Social interaction is like a dance between two or more individuals who communicate with each other by sending and receiving information. Social cues are multisensory, involving combinations of visual, olfactory, auditory, and tactile information. One individual transmits information through its behavior and the other interprets it, providing a behavioral response that contains further social information, and the dance continues. Unlike nonsocial behavior (e.g., a lone worm moving along a temperature gradient), social behavior is complex because the transmitter of the information changes the receiver’s subsequent behavior. From a statistical perspective, like individual behavior, variation in social behavior can be partitioned into its genetic and environmental contributions, and their interactions (1, 2). However, for social behavior, the social environment is a critical part of the equation (3, 4). Social interactions result in indirect genetic effects (IGEs), when the genetics of one individual affects another’s behavior (3, 4). IGEs are important for the evolution of social behavior; however, a mechanistic understanding of IGEs is lacking (5, 6). Which phenotypes should be measured when social interactions are an emergent property of the group? What genes and pathways are involved in IGEs, and are patterns of selection found in their DNA sequences? How can we validate and functionally investigate the role of genetic variants in a group setting? In PNAS, Avalos et al. (7) use genome-wide association studies (GWAS) in the eusocial honey bee to uncover genomic regions defined by colony allele frequencies that influence colony aggressive behavior.
Social behavior is exhibited by many organisms, from microbes to humans (8). Factors that affect social interactions include components of the social environment (e.g., density, space, and group social structure) and the physical environment (e.g., temperature and humidity) (9). Social experiences can affect different levels of biological organization (e.g., neural transmission, gene expression, and development) acting on different timescales (10). IGEs are found in many species of insects including beetles (11) and the fruit fly (12, 13), fire ant (7), and honey bee (6, 14); however, a link to group allele frequencies has not been established, and mechanistic studies of IGEs are lacking.
Social Insects as a Model to Study Mechanisms of IGEs
Why are eusocial insects an appealing model for the study of the mechanisms of IGEs, and what challenges exist (6, 7)?
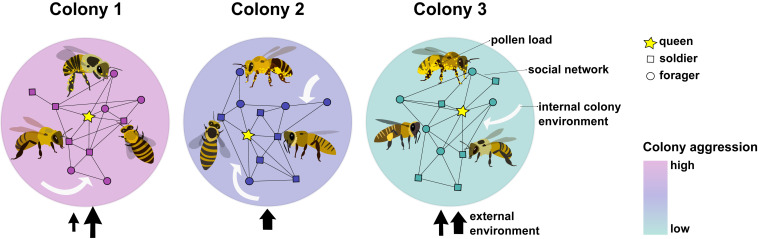
Like ants and termites, honey bees live in highly structured societies with a division of labor between the reproductive (the queen) and the nonreproductive workers (e.g., nurses, foragers, and soldiers) (15). Queen bees lay eggs and control their workers through chemical communication. Nurses tend the eggs and larvae, regulating their caste fate by controlling their diet. Caste members, each with their different tasks, cooperate and live in colony groups with overlapping generations. For a honey bee, the colony is its social context, and its behavior is a response to the colony’s needs. Behaviors of workers are highly stereotyped, and job specification is age-dependent (e.g., nurses transition to being foragers at around three weeks of age). It is well established that the brains of different worker castes (e.g., nurse and forager) show significant differences in gene expression that are very highly correlated with task (16). Honey bees also exhibit plastic changes in brain gene expression in response to modifications in the environment (e.g., the threat of an intruder) (17). Despite their structured societies, differences between colonies are the norm. Every colony is exposed to its myriad of internal and external environmental factors including factors such as outside temperature, humidity/rain, sunlight, day length, and colony size, number of individuals, density, space, the ratio of different castes, pheromones, intruders, foraging distances, and the quality and quantity of pollen and nectar available. Each colony has its microecology, which contributes to differences between groups and, consequently, differences in the colony-level social environment. In this context, allelic differences between colonies emerge over time, leading to some colonies exhibiting higher intensities of a behavior (e.g., defensive aggression) than others (Fig. 1). Eusocial insect colonies are a ripe source for group behavior studies, whether comparisons are made between groups of workers with different tasks or between colony-level group behaviors. The challenges in using social insects for studies of the mechanisms underlying IGE lie in the need for large sample sizes (a colony is an n = 1) and in applying ways to functionally validate candidate genes that affect the social environment at the colony level.
Fig. 1.
Pictorial representations of honey bee colonies. Soldiers (square), foragers (circle), and the queen (star) are connected through a social network. Colonies differ in their individual and group-level phenotypes. Some colonies are more aggressive (see color key for colony aggression); others have more successful foragers (see yellow pollen on foragers). Colonies are exposed to internal (white arrows) and external (black arrows) environments. Internal and external environmental factors differ between colonies, shown by the number and size of the white and black arrows. Examples of external environmental conditions are 1) the distance and quality of pollen and nectar and 2) the size and intensity of the black arrows representing colony intruders. Additional examples of external and internal conditions are discussed in the text. Image credit: Sydney Gram (University of Toronto, Toronto, Canada).
Measuring Defensive Aggression Behavior
Avalos et al. (7) use the IGE framework as a guide toward finding genes that affect colony-level aggression. Honey bee colonies were collected from Puerto Rican sites and transferred to the University of Puerto Rico field station for acclimation before experiments. Each colony had one queen and a brood and about 16,000 to 20,000 worker bees. Each queen was related to her workers and had mated with more than one drone. Individual and colony-level measures of aggression were performed. Individual-level aggression was measured by collecting soldier and forger bees after a disturbance. A string of 5- × 5-cm black suede patches was placed near the hive entrance, and the colony was disturbed with repeated strikes to the cover of the colony. Soldier bees left the hive and were collected in the act of stinging the black patch. Forager bees left the hive and returned with pollen and nectar. A colony-level aggression rank was produced 1) by counting the number of stingers or stings in the black patches and 2) by developing a combined behavioral score taken 2 wk postdisturbance which included two activity measures (running on the comb and hanging from the comb), a measure of arousal (flying around the hive), and a measure of the likelihood to sting. Both measures were significantly correlated and used in a multidimensional scaling (MDS) analysis to derive a colony-level phenotype dimensional vector called D1. The high quality of the behavioral phenotyping performed in the paper is notable.
Individual and Group GWAS
The authors found no significant associations in the individual GWAS based on a comparison of the DNA sequences of soldiers and foragers. The relatively small sample size (n = 177 individual bees with n = 10 soldiers n = 10 per colony) and the categorical phenotyping may have limited their power to detect associations (figure S1 in ref. 7). In contrast, and in line with expectations from IGEs, the colony-level GWAS showed particularly strong associations between colony allele frequencies and the quantitative composite phenotyping of colony aggression (D1), which accounted for 74% of the variance in the colony phenotype (figure 1C in ref. 7). The highly significant finding for the colony-level GWAS was surprising, given the n = 9 sample size. This is addressed in Avalos et al. (7).
What contributed to the highly significant associations found in the colony-level GWAS? The heritability of colony aggression was twice that of the individual aggression and high (0.63) for a behavioral phenotype, making it more likely to find significant GWAS hits in the colony GWAS data. The colony measures also had an unusually high genotype–phenotype correlation (P values of 10−10 to 10−50 for some loci), which may reflect the highly stereotyped behavior of castes in social insect colonies. The extent of phenotypic variance not explained by genotype was also very low, with genotype a continuous measure of colony allele frequency. Simulations revealed that the strong collinear relationships found can generate highly significant results even with small (n = 9) sample sizes. The high heritability measures found for a colony-level phenotype, colony-level genotype–phenotype correlations, and the capacity for deep behavioral phenotyping at the colony level lend support to the value of social insects for identifying mechanisms underlying IGEs.
Colony-Level GWAS and Candidate Genes for IGEs
How was colony-level genotype assessed? The genotypes of each bee were used to identify the minor allele at each polymorphic locus across the entire dataset. Samples were grouped by colony, and the frequency of that minor allele was calculated for each colony for each marker, resulting in a vector of nine (one per colony) allele frequencies for each single-nucleotide polymorphism (SNP) in the dataset. GWAS for the colony-level phenotype associated the per-colony minor allele frequency with the behavioral phenotype D1 from the MDS analysis. Significant associations were found for 1,172 SNPs located throughout the genome. A highly significant GWAS peak was found at LG07, a candidate for a large-effect-size locus affecting colony-level aggression. Many other significant SNPs were strongly correlated with one of the large-effect peaks (LG07). The LG07 peak had a strong selection signal and a high number of significant flanking SNPs in linkage disequilibrium.
Interestingly, the genes from the colony GWAS were enriched for Gene Ontology terms associated with the immunoglobulin (Ig) domain involved in an axon guidance pathway associated with brain development. Two of the honey bee genes centered on the locus LOC724823 on LG07 had fly orthologs in the dpr (defective proboscis extension response) family of Ig domain genes that encode sensory receptors and affect Drosophila courtship behavior (quicker to attempt mating and quicker to copulate). dpr acts downstream of fruitless, which genetically links aggression and courtship in Drosophila melanogaster (18). The GWAS peak that contained the dpr4 ortholog and other nearby loci had the strongest signature of selection.
How might the functional validation of candidate colony-level GWAS candidate genes proceed? In tightly socially integrated organisms such as the honey bee, colony-level phenotypes belong to the genomes of multiple interacting individuals. Consequently, it would not be appropriate to test the effect of a targeted CRISPR SNP alteration in a candidate gene at the individual level when addressing a colony-level phenotype. The question of how to use gene-editing technologies to alter colony-level phenotypes in social insect genomes is challenging. One way to begin might be to reconstruct colonies that carry CRISPR-edited genome alterations in the large-effect locus on LG07 that influences a high proportion of the variance on the colony-aggression phenotype (5). However, pleiotropies of the target genes and their consequences for each caste and overall colony functioning would need ample consideration.
In social insects, phenotypes belong to the genomes of multiple interacting individuals. Avalos et al. (7) provide a way forward for finding genes that affect the social environment, thereby expanding the conceptual framework of IGEs to studies of mechanism.
Footnotes
The author declares no competing interest.
See companion article, “Genomic regions influencing aggressive behavior in honey bees are defined by colony allele frequencies,” 10.1073/pnas.1922927117.
References
- 1.Sokolowski M. B., Levine J. D., “Nature-nurture interactions” in Social Behaviour, Genes, Ecology and Evolution, Szekely T., Moore A. J., Komdeur J., Eds. (Cambridge University Press, 2010), pp. 11–25. [Google Scholar]
- 2.Schneider J., Atallah J., Levine J. D., Social structure and indirect genetic effects: Genetics of social behaviour. Biol. Rev. Camb. Philos. Soc. 92, 1027–1038 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Moore A. J., Brodie E. D. III, Wolf J. B., Interacting phenotypes and the evolutionary process: I. Direct and indirect genetic effects of social interactions. Evolution 51, 1352–1362 (1997). [DOI] [PubMed] [Google Scholar]
- 4.Wolf J. B., Brodie E. D. III, Cheverud J. M., Moore A. J., Wade M. J., Evolutionary consequences of indirect genetic effects. Trends Ecol. Evol. (Amst.) 13, 64–69 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Linksvayer T. A., The molecular and evolutionary genetic implications of being truly social for social insects. Adv. Insect Physiol. 48, 271–293 (2015). [Google Scholar]
- 6.Weitekamp C. A., Libbrecht R., Keller L., Genetics and evolution of social behavior in insects. Annu. Rev. Genet. 51, 219–239 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Avalos A., et al. , Genomic regions influencing aggressive behavior in honey bees are defined by colony allele frequencies. Proc. Natl. Acad. Sci. U.S.A. 117, 17135–17141 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sokolowski M. B., Social interactions in “simple” model systems. Neuron 65, 780–794 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Rooke R., Rasool A., Schneider J., Levine J. D., Drosophila melanogaster behaviour changes in different social environments based on group size and density. Commun. Biol. 3, 304–310 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rittschof C. C., Hughes K. A., Advancing behavioural genomics by considering timescale. Nat. Commun. 9, 489 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carter M. J., Wilson A. J., Moore A. J., Royle N. J., The role of indirect genetic effects in the evolution of interacting reproductive behaviors in the burying beetle, Nicrophorus vespilloides. Ecol. Evol. 9, 998–1009 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kent C., Azanchi R., Smith B., Formosa A., Levine J. D., Social context influences chemical communication in D. melanogaster males. Curr. Biol. 18, 1384–1389 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Saltz J. B., Genetic composition of social groups influences male aggressive behaviour and fitness in natural genotypes of Drosophila melanogaster. Proc. Biol. Sci. 280, 20131926 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robinson G. E., Grozinger C. M., Whitfield C. W., Sociogenomics: Social life in molecular terms. Nat. Rev. Genet. 6, 257–270 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Wilson E. O., The Insect Societies (Belknap Press, Cambridge, MA, 1971). [Google Scholar]
- 16.Whitfield C. W., et al. , Genomic dissection of behavioral maturation in the honey bee. Proc. Natl. Acad. Sci. U.S.A. 103, 16068–16075 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shpigler H. Y., et al. , Behavioral, transcriptomic and epigenetic responses to social challenge in honey bees. Genes Brain Behav. 16, 579–591 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Vrontou E., Nilsen S. P., Demir E., Kravitz E. A., Dickson B. J., fruitless regulates aggression and dominance in Drosophila. Nat. Neurosci. 9, 1469–1471 (2006). [DOI] [PubMed] [Google Scholar]