Significance
Light and gravity are two key environmental factors that coordinately regulate plant gravitropism, but the underlying molecular mechanisms that drive this regulation are poorly understood. Here, we reveal that light represses expression of LAZY4, a positive regulator of gravitropism, by promoting degradation of PIF proteins to inhibit negative gravitropism of hypocotyls in Arabidopsis. Meanwhile, light induces LAZY4 expression by promoting HY5 accumulation to enhance positive gravitropism in roots. Our findings provide key insights regarding the mechanism by which light mediates organ-specific gravitropism in plants.
Keywords: light, gravitropism, PIFs, HY5, LAZY4
Abstract
Light and gravity are two key environmental factors that control plant growth and architecture. However, the molecular basis of the coordination of light and gravity signaling in plants remains obscure. Here, we report that two classes of transcription factors, PHYTOCHROME INTERACTING FACTORS (PIFs) and ELONGATED HYPOCOTYL5 (HY5), can directly bind and activate the expression of LAZY4, a positive regulator of gravitropism in both shoots and roots in Arabidopsis. In hypocotyls, light promotes degradation of PIFs to reduce LAZY4 expression, which inhibits the negative gravitropism of hypocotyls. LAZY4 overexpression can partially rescue the negative gravitropic phenotype of pifq in the dark without affecting amyloplast development. Our identification of the PIFs-LAZY4 regulatory module suggests the presence of another role for PIF proteins in gravitropism, in addition to a previous report demonstrating that PIFs positively regulate amyloplast development to promote negative gravitropism in hypocotyls. In roots, light promotes accumulation of HY5 proteins to activate expression of LAZY4, which promotes positive gravitropism in roots. Together, our data indicate that light exerts opposite regulation of LAZY4 expression in shoots and roots by mediating the protein levels of PIFs and HY5, respectively, to inhibit the negative gravitropism of shoots and promote positive gravitropism of roots in Arabidopsis.
Light and gravity are two critically important environmental signals for plant growth. Light regulates the development of plants throughout their life cycle from seed germination to flowering (1). Gravity controls the growth orientation and architecture of plants to ensure that shoots grow upward for efficient photosynthesis and reproduction and that roots grow downward for improved water absorption, nutrient acquisition, and anchorage (2–5). However, little is known about the molecular basis of how light and gravity coordinately regulate plant growth.
Upon illumination, the Arabidopsis transcriptome undergoes massive changes that are mediated by many transcription factors (6). PHYTOCHROME INTERACTING FACTORs (PIFs), a set of basic helix–loop–helix transcription factors that include PIF1, PIF3, PIF4, and PIF5, play important roles in phytochrome-signaling pathways (7). PIFs are negative regulators of photomorphogenesis, and the quadruple pif mutant (pifq) exhibits a photomorphogenic phenotype in darkness (8, 9). Upon interaction of photoactivated phytochromes with PIFs, PIF proteins are phosphorylated, ubiquitinated, and degraded, resulting in inhibition of PIF function and thus transcriptional reprogramming (10–14). All four PIFs (PIF1, PIF3, PIF4, and PIF5) were reported to preferentially bind to a core G-box DNA-sequence motif (CACGTG) and/or a PBE-box (CACATG) motif in the target promoters (15–18). In addition to their central role in photomorphogenesis, PIFs were also found to be involved in light inhibition of negative gravitropism of hypocotyls, possibly through targeting of REPRESSOR OF PHOTOSYNTHETIC GENES1 (RPGE1) to suppress conversion of endodermal gravity-sensing starch-filled amyloplasts into other plastids in darkness (19, 20).
ELONGATED HYPOCOTYL 5 (HY5), a well-known basic leucine zipper (bZIP) family transcription factor (21), promotes plant photomorphogenesis under diverse light conditions mainly through transcriptional regulation (22–25). Light stabilizes HY5 protein, and HY5 directly binds to ACGT-containing elements (ACEs) in the promoters of target genes to regulate expression of light-responsive genes (22, 23, 26, 27). HY5 was also reported to be involved in primary and lateral root formation and root gravitropism (22, 28, 29). Light signals perceived by the shoot also regulate root development (30–32). Under natural conditions, aboveground light can be stem-piped to roots to activate root phyB, which triggers HY5 accumulation to modulate root growth and gravitropism (31). Light-stabilized HY5 protein in shoots moves directly to roots to activate expression of HY5 and the nitrate transporter NRT2.1, which thereby regulate root growth and nitrate uptake (30).
Gravity provides a continuous directional environmental signal to plants. Plants are able to perceive the gravity vector mainly through specialized sensing cells called “statocytes,” which are located in shoot endodermal cells and root columella cells in Arabidopsis thaliana (3, 4, 33, 34). Statocytes contain high-density, starch-accumulating plastids known as amyloplasts that, under steady-state conditions, settle to the bottom of cells. Once the growth direction of the plant changes, amyloplasts sediment to the new cell bottom, which may convert the physical vector signal to the biochemical signal. Then, auxin is transported in a polar manner to form a gradient across the organ, which leads to the asymmetric growth of plant organs (35, 36).
LAZY family genes are a group of key factors that function in gravity signaling in several plant species (37–40). The rice LAZY1 gene was the first family member to be identified and was mapped using a rice cultivar line exhibiting a prostrate shoot phenotype (37, 38, 41, 42). A previous study identified a family of six LAZY1-like genes in Arabidopsis, with AtLAZY1 exhibiting the most similar sequence to OsLAZY1. The lateral shoots of lazy1 plants show a larger growth angle compared to the wild type (39). Moreover, loss of LAZY2 and LAZY4 function enhances the lazy1 gravitropic phenotype (39, 43, 44). Several studies have reported redundant function of LAZY2/LZY2/NGR1/DRO3, LAZY3/LZY4/NGR3/DRO2, and LAZY4/LZY3/NGR2/DRO1 in root gravitropism in Arabidopsis (40, 43, 44). Among these LAZY genes, the lazy4 (dro1) single mutant exhibits larger lateral root growth angles (43–45). Consistent with these findings, OsDRO1/OsLAZY4 controls rice root system architecture through positive regulation of the crown root angles (46). Recent studies showed that gravity-triggered polarly distributed LAZYs (LZYs) may recruit RLD family proteins to regulate asymmetric PIN3 distribution in the lateral root cap columella, which directs polar auxin transport to control bending growth in gravitropism (43, 47).
Here, we report that PIFs activate expression of LAZY4, a representative LAZY family member, in hypocotyls by directly binding to the G-box in the LAZY4 promoter in darkness. Light suppresses LAZY4 expression in hypocotyls by promoting degradation of PIFs, thus inhibiting negative gravitropism of hypocotyls. Increased LAZY4 expression in the pifq mutant is able to partially rescue the interrupted hypocotyl gravitropism phenotype of dark-grown pifq but not the development of endodermal amyloplasts. These data suggest the presence of another mechanism for regulation of hypocotyl negative gravitropism that is distinct from the previously reported PIFs-amyloplast pathway. We also demonstrate that HY5 is stabilized by light and binds to the LAZY4 promoter through ACEs, which results in induction of LAZY4 expression in roots and promotion of root positive gravitropism. Our study reveals a molecular basis for the coordination of light and gravity signaling in plants.
Results
PIFs Bind Directly to the LAZY4 Promoter In Vivo.
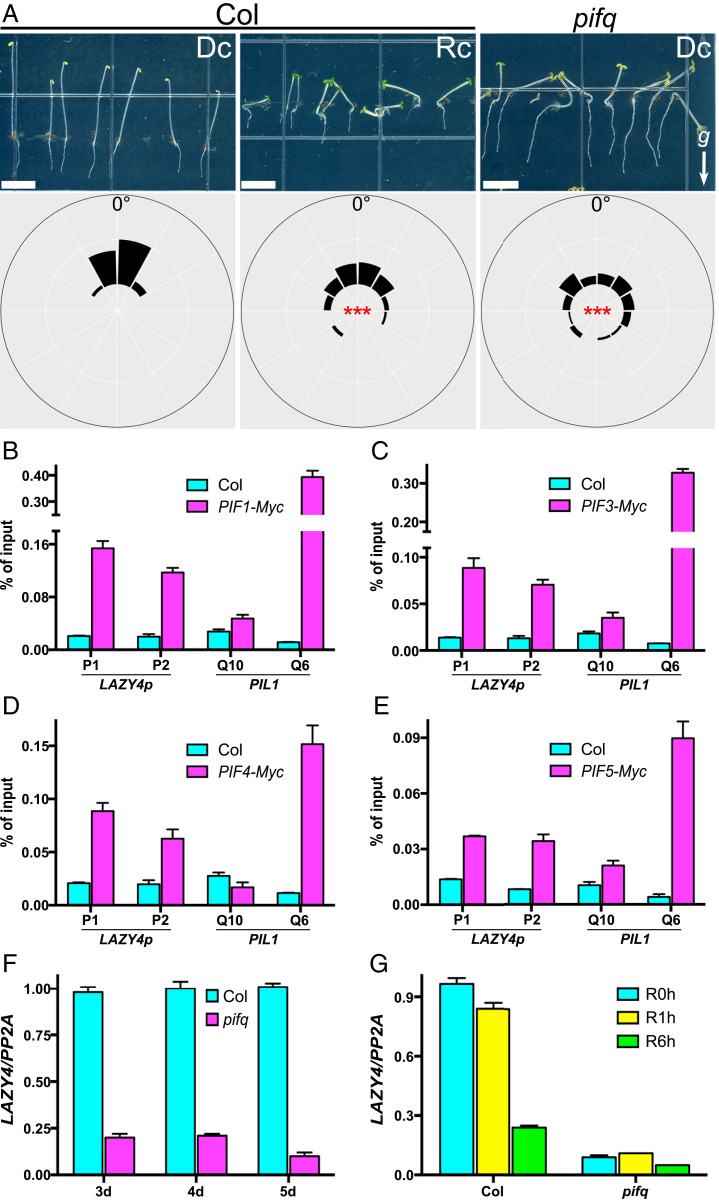
Previous studies proposed that light may inhibit the negative gravitropism of hypocotyls by promoting degradation of PIFs (9, 19). To test this model in our experimental condition, we grew seedlings on vertical plates in the dark or under red-light conditions. We observed that hypocotyls of dark-grown wild-type (Col) seedlings grew upward, whereas the hypocotyls of red-light–grown Col and dark-grown pifq exhibited interrupted negative gravitropism (Fig. 1A). A previous study suggested that PIFs maintain amyloplast development in the dark to promote negative gravitropism of Arabidopsis hypocotyls in the dark (19). To uncover specific molecular connections between the light signal and gravitropism, we searched for gravitropism-related genes in PIF targets by analyzing previously reported PIFs chromatin immunoprecipitation sequencing (ChIP-seq) data (15, 17, 18). We found that all four PIFs (PIF1, -3, -4, -5) bind to the LAZY4 promoter through the G-box located ∼2,000 bp upstream of the ATG start codon (SI Appendix, Figs. S1A and S2). As recent studies demonstrated that LAZY4 plays important roles in the gravitropism of both shoots and roots (40, 43–45), we speculated that LAZY4 may function downstream of PIFs, thus serving as a key node in light modulation of gravitropism. To confirm the ChIP-seq data described above, we performed chromatin immunoprecipitation (ChIP) assays using 4-d-old dark-grown Col and PIF1/3/4/5-Myc seedlings. Our analysis showed that all four of the PIF-Myc factors bound the G-box region of the LAZY4 promoter (Fig. 1 B–E).
Fig. 1.
PIFs directly activate LAZY4 expression in Arabidopsis hypocotyls. (A) Orientation of wild-type (Col) and pifq hypocotyls in red light or dark conditions. Seedlings were grown on the surface of vertical agar plates in dark or red light for 4 d (Top), and statistical analyses of the hypocotyls’ growth orientations are shown (Bottom). Frequencies of hypocotyl growth directions are shown in the corresponding angular position around a circle at intervals of 30° (n = 50 to 90). For statistical analyses, means of the absolute value of the angles between the growth direction and the upward vertical axis were compared. Asterisks indicate significant differences compared to Col in the dark, as determined by one-way ANOVA (***P < 0.001). The arrow labeled with a “g” represents the gravity direction (Scale bars, 5 mm.) (B–E) PIFs bind to the LAZY4 promoter in ChIP assays. Seedlings of 4-d-old dark-grown Col and PIF1-Myc (B), PIF3-Myc (C), PIF4-Myc (D), and PIF5-Myc (E) were collected for ChIP analyses using anti-Myc beads. LAZY4p-P1 and LAZY4p-P2 are two G-box–containing fragments within the LAZY4 promoter (see SI Appendix, Fig. S2, for their detailed locations). PIL1-Q10 and PIL1-Q6 were used as a negative and positive controls, respectively. (F and G) LAZY4 expression levels in Col and pifq hypocotyls. Total RNA was extracted from the hypocotyls of seedlings grown in the dark for 3, 4, or 5 d (F) or grown in the dark for 4 d and then transferred to red light for 1 or 6 h (G). Then, the levels of LAZY4 transcripts were examined by qRT-PCR. (B–G) Error bars represent the SD of three technical replicates. All experiments were repeated three times, and a representative result is shown.
Light Inhibits LAZY4 Expression in Hypocotyls by Promoting PIF Degradation.
In order to detect whether PIFs regulate LAZY4 transcript levels in hypocotyls, we compared the expression levels of LAZY4 in wild-type and pifq hypocotyls. Our analyses of seedlings grown in the dark for various time periods revealed that levels of LAZY4 transcripts were notably decreased in pifq mutant hypocotyls, indicating that PIFs positively regulate LAZY4 expression in Arabidopsis hypocotyls (Fig. 1F). As PIF proteins are rapidly degraded following light exposure, we next tested whether light inhibits LAZY4 transcription in hypocotyls. The expression of LAZY4 was dramatically decreased in the hypocotyls of wild type after light exposure, whereas the reduction of LAZY4 transcripts in the hypocotyls of the pifq mutant was not as obvious (Fig. 1G). Taken together, these results demonstrate that PIFs are the major transcriptional activators of LAZY4 in hypocotyls and that light-promoted PIF degradation leads to a dramatic decrease of LAZY4 expression in hypocotyls.
Overexpression of LAZY4 Partially Rescues the Gravitropic Phenotype of pifq without Affecting Amyloplast Development.
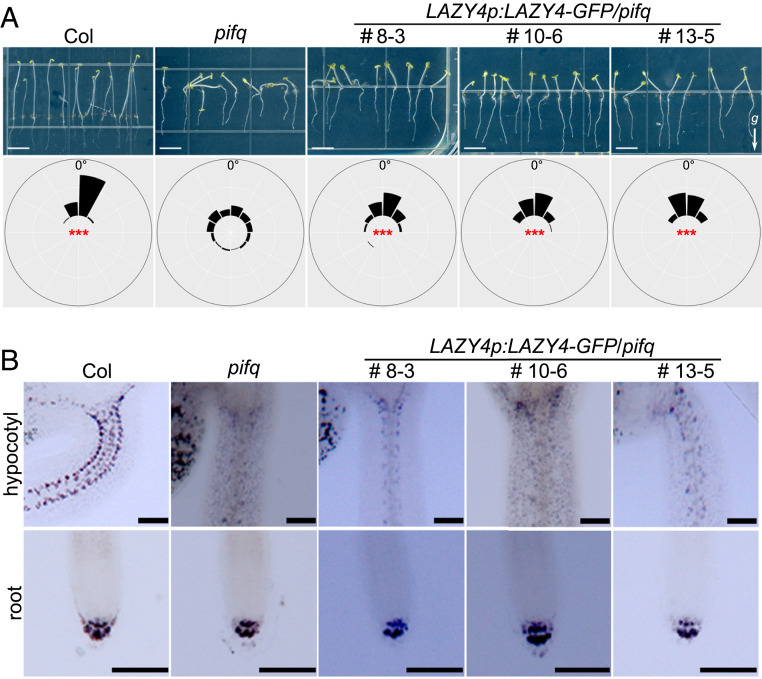
As previously reported, LAZY4 is a positive regulator of gravitropism in both shoots and roots (40, 43–45). To perform genetic analysis of the relationship between PIFs and LAZY4, we introduced a LAZY4-GFP fusion protein under the control of the native LAZY4 promoter into the pifq background and obtained three independent LAZY4p:LAZY4-GFP/pifq lines. In these three lines, the levels of LAZY4 transcripts were found to be higher than in the wild-type controls (SI Appendix, Fig. S3), indicating that these plants are LAZY4 overexpression lines. These results suggest the existence of other uncharacterized transcription factors that may activate LAZY4 expression in the dark. All three LAZY4p:LAZY4-GFP/pifq lines displayed improved negative gravitropism of their hypocotyls in the dark when compared with pifq (Fig. 2A). From these data, we concluded that LAZY4 acts downstream of PIFs and that the interrupted negative gravitropic phenotype of the pifq mutant can be partially rescued by increasing LAZY4 expression levels.
Fig. 2.
Expression of LAZY4-GFP partially rescues hypocotyl negative gravitropism but not amyloplast development in the pifq mutant. (A) Overexpression of LAZY4-GFP partially rescued the negative gravitropism of pifq hypocotyls. Seedlings of Col, pifq, and three LAZY4p:LAZY4-GFP/pifq transgenic lines were grown vertically in the dark for 4 d (Top). Frequencies of hypocotyl growth directions are shown in the corresponding angular position around a circle at intervals of 30° (n = 50 to 90; Bottom). For statistical analyses, means of the absolute value of the angles between the growth direction and upward vertical axis were compared. Asterisks indicate significant differences compared to pifq, as determined by one-way ANOVA (***P < 0.001). The arrow labeled with a “g” represents the gravity direction (Scale bars, 5 mm.) (B) Overexpression of LAZY4-GFP did not rescue amyloplast development in pifq hypocotyls. Amyloplasts in the hypocotyls (Top) and roots (Bottom) of 4-d-old dark-grown Col, pifq, and three LAZY4p:LAZY4-GFP/pifq transgenic lines were detected by I2-KI staining (Scale bars, 200 μm.)
A previous study reported that, in the dark, PIFs suppress the conversion of endodermal starch-filled amyloplasts into plastids with small starch granules, thereby enabling negative gravitropism of hypocotyls (19). Here, we found that PIFs positively regulate LAZY4 expression through direct binding of the LAZY4 promoter and ultimately promote hypocotyl negative gravitropism in the dark. Therefore, we investigated whether the PIFs-LAZY4 module also mediates amyloplast development. We examined and compared the endodermal amyloplasts in Col, pifq, and LAZY4:LAZY4-GFP/pifq lines by I2-KI staining. Amyloplast development is similar in the columella cells of all of the root caps. In contrast, hypocotyls of pifq exhibited fewer amyloplasts compared to wild-type hypocotyls, whereas LAZY4-GFP expression in the LAZY4:LAZY4-GFP/pifq lines did not rescue this reduced amyloplast development (Fig. 2B). Taken together, our data demonstrate that increasing LAZY4 transcript levels in the pifq mutant partially rescues hypocotyl negative gravitropism without altering endodermal amyloplast development. Thus, the PIFs-LAZY4 regulatory module represents another mechanism by which PIFs function in the coordination of light and gravity signaling, and this mechanism is independent from the previously reported PIF-amyloplast pathway (19).
HY5 Directly Binds to and Promotes Expression of LAZY4 in Arabidopsis Roots.
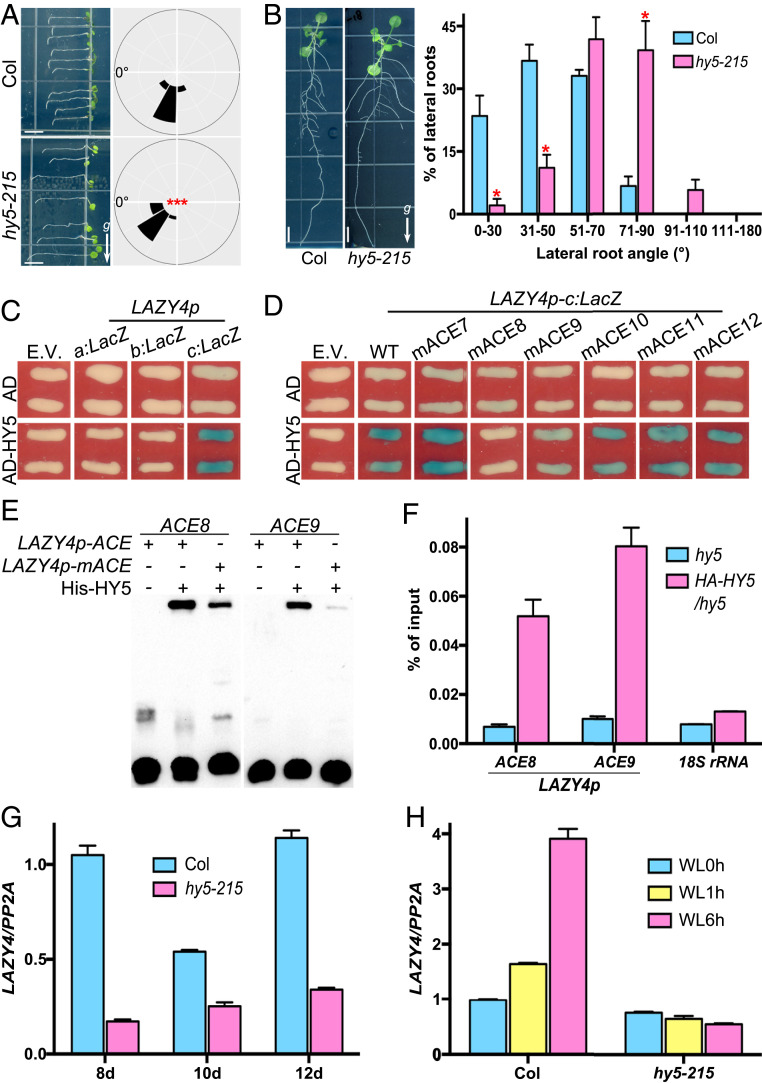
The investigation described above revealed a mechanism by which light inhibits the negative gravitropism of hypocotyls. We next asked whether light regulates the positive gravitropism of roots. Previous research reported that a mutation in HY5, a core positive transcription factor in photomorphogenesis, weakened gravitropic responses in both primary and lateral roots (22, 28, 29). We first examined the primary root gravitropism of the hy5 mutant and found it to be defective compared to the wild type in a gravistimulation test (Fig. 3A). The lateral root angles of the hy5-215 mutant were also found to be significantly larger when compared to those of the wild type (Fig. 3B). Taken together, these data suggest that HY5 plays roles in regulating the gravitropism of roots, and we next pursued study of the underlying molecular basis of the role of HY5 in this regulation.
Fig. 3.
HY5 directly activates LAZY4 expression in roots. (A and B) Mutation of HY5 disrupts Arabidopsis root gravitropism under light. (A) Seedlings of Col and hy5-215 were grown vertically in white light for 3 d and then rotated 90° for 12 h to induce gravistimulation. The angles between main root directions and the left horizontal axis were calculated. The frequencies of main root angles are shown in the corresponding angular position around a circle at intervals of 30° (n = 54). (B) Seedlings of Col and hy5-215 were grown vertically in white light for 12 d, and the distributions of lateral root angles were analyzed. Individual lateral root angle values were sorted into six categories, and percentages of each category were calculated. Bars represent SEM (n = 4 experiments; 40 to 50 lateral roots per experiment). In A and B, asterisks indicate significant differences, as determined by Student’s t test (***P < 0.001, *P < 0.05). Arrows with a “g” represent the gravity direction (Scale bars, 5 mm.) (C and D) HY5 binds to the LAZY4 promoter in the yeast one-hybrid assay. (C) The letters a, b, and c indicate three fragments of the LAZY4 promoter, as shown in SI Appendix, Fig. S2. Empty vector (E.V.) expressing the AD domain alone served as the negative control. (D) mACE7 to mACE12 represent fragment c with mutations (ACGT to gggg) in six different regions. (E) His-HY5 binds to LAZY4p-ACE8 and LAZY4p-ACE9 probes in EMSA. Recombinant His-HY5 was purified from Escherichia coli. LAZY4p-ACE and LAZY4p-mACE are wild-type and mutated probes, respectively. (F) HY5 binds to the LAZY4 promoter in ChIP assays. Seedlings of 4-d-old white light-grown Col and HA-HY5/hy5 were collected for ChIP analysis. LAZY4p-ACE8 and LAZY4p-ACE9 are two small fragments within fragment c of the LAZY4 promoter and were detected by qPCR. The 18S rRNA was used as a negative control. (G and H) HY5 positively regulates LAZY4 expression in Arabidopsis roots. (G) Seedlings of Col and hy5-215 were grown in white light for 8, 10, or 12 d or (H) in the dark for 4 d and then transferred to white light for 1 and 6 h, respectively. WL, white light. In G and H, total RNA was extracted from the roots, and the expression of LAZY4 was detected by qRT-PCR. PP2A was used as an internal control. (F–H) Error bars represent the SD of three technical replicates. These experiments were repeated three times, and a representative result is shown.
As HY5 is an extensively studied bZIP transcription factor, we searched for gravitropism-related genes in the targets of HY5. Analysis of existing HY5 ChIP-chip data (26, 27) revealed that HY5 can also bind to the LAZY4 promoter (SI Appendix, Fig. S1B). In order to confirm the binding of HY5 to the LAZY4 promoter and identify its binding region, we divided the LAZY4 promoter into three overlapping fragments, designated a, b, and c (SI Appendix, Fig. S2). Using a yeast one-hybrid assay, we found that HY5 was able to bind the c fragment of the LAZY4 promoter (Fig. 3C). Further mutagenesis analysis revealed that HY5 primarily binds to the ACE8 and/or ACE9 regions in the c fragment of the LAZY4 promoter (Fig. 3D and SI Appendix, Fig. S2). We next performed an electrophoretic mobility shift assay (EMSA), which further demonstrated the direct binding of HY5 to the ACE8 and ACE9 regions within the c fragment of the LAZY4 promoter (Fig. 3E). In addition, HY5 was shown to bind the LAZY4 promoter in vivo in our ChIP-qPCR analyses (Fig. 3F). Taken together, our data demonstrate that HY5 directly binds to the ACE8 and/or ACE9 regions in the c fragment of the LAZY4 promoter in vitro and in vivo.
To investigate the role of HY5 in regulating LAZY4 expression and root gravitropism, we compared the expression of LAZY4 in roots from the wild type and hy5 mutant using qRT-PCR analyses. Levels of LAZY4 transcripts decreased in hy5 mutant roots at all time points examined, suggesting that HY5 induces LAZY4 transcription in Arabidopsis roots (Fig. 3G). A previous study showed that LAZY4 expresses in both columella and endodermal cells of roots, but only the expression of LAZY4 in root columella cells plays a role in root gravitropic responses (43). Columella cells locate in the root tips, and we found that the expression of LAZY4 was actually reduced in the root tips when HY5 was mutated (SI Appendix, Fig. S4A). Since light promotes the accumulation of HY5 protein by repressing COP1 function (24, 48), we speculated that light may promote LAZY4 accumulation in roots, which we confirmed (Fig. 3H). Importantly, light-induced expression of LAZY4 was completely abolished in the hy5 mutant (Fig. 3H). Taken together, our data demonstrate that, in roots, HY5 positively regulates LAZY4 expression through direct binding to the LAZY4 promoter and that light increases accumulation of HY5 protein, which promotes LAZY4 expression.
The Defective Root Gravitropism of the hy5 Mutant Is Largely Rescued by Increased LAZY4 Expression.
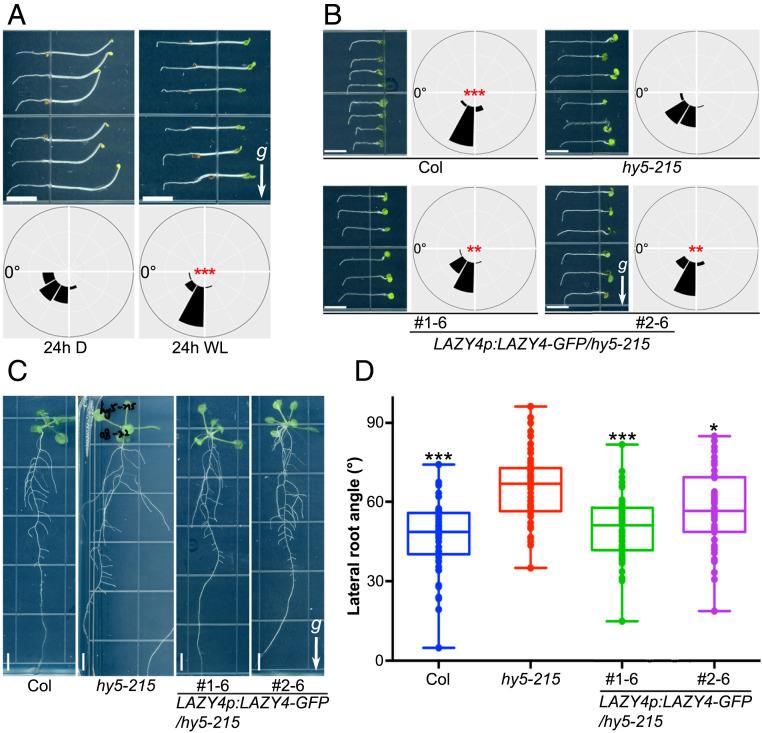
As light is capable of inducing LAZY4 expression via HY5 accumulation and LAZY4 plays positive roles in root gravitropism (40, 43–45), we speculated that light promotes positive gravitropism of roots. Our gravistimulation experiment indeed confirmed this expectation (Fig. 4A). This result is consistent with previous reports (49–52).
Fig. 4.
Expression of LAZY4-GFP largely rescues the positive gravitropism of hy5-215 roots. (A) Light promotes the gravitropism of Arabidopsis roots. Three-day-old dark-grown seedlings were either maintained in the dark or transferred to white light followed by a 90° rotation for 24 h to induce gravistimulation. Frequencies of root angles are shown in the corresponding angular position around a circle at intervals of 30° (n = 54). Asterisks indicate significant differences, as determined by Student’s t test (***P < 0.001). The arrow with a “g” represents the gravity direction (Scale bars, 5 mm.) (B) Expression of LAZY4-GFP partially rescued the gravitropism of the hy5 main roots. Seedlings of Col, hy5-215, and two LAZY4p:LAZY4-GFP/hy5-215 transgenic lines were grown in white light for 3 d and then rotated 90° for 12 h to induce gravistimulation. Frequencies of root angles are shown in the corresponding angular position around a circle at intervals of 30° (n = 54). Asterisks indicate significant differences compared to hy5-215, as determined by one-way ANOVA (***P < 0.001, **P < 0.01). The arrow with a “g” represents the gravity direction (Scale bars, 5 mm.) (C and D) Expression of LAZY4-GFP largely rescued the gravitropism of hy5 lateral roots. Seedlings of Col, hy5-215, and two LAZY4p:LAZY4-GFP/hy5-215 transgenic lines were grown in white light for 12 d (C), and the distribution of lateral root angles were analyzed (D, n = 40 to 50 for each genotype). Asterisks indicate significant differences compared to hy5-215, as determined by one-way ANOVA (***P < 0.001, *P < 0.05) (Scale bars, 5 mm.)
To perform a genetic analysis of the relationship between HY5 and LAZY4, we introduced a LAZY4-GFP fusion protein driven by the native LAZY4 promoter into the hy5 mutant and two independent LAZY4p:LAZY4-GFP/hy5-215 lines that were used for root gravitropism assays. The LAZY4 expression levels in these two lines were clearly higher than those in the hy5 mutant but less than that in the wild type (SI Appendix, Fig. S4B). We first examined the primary root gravitropism of the transgenic lines. LAZY4 expression driven by the native LAZY4 promoter is able to partially rescue the gravitropic phenotype of hy5 roots (Fig. 4B). The lateral roots of hy5-215 plants exhibit a larger growth angle, as previously reported (22, 28, 29), whereas the lateral root angles of the two transgenic lines are similar to that of wild-type plants (Fig. 4 C and D). Taken together, our data suggest that LAZY4 acts downstream of HY5 and that the abnormal gravitropism of hy5 roots can be largely rescued by increasing LAZY4 expression.
PIFs and HY5 Control LAZY4 Expression in Hypocotyls and Roots, Respectively, to Regulate Gravitropism.
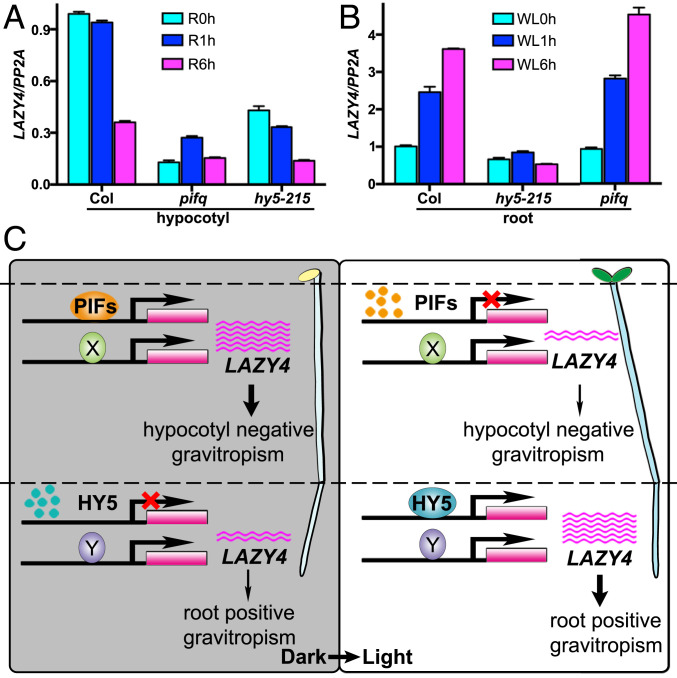
The study described above revealed that both PIFs and HY5 positively regulate the LAZY4 transcript levels. However, PIFs play an important role in regulation hypocotyl gravitropism, whereas HY5 functions mainly in root gravitropism. To uncover the specificity of these two groups of light-regulated transcription factors in gravitropism, we compared LAZY4 expression in hypocotyls and roots of Col, pifq, and hy5-215 in parallel during the dark-light transition. Although HY5 showed some positive regulation on LAYZ4 expression, PIFs predominantly mediated LAZY4 expression in hypocotyls (Fig. 5A). In contrast, HY5 predominantly mediated LAZY4 expression whereas PIFs did not show any regulation in roots (Fig. 5B). These expression patterns explain the observed organ-specific roles of PIFs and HY5 in gravitropism well.
Fig. 5.
PIFs and HY5 control the expression of LAZY4 in hypocotyls and roots, respectively, to regulate gravitropism. (A and B) LAZY4 expression levels in Col, pifq, and hy5-215 hypocotyls transferred from dark to red light (A) and in roots transferred from dark to white light (B). Total RNA was extracted from hypocotyls or roots from seedlings growing in the dark for 4 d and then transferred to light for 1 and 6 h, respectively. Transcripts were detected by qRT-PCR. Error bars represent the SD of three technical replicates. These experiments were repeated three times, and a representative result is shown. R, red light. WL, white light. (C) A model describing the roles of PIFs and HY5 in regulation of hypocotyl and root gravitropism via regulation of LAZY4 expression. In the dark, high accumulation of PIF proteins activates LAZY4 expression in hypocotyls to maintain their negative gravitropism. After light exposure, PIF proteins degrade quickly, and LAZY4 expression is dramatically reduced, which leads to inhibition of the negative gravitropism of hypocotyls. In roots, light exposure promotes accumulation of HY5 proteins, and LAZY4 expression increases, which promotes the positive gravitropism of roots. X and Y indicate speculated uncharacterized factors in the hypocotyls and roots, respectively.
From our findings, we propose a model to describe how light differentially mediates the expression of LAZY4 via PIFs and HY5 to regulate the gravitropism of hypocotyls and roots, respectively, in Arabidopsis (Fig. 5C). In hypocotyls, PIFs are stabilized in the dark, which activates LAZY4 expression and promotes the negative gravitropism of hypocotyls. Upon light exposure, phytochromes inhibit PIFs by targeting them for protein degradation, which results in reduced LAZY4 expression and inhibition of hypocotyl negative gravitropism. In roots, HY5 is degraded in darkness through the 26S proteasome pathway, leading to reduced LAZY4 expression in roots and relatively weak gravitropism. Upon light exposure, HY5 is stabilized to activate LAZY4 transcription in roots, which promotes root positive gravitropism (Fig. 5C).
Discussion
Light and gravity are constant environmental factors that coordinate to control plant development, but the mechanisms that underlie this coordinated regulation remain elusive. Red light inhibits the negative gravitropism of Arabidopsis hypocotyls, and the loss of phytochrome A and B function suppresses red-light inhibition of hypocotyl negative gravitropism (53, 54). Phytochromes promote the conversion of gravity-sensing amyloplasts to other plastids by mediating PIF degradation, which reduces the plant’s gravity-sensing capability and leads to inhibition of hypocotyl negative gravitropism (19). Subsequent studies revealed that endodermal PIF1 directly targets REPRESSOR OF PHOTOSYNTHETIC GENES1 (RPGE1) to inhibit conversion of amyloplasts to plastids with high expression of photosynthetic genes (20). Here, we revealed that LAZY4, a key factor in gravity signaling, is directly activated by PIFs in darkness to promote negative gravitropism of hypocotyls (Fig. 1). Previous studies showed that PIFs could function as either transcriptional activators or repressors (17, 18, 55–59), and PIFs clearly work as transcriptional activators in the regulation of LAZY4 in this study. Importantly, we demonstrated that this PIFs-LAZY4 module regulates gravitropism using a mechanism distinct from mediation of amyloplast development (Fig. 2). Together, these studies show that PIFs, the key transcriptional factors that function in light signaling, promote negative gravitropism of hypocotyls through at least two independent pathways: maintenance of amyloplast development and activation of LAZY4 gene expression. Since the lazy4 single mutant did not display reduced hypocotyl negative gravitropism (43, 44), and the possibility of the down-regulation of all of the LAZY family members simultaneously in the pifq mutant was excluded (SI Appendix, Fig. S5), how reduction of LAZY4 expression contributes to interrupted gravitropic response of pifq hypocotyls is unclear and needs further investigations in the future. PHYTOCHROME KINASE SUBSTRATE 4 (PKS4) and GRAVITROPIC IN THE LIGHT 1 (GIL1) have also been identified as negative regulators of light-inhibited negative gravitropism of Arabidopsis hypocotyls (60, 61). In the future, our understanding of the underlying molecular mechanism of light inhibition of hypocotyl negative gravitropism may be improved by studying the possible connections between these factors.
Light clearly promotes the gravitropic growth of roots. For example, maize roots and rice crown roots grew almost horizontally in the dark, and light exposure resulted in angled root growth (49–51). The roots of Arabidopsis phyA phyB double mutants exhibited a reduced gravitropic curvature (52). Mutation of HY5, a key transcription factor and positive regulator in photomorphogenesis, led to defective gravitropic responses in both primary and lateral roots (22, 28, 29). However, we still do not understand the molecular mechanisms that explain how these light-signaling factors regulate gravitropism. Our study shows that light-stabilized HY5 directly binds to the LAZY4 promoter and induces LAZY4 expression to promote root positive gravitropism (Figs. 3 and 4), revealing a molecular mechanism by which a key light-signaling factor regulates root gravitropism.
As a key environmental factor, light regulates many developmental processes of plants, and light-signaling pathways have been extensively studied. However, light-mediated processes differ in various plant organs. This phenomenon has been recognized for several decades, but the mechanisms underlying these differences have just begun to be revealed in recent years. Previously, we found that light promotes cotyledon expansion while inhibiting hypocotyl elongation by differentially mediating the expression of SAUR genes in Arabidopsis (62, 63). In this study, we demonstrated that light inhibits the gravitropism of hypocotyls but promotes the gravitropism of roots via differential regulation of LAZY4 gene expression to reveal a paradigm of organ-specific light response in plants. Although PIFs and HY5 are expressed in both hypocotyls and roots (22, 28, 30, 31, 64), they predominantly promote the expression of LAZY4 in hypocotyls and roots, respectively, in regulating gravitropic responses (Fig. 5). We speculate that unknown cofactors specifically expressed in hypocotyls or roots may work together with PIFs and HY5 to mediate the organ-specific regulation of LAZY4 expression, a hypothesis that will be addressed in future experiments.
Materials and Methods
Plant Materials and Growth Conditions.
The wild-type A. thaliana line used in this study was Columbia-0. The mutants and transgenic lines used in this study were described previously: hy5-215 (22); pif1 pif3 pif4 pif5 (pifq) (8); PIF1-Myc, PIF3-Myc, PIF4-Myc, and PIF5-Myc (65); and hy5 (SALK_096651) (66).
For all experiments, Arabidopsis seeds were surface-sterilized with 15% NaClO for 5 min followed by rinsing with sterile H2O three times. Seeds were sown onto Murashige and Skoog medium (MS, 1% sucrose and 0.8% agar) and cold-treated at 4 °C for 2 to 3 d in the dark. Subsequently, the seeds were irradiated with white light (100 μmol⋅m−2⋅s−1) for 3 h to induce germination and then grown under various light conditions.
Plasmid Construction.
The LAZY4 genomic sequence was fused with GFP and introduced into pJim19 (Bar) to generate the LAZY4p:LAZY4-GFP (Bar) construct. The LAZY4p:LAZY4-GFP (Bar) plasmid was digested with SbfI and EcoRI, and the LAZY4p:LAZY4-GFP cassette was inserted into the pCambia1300 plasmid to generate LAZY4p:LAZY4-GFP (Hyg). The LAZY4p:LAZY4-GFP (Bar) and LAZY4p:LAZY4-GFP (Hyg) plasmids were then introduced into Agrobacterium tumefaciens (strain GV3101) and transformed into the hy5-215 and pifq mutants via the floral dip method.
To generate the LAZY4p-a:LacZ, LAZY4p-b:LacZ, and LAZY4p-c:LacZ reporter constructs, the promoter fragments were amplified from the LAZY4p:LAZY4-GFP construct by PCR and cloned into the EcoRI-SalI site of pLacZi2μ (67). LacZ reporter genes driven by the various c fragments of the LAZY4 promoter with mutations in ACEs were amplified by overlap extension PCR using the LAZY4p-c:LacZ construct as the template. The AD-HY5 construct was described previously (68).
All primers used in this study are summarized in SI Appendix, Table S1.
Quantitative Real-Time PCR.
Total RNA from hypocotyls or roots was extracted using the Takara MiniBEST Plant RNA Extraction Kit (Takara). Reverse transcription was performed using ReverTra Ace qPCR RT Master Mix (TOYOBO) according to the manufacturer’s instructions. SYBR Premix ExTaq (Takara), complementary DNA templates, and primer sets were combined, and real-time qPCR analyses were performed on an Applied Biosystems 7500 Fast Real-Time PCR detection system (ABI). Each experiment was repeated with three independent samples, and qRT-PCR reactions were performed with three technical replicates for each sample. PP2A was used as an internal control. The primers used for qRT-PCR are listed in SI Appendix, Table S1.
Yeast One-Hybrid Assays.
The AD-fusion plasmid was cotransformed with the LacZ reporter genes under the control of various LAZY4 promoter fragments into the yeast strain EGY48. Transformants were selected and grown on proper dropout plates and then transferred to proper dropout medium containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside for interaction tests. Yeast transformation was conducted as described in the Yeast Protocols Handbook (Clontech).
EMSA.
EMSAs were performed using biotin-labeled probes and the Lightshift Chemiluminescent EMSA kit (Thermo Fisher). Briefly, 1 μg of His-HY5 fusion protein was incubated together with biotin-labeled wild type or mutated probes in 20-mL reaction mixtures containing 10 mM Tris-HCl, 150 mM KCl, 1 mM dithiothreitol, 50 ng/μL poly(dI-dC), 2.5% glycerol, 0.05% Nonidet P-40, 100 μM MgCl2, and 0.5 mg/mL bovine serum albumin for 20 min at room temperature and separated on 6% native polyacrylamide gels in Tris-glycine buffer. The labeled probes were detected according to the instructions provided with the EMSA kit. The sequences of the oligonucleotides used to generate the biotin-labeled probes are shown in SI Appendix, Table S1.
ChIP.
ChIP assays were performed as previously described (26). Seedlings were grown in the dark or in white light for 4 d, and then samples were harvested and treated with 1% formaldehyde under vacuum to cross-link the protein and DNA. Samples were ground into powder in liquid nitrogen, sonicated, and then incubated with anti-Myc agarose beads (Invitrogen) or anti-HA (Cell Signaling Technology) antibody to isolate chromatin complexes. Protein A magnetic beads (Invitrogen) were used to precipitate chromatin complexes from samples incubated with the anti-HA antibody. The precipitated DNA was recovered and analyzed by real-time PCR using the primers listed in SI Appendix, Table S1.
Gravitropism Analyses.
To investigate light-regulated gravitropic responses of hypocotyls, seeds were first irradiated with white light (100 μmol⋅m−2⋅s−1) for 3 h to induce germination and then incubated vertically in the dark or in red light (5 μmol⋅m−2⋅s−1) for 4 d. Growth orientations of hypocotyls were measured by degrees from the vertical axis.
For gravistimulation analyses of main roots, seedlings were grown vertically on MS plates for 3 d in darkness or white light. Then, seedlings of similar lengths were selected and transferred to new MS plates, with their roots vertically downward, and incubated for another 3 h. Next, the plates were rotated by 90° for gravitropic stimulation. Dim green light was used during manipulation of the seedlings in the dark. The curvature of the roots was measured from the digital images using ImageJ software (https://imagej.nih.gov/ij/).
To observe the lateral root angles, plants were grown vertically under constant white light for 12 d. The lateral root angles relative to the direction of gravity vector were measured using ImageJ at the position of 2 mm from the base of the lateral roots.
Staining of Amyloplasts.
Amyloplasts were stained with iodine as described previously (19). Four-day-old dark-grown seedlings were fixed with FAA (5% formaldehyde, 45% ethanol, 5% acetic acid) solution at 4 °C for 24 h. Then, the fixed seedlings were rinsed once in 50% (vol/vol) ethanol and stained in I2-KI solution (2% [wt/vol] iodine, 5% [wt/vol] potassium iodine, and 20% [wt/vol] chloral hydrate) for 1 to 2 min. Finally, the seedlings were de-stained in a 1:1:1 trichloroacetic acid:phenol:lactic acid solution for 5 min. Light microscopy was used to observe the amyloplasts.
Supplementary Material
Acknowledgments
We thank Dr. Peter Quail (University of California, Berkeley) for providing 35S:PIF-MYC transgenic seeds; Dr. Danmeng Zhu and Dr. Jian Li for providing UBQ10p:HA-HY5/hy5 transgenic seeds; Zhaoguo Deng for help with statistical analyses; and Zhaoxu Gao for bioinformatics analyses. This work was supported by grants from the National Key R&D Program of China (2017YFA0503800, 2018YFE0204700), the National Nature Science Foundation of China (31621001), the Peking-Tsinghua Center for Life Sciences, and the State Key Laboratory of Protein and Plant Gene Research.
Footnotes
The authors declare no competing interest.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2005871117/-/DCSupplemental.
Data Availability.
All data are included in the paper and SI Appendix. The materials described in the manuscript are freely available to readers upon request.
References
- 1.Sullivan J. A., Deng X. W., From seed to seed: The role of photoreceptors in Arabidopsis development. Dev. Biol. 260, 289–297 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Knight T. A., On the direction of the radicle and germen during the vegetation of seeds. Philos. Trans. R. Soc. Lond. B Biol. Sci. 96, 99–108 (1806). [Google Scholar]
- 3.Kiss J. Z., Mechanisms of the early phases of plant gravitropism. CRC Crit. Rev. Plant Sci. 19, 551–573 (2000). [DOI] [PubMed] [Google Scholar]
- 4.Morita M. T., Directional gravity sensing in gravitropism. Annu. Rev. Plant Biol. 61, 705–720 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Su S. H., Gibbs N. M., Jancewicz A. L., Masson P. H., Molecular mechanisms of root gravitropism. Curr. Biol. 27, R964–R972 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Tepperman J. M. et al., Expression profiling of phyB mutant demonstrates substantial contribution of other phytochromes to red-light-regulated gene expression during seedling de-etiolation. Plant J. 38, 725–739 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Leivar P., Quail P. H., PIFs: Pivotal components in a cellular signaling hub. Trends Plant Sci. 16, 19–28 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leivar P. et al., Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr. Biol. 18, 1815–1823 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shin J. et al., Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc. Natl. Acad. Sci. U.S.A. 106, 7660–7665 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bauer D. et al., Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell 16, 1433–1445 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiao Y., Lau O. S., Deng X. W., Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet. 8, 217–230 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Al-Sady B., Ni W., Kircher S., Schäfer E., Quail P. H., Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell 23, 439–446 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Al-Sady B., Kikis E. A., Monte E., Quail P. H., Mechanistic duality of transcription factor function in phytochrome signaling. Proc. Natl. Acad. Sci. U.S.A. 105, 2232–2237 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ni W. et al., Multisite light-induced phosphorylation of the transcription factor PIF3 is necessary for both its rapid degradation and concomitant negative feedback modulation of photoreceptor phyB levels in Arabidopsis. Plant Cell 25, 2679–2698 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hornitschek P. et al., Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 71, 699–711 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Oh E., Zhu J. Y., Wang Z. Y., Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 14, 802–809 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y. et al., A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLoS Genet. 9, e1003244 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pfeiffer A., Shi H., Tepperman J. M., Zhang Y., Quail P. H., Combinatorial complexity in a transcriptionally centered signaling hub in Arabidopsis. Mol. Plant 7, 1598–1618 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim K. et al., Phytochromes inhibit hypocotyl negative gravitropism by regulating the development of endodermal amyloplasts through phytochrome-interacting factors. Proc. Natl. Acad. Sci. U.S.A. 108, 1729–1734 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim K. et al., PIF1 regulates plastid development by repressing photosynthetic genes in the endodermis. Mol. Plant 9, 1415–1427 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Jakoby M. et al.; bZIP Research Group , bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7, 106–111 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Oyama T., Shimura Y., Okada K., The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 11, 2983–2995 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ang L. H. et al., Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol. Cell 1, 213–222 (1998). [DOI] [PubMed] [Google Scholar]
- 24.Osterlund M. T., Hardtke C. S., Wei N., Deng X. W., Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405, 462–466 (2000). [DOI] [PubMed] [Google Scholar]
- 25.Ulm R. et al., Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 101, 1397–1402 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee J. et al., Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19, 731–749 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang H. et al., Genome-wide mapping of the HY5-mediated gene networks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J. 65, 346–358 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Sibout R. et al., Opposite root growth phenotypes of hy5 versus hy5 hyh mutants correlate with increased constitutive auxin signaling. PLoS Genet. 2, e202 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song Y. H. et al., DNA-binding study identifies C-box and hybrid C/G-box or C/A-box motifs as high-affinity binding sites for STF1 and LONG HYPOCOTYL5 proteins. Plant Physiol. 146, 1862–1877 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen X. et al., Shoot-to-root mobile transcription factor HY5 coordinates plant carbon and nitrogen acquisition. Curr. Biol. 26, 640–646 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Lee H. J., Ha J. H., Park C. M., Underground roots monitor aboveground environment by sensing stem-piped light. Commun. Integr. Biol. 9, e1261769 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Gelderen K. et al., Far-red light detection in the shoot regulates lateral root development through the HY5 transcription factor. Plant Cell 30, 101–116 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blancaflor E. B., Fasano J. M., Gilroy S., Mapping the functional roles of cap cells in the response of Arabidopsis primary roots to gravity. Plant Physiol. 116, 213–222 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fukaki H. et al., Genetic evidence that the endodermis is essential for shoot gravitropism in Arabidopsis thaliana. Plant J. 14, 425–430 (1998). [DOI] [PubMed] [Google Scholar]
- 35.Morita M. T., Tasaka M., Gravity sensing and signaling. Curr. Opin. Plant Biol. 7, 712–718 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Nakamura M., Nishimura T., Morita M. T., Gravity sensing and signal conversion in plant gravitropism. J. Exp. Bot. 70, 3495–3506 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Li P. et al., LAZY1 controls rice shoot gravitropism through regulating polar auxin transport. Cell Res. 17, 402–410 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Yoshihara T., Iino M., Identification of the gravitropism-related rice gene LAZY1 and elucidation of LAZY1-dependent and -independent gravity signaling pathways. Plant Cell Physiol. 48, 678–688 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Yoshihara T., Spalding E. P., Iino M., AtLAZY1 is a signaling component required for gravitropism of the Arabidopsis thaliana inflorescence. Plant J. 74, 267–279 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Ge L., Chen R., Negative gravitropism in plant roots. Nat. Plants 2, 16155 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Abe K., Takahashi H., Suge H., Localization of cells containing sedimented amyloplasts in the shoots of normal and lazy rice seedlings. Biol. Sci. Space 8, 221–225 (1994). [DOI] [PubMed] [Google Scholar]
- 42.Abe K., Takahashi H., Suge H., Lazy gene (la) responsible for both an agravitropism of seedlings and lazy habit of tiller growth in rice (Oryza sativa L.). J. Plant Res. 109, 381–386 (1996). [DOI] [PubMed] [Google Scholar]
- 43.Taniguchi M. et al., The Arabidopsis LAZY1 family plays a key role in gravity signaling within statocytes and in branch angle control of roots and shoots. Plant Cell 29, 1984–1999 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoshihara T., Spalding E. P., LAZY genes mediate the effects of gravity on auxin gradients and plant architecture. Plant Physiol. 175, 959–969 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guseman J. M., Webb K., Srinivasan C., Dardick C., DRO1 influences root system architecture in Arabidopsis and Prunus species. Plant J. 89, 1093–1105 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Uga Y. et al., Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat. Genet. 45, 1097–1102 (2013). [DOI] [PubMed] [Google Scholar]
- 47.Furutani M. et al., Polar recruitment of RLD by LAZY1-like protein during gravity signaling in root branch angle control. Nat. Commun. 11, 76 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Osterlund M. T., Deng X. W., Multiple photoreceptors mediate the light-induced reduction of GUS-COP1 from Arabidopsis hypocotyl nuclei. Plant J. 16, 201–208 (1998). [DOI] [PubMed] [Google Scholar]
- 49.Mandoli D. F., Tepperman J., Huala E., Briggs W. R., Photobiology of diagravitropic maize roots. Plant Physiol. 75, 359–363 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feldman L. J., Briggs W. R., Light-regulated gravitropism in seedling roots of maize. Plant Physiol. 83, 241–243 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takano M. et al., Isolation and characterization of rice phytochrome A mutants. Plant Cell 13, 521–534 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Correll M. J., Kiss J. Z., The roles of phytochromes in elongation and gravitropism of roots. Plant Cell Physiol. 46, 317–323 (2005). [DOI] [PubMed] [Google Scholar]
- 53.Poppe C., Hangarter R. P., Sharrock R. A., Nagy F., Schäfer E., The light-induced reduction of the gravitropic growth-orientation of seedlings of Arabidopsis thaliana (L.) Heynh. is a photomorphogenic response mediated synergistically by the far-red-absorbing forms of phytochromes A and B. Planta 199, 511–514 (1996). [DOI] [PubMed] [Google Scholar]
- 54.Robson P. R. H., Smith H., Genetic and transgenic evidence that phytochromes A and B act to modulate the gravitropic orientation of Arabidopsis thaliana hypocotyls. Plant Physiol. 110, 211–216 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kidokoro S. et al., The phytochrome-interacting factor PIF7 negatively regulates DREB1 expression under circadian control in Arabidopsis. Plant Physiol. 151, 2046–2057 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Y., Liu Z., Chen Y., He J. X., Bi Y., PHYTOCHROME-INTERACTING FACTOR 5 (PIF5) positively regulates dark-induced senescence and chlorophyll degradation in Arabidopsis. Plant Sci. 237, 57–68 (2015). [DOI] [PubMed] [Google Scholar]
- 57.Zhang X. et al., Integrated regulation of apical hook development by transcriptional coupling of EIN3/EIL1 and PIFs in Arabidopsis. Plant Cell 30, 1971–1988 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Heng Y. et al., BBX4, a phyB-interacting and modulated regulator, directly interacts with PIF3 to fine tune red light-mediated photomorphogenesis. Proc. Natl. Acad. Sci. U.S.A. 116, 26049–26056 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shi H. et al., HFR1 sequesters PIF1 to govern the transcriptional network underlying light-initiated seed germination in Arabidopsis. Plant Cell 25, 3770–3784 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Allen T., Ingles P. J., Praekelt U., Smith H., Whitelam G. C., Phytochrome-mediated agravitropism in Arabidopsis hypocotyls requires GIL1 and confers a fitness advantage. Plant J. 46, 641–648 (2006). [DOI] [PubMed] [Google Scholar]
- 61.Schepens I., Boccalandro H. E., Kami C., Casal J. J., Fankhauser C., PHYTOCHROME KINASE SUBSTRATE4 modulates phytochrome-mediated control of hypocotyl growth orientation. Plant Physiol. 147, 661–671 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sun N. et al., Arabidopsis SAURs are critical for differential light regulation of the development of various organs. Proc. Natl. Acad. Sci. U.S.A. 113, 6071–6076 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dong J. et al., The transcription factors TCP4 and PIF3 antagonistically regulate organ-specific light induction of SAUR genes to modulate cotyledon opening during de-etiolation in Arabidopsis. Plant Cell 31, 1155–1170 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bai S. et al., PIF3 is involved in the primary root growth inhibition of Arabidopsis induced by nitric oxide in the light. Mol. Plant 7, 616–625 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dong J. et al., Arabidopsis DE-ETIOLATED1 represses photomorphogenesis by positively regulating phytochrome-interacting factors in the dark. Plant Cell 26, 3630–3645 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen H. et al., Integration of light and abscisic acid signaling during seed germination and early seedling development. Proc. Natl. Acad. Sci. U.S.A. 105, 4495–4500 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lin R. et al., Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science 318, 1302–1305 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li J. et al., Arabidopsis transcription factor ELONGATED HYPOCOTYL5 plays a role in the feedback regulation of phytochrome A signaling. Plant Cell 22, 3634–3649 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are included in the paper and SI Appendix. The materials described in the manuscript are freely available to readers upon request.