Significance
This study demonstrates that the extracellular protein human vitronectin (Vn) is able to bind both soluble calcium ions and mineralized hydroxyapatite with chemical specificity. The results have important implications for the normal functions of Vn and its role in the formation of plaques associated with age-related macular degeneration, Alzheimer’s disease, atherosclerosis, and other degenerative disorders. The molecular basis for plaque formation in these pathologies is unknown. The present findings shed light on this important process, advance our understanding of the mechanisms of abnormal plaque formation, and introduce a target with potential for developing diagnostic, preventive, or therapeutic approaches.
Keywords: macular degeneration, deposit, drusen, NMR, solid state
Abstract
The human blood protein vitronectin (Vn) is a major component of the abnormal deposits associated with age-related macular degeneration, Alzheimer’s disease, and many other age-related disorders. Its accumulation with lipids and hydroxyapatite (HAP) has been demonstrated, but the precise mechanism for deposit formation remains unknown. Using a combination of solution and solid-state NMR experiments, cosedimentation assays, differential scanning fluorimetry (DSF), and binding energy calculations, we demonstrate that Vn is capable of binding both soluble ionic calcium and crystalline HAP, with high affinity and chemical specificity. Calcium ions bind preferentially at an external site, at the top of the hemopexin-like (HX) domain, with a group of four Asp carboxylate groups. The same external site is also implicated in HAP binding. Moreover, Vn acquires thermal stability upon association with either calcium ions or crystalline HAP. The data point to a mechanism whereby Vn plays an active role in orchestrating calcified deposit formation. They provide a platform for understanding the pathogenesis of macular degeneration and other related degenerative disorders, and the normal functions of Vn, especially those related to bone resorption.
The extracellular deposits that accumulate under the retinal pigment epithelium (RPE) of the aging eye are a hallmark of age-related macular degeneration, Alzheimer’s disease, and dense deposit disease (1, 2) and share similarities with the plaques that form in atherosclerosis (3–10). These ectopic deposits are rich in blood proteins, lipids, and hydroxyapatite (HAP), the stable, mineralized form of hydroxylated calcium phosphate that makes up the bulk of bone and teeth (1, 10). Calcified deposits have been linked with progression of macular degeneration (11), but the process of deposit formation is poorly understood. Remarkably, subretinal deposits have been shown to consist of micrometer-size spherules, each composed of a cholesterol-rich lipid core, overlaid with a shell of HAP and a final top coat of protein (10). The spherules have been proposed to form by first mineralizing HAP shells around cholesterol-rich lipid droplets that accumulate in the sub-RPE space with age, and then, cloaking the shells with a protein layer that promotes spherule aggregation (10). In this model, HAP is thought to provide a generalized substrate for binding blood proteins, but protein association with calcified deposits appears to be specific (8–10). Notably, the protein Vn is unique in forming both an even coat on the spherule surface and a diffuse halo surrounding fully formed spherules (10). This is highly suggestive of a supersaturated solution of the spherule components that is phase-separated from the surrounding extracellular fluid and points to a potential, active role of Vn in coordinating HAP mineralization during retinal deposit formation.
Vn is a major blood protein that interacts with multiple ligands to regulate hemostasis, cell adhesion and migration, innate immunity, tissue remodeling, and bone remodeling (12–15). It is a lipid binding protein and a principal component of high density lipoprotein (16, 17), and pathogens bind Vn to acquire protection from complement-mediated lysis (18). Since Vn is also found in many types of abnormal deposits associated with degenerative disorders (6–10), understanding its role in deposit formation is important for advancing diagnostic and therapeutic approaches. Here, we show that Vn binds both soluble calcium ions and solid HAP with chemical specificity, and we ascribe the molecular basis for these interactions to its HX domain, whose three-dimensional (3D) structure was determined recently (19). The data, derived from solution NMR, solid-state NMR, sedimentation assays, DSF, and binding energy calculations, provide a view of this important macromolecular interaction. They indicate that Vn may be more than a passive component of sub-RPE deposits and suggest a model for its role in orchestrating HAP mineralization, as well as offer insights to the normal functions of Vn.
Results and Discussion
Vn Is a Selective Calcium-Binding Protein.
In blood, Vn circulates as an intact 75-kDa glycosylated molecule, or as two disulfide-linked 65-kDa and 10-kDa polypeptides (20, 21). The Vn sequence (SI Appendix, Fig. S1A) begins with a 44-residue somatomedin B (SMB) domain that is responsible for regulating plasminogen activation (22, 23), followed by an ArgGlyAsp motif that mediates binding to integrin receptors (24). These are linked to the HX domain by a 90-residue segment with predicted conformational disorder. The 325-residue HX domain comprises ∼70% of the sequence of mature Vn and contains binding sites for many functionally important ligands.
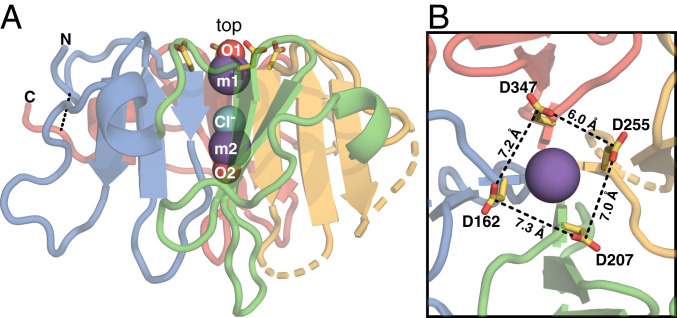
The structure of the HX domain (19), excluding two regions of relatively low conservation and low conformational order, is a four-bladed β-propeller, with each blade formed by one βββα HX repeat and the termini connected by a disulfide bond (Fig. 1 and SI Appendix, Fig. S1B). This fold is shared by all known HX domains (25). The propeller top – defined as the start of each β1 – forms a smooth surface, while longer flexible loops protrude from the bottom. The four β1 strands meet at the propeller center to form a channel that occludes a metal–chloride–metal ion triplet. Inside the channel, chloride is bound by four β1 amide hydrogens, and each metal ion is coordinated by four β1 carbonyl oxygens plus an oxygen from water or sulfate (Fig. 1A and SI Appendix, Fig. S2). The metal ions were assigned to sodium, based on the temperature B factors and considerations of the coordination sphere geometry (19, 26), but we note that X-ray diffraction cannot readily distinguish between sodium and calcium.
Fig. 1.
Structure of the Vn-HX propeller. Shown for molecule A of the asymmetric unit (Protein Data Bank ID code 6O5E). Backbone colors denote the repeat units corresponding to each propeller blade: HX1 (blue), HX2 (green), HX3 (yellow), and HX4 (red). Broken lines denote gaps in the protein chain due to missing electron density. The rim-Asp side chains (yellow/red) project from the top. (A) Side view. Spheres represent Na+ (m1 and m2; slate) and Cl− (aqua) ions occluded in the channel and water oxygen atoms (O1 and O2; red). The N- and C-termini are connected by a disulfide bond (dashed line). (B) Top view. The rim-Asp carboxylates form a ∼7 Å x 7 Å corral around the channel opening.
In proteins, calcium ligation is typically mediated by carboxylate oxygens (27), and carboxylate-rich matrix proteins – such as osteocalcin, osteopontin, and collagen – as well as the tricarboxylate molecule citrate, have all been shown to bind HAP selectively and mediate HAP crystal growth in bone and tooth enamel (28–31). In the Vn HX domain, four Asp sidechains (D162, D207, D255, D347), each at the start of a β1 strand, project from the top of the channel, forming a 7 × 7 Å corral around its opening (Fig. 1B). The carboxylate groups of these “rim-Asp” are oriented with their planes perpendicular to each other, in a chiral arrangement that appears to minimize repulsive forces while focusing the negative charge at the channel opening (SI Appendix, Fig. S1C). These structural features are highly suggestive of a calcium binding site and led us to ask whether the association of Vn with HAP is related to the interaction of calcium with its HX domain. NMR is ideally suited for addressing this question: Collectively, solution and solid-state NMR methods provide access to a wide range of molecular dynamics and distance scales, enabling proteins to be studied in both their freely soluble and sedimented states.
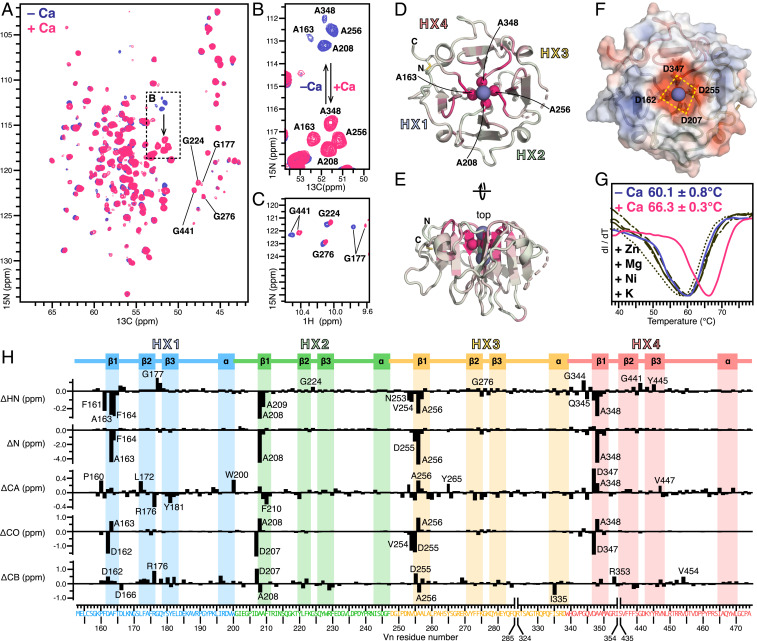
The NMR spectra of 15N/13C-labeled Vn-HX in calcium-free aqueous solution (Fig. 2 A–C and SI Appendix, Fig. S3A) have highly dispersed 1H, 15N, and 13C signals, as expected for a conformationally ordered structure. Analysis of the chemical shifts (SI Appendix, Fig. S3B), which we assigned by triple resonance experiments, demonstrates that the protein in solution has the same βββα-repeat structure and conformational order profile as observed in the crystalline state. Notably, four Gly (G177, G224, G276, G441) that form the β2-β3 turns at the propeller top, and stabilize the conformations of the rim-Asp carboxylates via hydrogen bonds, have highly deshielded amide 1HN and 15N chemical shifts (Fig. 2C, blue). Moreover, four “rim-Ala” (A163, A208, A256, A348), lining the channel one level below the rim-Asp, have relatively shielded 1HN and 15N chemical shifts, as well as a striking pattern of 15N/13CA cross peaks (Fig. 2 A and B, blue) that mirrors the positions of the corresponding backbone atoms in the structure (Fig. 2D). These features reflect a strong coupling of these atomic sites with the electric field generated by the rim-Asp carboxylate groups.
Fig. 2.
Effects of ionic calcium on Vn-HX. (A–C) Selected regions of 2D planes taken from 3D HNCA NMR spectra (A and B) and 2D HSQC NMR spectra (C) of 13C/15N-labeled Vn-HX in 300 mM NaCl, without (blue) or with (pink) 2 mM CaCl2. (D and E) Cartoon representations of the structure of Vn-HX. The color scale reflects the magnitude of calcium-induced 15N chemical shift perturbation from 0 ppm (gray) to 5 ppm (pink). Highly perturbed amide N atoms of four rim-Ala near the top of the channel are shown as spheres. Broken lines denote gaps in the protein chain due to missing electron density in the structure. HX1–HX4 denote the HX repeats. (F) Surface representation showing the four rim-Asp carboxylates the top of the channel. Colors denote surface electrostatic potential from −5 kT/e (red) to +5 kT/e (blue). (G) DSF traces of Vn-HX in 300 mM NaCl, without added CaCl2 (blue), with 2 mM CaCl2 (pink), or with 2 mM of the metal salts (black): KCl (––), ZnCl2 (・・・), NiCl2 (–・–), or MgCl2 (– –). Values of the melting temperature represent the mean and standard deviation of triplicate independent experiments. (H) Residue-specific perturbations (Δ) induced by 2 mM CaCl2 for 1HN, 15N, 13CA, 13CO, and 13CB chemical shifts (Δ < 0 reflects deshielding). The amino acid sequence is colored by HX repeat unit: HX1 (blue), HX2 (green), HX3 (yellow), and HX4 (red). The protein secondary structure is outlined at the top.
Addition of CaCl2 induces the growth of a second set of NMR signals that gain intensity with increasing CaCl2 concentration and dominate the spectra at 8 mM CaCl2, but does not alter the overall protein structure (SI Appendix, Figs. S3C and S4). The data reflect slow exchange between calcium-bound and calcium-free protein, relative to the chemical shift time scale. We derived binding curves from the change in signal height with calcium addition for each of six well-resolved peaks (SI Appendix, Fig. S4). The curves yield an average apparent equilibrium dissociation constant (Kd) of 27±1.5 µM (assuming 1:1 binding), suggesting that a subtle conformational rearrangement may contribute to the observation of slow exchange. Importantly, the apparent Kd, which is ∼10 times higher than the 2.5–5 µM concentration of Vn in blood (12–14) and ∼30 times lower than the 1.3–1.5 mM concentration of ionic calcium in blood (27), indicates that all Vn is calcium-bound in vivo.
Size exclusion chromatography demonstrates that Vn-HX remains monomeric in the presence of calcium, excluding the possibility that the NMR perturbations reflect protein oligomerization (SI Appendix, Fig. S3D). However, calcium binding appears to be associated with increased conformational order, as it enables the detection of additional NMR peaks. In line with this observation, DSF analysis shows that calcium increases the protein melting temperature by ∼6 °C (Fig. 2G). A similar calcium-related thermal stabilization has been reported for the HX domain of matrix metalloprotease-19, whose structure remains undetermined (32). For Vn-HX, the effect is specific to calcium since other metal ions (Zn2+, Mg2+, Ni2+, and K+) – among which Zn2+ is also enriched in sub-RPE deposits (33) – produce no such change.
Calcium-induced perturbations of 1HN, 15N, 13CA, 13CO, and 13CB chemical shifts track the repeating elements of the Vn-HX propeller structure (Fig. 2H) and map primarily to the top of the channel (Fig. 2 D and E). A dramatic effect is seen for the amide nitrogen atoms of the rim-Ala: The entire group of four 15N signals shifts 5 ppm downfield while preserving its striking structure-mirroring 15N/13CA peak pattern (Fig. 2 A and B, pink). The carbonyl carbons of both rim-Ala and rim-Asp are also highly affected, as are the amide hydrogens and nitrogens of four residues that line the channel one level below them (F164, A209, A257, and A349), and of the four β2-β3 Gly that stabilize the rim-Asp (Fig. 2C, pink). The effects of calcium extend to atomic sites connected to the channel via the extensive hydrogen bond network of the propeller structure (SI Appendix, Fig. S5). Overall, the data indicate that ionic calcium binds at the top of the channel opening, where the rim-Asp carboxylates generate a focused electric field (Fig. 2F), rather than inside the channel.
The metal ions occluded in the channels of other HX domains have been variably assigned to sodium or calcium, in different and sometimes conflicting arrangements (reviewed in ref. 34). To assess the potential of calcium occlusion inside the Vn channel, we compared the free energies associated with transferring sodium or calcium from bulk aqueous solution to the internal metal binding sites, m1 and m2 (Fig. 1 and SI Appendix, Fig. S2). The energies, calculated with an approach (35) based on molecular mechanics and Poisson–Boltzmann continuum electrostatics (36, 37), reveal a distinct selectivity for sodium (Table 1). Sodium selectivity is maintained whether the channel-capping oxygen in the coordination sphere of m2 comes from water or sulfate, although these cases have some energetic differences, and the m2 site becomes more permissive for calcium with sulfate oxygen coordination. In the case of water coordination, the energetic penalty for replacing sodium with calcium is 34 kcal/mol for site m1, 30 kcal/mol for site m2, and 78 kcal/mol for both sites combined, while these values are 29 kcal/mol, 8 kcal/mol, and 53 kcal/mol in the case of sulfate coordination at m2. Unlike ion-conducting channels, the Vn channel is constrained by the propeller structure, which is extensively hydrogen-bonded and held together by favorable ring stacking and hydrophobic interactions, and the occluded metal ions appear to be structural. Based on the NMR and computational data, we propose that the principal binding site for aqueous calcium is not occluded and involves the rim-Asp carboxylate groups at the top of the propeller.
Table 1.
Interaction energy of Na+ and Ca2+ ions with the Vn-HX channel interior
| O1 – m1 – Cl− – m2 – O2 | ΔEVDW | ΔEELEC | ΔETOT | |
| Molecule A | water – Na+ – Cl− – Na+ – water | 29.42 | −248.50 | −219.08 |
| water – Na+ – Cl− – Ca2+ – water | 33.78 | −223.29 | −189.51 | |
| water – Ca2+ – Cl− – Na+ – water | 34.46 | −220.02 | −185.56 | |
| water – Ca2+ – Cl− – Ca2+ – water | 38.81 | −179.90 | −141.09 | |
| Molecule B | water – Na+ – Cl− – Na+ – SO42− | 29.86 | −274.86 | −245.00 |
| water – Na+ – Cl− – Ca2+ – SO42− | 33.74 | −271.17 | −237.43 | |
| water – Ca2+ – Cl− – Na+ – SO42− | 35.26 | −251.56 | −216.30 | |
| water – Ca2+ – Cl− – Ca2+ – SO42− | 39.15 | −231.11 | −191.96 |
Values of ΔEVDW, ΔEELEC, and their sum ETOT (kcal/mol) reflect the van der Waals and electrostatic energy differences associated with transferring each metal ion from bulk aqueous solution to the occluded m1 and m2 channel sites. Energies were calculated for molecules A and B of the crystal structure (Protein Data Bank ID code 6O5E).
Vn Is a Selective HAP Binding Protein.
The protein binding capacity of HAP is well known (38), and its interaction with blood proteins has been explored in the context of macular degeneration (39). Our investigation of Vn was motivated by its unique pattern of association with retinal HAP spherules – Vn forms both an even coat on the spherule surface and a diffuse phase enveloping fully formed spherules (10) – but Vn is also critical for osteoclast bone resorption (15) where affinity for HAP is also important.
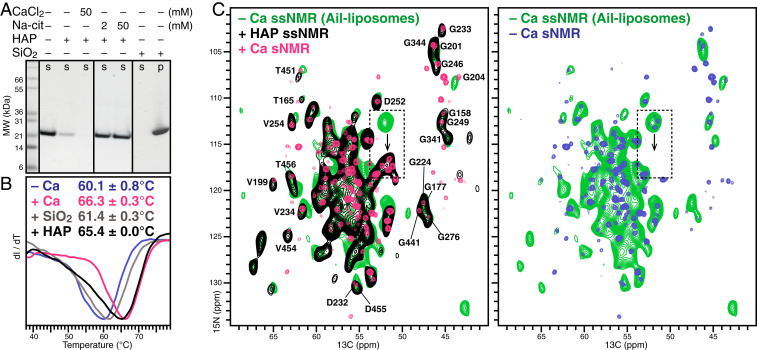
To test whether the HX domain contributes to the association of Vn with HAP, we incubated the protein with HAP microspheres (2.5 µm diameter) and performed polyacrylamide gel electrophoresis in sodium dodecyl sulfate (SDS-PAGE) on the supernatant obtained after centrifugation (Fig. 3A). The results show that HAP avidly pulls Vn from solution. Notably, cosedimentation is completely blocked by citrate, a known chelate and HAP binder (29). However, inclusion of CaCl2 in the protein solution does not inhibit, but rather enhances, Vn sedimentation with HAP, an effect also reported for osteocalcin (30, 31).
Fig. 3.
Vn-HX binds HAP and ionic calcium via the same calcium binding site. (A) Cosedimentation of Vn-HX with HAP microspheres. Supernatant (s) and sedimented (p) fractions were obtained after incubation of Vn-HX with either HAP or SiO2, followed by centrifugation, and analyzed by Coomassie-stained SDS/PAGE. (B) DSF traces of Vn-HX in 300 mM NaCl, without added CaCl2 (blue), with 2 mM CaCl2 (pink), with HAP (black), or with SiO2 (gray). Values of the melting temperature represent the mean and standard deviation of triplicate independent experiments. (C) Solid-state NMR 2D NCA spectra obtained for Vn-HX bound to HAP nanoparticles (black) or bound to liposomes reconstituted with the Vn-binding outer membrane protein Ail (green). The solution NMR HNCA spectra of soluble Vn-HX, without (blue) or with (pink) 2 mM CaCl2, are overlaid. The dashed-line box highlights the calcium-sensitive signals of the four rim-Ala.
DSF experiments performed by adding soluble Vn to HAP microspheres show that HAP binding increases the melting temperature by ∼6 °C (Fig. 3B) and, thus, provides the same degree of stabilization as ionized calcium. Since Vn was named after its ability to bind glass (40), we asked whether the thermal stability might reflect nonspecific surface adsorption, or even precipitation, rather than a specific chemical effect. As expected, incubation of Vn with silica gel (60-nm diameter) results in complete protein sedimentation from solution (Fig. 3A). In contrast to HAP, however, silica-adsorbed Vn has essentially the same melting transition temperature as in calcium-free solution (Fig. 3B). The combined data, therefore, reflect a specific, calcium-mediated interaction of Vn with HAP.
Lastly, we examined the conformation of HAP-bound Vn. While the Vn–HAP complex is prohibitive for solution NMR studies, solid-state NMR has no physical size limitation and can be used to examine large molecular assemblies that are immobilized on the NMR time scale (41, 42). We prepared the NMR sample by incubating 13C/15N-labeled, calcium-free Vn-HX with HAP nanoparticles (50 nm diameter) and then transferred the bound complex into the magic angle spinning (MAS) rotor by centrifugation. This preparation gave excellent one-dimensional (1D) 15N and 13C MAS NMR spectra by through-space dipolar cross-polarization (CP) from 1H, but essentially no signal in the spectra obtained by through-bond polarization transfer, confirming that association with HAP effectively immobilizes Vn on the microsecond time scale (SI Appendix, Fig. S6).
The two-dimensional (2D), CP-based 15N/13CA and 1H/15N heteronuclear correlation spectra, obtained for HAP-bound Vn at 500 MHz, have very good signal-to-noise ratio and resolution (Fig. 3C and SI Appendix, Fig. S6). Both display resonance patterns similar to their counterparts in solution (the solution NMR 1H/15N/13CA spectrum has more signals because it correlates HN of residue i to CA of both residue i and i-1), notwithstanding the different temperatures utilized for solution (30 °C) and solid-state (5 °C) NMR experiments, and the widely different physical states of the samples. The similarities are especially evident among the Gly and Ala signals, which are relatively well resolved in the solid state. The combined data show that the HX domain retains its β-propeller fold upon binding crystalline HAP.
Comparison with the solution NMR spectra reveals a striking feature in the 15N/13CA spectrum of HAP-bound Vn: The rim-Ala signal quartet coincides with its calcium-bound position in solution (Fig. 3C, black/pink, dashed box). Although the overall broader lines of the solid-state NMR spectrum do not provide resolution among the four peaks, the ∼5 ppm 15N shift downfield is so large that it can be readily detected. To examine the source of this effect in the solid state, we acquired 15N/13CA spectra of the protein sedimented with the outer membrane protein Ail in calcium-free liposomes (Fig. 3C and SI Appendix, Fig. S6, green). Ail binds serum Vn as part of its function to promote resistance to host immune defenses in the pathogenic bacterium Yersinia pestis (43), and we have shown that the HX domain is central for this interaction (19). The 15N/13CA spectrum for this complex is similar to that of HAP-bound Vn with the notable exception of the four rim-Ala signals, which now coincide with the signal of calcium-free Vn in the solution NMR spectrum (Fig. 3C, blue). The combined solution and solid-state NMR data, therefore, demonstrate that calcium association is responsible for the downfield 15N shift in both the solution and solid-state NMR spectra. This implies that the same calcium-binding site is implicated in the association of Vn with either soluble ionic calcium or HAP.
Possible Roles of Vn in Calcified Deposit Formation.
In summary, we have demonstrated that the HX domain of Vn is capable of binding both soluble ionic calcium and crystalline HAP with high affinity and chemical specificity. NMR estimates of the calcium-binding affinity indicate that circulating Vn is calcium-bound in vivo. The calcium binding site maps to the top of the Vn-HX propeller, where four Asp generate a highly focused electronegative potential above the channel opening. Calcium is unlikely to be occluded inside the channel, in line with computational energy estimates of the channel’s preferred selectivity for sodium, although the precise molecular basis of calcium binding will have to be verified. Importantly, the same site is involved in binding both ionic calcium and HAP, and ionic calcium cooperatively enhances the affinity of Vn for HAP.
The ability to bind calcium is a newly recognized property of Vn, which has important consequences for its roles in abnormal deposit formation as well as its multiple normal functions in blood, most notably, as a regulator of osteoclast bone resorption (15). These and other functions of Vn have been incompletely understood, owing to the lack of structural information. The structure of the HX domain provides a useful molecular platform for rectifying this knowledge gap, and the present results offer insights about the potential roles of Vn in the formation of calcified protein-lipid deposits (Fig. 4).
Fig. 4.
Models for the roles of Vn in calcified deposit formation. Vn (cartoon representation) is colored by repeat units: HX1 (blue), HX2 (green), HX3 (yellow), and HX4 (red). Rim-Asp, peripheral acidic residues and lipid acyl tails are shown as sticks (yellow). Calcium (green), chloride (aqua), sodium (slate), oxygen (red), and phosphorus (orange) are shown as spheres. (A) Alignment of the propeller top surface with the calcium ions in the HAP (001) lattice (dashed lines). Red circles mark HAP calcium ion positions that coincide with the rim-Asp and other peripheral acidic residues. (B) Circulating Vn may catalyze nucleation of calcium-phosphate ion clusters. (C and D) Exchange of ionic calcium between Vn and the phospholipid headgroups of a lipid droplet (yellow) or the surface of HAP (pink).
How might ionic calcium mediate the interaction of Vn with HAP? The properties of HAP are relevant to this question. At neutral pH, HAP is the most thermodynamically stable form of calcium phosphate, but in vivo, the HAP surface is highly dynamic and its composition and structure are constantly fluctuating in response to physicochemical conditions (44–47). Similarly, in vitro, the surface of nanocrystalline HAP undergoes continuous exchange of calcium and phosphate with its environment and is substantially less ordered than the crystalline lattice found in the core (48). Moreover, recent studies highlight the role of protein-based macromolecular crowding in the nucleation of phase-separated calcium-phosphate ion clusters at a templating surface (44–47). The diffuse immunofluorescence observed for Vn and calcium-phosphate around retinal spherules (10) is visually suggestive of such an amorphous prenucleation phase.
The carboxylate groups of the rim-Asp, which drive the association of ionic calcium with Vn, may also provide a template for ion cluster nucleation and HAP mineralization (Fig. 4 A and B). Their positions on the Vn propeller surface, as well as those of four additional Asp and Glu (E154, E203, D252, D442) that surround them, match the interatomic spacing of calcium ions in the HAP 001 lattice (Fig. 4A), suggesting that they may play both templating and surface-anchoring roles that protect mineralized HAP from dissolution. In the presence of lipid droplets, the calcium-mediated interaction of Vn with lipid phosphate groups is likely to be important (Fig. 4C). The affinity of phospholipids for calcium is well known (49), and phospholipids have been shown to nucleate calcium-phosphate clusters on membrane surfaces (50). Lipid phosphate groups may thus be expected to provide a template for Vn-mediated epitaxial mineralization of HAP on the surface of lipid droplets. Finally, we note that our estimate for the calcium binding affinity of Vn is sufficiently high to maintain circulating Vn in a calcium-bound state, yet sufficiently low for exchange of Vn-bound calcium with the surface of HAP or lipid droplets (Fig. 4D). Such calcium exchange interactions could promote the accumulation of a Vn surface layer that regulates HAP crystal growth and stabilizes it against dissolution. This also provides an explanation for the enhanced affinity of Vn for HAP observed in the presence of ionic calcium (Fig. 3A) and may be related to the normal function of Vn in bone remodeling.
In the context of calcified deposit formation, we propose that Vn may act as a buffer for ionic calcium in blood, a coordinator of calcium-phosphate deposition and mineralization on the surface of lipid droplets, and a regulator of HAP crystal growth. The present data provide a platform for addressing the role of Vn in the formation of ectopic deposits that are associated with age-related macular degeneration and other degenerative pathologies, as well as understanding the consequences of calcium binding for the normal functions of Vn in blood.
Materials and Methods
Protein Preparation.
Vn-HX was prepared from Escherichia coli as described (19). All buffer solutions were prepared with Milli-Q deionized water. For calcium-free preparations, the protein was folded by dropwise dilution from buffer T1 (20 mM Tris⋅HCl pH 8, 6 M guanidine, 10 mM dithiothreitol) into buffer T2 (20 mM Tris⋅HCl pH 8, 500 mM ArgCl, 300 mM NaCl, 5 mM β-mercaptoethanol, 1 mM hydroxyethyldisulfide), followed by dialysis into buffer M1 (20 mM MES, pH 6.5, 300 mM NaCl) and size exclusion chromatography (Superdex 200 10/300 GL, GE Healthcare). Calcium-containing samples were prepared by supplementing buffer M1 with CaCl2.
Uniformly 15N/13C-labeled protein for NMR studies was prepared by growing bacteria on minimal M9 medium containing (15NH4)2SO4 and 13C-glucose (Cambridge Isotopes). The samples for solution NMR contained 64–90 µM Vn-HX in buffer M1, with 5% D2O, in a volume of 160 μL. The samples were supplemented with 3–4.5 mM 1,2-diheptanoyl-sn-glycero-3-phosphocholine (1:50 protein-to-lipid ratio).
HAP-associated Vn for solid-state NMR was prepared by gently mixing 850 µL of 192 µM Vn-HX in buffer M1 with 200 mg of HAP nano-powder (50 nm diameter, 80 m2/g surface area; Sigma-Aldrich catalog no. 900194), removing the supernatant by centrifugation, washing the sediment with 20 mM MES pH 6.5, and loading the sediment into the 36 µL of volume of a 4-mm MAS rotor.
Liposomes reconstituted with the outer membrane protein Ail were prepared with a 7.5/2.5 molar ratio of the lipids dimyristoyl-phosphatidyl-choline and dimyristoyl-phosphatidyl-glycerol (Avanti), as described (51). Liposome-associated Vn for solid-state NMR studies was prepared by mixing 2.5 mL of 179 µM Vn-HX with 2 mL of 143 µM Ail liposomes in buffer P1 (20 mM NaH2PO4, pH 6.5, 300 mM NaCl). The mixture was dialyzed overnight at 4 °C against buffer P2 (10 mM NaH2PO4, pH 6.5) supplemented with 150 mM NaCl, followed by stepwise dialysis into buffer P2 with 100, 50, and 0 mM NaCl. The resulting liposomes were sedimented by centrifugation (326,000 × g, 16 h, 4 °C) and transferred to the MAS rotor.
DSF Experiments.
Protein melting experiments were performed in a 96-well plate format, using a LightCycler 480 instrument (Roche), with a linear temperature gradient of 0.03 °C/sec from 20 °C to 95 °C. Fluorescence intensity was measured at 0.3 °C intervals, with excitation and emission wavelengths of 465 nm and 580 nm. Melting curves were analyzed using Roche Protein Melt software. Samples for analysis contained 1.2 µM protein mixed with a 500-fold dilution of the dye SYPRO Orange (5000× in DMSO; Invitrogen). Additives were mixed in to maintain constant composition and volume (15 µL) of buffer M1. Experiments with solid HAP and SiO2 were performed by adding 840 µg of either solid directly into the plate wells.
Cosedimentation Assays.
HAP (2.5 µm diameter, 100 m2/g surface area; Sigma-Aldrich catalog no. 900195) and SiO2 gel (60 nm diameter; Sigma-Aldrich catalog no. 288594) were washed three times by suspension in buffer M1 and centrifugation (5,000 × g, 5 min, 4 °C). Sedimentation of Vn-HX was tested by gently mixing 200 µL of 10 µM Vn-HX in buffer M1 with 10 mg of HAP or SiO2, then separating the solid from the supernatant by centrifugation (5,000 × g, 5 min, 4 °C), washing the solid with 200 µL of fresh buffer, and collecting it by centrifugation. Protein in the centrifuge fractions was analyzed by Coomassie-stained SDS/PAGE. Buffer M1 was supplemented with CaCl2 or sodium citrate to examine the effects of these compounds on Vn-HX sedimentation.
NMR Spectroscopy.
Solution NMR experiments were performed at 30 °C, on a Bruker Avance 600 MHz spectrometer, equipped with a 1H/13C/15N triple-resonance cryoprobe. The NMR data were processed using TopSpin (Bruker) and analyzed using CCPN 2.4.2 (52). The 1HN, 15N, 13CO, 13CA and 13CB chemical shifts were assigned using 2D 1H/15N heteronuclear single quantum coherence (HSQC) experiments, and 3D 1H/15N/13C HNCA, HNCO, CBCACONH experiments (53). The assigned chemical shifts have been deposited in the BMRB database (ID codes 50241 and 50261).
Solid-state NMR studies were performed at 5 °C, on a Bruker Avance 500 MHz spectrometer equipped with a Bruker 1H/15N/13C, 4 mm, Efree MAS probe, operating at a spinning rate of 10 kHz ± 5 Hz. Typical π/2 pulse lengths for 1H, 13C, and 15N were 2.5 μs, 2.5 μs, and 5 μs. SPINAL64 (54), implemented with a 90-kHz RF field strength, was used for 1H decoupling during data acquisition. The 1D 13C and 15N spectra were acquired using either dipolar-based CP (55) or J-coupling–based polarization transfer with the INEPT sequence (56). The respective delays in the INEPT and refocused INEPT blocks were set to 1/4 × J (1.7 ms) and 1/6 × J (1.1 ms), to obtain positive phase carbon signals. The 2D heteronuclear 13C/15N correlation spectra were acquired using SPECIFIC-CP (57) for band selective polarization transfer, during which continuous wave 1H decoupling (100 kHz RF field strength) was applied, and a tangent ramp was applied on the 15N channel with contact times of 5 ms for N-CA transfer.
Binding Energy Calculations.
Molecules A and B, in the asymmetric unit of the crystal structure of Vn-HX (Protein Data Bank ID code 6O5E) were each prepared using PBD Reader in CHARMM-GUI (58), with bound water at the O1 site above the channel, and bound water (molecule A) or bound sulfate (molecule B) at the bottom, O2 site (Fig. 2). While one can, in principle, use the molecular dynamics free energy perturbation approach (35) to calculate the binding energy differences of metal ions in the two sites, m1 and m2 (Fig. 2), such calculations would inevitably have large errors associated with the large solvation energy difference between Na+ (∼98 kcal/mol) and Ca2+ (∼380 kcal/mol). To avoid this problem, we used the molecular mechanics and Poisson–Boltzmann continuum electrostatics approach to calculate the binding energies of Na+ and Ca2+ in m1 and m2. For each protein molecule, A and B, and each of four combinations of Na+ and Ca2+ in the m1 and m2 sites, we calculated the electrostatic solvation energies of Vn-HX with m1 and m2 occupied [EELEC(Vn,m1m2)], with m1 and m2 unoccupied [EELEC(Vn)], with m1 only occupied [EELEC(Vn,m1)], and with m2 only occupied [EELEC(Vn,m2)], using the PDBEQ module (36) in CHARMM (59) and the PDBEQ Solver function (60) in CHARMM-GUI. The protein, and water dielectric constants were set to 1 and 80, respectively, and a grid spacing of 0.25 Å was used for finite-difference Poisson–Boltzmann calculations. The electrostatic contribution to the binding energy was calculated as: ΔEELEC = EELEC(Vn,m1m2) − EELEC(Vn) − EELEC(Vn,m1) − EELEC(Vn,m2). The van der Waals energy contribution was similarly calculated.
Data and Materials Availability.
All data needed to evaluate the conclusions are present in the paper and/or SI Appendix. NMR chemical shifts have been deposited in Biological Magnetic Resonance Data Bank, http://www.bmrb.wisc.edu (ID codes 50241 and 50261).
Supplementary Material
Acknowledgments
We thank Bernhard Lechtenberg and Ian Pass for useful discussion. This study was supported by NIH Grant GM 118186 (to F.M.M.) and from a Canadian Institutes of Health Research Postdoctoral Fellowship (to K.S.). It utilized the Cancer Center Structural Biology Resource supported by Grant P30 CA030199.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: NMR data have been deposited in Biological Magnetic Resonance Data Bank (ID codes 50241 and 50261).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2007699117/-/DCSupplemental.
References
- 1.Bergen A. A. et al., On the origin of proteins in human drusen: The meet, greet and stick hypothesis. Prog. Retin. Eye Res. 70, 55–84 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Curcio C. A., Soft drusen in age-related macular degeneration: Biology and targeting via the oil spill strategies. Invest. Ophthalmol. Vis. Sci. 59, AMD160-AMD181 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hageman G. S. et al., An integrated hypothesis that considers drusen as biomarkers of immune-mediated processes at the RPE-Bruch’s membrane interface in aging and age-related macular degeneration. Prog. Retin. Eye Res. 20, 705–732 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Khan K. N. et al., Differentiating drusen: Drusen and drusen-like appearances associated with ageing, age-related macular degeneration, inherited eye disease and other pathological processes. Prog. Retin. Eye Res. 53, 70–106 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Csincsik L. et al., Peripheral retinal imaging biomarkers for Alzheimer’s disease: A pilot study. Ophthalmic Res. 59, 182–192 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mullins R. F., Russell S. R., Anderson D. H., Hageman G. S., Drusen associated with aging and age-related macular degeneration contain proteins common to extracellular deposits associated with atherosclerosis, elastosis, amyloidosis, and dense deposit disease. FASEB J. 14, 835–846 (2000). [PubMed] [Google Scholar]
- 7.Shin T. M. et al., Formation of soluble amyloid oligomers and amyloid fibrils by the multifunctional protein vitronectin. Mol. Neurodegener. 3, 16 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hageman G. S., Mullins R. F., Russell S. R., Johnson L. V., Anderson D. H., Vitronectin is a constituent of ocular drusen and the vitronectin gene is expressed in human retinal pigmented epithelial cells. FASEB J. 13, 477–484 (1999). [DOI] [PubMed] [Google Scholar]
- 9.Johnson L. V. et al., Cell culture model that mimics drusen formation and triggers complement activation associated with age-related macular degeneration. Proc. Natl. Acad. Sci. U.S.A. 108, 18277–18282 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thompson R. B. et al., Identification of hydroxyapatite spherules provides new insight into subretinal pigment epithelial deposit formation in the aging eye. Proc. Natl. Acad. Sci. U.S.A. 112, 1565–1570 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan A. C. S. et al., Calcified nodules in retinal drusen are associated with disease progression in age-related macular degeneration. Sci. Transl. Med. 10, eaat4544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomasini B. R., Mosher D. F., Vitronectin. Prog. Hemost. Thromb. 10, 269–305 (1991). [PubMed] [Google Scholar]
- 13.Preissner K. T., Reuning U., Vitronectin in vascular context: Facets of a multitalented matricellular protein. Semin. Thromb. Hemost. 37, 408–424 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Leavesley D. I. et al., Vitronectin–Master controller or micromanager? IUBMB Life 65, 807–818 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Chambers T. J., Fuller K., How are osteoclasts induced to resorb bone? Ann. N. Y. Acad. Sci. 1240, 1–6 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Yoneda A., Ogawa H., Kojima K., Matsumoto I., Characterization of the ligand binding activities of vitronectin: Interaction of vitronectin with lipids and identification of the binding domains for various ligands using recombinant domains. Biochemistry 37, 6351–6360 (1998). [DOI] [PubMed] [Google Scholar]
- 17.Vaisar T. et al., Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL. J. Clin. Invest. 117, 746–756 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh B., Su Y. C., Riesbeck K., Vitronectin in bacterial pathogenesis: A host protein used in complement escape and cellular invasion. Mol. Microbiol. 78, 545–560 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Shin K. et al., Structure of human Vitronectin C-terminal domain and interaction with Yersinia pestis outer membrane protein Ail. Sci Adv. 5, eaax5068 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Conlan M. G., Tomasini B. R., Schultz R. L., Mosher D. F., Plasma vitronectin polymorphism in normal subjects and patients with disseminated intravascular coagulation. Blood 72, 185–190 (1988). [PubMed] [Google Scholar]
- 21.Kubota K., Katayama S., Matsuda M., Hayashi M., Three types of vitronectin in human blood. Cell Struct. Funct. 13, 123–128 (1988). [DOI] [PubMed] [Google Scholar]
- 22.Seiffert D., Ciambrone G., Wagner N. V., Binder B. R., Loskutoff D. J., The somatomedin B domain of vitronectin. Structural requirements for the binding and stabilization of active type 1 plasminogen activator inhibitor. J. Biol. Chem. 269, 2659–2666 (1994). [PubMed] [Google Scholar]
- 23.Zhou A., Huntington J. A., Pannu N. S., Carrell R. W., Read R. J., How vitronectin binds PAI-1 to modulate fibrinolysis and cell migration. Nat. Struct. Biol. 10, 541–544 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Pytela R., Pierschbacher M. D., Ruoslahti E., A 125/115-kDa cell surface receptor specific for vitronectin interacts with the arginine-glycine-aspartic acid adhesion sequence derived from fibronectin. Proc. Natl. Acad. Sci. U.S.A. 82, 5766–5770 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piccard H., Van den Steen P. E., Opdenakker G., Hemopexin domains as multifunctional liganding modules in matrix metalloproteinases and other proteins. J. Leukoc. Biol. 81, 870–892 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Zheng H. et al., CheckMyMetal: A macromolecular metal-binding validation tool. Acta Crystallogr. D Struct. Biol. 73, 223–233 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carafoli E., Krebs J., Why calcium? How calcium became the best communicator. J. Biol. Chem. 291, 20849–20857 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boskey A. L., Roy R., Cell culture systems for studies of bone and tooth mineralization. Chem. Rev. 108, 4716–4733 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu Y. Y., Rawal A., Schmidt-Rohr K., Strongly bound citrate stabilizes the apatite nanocrystals in bone. Proc. Natl. Acad. Sci. U.S.A. 107, 22425–22429 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoang Q. Q., Sicheri F., Howard A. J., Yang D. S., Bone recognition mechanism of porcine osteocalcin from crystal structure. Nature 425, 977–980 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Scudeller L. A. et al., Orientation and conformation of osteocalcin adsorbed onto calcium phosphate and silica surfaces. Biointerphases 12, 02D411 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mysliwy J., Dingley A. J., Sedlacek R., Grötzinger J., Structural characterization and binding properties of the hemopexin-like domain of the matrixmetalloproteinase-19. Protein Expr. Purif. 46, 406–413 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Flinn J. M., Kakalec P., Tappero R., Jones B., Lengyel I., Correlations in distribution and concentration of calcium, copper and iron with zinc in isolated extracellular deposits associated with age-related macular degeneration. Metallomics 6, 1223–1228 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Cha H., Kopetzki E., Huber R., Lanzendörfer M., Brandstetter H., Structural basis of the adaptive molecular recognition by MMP9. J. Mol. Biol. 320, 1065–1079 (2002). [DOI] [PubMed] [Google Scholar]
- 35.Noskov S. Y., Bernèche S., Roux B., Control of ion selectivity in potassium channels by electrostatic and dynamic properties of carbonyl ligands. Nature 431, 830–834 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Im W., Beglov D., Roux B., Continuum solvation model: Computation of electrostatic forces from numerical solutions to the Poisson-Boltzmann equation. Comput. Phys. Commun. 111, 59–75 (1998). [Google Scholar]
- 37.Roux B., Bernèche S., Im W., Ion channels, permeation, and electrostatics: Insight into the function of KcsA. Biochemistry 39, 13295–13306 (2000). [DOI] [PubMed] [Google Scholar]
- 38.Cummings L. J., Snyder M. A., Brisack K., Protein chromatography on hydroxyapatite columns. Methods Enzymol. 463, 387–404 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Arya S. et al., Quantitative analysis of hydroxyapatite-binding plasma proteins in genotyped individuals with late-stage age-related macular degeneration. Exp. Eye Res. 172, 21–29 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Hayman E. G., Pierschbacher M. D., Ohgren Y., Ruoslahti E., Serum spreading factor (vitronectin) is present at the cell surface and in tissues. Proc. Natl. Acad. Sci. U.S.A. 80, 4003–4007 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marassi F. M., Ramamoorthy A., Opella S. J., Complete resolution of the solid-state NMR spectrum of a uniformly 15N-labeled membrane protein in phospholipid bilayers. Proc. Natl. Acad. Sci. U.S.A. 94, 8551–8556 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bertini I. et al., Solid-state NMR of proteins sedimented by ultracentrifugation. Proc. Natl. Acad. Sci. U.S.A. 108, 10396–10399 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bartra S. S. et al., Yersinia pestis uses the Ail outer membrane protein to recruit vitronectin. Microbiology 161, 2174–2183 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dorozhkin S. V., Epple M., Biological and medical significance of calcium phosphates. Angew. Chem. Int. Ed. Engl. 41, 3130–3146 (2002). [DOI] [PubMed] [Google Scholar]
- 45.Gower L. B., Biomimetic model systems for investigating the amorphous precursor pathway and its role in biomineralization. Chem. Rev. 108, 4551–4627 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Uskoković V., The role of hydroxyl channel in defining selected physicochemical peculiarities exhibited by hydroxyapatite. RSC Adv. 5, 36614–36633 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rao A., Cölfen H., Mineralization and non-ideality: On nature’s foundry. Biophys. Rev. 8, 309–329 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jäger C., Welzel T., Meyer-Zaika W., Epple M., A solid-state NMR investigation of the structure of nanocrystalline hydroxyapatite. Magn. Reson. Chem. 44, 573–580 (2006). [DOI] [PubMed] [Google Scholar]
- 49.McLaughlin S., Mulrine N., Gresalfi T., Vaio G., McLaughlin A., Adsorption of divalent cations to bilayer membranes containing phosphatidylserine. J. Gen. Physiol. 77, 445–473 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dey A. et al., The role of prenucleation clusters in surface-induced calcium phosphate crystallization. Nat. Mater. 9, 1010–1014 (2010). [DOI] [PubMed] [Google Scholar]
- 51.Ding Y., Fujimoto L. M., Yao Y., Marassi F. M., Solid-state NMR of the Yersinia pestis outer membrane protein Ail in lipid bilayer nanodiscs sedimented by ultracentrifugation. J. Biomol. NMR 61, 275–286 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vranken W. F. et al., The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins 59, 687–696 (2005). [DOI] [PubMed] [Google Scholar]
- 53.Cavanagh J., Fairbrother W. J., Palmer A. G., Skelton N. J., Protein NMR Spectroscopy: Principles and Practice, Cavanagh J., Fairbrother W. J., Palmer A. G., Rance M., Skelton N. J., Eds. (Academic Press, Burlington, ed. 2, 2007). [Google Scholar]
- 54.Fung B. M., Khitrin A. K., Ermolaev K., An improved broadband decoupling sequence for liquid crystals and solids. J. Magn. Reson. 142, 97–101 (2000). [DOI] [PubMed] [Google Scholar]
- 55.Pausak S., Pines A., Gibby M. G., Waugh J. S., Carbon-13 shielding tensors in single crystal durene. J. Chem. Phys. 58, 591–595 (1973). [Google Scholar]
- 56.Morris G. A., Freeman R., Enhancement of nuclear magnetic resonance signals by polarization transfer. J. Am. Chem. Soc. 101, 760–762 (1979). [Google Scholar]
- 57.Baldus M., Petkova A., Herzfield J., Griffin R., Cross polarization in the tilted frame: Assignment and spectral simplification in heteronuclear spin systems. Mol. Phys. 95, 1197–1207 (1998). [Google Scholar]
- 58.Jo S., Kim T., Iyer V. G., Im W., CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 (2008). [DOI] [PubMed] [Google Scholar]
- 59.Brooks B. R. et al., CHARMM: The biomolecular simulation program. J. Comput. Chem. 30, 1545–1614 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jo S., Vargyas M., Vasko-Szedlar J., Roux B., Im W., PBEQ-Solver for online visualization of electrostatic potential of biomolecules. Nucleic Acids Res. 36, W270–W275 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.