic monkeys is mediated by neutralization, while 21% is mediated by Fc-related effects. These findings have implications for the use of NAbs as an adjunct to treatment with ARVs.
Though many studies have addressed the role of Fc function in protection against SHIV infection in macaques (13, 25–27) or HIV-1 in mouse models (9–12), fewer studies have quantified Fc function in a therapeutic setting. A study in pig-tail macaques using PGT121 and PGT121/LALA suggested that Fc functions were redundant for therapy, irrespective of NAb dose (13). Another study in chronically infected humanized mice demonstrated that at 5 h after infusion of a wild-type (WT) compared to a G236R/L328R (GRLR) (Fc-null) NAb, there was a statistically greater loss of Gag+ cells from the spleen (12). Three other studies using humanized mice demonstrated that Fc modifications of NAbs have differential effects on plasma-virus rebound; however, there were only limited assessments of the effects on PVL kinetics in these studies. A recent study (10) tested the NAbs 10-1074 and 10-1074/LALA in a novel splenic-injected primary HIV-infected reservoir mouse model during coinfusion of infected cells and NAbs. The NAb groups differed in time to rebound, but neither the relative contribution of neutralization versus Fc-mediated effects nor the size of the infected cell population were quantified. Halper-Stromberg et al. (11) used a combination of three NAbs with either WT or null Fc function, administered as three to five doses at 4 d postvirus challenge in humanized non-obese diabetic (NOD).Rag1KO.IL2RγcKO mice. The NAb groups showed similar initial rate of plasma-virus clearance, and differences between the groups arose only several weeks after NAb infusion during the rebound period. Bournazos et al. (9) tested Fc-modified combination NAbs in a multidose therapeutic NRG mouse model. Mice were challenged with HIV-1YU2 and, 3 wk later, were treated with a combination of Fc-modified 3BNC117, 10-1074, and PG16. NAbs were dosed every 5 to 7 d for 6 wk, and PVL was measured weekly. A comparison between the Fc-null and Fc-enhanced NAb variants demonstrated a significantly greater frequency of viremic animals in the Fc-null group at 3 wk postinitiation of treatment, during the rebound phase. Differences in the rate of early phase PVL decline were not quantified. In contrast, a nonfucosylated version of the NAb b12 with enhanced effector activity in vitro did not improve efficacy in a SHIV challenge model in rhesus macaques (26).
Our study applies mathematical modeling to accurately quantify the role of Fc function on the antiviral activity of an NAb within the first few days after infusion. We specifically show a role for Fc-mediated function in acute PVL kinetics, but that this does not contribute to differences in time to, or level of, PVL rebound, which appear to be dependent on pretreatment PVL. The results presented here, in concert with those of Wang et al. (28), provide clear, quantitative evidence for the timing and degree of Fc-mediated function in the therapeutic antiviral activity of HIV NAbs. The additional contribution of Fc-mediated functions to the antiviral activity by NAbs, in contrast to ARVs, suggests that Fc-enhanced NAbs should provide earlier and faster virus clearance. Improving therapeutic efficacy of NAbs is especially important in light of recent findings that the majority of the latent reservoir is seeded at the time of ARV initiation (29, 30), and future studies must address the relevance of ARVs versus NAbs for treatment in conjunction with the seeding of latent reservoirs at the time of treatment initiation.
Paradoxically, the LS/DEL and LS/LALA NAb variants both showed identical diminished antiviral kinetics in vivo, suggesting that the mechanism(s) underlying the Fc-mediated antiviral activity may be more complicated than initially assumed. Our results point to several possibilities for this paradoxical effect. Firstly, the VRC07-523LS/DEL infusion in monkeys cross-linked the FcγRIII on effector cells, leading to NK-cell death. Similar effects of NK-cell loss have been observed in a phase I trial with a CD19 antibody containing the S239D and I332E mutations (31). Secondly, given the large excess of antibody over antigen in vivo, VRC07-523LS/DEL likely saturates both targets and effector cells with antibody, thereby leading to a situation analogous to the prozone effect observed for in vitro ADCC assays (32). Differences in saturation of cell-surface FcγR is also a potential explanation for a prior study reporting comparable pharmacokinetics and difference in efficacy between the GRLR and G236A/S239D/A330L/I332E variants, which were coadministered with an excess of intravenous Ig (9). Thirdly, Env is sparse on the virion surface (33), and the affinity of the VRC07-523LS or LS/DEL antibodies may not compensate for the avidity required for phagocytosis. Lastly, both the LS/LALA and LS/DEL antibodies are ablated for complement binding, suggesting that complement may play a role in antibody-mediated virus clearance, either through virolysis or virus-specific cytolysis. Whether the low density of Env trimers on the virion surface and the maximum of three binding sites per trimer for a single NAb can trigger complement fixation is not clear, and there are likely to be strain-specific differences in Env incorporation in virions. The delay in PVL after NAb infusion further supports the evidence that virolysis may not be the primary antiviral mechanism. It appears more likely that complement may act through lysis of infected cells, and the lag phase of antiviral effects may come from the time taken for the NAb to reach sites of viral replication. We did not observe any difference in the long-term effects of NAb treatment. However, these data using a human NAb in macaques must be interpreted with caution. Beyond the first week post-NAb therapy, most animals generate antidrug antibodies (ADAs) against the infused NAb. Even animals within the same NAb group are expected to differ in the quantity and quality of ADAs. This would impact the duration of viral suppression as well as active or passive decay of infected cells. Viral evolution over time varies across animals, and the in vivo fitness of escape variants remains unknown. It is also unlikely that a single dose of NAb will have a prolonged effect on the virus, as borne out in multiple human trials of short-term, broadly neutralizing antibody treatment. These findings highlight the role of NAb effector function in vivo and have implications in the design and clinical testing of antibodies which have been altered with the goal of augmenting Fc-mediated effector functions and the importance of addressing complement-mediated effects.
While there are limitations to animal models of human virus infection, there are similarities between human and macaque immune biology, as well as HIV and SHIV infection (20, 34), suggesting that the findings from this study could be extrapolated to NAb therapy in HIV-infected people. In particular, we show that the SHIV-macaque model recapitulates both the 1- to 2-d lag phase before PVL decline begins to appear, as well as the monophasic decay of PVL after day 1, which corresponds to the passive loss of actively infected cells. Assuming ADCC is one of the operative antiviral Fc functions, the contribution of neutralization versus ADCC for any given NAb will be dependent upon the amount of free virus available for neutralization compared to virus-expressing cells that would be available for killing by ADCC. We performed these experiments in viremic monkeys, in which there was presumably more cell-free virus than virus-expressing cells. The contribution of ADCC may, of course, be much greater in an ART-suppressed situation, where there may be greater relative availability of virus-expressing cells than free virus, and therefore greater contribution of ADCC. Future studies must address other strategies to enhance the clearance of infected cells.
Methods
Antibodies.
Point mutations were introduced in antibody heavy-chain plasmids by using site-directed mutagenesis (GenScript). Heavy and light chains were cotransfected into Expi293 cells per manufacturer instructions, and antibodies were purified from culture supernatants by Protein A-Sepharose affinity chromatography. Of the 12 animals treated with VRC07-523LS, 3 of the animals used for the transcriptomics analysis were treated with antibody made and purified from Chinese hamster ovary cells under good manufacturing process guidelines.
Neutralization Assays.
Single-round-of-replication Env pseudoviruses were prepared, titers were determined, and the pseudoviruses were used to infect TZM-bl target cells, as described (35). Neutralization of monoclonal antibodies was determined by using replication-competent SHIVSF162P3 and a panel of six HIV-1 Env-pseudoviruses. Each NAb was assayed at fivefold dilutions, starting at a concentration of 50 μg/mL. The neutralization titers were calculated as a reduction in luminescence units compared with control wells and reported as either IC50 or IC80 in micrograms per milliliter.
ADCC Assays.
CEM-NKr cells were infected with SHIVSF162P3 and sorted for Env-expressing cells by using VRC07-523LS and labeled anti-human IgG secondary antibody. Sorted cells were monitored for Env expression and used in assays after complete loss of signal from the VRC07-523LS antibody used in the sort. For ADCC assays, infected CEM cells were labeled with 0.05 to 0.1 μM Cell-Trace Violet (CTV) dye (Thermofisher, catalog no. C34557) for 10 min at 37 °C, washed, and resuspended in RPMI medium with 10% serum, 100 U/mL penicillin, and 100 μg/mL streptomycin. NK cells were sorted from fresh rhesus macaque peripheral blood mononuclear cells (PBMCs) (Live CD3− CD20− CD14− HLADR− CD41− CD8+). Sorted cells were >95% CD159a+ and used as effectors. CTV-labeled target cells were incubated with antibody for 10 min at room temperature and combined with effector cells at a 1:10 ratio in 96-well plates. Following a 1-h incubation at 37 °C, cells were stained with a viability dye and assessed for target-cell death. For the prozone assays, CTV-labeled target cells and effector cells (KHYG-Rh cells expressing rhesus macaque FcγRIII) were individually incubated with antibodies for 10 min at room temperature followed by a wash and were then mixed together with the other cell type, either in the presence or absence of added antibody for 3 h at 37 °C. At the end of the assay, cells were washed and stained for viability, followed by staining for the bound antibody using anti-human IgG.
Phagocytosis Assays.
Phagocytosis assays were performed as described (36) with minor modifications. Biotinylated resurfaced stabilized core 3 (RSC-3) antigen was incubated overnight and at 4 °C with 1 μM fluorescent neutravidin beads. Following two washes in phosphate-buffered saline (PBS)–1% bovine serum albumin (BSA), 9 × 105 RSC-3–coated beads were incubated for 2 h at 37 °C/5% CO2 with the antibodies of interest at the indicated concentrations. Subsequently, 2 × 104 THP-1 cells were added for 4 h (bead:cell ratio of 45:1). At the end of the incubation period, cells were fixed in 1% paraformaldehyde (PFA) before acquisition in the flow cytometer.
Enzyme-Linked Immunosorbent Assay for Complement Binding.
Antibodies were tested for complement fixation as described in ref. 25.
FcγR and FcRn Binding.
His-tagged FcγRs were expressed in 293F or Expi293 cells and purified from culture supernatants by using Ni-nitriloacetic acid (Ni-NTA) chromatography. FcRn was purchased from Acro Biosystems (catalog no. FCM-C5284). Binding affinities were measured by biolayer interferometry. All receptors were loaded on HIS1K biosensors (ForteBio, catalog no. 18-5122) and probed into antibodies in 1× kinetics buffer. Loading time and concentration, antibody concentration, and association and dissociation times were optimized individually for each receptor. For pH 6 measurements, antibodies were diluted into phosphate buffer at pH 6. All data were fit by using a 1:1 model with R2 > 0.98 and χ2 < 3.
Animal Studies.
All animals were housed and cared for in accordance with the Guide for the Care and Use of Laboratory Animals (37) in a Biosafety Level-2 National Institute of Allergy and Infectious Diseases (NIAID) facility. All animal procedures and experiments were performed according to protocols approved by the Institutional Animal Care and Use Committee of the Vaccine Research Center, NIAID, NIH. Indian-origin rhesus macaques were challenged with 20 median tissue culture infectious-dose units of SHIV SF162P3 (stock VRCp2) i.v. and monitored weekly for PVL. Animals were assigned to antibody-treatment groups to obtain equivalent distribution based on pretreatment viral load. Animals were treated with a single dose of 20 mg/kg antibody i.v. bolus injection at 6 wk following virus challenge. Plasma viremia was quantitated by using a qRT-PCR–based method to quantify simian immunodeficiency virus gag RNA levels with a detection limit of 15 copies per mL, as described in ref. 38. Three additional animals in the VRC07-523LS group used for transcriptomics were treated with antibody at 12 to 14 wk postchallenge. These animals controlled plasma virus to undetectable levels by day 5 after antibody infusion and did not have sufficient sampling and were therefore excluded from virus-clearance kinetics analyses.
Antibody Pharmacokinetics.
Antibody levels in plasma were measured by a capture enzyme-linked immunosorbent assay (ELISA). High-binding ELISA plates (Corning, catalog no. 3690) were coated with 2 μg/mL antigen (RSC-3) in PBS (pH 7.4) at 37 °C for 1 h and blocked with blocking buffer containing 2% BSA and 5% skim milk for 30 min at room temperature. After washing, serial dilutions of heat-inactivated plasma in blocking buffer were added to the wells and incubated for 1 h at room temperature. Plates were then washed in PBS with 0.1% Tween-20, probed with anti-human secondary antibody (Southern Biotech, catalog no. 4029-05) for 30 min at room temperature, washed again, and developed with 3,3′,5,5′-tetramethylbenzidine substrate for 10 min. Reactions were stopped by the addition of 1 N sulfuric acid and read at 450 nm. Standard curves of purified antibody and control samples with known amounts of NAb spiked into naïve monkey plasma were used in every plate.
Flow Cytometry for Cell-Bound Infused NAb and Cell-Subset Frequency.
Reagents.
The 5C9 (anti-Idiotype for VRC07-523LS and Fc variants) with a C-terminal His-tag and Avi-tag was expressed as a Fab in Expi293 cells and purified through an Ni-NTA affinity column, followed by size-exclusion chromatography. Purified Fabs were biotinylated by using BirA ligase (Avidity, catalog no. BirA500) and then tetramerized on fluorescently labeled streptavidin.
Staining.
A total of 100 μL of ethylenediaminetetraacetic acid-anticoagulated whole blood was washed twice in 2 mL of ammonium–chloride–potassium lysis buffer (Quality Biological, catalog no. 118-156-101), followed by one wash in 3 mL of 1× PBS. Cells were incubated on ice for 20 min with fluorescently labeled antibodies, washed once with 3 mL of 2% BSA, and fixed in 1% PFA before acquisition on a flow cytometer. Antibodies in the staining panel were 5C9 BV421 (described above), XCR1-fluorescein isothiocyanate (FITC) (Biolegend, catalog no. 372612), CD3-BB660 (BD custom conjugate), CD66abce PerCP Vio700 (Miltenyi, catalog no. 130-103-794), CD14 BV510 (Biolegend, catalog no. 301842), CD64 BV605 (Biolegend, catalog no. 305034), HLA-DR BV655 (Biolegend, catalog no. 307650), CD16 BV711 (Biolegend, catalog no. 302044), CD123 BV786 (BD, catalog no. 564196), Clec9A Alexa 647 (BD, catalog no. 564267), CD11c (APC Fire750, Biolegend, catalog no. 371510), CD32 PE (Biolegend, catalog no. 303206), CD20 PECF594 (BD, catalog no. 562295), CD159a Cy7PE (Beckman, catalog no. B10246), CD4 BUV395 (BD, catalog no. 563550), CD41 BUV496 (BD custom conjugate), CD141 BUV737 (BD custom conjugate), and CD8 BUV805 (BD, catalog no. 564912).
Transcriptomics.
Cell sorting.
For transcriptomics, frozen PBMCs were thawed and stained for viability, followed by a panel of antibodies—XCR1 FITC (Biolegend, catalog no. 372612), CD20 PerCPCy5.5 (Biolegend, catalog no. 302326), 5C9 (custom in-house, described above), CD8 BV605 (BD, catalog no. 564115), HLA-DR BV650 (BD, catalog no. 307650), CD41 BV711 (BD, catalog no. 740778), Clec9A Alexa 647 (BD, catalog no. 564267), CD14 Alexa 700 (BD, catalog no. 557923), CD11c APC Fire750 (Biolegend, catalog no. 371510), CD282 PE (BD, catalog no. 565349), CD3 PECF594 (BD, catalog no. 562406), and CD159a Cy7PE (Beckman, catalog no. B10246)—for 20 min on ice, washed, and resuspended in 2% BSA. NK cells were identified as CD8+ CD159a+ and negative for all other markers in the panel. Samples were stored on ice until sorting on a flow cytometer. Sorted cells were lysed in RNAzol RT (MRC, catalog no. RN190) and stored frozen at −80 °C until use.
Sequencing.
Sequencing libraries were prepared and sequenced as described in ref. 39. In brief, total RNA was extracted from sorted NK cells and enriched for polyadenylated species by two sequential rounds of binding to oligo-dT Dynabeads (Life Technologies), chemically fragmented in the presence of Mg2+, and reverse-transcribed by using SuperScript III reverse transcriptase (Life Technologies). Second-strand complementary DNA (cDNA) synthesis, end repair, A-tailing, and sequencing-adaptor ligation were performed by using NEBNext enzyme modules (New England Biolabs). Libraries were amplified by using universal and indexed primers from the NEBNext system with Kapa 2x Hot Start Readymix (Kapa Biosystems). Amplified libraries were size-selected by using Beckman-Coulter Ampure XP beads, quantified by qPCR using the Kapa Library Quantification Kit for Illumina (Kapa Biosystems), and checked for sizing by electrophoresis on a BioAnalyzer (Agilent). Completed libraries were loaded on Illumina Truseq Paired-End v2 Cluster Kits and sequenced in 2 × 150 base-paired-end runs on an Illumina HiSeq 4000 sequencer with an average of 20 million reads per sample.
Data analysis.
Raw reads were trimmed for low-quality reads and miscalled nucleotides by using Trimmomatic (Version 0.39). Reads that passed the trimming step were aligned to the rhesus macaque genome (Ensembl Mmul_8.0.1, Release 88) by using the STAR aligner (Version 2.7.0e). Aligned reads were counted by using HTSeq (Version 0.9.1). Differential expression analysis between preinfusion and the two postinfusion timepoints was performed by using the R package edgeR. For each subset and Nab, a generalized linear model was fitted with the animals and timepoints as independent variables and the gene counts as a dependent variable. A likelihood-ratio test and Benjamini–Hochberg adjustment was used to test statistically for differential expression. Adjusted P value below or equal to 0.05 was used to defined differentially regulated genes. Gene Set Enrichment Analysis (preranked by edge R -log10 [adjusted P value]) was used to identify Kyoto Encyclopedia of Genes and Genomes pathways enriched among genes regulated postinfusion.
Data availability.
R code used for the transcriptomic analysis is available at https://github.com/sekalylab/vrc07. Raw RNA-sequencing data has been deposited in the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/) with accession no. GSE136938.
Env Sequencing from Plasma.
Sequencing.
Virions were concentrated from SHIV-infected macaque plasma by centrifugation at 21,100 × g for 1.5 h at 4 °C. RNA was extracted from virion pellets by using QIAmp Viral RNA Mini kits (Qiagen) and immediately reverse transcribed. Reverse-transcription (RT) reactions included 100 nM RT primer (TRTAATAAATCCCTTCCAGTCCCCCC), 100 units of SuperScript III Reverse Transcriptase, 500 mM deoxynucleoside triphosphate, 5 mM dithiothreitol, 20 units of RNAseOUT, and 1× first Strand Buffer (all from ThermoFisher Scientific). Reverse-transcription reaction conditions were 50 °C for 50 min, 85 °C for 10 min, and 4 °C hold. Copy numbers of resulting cDNAs were determined by limiting-dilution PCR using fluorescence-assisted clonal amplification (40). An estimated 600 cDNA templates were then subjected to near-full-length env PCR by using forward primer GAGCAGAAGACAGTGGCAATGA (41) and reverse primer CCCACGCGCYCGCAAGAG. PCR-amplified cDNA was prepared for Pacific Biosciences single-molecule real-time (SMRT) sequencing by using the SMRTbell Barcoded Adapter Complete Prep Kit and the SMRTbell DNA Damage Repair Kit for multiplex SMRT sequencing and then sequenced on a Pacific Biosciences Sequel with a 20-h movie time.
Data analysis.
Pacific Biosciences circular consensus sequence reads were generated at 99.9% identity, using the Quiver framework for consensus generation. The resulting fastq files were clustered by dereplicating reads and keeping a count of read abundance using Collapser command from FASTX-Toolkit (hannonlab.cshl.edu/fastx_toolkit). The clustered species were trimmed for primer sequences, and sequence reads under 1,000 base pairs in length were discarded. These species were further translated into amino acid sequences, and the dereplication step was repeated to determine unique occurrence of each amino acid sequence. Species with fewer than five reads were discarded at this step to get high-confidence subsampled sequences. MAFFT (42) was used to align the sequences, and the alignments were improved manually, with assessment of indels and removal of out-of-frame sequences. Mutational selection by NAbs at each position in Env was assessed by Fisher’s exact test for nonrandom differential distribution of character states between preinfusion and postinfusion amino acid sequence alignments, with dereplicated data weighted by read count. False discovery rate correction was performed in this analysis by using Bonferroni correction. Sequence-alignment patterns were visualized by using logo plots.
Cell-Associated DNA Load.
Frozen PBMCs were thawed, stained for viability (Thermofisher, catalog no. L34955) and then with a mixture of antibodies—CD14 BV510 (Biolegend, catalog no. 301842), CD8 BV605 (BD, catalog no. 564116), CD56 BV650 (BD, catalog no. 740579), CD4 Alexa 700 (Biolegend, catalog no. 317426), CD3 Cy7APC (BD, catalog no. 557757), TCRgd PE (Thermofisher, catalog no. MHGD04), CD20 PECF594 (BD, catalog no. 562295), and CD159a Cy7PE (Beckman, catalog no. B10246)—for 20 min on ice, followed by two washes. Cells were resuspended in 2% BSA and sorted for live CD14− CD20− CD159a− CD56− TCRgd− CD3+ CD8− CD4+/− cells. Sorted cells were pelleted by centrifugation, snap-frozen in liquid nitrogen, and stored a −80 °C until further use. Cell pellets were processed and tested for viral DNA, as described in ref. 43.
Statistical Analysis.
Statistical analysis was performed by using GraphPad Prism (Version 7.0d). Comparisons of pretreatment baseline viral load, slope of plasma-virus clearance, and fold changes in PVL between baseline and day 1.5 across NAb treatment groups were calculated by using one-way ANOVA. All analyses were corrected for multiple comparisons by the Tukey method.
Supplementary Material
Acknowledgments
We thank Elizabeth McCarthy, Jumagul Noor, Alida Taylor, and Hana Bao in the Translational Research Program of the Vaccine Research Center (VRC) and Megan DeMouth, Michelle Cully, and Keyun Wang in the Virology Laboratory, VRC, for help in organizing the animal experiments; Nancy Sullivan and John Misasi in the Biodefense Research Section, VRC, for providing the mAb114 plasmid; Farida Laboune and Amy Ransier in the Sequencing Core, VRC for processing samples for the transcriptomics analysis; Richard Nguyen, Julie Hill, Erica Smit, and Steve Perfetto in the Flow Cytometry Core, VRC, for help with flow cytometry; Kathy Foulds, Amy Noe, Shing-Fen Kao, Nadesh Nji, Dillon Flebbe, and Evan Lamb in the Nonhuman Primate Immunogenicity Core, VRC, for generous help in providing samples for optimizing assays; Nicole Doria-Rose and Stephen Schmidt in the Humoral Immunology Section, VRC, for neutralization assays; Margaret Beddal in the ImmunoTechnology Section, VRC, for custom conjugation of the anti-human IgG-Ax680; Guido Ferrari at the Duke Human Vaccine Institute for advice on the ADCC assays; and David Evans at the Department of Microbiology and Immunobiology, Harvard Medical School, New England Primate Research Center, for providing the KHYG-Rh cell line. We thank all members of the VRC, particularly the Immunology Laboratory and Virology Laboratory, for helpful comments and suggestions. We also thank the Quantitative Molecular Diagnostics Core of the AIDS and Cancer Virus Program, Frederick National Laboratory, for PVL analyses. This work was supported in part with federal funds from the National Cancer Institute, NIH, under Contract HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government. This work also made use of the High-Performance Computing Resource in the Core Facility for Advanced Research Computing at Case Western Reserve University. Portions of this work were performed under the auspices of the U.S. Department of Energy under Contract 89233218CNA000001 and funded by NIH Grants R01-AI028433 (to A.S.P.), R01-OD011095 (to A.S.P.), and P01-AI131365. S.F., R.P.S., and R.A.K. were supported by Bill and Melinda Gates Foundation Grant OPP1147555. Support for this work was provided by the Intramural Research Program of the VRC, NIAID, NIH.
Footnotes
Competing interest statement: J.R.M. and W.S. are inventors on a patent titled “Broadly neutralizing HIV-1 VRC07 antibodies that bind to the CD4-binding site of the envelope protein” (E-051-2012) for the VRC07-523LS antibody used in this study.
This article is a PNAS Direct Submission.
Data deposition: R code used for the transcriptomic analysis is available at GitHub (https://github.com/sekalylab/vrc07). Raw RNA-sequencing data have been deposited in the Gene Expression Omnibus database (accession no. GSE136938).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2008236117/-/DCSupplemental.
References
- 1.Bar-On Y. et al., Safety and antiviral activity of combination HIV-1 broadly neutralizing antibodies in viremic individuals. Nat. Med. 24, 1701–1707 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caskey M. et al., Viraemia suppressed in HIV-1-infected humans by broadly neutralizing antibody 3BNC117. Nature 522, 487–491 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caskey M. et al., Antibody 10-1074 suppresses viremia in HIV-1-infected individuals. Nat. Med. 23, 185–191 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lynch R. M. et al.; VRC 601 Study Team , Virologic effects of broadly neutralizing antibody VRC01 administration during chronic HIV-1 infection. Sci. Transl. Med. 7, 319ra206 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Bar K. J. et al., Effect of HIV antibody VRC01 on viral rebound after treatment interruption. N. Engl. J. Med. 375, 2037–2050 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mendoza P. et al., Combination therapy with anti-HIV-1 antibodies maintains viral suppression. Nature 561, 479–484 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scheid J. F. et al., HIV-1 antibody 3BNC117 suppresses viral rebound in humans during treatment interruption. Nature 535, 556–560 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilbert P. B. et al., Basis and statistical design of the passive HIV-1 antibody mediated prevention (AMP) test-of-concept efficacy trials. Stat. Commun. Infect. Dis. 9, 20160001 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bournazos S. et al., Broadly neutralizing anti-HIV-1 antibodies require Fc effector functions for in vivo activity. Cell 158, 1243–1253 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flerin N. C. et al., Establishment of a novel humanized mouse model to investigate in vivo activation and depletion of patient-derived HIV latent reservoirs. J. Virol. 93, e02051-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Halper-Stromberg A. et al., Broadly neutralizing antibodies and viral inducers decrease rebound from HIV-1 latent reservoirs in humanized mice. Cell 158, 989–999 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu C. L. et al., Enhanced clearance of HIV-1-infected cells by broadly neutralizing antibodies against HIV-1 in vivo. Science 352, 1001–1004 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parsons M. S. et al., Fc-dependent functions are redundant to efficacy of anti-HIV antibody PGT121 in macaques. J. Clin. Invest. 129, 182–191 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perelson A. S. et al., Decay characteristics of HIV-1-infected compartments during combination therapy. Nature 387, 188–191 (1997). [DOI] [PubMed] [Google Scholar]
- 15.Perelson A. S., Neumann A. U., Markowitz M., Leonard J. M., Ho D. D., HIV-1 dynamics in vivo: Virion clearance rate, infected cell life-span, and viral generation time. Science 271, 1582–1586 (1996). [DOI] [PubMed] [Google Scholar]
- 16.Ho D. D. et al., Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature 373, 123–126 (1995). [DOI] [PubMed] [Google Scholar]
- 17.Markowitz M. et al., A novel antiviral intervention results in more accurate assessment of human immunodeficiency virus type 1 replication dynamics and T-cell decay in vivo. J. Virol. 77, 5037–5038 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei X. et al., Viral dynamics in human immunodeficiency virus type 1 infection. Nature 373, 117–122 (1995). [DOI] [PubMed] [Google Scholar]
- 19.Rudicell R. S. et al.; NISC Comparative Sequencing Program , Enhanced potency of a broadly neutralizing HIV-1 antibody in vitro improves protection against lentiviral infection in vivo. J. Virol. 88, 12669–12682 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harouse J. M. et al., Mucosal transmission and induction of simian AIDS by CCR5-specific simian/human immunodeficiency virus SHIV(SF162P3). J. Virol. 75, 1990–1995 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zalevsky J. et al., Enhanced antibody half-life improves in vivo activity. Nat. Biotechnol. 28, 157–159 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hezareh M., Hessell A. J., Jensen R. C., van de Winkel J. G., Parren P. W., Effector function activities of a panel of mutants of a broadly neutralizing antibody against human immunodeficiency virus type 1. J. Virol. 75, 12161–12168 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lazar G. A. et al., Engineered antibody Fc variants with enhanced effector function. Proc. Natl. Acad. Sci. U.S.A. 103, 4005–4010 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corti D. et al., Protective monotherapy against lethal Ebola virus infection by a potently neutralizing antibody. Science 351, 1339–1342 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Hessell A. J. et al., Fc receptor but not complement binding is important in antibody protection against HIV. Nature 449, 101–104 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Moldt B. et al., A nonfucosylated variant of the anti-HIV-1 monoclonal antibody b12 has enhanced FcγRIIIa-mediated antiviral activity in vitro but does not improve protection against mucosal SHIV challenge in macaques. J. Virol. 86, 6189–6196 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hessell A. J. et al., Effective, low-titer antibody protection against low-dose repeated mucosal SHIV challenge in macaques. Nat. Med. 15, 951–954 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang P. et al., Quantifying the contribution of Fc-mediated effector functions to the antiviral activity of anti–HIV-1 IgG1 antibodies in vivo. Proc. Natl. Acad. Sci. U.S.A. 117, 18002–18009 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abrahams M. R. et al., The replication-competent HIV-1 latent reservoir is primarily established near the time of therapy initiation. Sci. Transl. Med. 11, eaaw5589 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pankau M. D. et al., Dynamics of HIV DNA reservoir seeding in a cohort of superinfected Kenyan women. PLoS Pathog. 16, e1008286 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woyach J. A. et al., A phase 1 trial of the Fc-engineered CD19 antibody XmAb5574 (MOR00208) demonstrates safety and preliminary efficacy in relapsed CLL. Blood 124, 3553–3560 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lü F. X. et al., Immunoglobulin concentrations and antigen-specific antibody levels in cervicovaginal lavages of rhesus macaques are influenced by the stage of the menstrual cycle. Infect. Immun. 67, 6321–6328 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu P. et al., Electron tomography analysis of envelope glycoprotein trimers on HIV and simian immunodeficiency virus virions. Proc. Natl. Acad. Sci. U.S.A. 100, 15812–15817 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manickam C., Shah S. V., Nohara J., Ferrari G., Reeves R. K., Monkeying around: Using non-human primate models to study NK cell biology in HIV infections. Front. Immunol. 10, 1124 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sarzotti-Kelsoe M. et al., Optimization and validation of the TZM-bl assay for standardized assessments of neutralizing antibodies against HIV-1. J. Immunol. Methods 409, 131–146 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ackerman M. E. et al., A robust, high-throughput assay to determine the phagocytic activity of clinical antibody samples. J. Immunol. Methods 366, 8–19 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.National Research Council , Guide for the Care and Use of Laboratory Animals, (National Academies Press, Washington, DC, ed. 8, 2011). [Google Scholar]
- 38.Li H. et al., Envelope residue 375 substitutions in simian-human immunodeficiency viruses enhance CD4 binding and replication in rhesus macaques. Proc. Natl. Acad. Sci. U.S.A. 113, E3413–E3422 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu J., Boritz E., Wylie W., Douek D. C., Stochastic principles governing alternative splicing of RNA. PLOS Comput. Biol. 13, e1005761 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boritz E. A. et al., Multiple origins of virus persistence during natural control of HIV infection. Cell 166, 1004–1015 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laird Smith M. et al.; IAVI Protocol C Investigators & The IAVI African HIV Research Network , Rapid sequencing of complete env genes from primary HIV-1 samples. Virus Evol. 2, vew018 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Katoh K., Misawa K., Kuma K., Miyata T., MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hansen S. G. et al., Addendum: Immune clearance of highly pathogenic SIV infection. Nature 547, 123–124 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
R code used for the transcriptomic analysis is available at https://github.com/sekalylab/vrc07. Raw RNA-sequencing data has been deposited in the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/) with accession no. GSE136938.