Significance
Vps13, a conserved phospholipid transporter, localizes to contact sites between the endoplasmic reticulum (ER) and different organelles. While budding yeast has one VPS13 gene, humans have four. The yeast Vps13 protein is most closely related to VPS13A and VPS13C. VPS13A localizes to ER–mitochondria contact sites, while VPS13C resides at ER–late endosome contact sites. Here we report that the vps13Δ mutant is defective in the selective autophagy of ER (ER-phagy). In the absence of Vps13, the ER accumulates at late endosomes and fails to be packaged into autophagosomes. These findings highlight the importance of Vps13 function at late endosomes in ER-phagy, and may have implications for understanding the role of VPS13C, which has been linked to early-onset Parkinson’s disease.
Keywords: ER-phagy, lipid transporter, contact site, autophagy, Vps13
Abstract
Endoplasmic reticulum (ER) macroautophagy (hereafter called ER-phagy) uses autophagy receptors to selectively degrade ER domains in response to starvation or the accumulation of aggregation-prone proteins. Autophagy receptors package the ER into autophagosomes by binding to the ubiquitin-like yeast protein Atg8 (LC3 in mammals), which is needed for autophagosome formation. In budding yeast, cortical and cytoplasmic ER-phagy requires the autophagy receptor Atg40. While different ER autophagy receptors have been identified, little is known about other components of the ER-phagy machinery. In an effort to identify these components, we screened the genome-wide library of viable yeast deletion mutants for defects in the degradation of cortical ER following treatment with rapamycin, a drug that mimics starvation. Among the mutants we identified was vps13Δ. While yeast has one gene that encodes the phospholipid transporter VPS13, humans have four vacuolar protein-sorting (VPS) protein 13 isoforms. Mutations in all four human isoforms have been linked to different neurological disorders, including Parkinson’s disease. Our findings have shown that Vps13 acts after Atg40 engages the autophagy machinery. Vps13 resides at contact sites between the ER and several organelles, including late endosomes. In the absence of Vps13, the cortical ER marker Rtn1 accumulated at late endosomes, and a dramatic decrease in ER packaging into autophagosomes was observed. Together, these studies suggest a role for Vps13 in the sequestration of the ER into autophagosomes at late endosomes. These observations may have important implications for understanding Parkinson’s and other neurological diseases.
The endoplasmic reticulum (ER) forms a continuous network of interconnected sheets and tubules that undergoes dynamic rearrangements (1). The ER mediates fundamental cellular processes that include calcium storage, protein and lipid biosynthesis, as well as interorganelle communication (1). To maintain cellular homeostasis, the ER has evolved quality control mechanisms. In response to unfolded or misfolded proteins, the unfolded protein response (UPR) is up-regulated. This leads to an increase in chaperone levels and the degradation of misfolded proteins via ER-associated protein degradation (ERAD). ERAD retrotranslocates terminally unfolded or misfolded proteins across the ER membrane into the cytosol, where they are degraded by the ubiquitin-proteasome system (2). Certain proteins, such as aggregation-prone proteins, do not induce the UPR, while other proteins are resistant to ERAD. These proteins must be disposed of by alternate degradation pathways that are regulated independent of the UPR (3, 4).
Macro-ER-phagy (herein called ER-phagy) is an alternate ER degradation pathway that sequesters ER domains into autophagosomes and delivers them to lysosomes (mammals) or vacuoles (yeast), where they are degraded (5). Unlike bulk autophagy, which nonselectively scavenges cytosolic components for nutrients, ER-phagy is mediated by receptors that selectively package ER fragments into autophagosomes (5). These receptors contain at least one motif that binds to LC3 (mammals) or Atg8 (yeast) to connect the ER to core autophagy (Atg) machinery (5, 6). Currently, seven receptors (FAM134B, RTN3, SEC62, CCPG1, ATL3, TEX264, and CALCOCO1) have been identified in mammals (5, 7), and two (Atg39 and Atg40) in budding yeast (8). Atg39 is required for nuclear ER degradation, while Atg40 is predominately needed for the degradation of cortical and cytoplasmic ER. The cortical ER in yeast consists of sheets and tubules (1). Consistent with the observation that a small fraction of Atg40 also resides on the perinuclear ER, Atg40 mediates nucleophagy as well (8). Atg40 has a similar domain structure to the mammalian ER-phagy sheet receptor FAM134B (8, 9), yet it localizes to the tubular ER like the ER-phagy receptor RTN3 (10). Atg40, FAM134B, and RTN3 contain tandem reticulon homology domains (RHDs) or putative RHDs that anchor these receptors to the cytosolic face of the ER (11).
While autophagy receptors localize throughout the contiguous ER network, ER-phagy occurs at discrete sites. We recently showed that a cytosolic coat complex, Lst1–Sec23, works with Atg40 to target domains of the ER for vacuolar degradation (3). The role of Lst1 appears to be conserved as its mammalian homolog, SEC24C, is also required for ER-phagy (3). Once a domain on the ER is marked for degradation, it must interact with the Atg machinery. In budding yeast, ER-phagy occurs at perivacuolar sites, where the Atg machinery assembles upon the induction of ER autophagy (8). We previously showed that the association of Atg40 with core Atg proteins requires actin polymerization and Lnp1 (12). Lnp1 is a membrane protein that resides at the three-way junctions of the ER network, where it stabilizes the network as it is being remodeled (13). In the absence of Lnp1, Atg40 lines the ER at the cell periphery and fails to colocalize with Lst1 and Atg11, the scaffold for the assembly of the Atg proteins needed for selective autophagy (12).
While many autophagy receptors have been identified, little is known about other components required for ER-phagy. In an effort to identify new components and gain insight into the mechanism of ER-phagy, we screened the yeast genomic library of viable deletion mutants for defects in the vacuolar delivery of a fluorescently tagged ER reporter, Sec61. In addition to identifying known components required for cortical ER-phagy, such as Atg40, Lst1, and Lnp1 (3, 8, 12), we identified vacuolar protein-sorting (Vps) protein 13 as a key player in ER-phagy. Vps13 is a protein that transports lipids between the ER and adjacent organelles. These organelles include endosomes, vacuoles, the nucleus, and mitochondria (14–16). Mutations in the two closest mammalian Vps13 homologs, VPS13A and VPS13C, have been linked to neurodegenerative disorders that include chorea acanthocytosis and Parkinson’s disease (16). Here we identify the stage of the ER-phagy pathway at which Vps13 functions and discuss its possible role in this pathway.
Results
Identification of Mutants Defective in Rapamycin-Induced ER-Phagy.
To identify new genes whose products act in ER-phagy, we used fluorescence microscopy to screen a genome-wide yeast library of viable deletion mutants and scored the strains displaying a defect in the delivery of the ER to the vacuole. We chose to monitor two different ER markers that have distinct localizations, Sec61 and Rtn1. Sec61 localizes to the nuclear ER, cortical ER, and cytoplasmic elements connecting the cortical and nuclear ER, while Rtn1 primarily resides on the cortical ER (17). To perform this screen, we induced ER-phagy as previously described (12), with rapamycin, a drug that mimics starvation by inhibiting TORC1 activity (6). Mutant cells were first assessed for their ability to transport Sec61-mCherry (expressed from a CEN plasmid) to the vacuole 16 h after rapamycin treatment. To eliminate false positives that may have resulted from poor transformation of the plasmid, Sec61-GFP (green fluorescent protein) was integrated into the genome of the 200 identified candidates (SI Appendix, Table S1) and these mutants were retested. The 87 mutants deemed as defective in the delivery of Sec61-GFP to the vacuole after retesting were then counterscreened with Rtn1-GFP to exclude strains exclusively affecting Sec61-GFP. To eliminate mutants defective in bulk autophagy, the 60 mutants found to be defective in the delivery of both Sec61 and Rtn1 were screened for defects in the vacuolar delivery of GFP-Atg8, which is mainly transported to the vacuole by bulk autophagosomes during starvation (6). This analysis revealed that 39 of the 60 mutants identified in the screen were also defective in bulk autophagy and consequently excluded (SI Appendix, Table S1). Among the eliminated mutants were strains lacking a known ATG gene, validating the robustness of our approach.
While many of the Atg proteins needed for bulk autophagy were found to be needed for ER-phagy (SI Appendix, Table S1), several key ATG genes were not identified (SI Appendix, Table S2). When we reassayed mutants lacking these Atg proteins for defects in the cleavage of Sec61-GFP to GFP, a defect was observed in all cases (SI Appendix, Fig. S1B). Therefore, in addition to the ATG genes listed in SI Appendix, Table S2, ATG2, ATG16, ATG29, and ATG31 are required for ER-phagy (SI Appendix, Table S3). Other proteins needed for bulk autophagy such as Rab GTPases (Ypt5 and Ypt7) and Rab activators (Ccz1 and Trs85) were also identified in the screen (SI Appendix, Table S2) (18). Surprisingly, however, while Atg21 and Atg24 are needed for nitrogen-induced ER-phagy in fission yeast (19), we did not identify a requirement for these proteins in the screen (SI Appendix, Table S2). This result was confirmed using a cleavage assay (SI Appendix, Fig. S1A).
Most importantly, our screen also identified proteins whose loss disrupted ER degradation but not GFP-Atg8 traffic to the vacuole (SI Appendix, Table S2). Consistent with previous studies, these proteins included Atg40, Lnp1, and Lst1 (3, 8, 12). Other examples included proteins required for the organization of the cytoskeleton (Bem2, End2, and Gic2), ubiquitin system (Doa4), and two vacuolar protein-sorting proteins (Vps13 and Vps38) (https://www.yeastgenome.org/). Using fluorescence microscopy and biochemical cleavage assays, a requirement in ER degradation was confirmed for both Vps13 and Vps38 in our strain background (Figs. 1 and 2 A and B and SI Appendix, Fig. S2). We have focused on Vps13 for the remainder of this study because its human homologs have been associated with devastating neurobiological diseases. The analysis of the remaining mutants will be discussed elsewhere.
Fig. 1.
Loss of Vps13 leads to a defect in the degradation of Sec61-GFP. (A) The translocation of Sec61-GFP to the vacuole is disrupted in the vps13Δ mutant. (A, Left) Fluorescence images of yeast cells expressing Sec61-GFP. Cells were treated with rapamycin for 16 h. Vacuoles were stained with FM4-64. (A, Right) The percentage of cells with GFP in the vacuole was quantified. Error bars represent SEM; n = 3 independent experiments. **P < 0.01, Student’s t test. (B) The degradation of Sec61-GFP to the vacuole is disrupted in the vps13Δ mutant. (B, Left) Cells expressing Sec61-GFP were treated with rapamycin for 0 or 16 h. Cell lysates were prepared as described in Materials and Methods and immunoblotted using an anti-GFP antibody. Adh1 was used as a loading control. (B, Right) The cleavage of Sec61-GFP to GFP was quantified by calculating the percentage of free GFP divided by total GFP (GFP + Sec61-GFP). The data were normalized to the WT control. Error bars represent SEM; n = 3 independent experiments. **P < 0.01, Student’s t test. (Scale bar, 2 μm.)
Fig. 2.
Vps13 is required for cortical ER-phagy but not nuclear ER-phagy. (A) The translocation of Rtn1-GFP to the vacuole is disrupted in the vps13Δ mutant. (A, Left) Fluorescence images of yeast cells expressing Rtn1-GFP. Cells were treated with rapamycin for 16 h. Vacuoles were stained with FM4-64. (A, Right) The percentage of cells with GFP in the vacuole was quantified. Error bars represent SEM; N = 3 independent experiments. **P < 0.01, ***P < 0.001, Student’s t test. (B) The degradation of Rtn1-GFP in the vacuole is disrupted in the vps13Δ mutant. (B, Left) Cells expressing Rtn1-GFP were treated with rapamycin for 0 or 16 h. Cell lysates were immunoblotted using an anti-GFP antibody. Adh1 was used as a loading control. (B, Right) The percentage of free GFP divided by total GFP was quantified. The data were normalized to the WT control. Error bars represent SEM; N = 3 independent experiments. **P < 0.01, ****P < 0.0001, Student’s t test. (C, Left) Western blot analysis of the cleavage of Hmg1-GFP to GFP after rapamycin treatment for 0 or 16 h. Adh1 was used as a loading control. (C, Right) The percentage of free GFP divided by total GFP was quantified. The data were normalized to the WT control. Error bars represent SEM; N = 3 independent experiments. NS, nonsignificant (P ≥ 0.05), ***P < 0.001, Student’s t test. (Scale bar, 2 μm.)
Vps13 Specifically Acts in Cortical ER-Phagy.
Vps13 is related to Atg2, a phospholipid transporter that establishes membrane contact sites between the ER and growing autophagosome to transport lipids between these two organelles (20–24). Vps13 resides at ER–nuclear, ER–vacuole, and ER–mitochondria contact sites (15). While an ER degradation defect was found in the vps13Δ mutant, deletion mutants lacking other ER contact-site proteins, such as Vac8, Nvj1, Nvj2, Nvj3, Osh1, Mdm1, Lam5, and Lam6 (25), were not identified in the screen (SI Appendix, Table S2). This observation was validated when we examined Sec61-GFP cleavage in cells lacking these ER–vacuole contact-site proteins (SI Appendix, Fig. S3). Together, these findings suggest that Vps13 may perform a unique function in ER-phagy at a specific ER contact site.
Interestingly, although Vps13 was found to be needed for cortical ER-phagy (Fig. 2 A and B), it was, like Lst1 (3), dispensable for the degradation of the nuclear marker Hmg1 (Fig. 2C and SI Appendix, Fig. S4A). Therefore, unlike Atg40, which functions in both ER-phagy pathways (8), Lst1 and Vps13 appear to specifically function in cortical ER-phagy. We also did not observe a cleavage defect for the piecemeal autophagy nuclear ER protein Nvj1-GFP (26) in the vps13Δ mutant, while the vps38Δ mutant displayed a defect (SI Appendix, Fig. S4B). Piecemeal autophagy is a non–autophagosome-mediated microautophagy pathway (26).
Because a partial defect in bulk autophagy was previously reported in mammalian cells lacking VPS13A (27), we sought to use more sensitive assays to evaluate whether yeast Vps13 is required for bulk autophagy. Pho8∆60, a vacuolar alkaline phosphatase lacking its N-terminal 60 amino acids, provides a quantitative method for measuring autophagy. During starvation, Pho8∆60 is delivered from the cytosol to the vacuole and activated (28). We observed a 24% reduction in Pho8∆60 activity in the vps13∆ mutant when compared with its isogenic wild-type (WT) strain (SI Appendix, Fig. S5A). This small defect was confirmed when Western blot analysis was used to monitor GFP-Atg8 flux to the vacuole in rapamycin-treated cells (SI Appendix, Fig. S5 B and C) (29). Based on these findings, we concluded that the loss of Vps13 has a minor effect on bulk autophagy; however, this defect was too small to explain the magnitude of the ER-phagy defect in vps13∆ cells. Interestingly, when we examined the cleavage of the mitochondrial outer-membrane protein OM45-GFP in the vps13∆ mutant (SI Appendix, Fig. S6), we observed an increase and not a decrease in mitophagy. This observation is consistent with a previous study that used a different reporter to measure mitophagy in the vps13Δ mutant (15). In contrast to yeast Vps13, the loss of VPS13A was recently reported to lead to a defect in mitophagy (30).
Vps13 is recruited to different intracellular membrane-bound compartments by organelle-specific adaptors (14, 15, 31). These adaptors include Ypt35 (endosome and vacuole), Mcp1 (mitochondria), and Spo71 (prospore membrane). Vps13 might work in conjunction with one of these adaptors to mediate ER-phagy. To address this possibility, we assayed for the cleavage of Rtn1-GFP to GFP in mutants lacking Ypt35, Mcp1, or Spo71 (SI Appendix, Fig. S7). Interestingly, none of the mutants in the known adaptors displayed a defect (SI Appendix, Fig. S7). This observation is consistent with the fact that these adaptors were not identified in our screen (SI Appendix, Table S2). It also raises the possibility that Vps13 may work with a currently unidentified adaptor to mediate ER-phagy.
Vps13 Acts after Atg40.
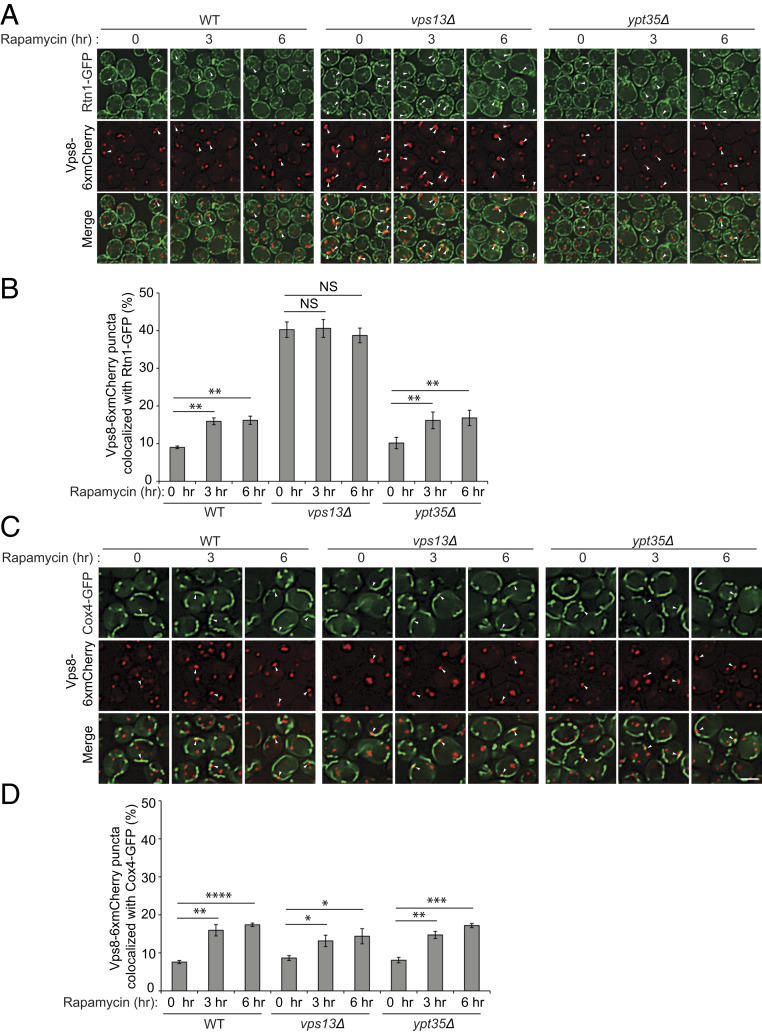
To begin to understand the role of Vps13 in ER-phagy, we localized Vps13 in rapamycin-treated cells. Earlier studies revealed that the majority of Vps13 puncta reside at the late endosome under normal growth conditions (15, 31). Using a previously described yeast strain expressing Vps13-GFP1360 (hereafter referred to as Vps13-GFP) (15), we observed that perivacuolar Vps13-GFP puncta decreased in the presence of rapamycin and instead lined the vacuolar membrane (Fig. 3A). In the absence of rapamycin, Vps13-GFP puncta largely colocalized (57%) with Vps8-6xmCherry, a marker protein for the late endosome (Fig. 3 B–D). Upon the addition of rapamycin, the perivacuolar Vps13-GFP puncta that colocalized with Vps8-6xmCherry dramatically decreased (Fig. 3C). While the punctate pattern of Vps13-GFP localization changed in the presence of rapamycin, Vps8 puncta did not (Fig. 3 E and F). This observation indicates that the rapamycin-induced change in Vps13 distribution reflects a relocation of Vps13 from late endosomes rather than a change in late-endosome structure.
Fig. 3.
Colocalization of Vps8 with Vps13 decreases in response to rapamycin treatment. (A) Fluorescence images of cells expressing Vps13-GFP 0 and 3 h after rapamycin treatment. The vacuolar membrane was stained with FM4-64. (B) Fluorescence images of cells expressing Vps13-GFP and Vps8-6xmCherry at the indicated time points in response to rapamycin treatment. The arrowheads point to Vps13 puncta colocalized with Vps8 puncta. (C) Quantification of Vps8-6xmCherry puncta that colocalize with Vps13-GFP puncta for cells in B. Error bars represent SEM; N = 3 independent experiments. **P < 0.01, Student’s t test. (D) Quantification of Vps13-GFP puncta that colocalize with Vps8-6xmCherry in cells shown in B. Error bars represent SEM; n = 3 independent experiments. NS, nonsignificant (P ≥ 0.05), Student’s t test. (E) Fluorescence images of yeast cells expressing Vps8-6xmCherry at the indicated time points in response to rapamycin treatment. (F) Quantification of Vps8-6xmCherry puncta that localize to the vacuolar membrane for cells in E. Error bars represent SEM; N = 3 independent experiments. NS, nonsignificant (P ≥ 0.05), Student’s t test. (Scale bars, 2 μm.) DIC, differential interference contrast.
Next, we addressed whether Vps13 is degraded during ER-phagy. We determined if Vps13 traffics to the lumen of the vacuole in WT, atg14Δ, and atg40Δ cells upon rapamycin treatment. The ypt35Δ mutant was also examined as a control. As mentioned above, cortical ER-phagy is disrupted in the atg40Δ mutant, while autophagosomes fail to form in atg14Δ cells (6, 8). As expected, Vps13-GFP was observed in the vacuole of WT and ypt35Δ cells (SI Appendix, Fig. S8 A and B). Unexpectedly, however, we found that the delivery of Vps13-GFP to the vacuole was disrupted in the atg14Δ, but not atg40Δ, mutant (SI Appendix, Fig. S8 A and B). Similar results were obtained when we monitored the cleavage of Vps13-GFP to GFP by Western blot analysis (SI Appendix, Fig. S8 C–E). Thus, these data imply that during rapamycin treatment, Vps13 is delivered to the vacuole via autophagosomes independent of specific components of the ER-phagy machinery. Therefore, the vacuolar delivery and degradation of Vps13 during rapamycin treatment may reflect the degradation of a cytosolic pool of Vps13 that occurs via bulk autophagy.
Next, we employed double-mutant analysis to ask if Vps13 and Atg40 act on the same ER-phagy pathway. We found that the Sec61-GFP degradation defect in the atg40Δ and vps13Δ single knockouts was comparable to that observed in the atg40Δvps13Δ double mutant (Fig. 4), implying that Vps13 and Atg40 act on the same cortical ER-phagy pathway and not on parallel or converging pathways.
Fig. 4.
Vps13 and Atg40 function in the same ER-phagy pathway. (A) Atg40 and Vps13 act on the same pathway. Fluorescence images of yeast cells expressing Sec61-GFP after rapamycin treatment. The vacuolar membrane was stained with FM4-64. (B) The percentage of cells with GFP in the vacuole was quantified. Error bars represent SEM; n = 3 independent experiments. NS, nonsignificant (P ≥ 0.05), Student’s t test. (C) Western blot analysis of Sec61-GFP cleavage to GFP before and after rapamycin treatment. Adh1 was used as a loading control. (D) The percentage of free GFP divided by total GFP was quantified. The data were normalized to the WT control. Error bars represent SEM; n = 3 independent experiments. NS, nonsignificant (P ≥ 0.05), Student’s t test. (Scale bar, 2 μm.)
To ask if Vps13 plays a role in the assembly of the Atg machinery during ER-phagy, we assessed the colocalization of Atg8 with Atg40 and Lst1, as well as the colocalization of Atg40 with Atg11, in the vps13Δ mutant. Atg8 is the binding partner for Atg40, and Atg11 is the scaffold protein for the assembly of the Atg machinery during selective autophagy (6, 8). We observed that the distribution of Atg40 puncta was unaltered in the vps13Δ mutant (SI Appendix, Fig. S9A; compare Atg40 puncta in the vps13Δ and lnp1Δ mutants). This is unlike what was observed in the lnp1Δ mutant, where Atg40 puncta line the cell periphery and fail to access the perivacuolar autophagy machinery (13). Consistent with the finding that Atg40 puncta are unaltered in the vps13Δ mutant, the loss of Vps13 did not disrupt the colocalization of Atg40 with Atg8 (SI Appendix, Fig. S9B) or Atg11 (SI Appendix, Fig. S9C). Furthermore, the colocalization of Atg8 with Lst1 appeared to be similar in WT and the vps13Δ mutant (SI Appendix, Fig. S9D). We conclude that Atg40 remains accessible to the Atg machinery in the absence of Vps13.
Vps13 Is Required for the Sequestration of the ER into Autophagosomes.
As yeast Vps13 largely localizes to late endosomes, we asked whether the cortical ER protein Rtn1 accumulates at late endosomes in response to rapamycin. Under normal growth conditions, we observed a small pool of Vps8-6xmCherry puncta (∼10%) colocalizing with Rtn1-GFP. This pool increased following rapamycin treatment in WT and ypt35Δ cells (Fig. 5 A and B). In the vps13Δ mutant, however, late endosomes were somewhat enlarged and the colocalization of Vps8 with Rtn1 was dramatically elevated relative to WT. This occurred even in the absence of rapamycin (Fig. 5 A and B). Interestingly, although rapamycin also increased the colocalization of Vps8 puncta with the mitochondrial marker Cox4 (https://www.yeastgenome.org/), the loss of Vps13 did not alter the colocalization of these two proteins (Fig. 5 C and D).
Fig. 5.
ER, but not mitochondria, accumulates at the late endosome in the vps13∆ mutant. (A) Rtn1 accumulates at the late endosome in the vps13∆ mutant. Fluorescence images of cells expressing Rtn1-GFP and Vps8-6xmCherry at the indicated time points in response to rapamycin treatment. The arrowheads mark Vps8 puncta that colocalize with Rtn1. (B) The percentage of Vps8 puncta colocalizing with Rtn1 was quantified. Error bars represent SEM; N = 3 independent experiments. NS, nonsignificant (P ≥ 0.05), **P < 0.01, Student’s t test. (C) Mitochondria do not accumulate at the late endosome in the vps13∆ mutant. Fluorescence images of cells expressing Cox4-GFP and Vps8-6xmCherry at the indicated time points in response to rapamycin treatment. The arrowheads mark Vps8 puncta that colocalize with Cox4. (D) The percentage of Vps8 puncta colocalizing with Cox4 was quantified. Error bars represent SEM; N = 3 independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, Student’s t test. (Scale bars, 2 μm.)
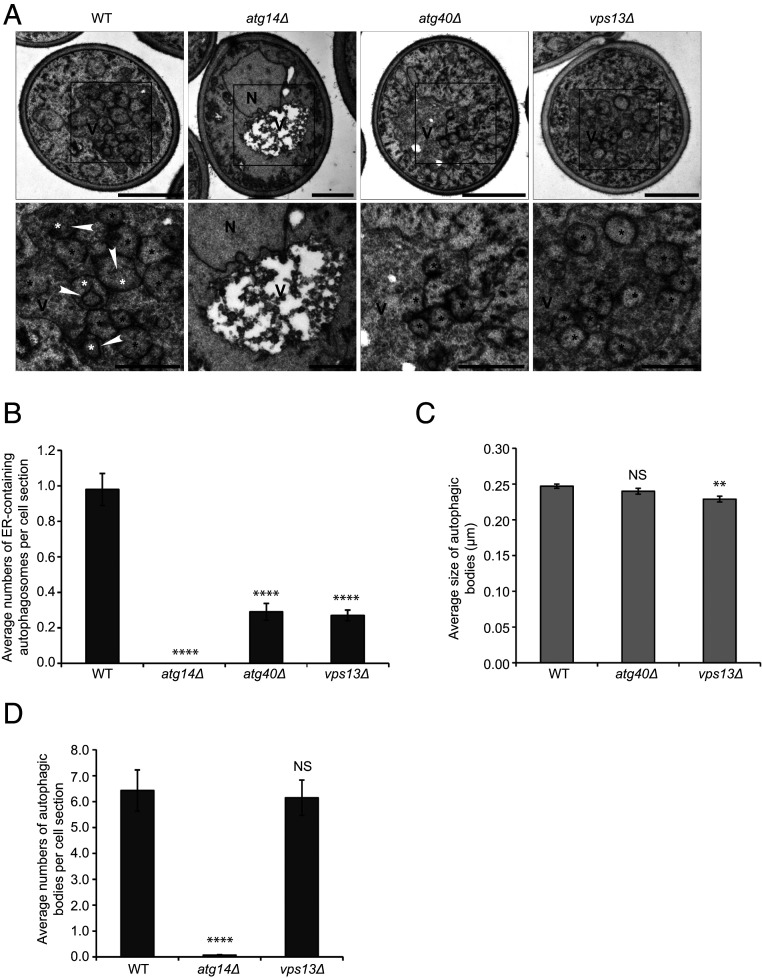
A possible explanation for our data is that Vps13 regulates the packaging of the ER into autophagosomes at late endosomes. To address this possibility, we performed transmission electron microscopy (EM). Transmission EM analysis was performed with cells depleted of the vacuolar protease Pep4, which allowed for the accumulation of autophagic bodies in the vacuolar lumen. We found that like atg40Δ, the vps13Δ mutant exhibited a 70% reduction in the packaging of the ER into autophagosomes when compared with the WT (Fig. 6 A and B). Consistent with the finding that the vps13Δ mutant displayed only a minor delay in autophagic flux (SI Appendix, Fig. S5), we found a slight reduction in autophagosome size (Fig. 6C) and no reduction in autophagosome number (Fig. 6D). In contrast, no autophagosomes were found in the atg14Δ mutant, which is known to block autophagosome formation (Fig. 6D) (6). As the loss of Vps13 did not alter autophagosome number, the reduced number of ER-containing autophagosomes in the vps13Δ mutant is the consequence of a defect in ER packaging.
Fig. 6.
Packaging of the ER into autophagosomes is impaired in the vps13∆ mutant. (A, Top) Representative electron micrographs of yeast cells treated for 12 h with rapamycin. (A, Bottom) The boxed areas (Top) are enlarged. White asterisks mark an autophagic body containing an ER fragment; black asterisks mark an autophagic body lacking an ER fragment. Arrowheads point to an ER fragment inside an autophagic body. N, nucleus; V, vacuole. The darker tone of the vacuole lumen in WT, atg40Δ, and vps13Δ cells, relative to atg14Δ, is a consequence of continued autophagic flux in these strains. (B) Average number of ER-containing autophagosomes per cell section. (C) Average size of autophagic bodies. (D) Average number of autophagosomes per cell section. Error bars represent SEM; N = 100 cells. NS, nonsignificant (P ≥ 0.05), **P < 0.01, ****P < 0.0001, Student’s unpaired t test. (Scale bars, 1 μm [A, Top] and 500 nm [A, Bottom].)
Discussion
While many ER-phagy receptors have been identified, little is known about the machinery that works with these receptors or acts downstream of these receptors. We previously reported that a COPII coat subcomplex containing Lst1 and Sec23 works with Atg40 and Atg8 to target domains of the ER for degradation (3). This noncanonical form of the coat, that mediates ER-phagy, does not appear to contain other COPII coat subunits. We have named these Lst1- and Atg40-containing domains of ER-phagy “ERPHSs” (ER-phagy sites). The role of Lst1 in ER-phagy is conserved from yeast to man as its homolog, SEC24C, is also required for ER-phagy in U2OS cells (3). We have also shown that Lnp1, a protein that stabilizes nascent three-way junctions of the ER (13), is required for the formation of ERPHSs (3).
In search of additional components of the ER-phagy machinery, we screened a yeast deletion library for mutants displaying a defect in the vacuolar degradation of the ER during rapamycin treatment. Mutants found to be defective in the degradation of two ER markers, Sec61 and the cortical ER marker Rtn1, were then counterscreened for the impairment of bulk autophagy. This analysis resulted in the identification of 21 mutants that were specifically defective in ER degradation but not bulk autophagy. In addition to known players such as Atg40, Lnp1, and Lst1, we identified genes encoding components that impact the cytoskeleton, the ubiquitin system, and two Vps proteins.
In this report, we have focused on Vps13, a conserved lipid transporter that is related to Atg2 (16, 20). Vps13 is required for Atg40-mediated cortical ER-phagy. Unlike Atg2, Vps13 is not needed for autophagosome formation, nor does it appear to be needed for nucleophagy or mitophagy. While yeast has one Vps13 protein, humans have four: VPS13A, VPS13B, VPS13C, and VPS13D (16, 32, 33). Yeast Vps13 is most closely related to VPS13A and VPS13C (15). Loss-of-function mutations in VPS13A, which localizes to ER–mitochondria contact sites, have been linked to the genetic disorder chorea acanthocytosis (16, 34, 35). In contrast, loss-of-function mutations in VPS13C, which localizes to ER–late endosome contact sites, have been associated with early-onset Parkinson’s disease (16, 36). Interestingly, the loss of SEC24C, FAM134B, LNP1, and ATL3 (another ER-phagy receptor) has also been linked to neurodegenerative disorders, specifically hereditary spastic paraplegias (9, 37, 38).
Here we present evidence that Vps13 may mediate ER-phagy at late endosomes. This proposal is primarily based on two observations. In the absence of Vps13, the cortical ER marker Rtn1 accumulates at or near the late endosome, and the packaging of the ER into autophagosomes is dramatically reduced in vps13Δ mutant cells during ER-phagy. Our findings also indicate that Vps13 does not play a major role in autophagosome biogenesis. The loss of Vps13 did not decrease autophagosome number and only resulted in a very minor decrease in autophagosome expansion. The observed decrease in autophagosome expansion was too small to account for the defect in ER-phagy.
ER packaging into autophagosomes also requires that Atg40 and Lst1 work in conjunction with the autophagy machinery (3, 8). We have found that Atg40 and Lst1 still colocalize with Atg8 in vps13Δ cells. Atg40 also continues to colocalize with the scaffold protein for selective types of autophagy, Atg11, in the vps13Δ mutant. Together, these findings imply that Vps13 is needed after Atg40 interacts with Lst1 and the autophagy machinery but before the ER is packaged into autophagosomes.
Recent cryoelectron microscopy analysis indicates that Vps13 forms an elongated structure with a hydrophobic groove along its length (39). This groove is thought to channel phospholipids across the cytoplasmic space between the membranes of the two organelles that are linked by Vps13 (16). Based on the prominent localization of Vps13 to the late endosome and the accumulation of the ER at the late endosome in vps13 mutant cells, we propose that within the context of ER-phagy, Vps13 acts to exchange phospholipid between the ER and late endosome. This function may be needed to allow the packaging of ER fragments into autophagosomes. Since the ER is the major site of lipid synthesis, it seems likely that the direction of Vps13-mediated lipid flux is from the ER to the late endosome. The Vps13 requirement in ER-phagy could reflect a need to deplete phospholipid from ER fragments to facilitate their autophagosomal engulfment. Alternatively, Vps13 may be needed to incorporate additional lipid or a specific lipid species into endosomal membranes during ER-phagy. Additional experiments will be required to address these possibilities. As these results have heightened the importance of Vps13 at late endosomes in ER-phagy, they may have implications for understanding the role of VPS13C in early-onset Parkinson’s disease.
Materials and Methods
Strains and Growth Conditions.
Yeast strains used in this study are listed in SI Appendix, Table S4. Unless otherwise stated, the growth of yeast in liquid culture was monitored by measuring the optical density at 600 nm (OD600). All yeast deletion strains were constructed using the Longtine toolbox (40). Integration or CEN vectors were used to generate strains that express GFP- or mCherry-tagged proteins. The strain expressing Vps13-GFP, described in Park et al. (15), was a gift from A.M.N.
ER-phagy was induced as described previously (12). Briefly, yeast cells were grown to exponential phase (OD600 0.2) in synthetic complete or dropout medium containing 2% glucose (SD). Rapamycin was added to a final concentration of 200 ng/mL and the cells were incubated for 12 to 24 h at 30 °C.
To induce mitophagy, yeast cells expressing OM45-GFP were grown in rich medium (YPD; 1% yeast extract, 2% peptone, 2% glucose) to exponential phase (OD600 0.5). Subsequently, the cells were shifted to YPL (1% yeast extract, 2% peptone, 2% lactate) medium for 16 to 24 h, until they reached a final OD600 of 1.5. Cells were then pelleted and resuspended in SD medium without ammonium sulfate and amino acids (SD-N) and cultured for 6 h.
Screen for Mutants Defective in ER-Phagy.
Fluorescence microscopy was used to screen the yeast deletion library for mutants defective in ER-phagy. To perform this screen, cells were transformed in a 96-well plate with Sec61-mCherry (CEN LEU2) and grown overnight in SD-Leu. The transformed cells were then transferred into special optics 96-well plates and grown overnight at 30 °C. The next day, the transformants were diluted 1:250 and incubated with 400 ng/mL rapamycin for 16 h. Mutants were considered to be defective if at least half of the analyzed cells failed to deliver Sec61-mCherry to the vacuole. Sec61-GFP was then integrated into the genome of the defective mutants and they were reexamined. Mutants deemed defective were also screened for defects in the delivery of Rtn1-GFP to the vacuole. Mutants displaying defects with both markers were considered to be defective in ER degradation. These mutants were assessed for defects in bulk autophagy by measuring the translocation of GFP-Atg8 to the vacuole in 50 to 100 cells that were treated for 1 h in SD-N medium at 30 °C. Mutants found to be defective in the delivery of Sec61-GFP and Rtn1-GFP to the vacuole are listed in SI Appendix, Table S2.
FM4-64 Staining.
To stain the vacuolar membrane, 0.5 OD600 units of cells were centrifuged at 3,000 × g for 5 min. The cell pellet was then resuspended in 100 μL of YPD medium containing FM4-64 (1.6 μM final concentration) and incubated at 30 °C for 30 to 60 min. The cells were centrifuged again and the pellet was washed twice with YPD medium to remove excess FM4-64. Vacuoles were viewed using an Axio Imager Z1 fluorescence microscope as described below.
Fluorescence Microscopy.
Approximately 0.2 to 0.5 OD600 units of cells were pelleted and imaged on an Axio Imager Z1 fluorescence microscope. This microscope is equipped with an A-Plan 10× 0.25 Ph1 objective lens (Zeiss), Compact Light Source HXP 120V, and Axiocam 506 mono digital camera. Acquisition was performed with ZEN software and deconvoluted using Open Lab (Improvision) using the manufacturer’s parameters. Subsequent analysis was performed using Fiji ImageJ or Photoshop CS4 (Adobe).
GFP Cleavage Assays.
Approximately 5.0 OD600 units of cells expressing GFP-tagged proteins (Sec61, Rtn1, Hmg1, Nvj1, Atg8, OM45, and Vps13) were harvested by centrifugation (3,000 × g, 5 min). The cells were washed twice with distilled H2O and treated for at least 30 min with 1.0 mL of ice-cold 10% trichloroacetic acid (TCA), and then pelleted at 13,000 × g for 5 min. The pellet was washed twice with 1.0 mL of ice-cold acetone to remove residual TCA, dried at room temperature, and resuspended in 50 μL of buffer (50 mM Tris⋅HCl, 1 mM ethylenediaminetetraacetate, 1% sodium dodecyl sulfate [SDS], 6 M urea, pH 7.5) that was heated to 100 °C. Silica beads (equal to the volume of the sample) were added and the samples were vortexed for 5 min and then incubated at 55 °C for 5 min. Sample buffer (150 mM Tris⋅HCl, 6% SDS, 6 M urea, 10% 2-mercaptoethanol, 0.002% bromophenol blue, pH 6.8) was added to a final volume of 100 μL, before the sample was vortexed (5 min) and incubated at 55 °C for an additional 5 min. All samples were centrifuged at 13,000 × g for 10 min to remove debris and subjected to SDS/polyacrylamide-gel electrophoresis. All GFP-tagged fusion proteins and free GFP were detected by Western blot analysis using an anti-GFP mouse monoclonal antibody (1:3,000 dilution; Roche; clones 7.1 and 13.1). Adh1 was used as a loading control and detected with an Adh1 rabbit polyclonal antibody (1:10,000 dilution; AB1202; EMD Millipore).
Vacuolar Alkaline Phosphatase Activity.
Alkaline phosphatase (ALP) activity assays were performed as described previously (28) with minor modifications. Briefly, cells expressing the cytoplasmic form of Pho8 (Pho8∆60) were grown in SD complete medium to log phase (OD600 0.5 to 1.0). The cells were centrifuged, washed twice with sterile H2O, and shifted to starvation medium (SD-N) for 2 h at 30 °C to induce autophagy. The equivalent of 5.0 OD600 units of cells was harvested and washed with 1 mL of ice‐cold 0.85% NaCl containing 1 mM phenylmethylsulfonyl fluoride (PMSF) and pelleted at 3,000 × g for 5 min. Ice-cold lysis buffer (20 mM Pipes, pH 7.2, 0.5% Triton X-100, 50 mM KCl, 100 mM potassium acetate, 10 mM MgSO4, 10 μM ZnSO4, and 1 mM PMSF) and an equivalent volume (300 μL) of silica beads were added to the pelleted cells, and the samples were vortexed for 1 min at 4 °C. This step was repeated five times (1 min each time with 1-min intervals), followed by centrifugation at 14,000 × g for 5 min at 4 °C to remove debris. The supernatant was collected and the protein concentration was measured using Bradford reagent (Bio-Rad).
To analyze ALP activity, 100 μL of the supernatant was mixed thoroughly with 400 μL of the ALP substrate solution (1.25 mM p-nitrophenyl phosphate, 250 mM Tris⋅HCl, pH 8.5, 0.4% Triton X-100, 10 mM MgSO4, and 10 μM ZnSO4). Then the mixture was incubated in a water bath for 10 min at 37 °C, allowing for the production of p-nitrophenol. Stop buffer (1 M glycine/KOH, pH 11.0) was added (500 μL) to the samples to terminate the reaction, and p-nitrophenol was measured at 400 nm using a spectrophotometer. ALP activity was normalized to equal protein concentration.
Electron Microscopy.
Cells were grown in synthetic medium before inducing autophagy for 12 h by the addition of 200 ng/mL rapamycin. Subsequently, the cells were collected by centrifugation and processed for electron microscopy as previously described (3). The average number of autophagic bodies per cell section and the average number of autophagic bodies containing ER fragments were statistically assessed by counting 100 randomly selected cell profiles over two grids for each analyzed condition. ER fragments within autophagic bodies were identified by their morphology and were at least 25 nm in length. The cortical ER consisted of well-defined electron-dense tubules that were 3 to 5 nm in width.
Statistical Analysis.
At least three independent replicates were performed for all experiments. For fluorescence microscopy, a minimum number of 100 yeast cells were quantified for each replicate and representative images are shown. Data are presented as SEM. Probability values (P value) were calculated using the Student’s t test, and all comparisons with a P value < 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
We thank Serena Cervantes for her efforts in the early stages of this project. Salary support for S.F.-N., S.C., S.P., and Y.C. was provided by 1R01GM114111, 1R01GM115422, and 5R35GM131681. Salary support for P.J.N. and D.L. was provided by 1R01GM35370 and 1R01GM082861. F.R. is supported by Netherlands Organization for Health Research and Development TOP (ZonMW TOP) (91217002), Earth and Life Sciences (ALW) Open Programme (ALWOP.310), Marie Skłodowska-Curie Cofund (713660), and Marie Skłodowska-Curie European Training Networks (765912) grants. M.M. is supported by an ALW Open Programme (ALWOP.355).
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2008923117/-/DCSupplemental.
Data Availability.
All data are made available in this paper.
References
- 1.Chen S., Novick P., Ferro-Novick S., ER structure and function. Curr. Opin. Cell Biol. 25, 428–433 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walter P., Ron D., The unfolded protein response: From stress pathway to homeostatic regulation. Science 334, 1081–1086 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Cui Y. et al., A COPII subunit acts with an autophagy receptor to target endoplasmic reticulum for degradation. Science 365, 53–60 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Forrester A. et al., A selective ER-phagy exerts procollagen quality control via a calnexin-FAM134B complex. EMBO J. 38, e99847 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hübner C. A., Dikic I., ER-phagy and human diseases. Cell Death Differ. 27, 833–842 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mizushima N., Yoshimori T., Ohsumi Y., The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27, 107–132 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Stefely J. A. et al., Mass spectrometry proteomics reveals a function for mammalian CALCOCO1 in MTOR-regulated selective autophagy. Autophagy, 1–19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mochida K. et al., Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature 522, 359–362 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Khaminets A. et al., Regulation of endoplasmic reticulum turnover by selective autophagy. Nature 522, 354–358 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Grumati P. et al., Full length RTN3 regulates turnover of tubular endoplasmic reticulum via selective autophagy. eLife 6, e25555 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilkinson S., Emerging principles of selective ER autophagy. J. Mol. Biol. 432, 185–205 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen S., Cui Y., Parashar S., Novick P. J., Ferro-Novick S., ER-phagy requires Lnp1, a protein that stabilizes rearrangements of the ER network. Proc. Natl. Acad. Sci. U.S.A. 115, E6237–E6244 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen S. et al., Lunapark stabilizes nascent three-way junctions in the endoplasmic reticulum. Proc. Natl. Acad. Sci. U.S.A. 112, 418–423 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.John Peter A. T. et al., Vps13-Mcp1 interact at vacuole-mitochondria interfaces and bypass ER-mitochondria contact sites. J. Cell Biol. 216, 3219–3229 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park J. S. et al., Yeast Vps13 promotes mitochondrial function and is localized at membrane contact sites. Mol. Biol. Cell 27, 2435–2449 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar N. et al., VPS13A and VPS13C are lipid transport proteins differentially localized at ER contact sites. J. Cell Biol. 217, 3625–3639 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen S., Novick P., Ferro-Novick S., ER network formation requires a balance of the dynamin-like GTPase Sey1p and the Lunapark family member Lnp1p. Nat. Cell Biol. 14, 707–716 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lamb C. A., Longatti A., Tooze S. A., Rabs and GAPs in starvation-induced autophagy. Small GTPases 7, 265–269 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao D. et al., Atg20- and Atg24-family proteins promote organelle autophagy in fission yeast. J. Cell Sci. 129, 4289–4304 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Osawa T. et al., Atg2 mediates direct lipid transfer between membranes for autophagosome formation. Nat. Struct. Mol. Biol. 26, 281–288 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Gómez-Sánchez R. et al., Atg9 establishes Atg2-dependent contact sites between the endoplasmic reticulum and phagophores. J. Cell Biol. 217, 2743–2763 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chowdhury S. et al., Insights into autophagosome biogenesis from structural and biochemical analyses of the ATG2A-WIPI4 complex. Proc. Natl. Acad. Sci. U.S.A. 115, E9792–E9801 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kotani T., Kirisako H., Koizumi M., Ohsumi Y., Nakatogawa H., The Atg2-Atg18 complex tethers pre-autophagosomal membranes to the endoplasmic reticulum for autophagosome formation. Proc. Natl. Acad. Sci. U.S.A. 115, 10363–10368 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valverde D. P. et al., ATG2 transports lipids to promote autophagosome biogenesis. J. Cell Biol. 218, 1787–1798 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eisenberg-Bord M., Shai N., Schuldiner M., Bohnert M., A tether is a tether is a tether: Tethering at membrane contact sites. Dev. Cell 39, 395–409 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Roberts P. et al., Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol. Biol. Cell 14, 129–141 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muñoz-Braceras S., Calvo R., Escalante R., TipC and the chorea-acanthocytosis protein VPS13A regulate autophagy in Dictyostelium and human HeLa cells. Autophagy 11, 918–927 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noda T., Klionsky D. J., The quantitative Pho8Delta60 assay of nonspecific autophagy. Methods Enzymol. 451, 33–42 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Guimaraes R. S., Delorme-Axford E., Klionsky D. J., Reggiori F., Assays for the biochemical and ultrastructural measurement of selective and nonselective types of autophagy in the yeast Saccharomyces cerevisiae. Methods 75, 141–150 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Yeshaw W. M. et al., Human VPS13A is associated with multiple organelles and influences mitochondrial morphology and lipid droplet motility. eLife 8, e43561 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bean B. D. M. et al., Competitive organelle-specific adaptors recruit Vps13 to membrane contact sites. J. Cell Biol. 217, 3593–3607 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kolehmainen J. et al., Cohen syndrome is caused by mutations in a novel gene, COH1, encoding a transmembrane protein with a presumed role in vesicle-mediated sorting and intracellular protein transport. Am. J. Hum. Genet. 72, 1359–1369 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anding A. L. et al., Vps13D encodes a ubiquitin-binding protein that is required for the regulation of mitochondrial size and clearance. Curr. Biol. 28, 287–295.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rampoldi L. et al., A conserved sorting-associated protein is mutant in chorea-acanthocytosis. Nat. Genet. 28, 119–120 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Ueno S. et al., The gene encoding a newly discovered protein, chorein, is mutated in chorea-acanthocytosis. Nat. Genet. 28, 121–122 (2001). [DOI] [PubMed] [Google Scholar]
- 36.Lesage S. et al.; French Parkinson’s Disease Genetics Study (PDG); International Parkinson’s Disease Genomics Consortium (IPDGC) , Loss of VPS13C function in autosomal-recessive parkinsonism causes mitochondrial dysfunction and increases PINK1/parkin-dependent mitophagy. Am. J. Hum. Genet. 98, 500–513 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen Q. et al., ATL3 is a tubular ER-phagy receptor for GABARAP-mediated selective autophagy. Curr. Biol. 29, 846–855.e6 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Wang B. et al., The COPII cargo adapter SEC24C is essential for neuronal homeostasis. J. Clin. Invest. 128, 3319–3332 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li P., Lees J. A., Lusk C. P., Reinisch K. M., Cryo-EM reconstruction of a VPS13 fragment reveals a long groove to channel lipids between membranes. J. Cell Biol. 219, e202001161 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Longtine M. S. et al., Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 (1998). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are made available in this paper.