Significance
The mechanism of eukaryotic transcription-coupled nucleotide excision repair (TC-NER) is poorly understood. Here, we studied the genome-wide role of yeast Rad26, a homolog of the human Cockayne syndrome group B (CSB) protein, in the repair of UV-induced DNA damage. We found that Rad26 is not uniformly required for TC-NER in transcribed chromatin. Instead, nucleosome organization and the distribution of transcription regulators such as TFIIH and Spt4/Spt5 significantly modulate the requirement for Rad26 in TC-NER. Importantly, TC-NER is conserved from yeast to humans. Our data provide insights into the mechanism of TC-NER in eukaryotes.
Keywords: DNA repair, nucleosome, TFIIH, Spt4/Spt5, Cockayne syndrome
Abstract
Transcription-coupled nucleotide excision repair (TC-NER) is an important DNA repair mechanism that removes RNA polymerase (RNAP)-stalling DNA damage from the transcribed strand (TS) of active genes. TC-NER deficiency in humans is associated with the severe neurological disorder Cockayne syndrome. Initiation of TC-NER is mediated by specific factors such as the human Cockayne syndrome group B (CSB) protein or its yeast homolog Rad26. However, the genome-wide role of CSB/Rad26 in TC-NER, particularly in the context of the chromatin organization, is unclear. Here, we used single-nucleotide resolution UV damage mapping data to show that Rad26 and its ATPase activity is critical for TC-NER downstream of the first (+1) nucleosome in gene coding regions. However, TC-NER on the transcription start site (TSS)-proximal half of the +1 nucleosome is largely independent of Rad26, likely due to high occupancy of the transcription initiation/repair factor TFIIH in this nucleosome. Downstream of the +1 nucleosome, the combination of low TFIIH occupancy and high occupancy of the transcription elongation factor Spt4/Spt5 suppresses TC-NER in Rad26-deficient cells. We show that deletion of SPT4 significantly restores TC-NER across the genome in a rad26∆ mutant, particularly in the downstream nucleosomes. These data demonstrate that the requirement for Rad26 in TC-NER is modulated by the distribution of TFIIH and Spt4/Spt5 in transcribed chromatin and Rad26 mainly functions downstream of the +1 nucleosome to remove TC-NER suppression by Spt4/Spt5.
Nucleotide excision repair (NER) is critical for the repair of cytotoxic and mutagenic DNA lesions, including ultraviolet (UV)-induced cyclobutane pyrimidine dimers (CPDs) and (6, 4) photoproducts (6-4PPs). NER consists of two subpathways: global genomic nucleotide excision repair (GG-NER) and transcription-coupled nucleotide excision repair (TC-NER), which mainly differ in how the lesion is recognized (1). CPDs located in the transcribed strand (TS) of active genes are rapidly removed by TC-NER, initiated when elongating RNA polymerase (RNAP) stalls at a CPD lesion (2). CPDs located elsewhere in the genome (e.g., the nontranscribed strand [NTS] of genes or intergenic regions) are recognized by damage recognition factors (e.g., UV-DDB, XPC) and repaired by GG-NER, which is generally less efficient than TC-NER (2). Due to the different efficiency between the two NER subpathways, transcribed genes exhibit strong strand asymmetry in the repair of CPDs, with preferential repair in the TS relative to the NTS (3), which is mirrored by the reported strand-biased somatic mutations in skin cancers (4).
The initiation of TC-NER upon RNAP stalling requires specific factors. These factors respond rapidly to transcription-blocking DNA damage, binding to stalled RNAP to coordinate assembly of downstream NER factors (5). Several TC-NER factors have been identified, including bacterial Mfd (6), yeast Rad26 (7), and mammalian CSB (8). The bacterial Mfd protein functions as a DNA translocase and can displace RNAP stalled at a lesion (9, 10). Mfd then recruits general NER factors such as the UvrAB complex to repair the exposed damage (10, 11). Although the function of Mfd is well characterized, the mechanism for eukaryotic TC-NER factors (e.g., Rad26 and CSB) is less understood (5). Both CSB and Rad26 are ATP-dependent DNA translocases and can promote RNA polymerase II (Pol II) forward movement (12, 13), similar to Mfd; however, neither of them can displace stalled Pol II from DNA in vitro (12, 14). This raises important questions about whether the forward translocation of Pol II promoted by CSB/Rad26 is required for lesion recognition and TC-NER initiation, and how eukaryotic cells deal with the stalled Pol II to avoid repair inhibition.
The mechanism of CSB/Rad26 is further complicated by the packaging of genomic DNA into chromatin, which may influence TC-NER in different ways. For example, nucleosomes around stalled Pol II may restrict the access of repair factors and/or Pol II backtracking (2), which may require chromatin remodeling to overcome this physical barrier. Additionally, CSB/Rad26 and some downstream NER factors play important roles in transcription regulation (12, 13, 15), and their distribution may be affected by chromatin. In this regard, recent studies have shown that nucleosomes significantly modulate distribution of the transcription initiation/NER factor TFIIH, which exhibits highest occupancy near the first (+1) nucleosome in the coding region (16, 17). Furthermore, occupancy of the transcription elongation factor Spt4/Spt5, a known TC-NER suppressor in yeast (18, 19), also fluctuates along the transcribed gene (20). Hence, the availability of these transcription/repair factors varies significantly in different chromatin regions. However, how the variable occupancy of these factors affects TC-NER has not been characterized.
High-resolution methods for mapping CPD repair across the genome (3, 21) provide a unique opportunity to elucidate how CSB and Rad26 promote TC-NER in chromatin. Here, we employed a high-throughput CPD mapping method known as CPD-seq (21) to generate base-resolution repair maps among ∼5,200 Pol II-transcribed yeast genes (22) in WT and Rad26-deficient yeast strains. Our data reveals a varying requirement for Rad26 in activating TC-NER in different chromatin regions. We further show that the differential requirement for Rad26 is correlated with the distribution of TFIIH and Spt4/Spt5 along the transcribed gene.
Results
TC-NER Is Generally Impaired in Rad26-Deficient Yeast Strains across the Genome.
We recently developed CPD-seq for precise mapping of CPD lesions (21). CPD-seq employs DNA repair enzymes T4 endonuclease V (T4 endo V) and apurinic/apyrimidinic endonuclease (APE1) to generate a new ligatable 3′-OH group immediately upstream of the CPD lesion, which is ligated to an adaptor DNA for next-generation sequencing (SI Appendix, Fig. S1A).
To investigate the genome-wide role of Rad26 in TC-NER, we conducted CPD-seq experiments in Rad26-proficient (i.e., WT) and Rad26-deficient (i.e., rad26∆) Saccharomyces cerevisiae strains. Counting the two nucleotides immediately upstream of the 5′ end of CPD-seq reads revealed a high enrichment of dipyrimidines (i.e., TT, TC, CT, and CC dinucleotides) in UV-irradiated cells, but not in “No UV” control (SI Appendix, Fig. S1 B and C), which is indicative of specific mapping of CPD lesions, as shown in our previous studies (21, 23). We used CPD-seq data to analyze the distribution of CPDs both immediately after UV irradiation (0 h) and following 2 h of repair incubation across ∼5,200 Pol II-transcribed yeast genes (22). After normalizing CPD-seq counts at 2 h by the initial counts at 0 h, we found that there were significantly lower remaining CPDs (or unrepaired CPDs) in the TS relative to the nontranscribed strand (NTS) in WT cells (Fig. 1A), consistent with previous results showing more rapid repair on the TS by TC-NER (3, 21). In contrast, TC-NER was significantly reduced in a RAD26 full deletion yeast strain (i.e., rad26∆), as shown by high remaining CPDs in the TS in the mutant strain (Fig. 1B). Residual TC-NER was still seen in the rad26∆ mutant (Fig. 1B), suggesting the existence of Rad26-independent TC-NER in some genes (24).
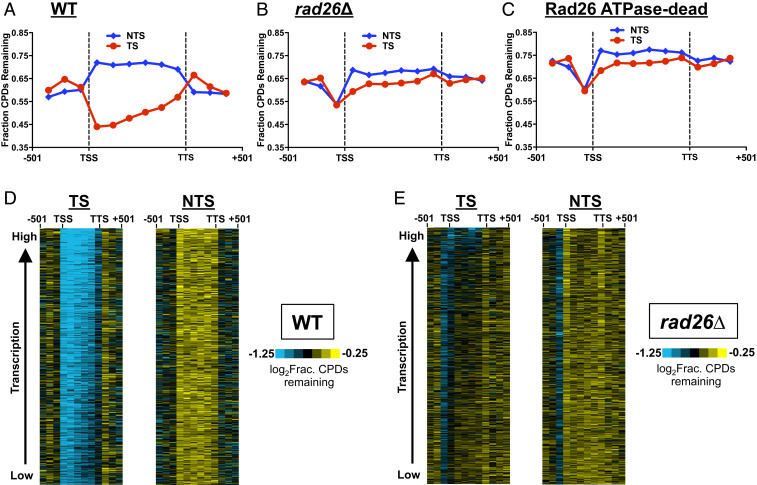
Fig. 1.
Rad26 is generally required for TC-NER in the yeast genome. (A) CPD repair is faster in the TS relative to the NTS in WT cells. The coding region of each gene (n = 5,205) was separated into six equal-sized bins. Three additional bins in the promoter and terminator were also presented. The number of CPD-seq reads at 2 h was counted in each bin and normalized by CPD-seq reads at 0 h. The resulting fraction of remaining CPDs was plotted. Repair in the flanking sequences (e.g., upstream of TSS and downstream of TTS) appears to be faster than repair in the NTS of the coding region, likely because many of these flanking sequences overlap with the transcribed regions of neighboring genes and, thus, are partially repaired by TC-NER. (B) Deletion of RAD26 significantly reduces repair in the TS. CPD-seq data in the rad26∆ mutant was analyzed. (C) The ATPase activity of Rad26 is essential for TC-NER. Same as A, but CPD-seq data in the Rad26 ATPase-dead mutant was analyzed. (D) Gene cluster plot of fraction of remaining CPDs in the TS (Left) and NTS (Right) in WT cells. Each column represents one bin, from promoter to terminator of each gene, and rows indicate fraction of remaining CPDs at 2 h. Median levels of CPDs are depicted in black. Blue and yellow indicate low and high levels of CPDs, respectively (see color bar). Rows were sorted by gene transcription frequency (28), from lowest to highest. (E) Same as in D, except analysis of rad26∆ CPD-seq data.
Both Rad26 and CSB are ATP-dependent DNA translocases that enhance Pol II forward translocation (12, 13); however, the importance of the translocase activity in repair is not fully understood (5). To investigate the genome-wide function of the Rad26 translocase activity in TC-NER, we used CRISPR-Cas9 (25) to generate a Rad26 ATPase-dead strain by mutating the catalytic lysine residue to an arginine (i.e., Rad26-K328R) in its chromosomal location. Previous studies have shown that the K328R mutation abolishes the DNA translocase activity of Rad26 in promoting Pol II transcription over DNA barriers (12). Additionally, mutations in the ATPase domain do not affect cellular Rad26 protein stability (26). Analysis of CPD-seq data revealed generally impaired TC-NER in the Rad26 ATPase-dead strain (Fig. 1C), similar to rad26∆. Hence, our data indicates that the ATP-dependent DNA translocase activity of Rad26 is critical for TC-NER across the yeast genome.
Although Rad26-independent TC-NER has been reported in specific genes (24, 27), it remains unclear to what extent Rad26 is required for TC-NER at the genome scale. To address this question, we sorted yeast genes by transcription frequency (28) and generated gene cluster plots of remaining CPDs following 2 h repair. As expected, the remaining CPD levels were generally lower in the TS (Fig. 1 D, Left) relative to the NTS (Fig. 1 D, Right) in WT cells, regardless of gene expression levels, suggesting that the TS is rapidly repaired by TC-NER. RAD26 deletion significantly weakened TC-NER in many genes, resulting in comparable remaining CPDs between the TS and NTS (Fig. 1E), consistent with our data shown in Fig. 1B. Faster repair in the TS relative to the NTS was observed in a small number of highly transcribed genes in the rad26∆ mutant (see the top part of Fig. 1 E, Left), suggesting that Rad26-independent TC-NER is mainly enriched in a subset of actively transcribed genes. Further pathway analysis of these Rad26-independent genes revealed that they are involved in various cellular processes such as protein folding, glycolysis, and ribosomal functions (SI Appendix, Table S1).
Taken together, these data indicate that Rad26 is generally required for TC-NER in most yeast genes, and Rad26-independent TC-NER mainly occurs in a few highly transcribed genes.
Rad26 Is Specifically Required for TC-NER in Transcribed Regions Downstream of the +1 Nucleosome.
We next sought to understand how nucleosomes affect the requirement for Rad26 in TC-NER. Previous studies have shown that transcribed genes in eukaryotes are organized into regularly spaced nucleosomes downstream of the transcription start site (TSS), starting from the +1 nucleosome, which is nearest the TSS, followed by the +2, +3, +4 nucleosomes and so on ref. 29. While the nucleosome organization is known to be important for proper gene transcription, how it impacts the function of Rad26 in TC-NER has not been analyzed.
We aligned the TSS of ∼5,200 genes and analyzed CPD-seq reads at 2 h (normalized to 0 h) at each position from 200 bp upstream to 650 bp downstream of the TSS. Notably, this single-nucleotide resolution analysis revealed faster repair in the TS relative to the NTS in WT cells (Fig. 2A). This analysis also showed a periodic pattern of remaining CPDs in the NTS, but not in the TS (Fig. 2A), which is consistent with our recent report (30). The peaks of remaining CPDs were indicative of positions with slow repair, and they generally overlapped with the central dyad locations of nucleosomes (Fig. 2A, gray background), indicating that GG-NER (i.e., repair of the NTS) is inhibited by nucleosomes at their central dyad positions. In contrast, our data showed no periodic CPD peaks in the TS in WT cells (Fig. 2A), indicating that TC-NER is not inhibited by nucleosomes, presumably due to transient disruption of the nucleosome structure during Pol II elongation (2).
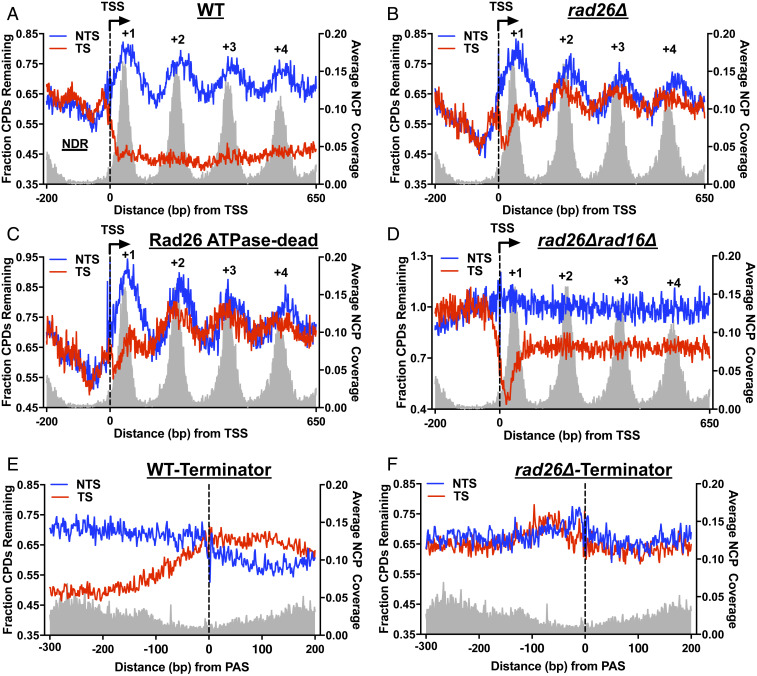
Fig. 2.
Rad26 is specifically required for TC-NER downstream of the +1 nucleosome in the coding region. (A) Nucleosome organization modulates GG-NER but not TC-NER in WT cells. Yeast genes (n = 5,205) were aligned at their TSS (22) and the fraction of remaining CPDs at 2 h was plotted. TS and NTS were analyzed separately. The average nucleosome coverage (right y axis), shown as the gray background, was derived from a published MNase nucleosome map (43). NCP, nucleosome core particle. (B) Rad26 is important for TC-NER in regions downstream of the +1 nucleosome. Same as A, except CPD-seq data were obtained from the rad26∆ mutant. (C) The Rad26 ATPase activity is required for TC-NER downstream of the +1 nucleosome. CPD-seq data in the Rad26 ATPase-dead mutant was analyzed. (D) Deletion of the GG-NER factor Rad16 abolished the periodic repair pattern in both strands. CPD-seq data from the rad16∆rad26∆ double mutant was analyzed. (E) High-resolution repair analysis around the termination site in WT cells. Annotated yeast PAS were obtained from the published study (22). Genes were aligned at the PAS, and the remaining CPDs were analyzed and plotted. Gene is transcribed from left to right. (F) Rad26 is essential for TC-NER in the termination region. Same as E, except CPD-seq data in the rad26∆ mutant were analyzed.
Analysis of CPD-seq data in the rad26∆ mutant revealed periodic CPD peaks in both TS and NTS (Fig. 2B and SI Appendix, Fig. S2A). This indicates that repair in both strands is inhibited by nucleosomes when Rad26 is absent. However, CPD peaks in the TS were only seen in regions downstream of the +1 nucleosome, including the dyad positions of the +2, +3, and +4 nucleosomes (Fig. 2B and SI Appendix, Fig. S2A). Similarly, CPD peaks in the TS were also found in the Rad26 ATPase-dead mutant in the downstream nucleosomes, but not in the +1 nucleosome, following 1 h and 2 h repair (Fig. 2C and SI Appendix, Fig. S2B). Fast repair was observed on both strands in the nucleosome-depleted region (NDR) immediately upstream of the TSS in Rad26-deficient strains (Fig. 2 B and C), which is likely due to increased damage accessibility to GG-NER factors when nucleosomes are depleted. Further analysis of CPDs after 2 h repair in rad26∆ among gene groups with different expression levels (i.e., high, medium, and low) (28) showed similar CPD peaks in the TS, in an expression-independent manner (SI Appendix, Fig. S2 C–E), although highly expressed genes had higher residual TC-NER in the +1 nucleosome.
The periodic peaks of remaining CPDs in the TS in rad26∆ suggest that damage in this strand is repaired in the context of nucleosomes, likely by GG-NER. To test this hypothesis, we conducted CPD-seq in a rad26∆rad16∆ double mutant that is defective in both GG-NER and TC-NER. Deletion of the GG-NER gene RAD16 (31) abolished the periodic CPD pattern in both strands (Fig. 2D), suggesting that damage in the downstream nucleosomes of the TS was indeed repaired by GG-NER in rad26∆ cells. While GG-NER was severely impaired, residual TC-NER was seen in the rad26∆rad16∆ double mutant in the +1 nucleosome as well as the downstream nucleosomes (Fig. 2D). The TC-NER pattern in the double mutant is different from the pattern in the rad26∆ single mutant, which mainly exhibits TC-NER in the +1 nucleosome (Fig. 2B). However, the detailed mechanism for the difference is unclear.
A closer examination of the CPD-seq data in rad26∆ revealed that the residual TC-NER is particularly apparent for damage located within ∼30 bp downstream of the TSS and is detectable for most of the +1 nucleosome (SI Appendix, Fig. S2F). Previous studies showed Rad26-independent TC-NER immediately downstream of the TSS in specific yeast genes (32) and CSB-independent TC-NER near the TSS in a human gene (33). Our genomic data, consistent with these early observations, further demonstrate that the Rad26-independent TC-NER near the TSS is widespread for many genes and is associated with the +1 nucleosome.
Rad26 Is Essential for TC-NER in the Transcription Termination Region.
How TC-NER functions in transcription termination regions has been largely uncharacterized. To examine if there is active TC-NER in the termination region and the role of Rad26 in this region, we analyzed repair at gene 3′ ends in WT and rad26∆ cells. Pol II termination occurs in a short DNA window downstream of the polyadenylation site (PAS) where Pol II is released from DNA (20). We aligned yeast genes at the annotated PAS (22) and analyzed remaining CPDs at 2 h at each position around the PAS (i.e., from −300 bp to +200 bp relative to the PAS). Our data showed apparent TC-NER upstream of the PAS in WT cells (Fig. 2E). TC-NER activity decreased gradually as Pol II approaches the PAS, starting from ∼150 bp upstream of the PAS, and no TC-NER was seen at the PAS (Fig. 2E), likely due to paused Pol II elongation (20). Repair in the NTS was slightly faster than the TS downstream of the PAS (Fig. 2E), which may be caused by TC-NER activity from the adjacent gene. No TC-NER was seen near the PAS in rad26∆ (Fig. 2F and SI Appendix, Fig. S3A) or in the Rad26 ATPase-dead mutant (SI Appendix, Fig. S3B), indicating that Rad26 and its ATPase activity is essential for TC-NER in the termination region.
Rad26 Requirement in TC-NER Is Correlated with TFIIH and Spt4/Spt5 Distribution.
To understand why Rad26 is specifically required for TC-NER downstream of the +1 nucleosome, we analyzed distribution of transcription initiation factor TFIIH and elongation factor Spt4/Spt5 in Pol II-transcribed genes. We focused on TFIIH and Spt4/Spt5 because they both play dual roles in transcription and NER. TFIIH is a general transcription factor that functions in NER by unwinding DNA to facilitate repair incision (15). Spt4/Spt5 is an essential transcription elongation factor that suppresses TC-NER in yeast (18, 19).
We first analyzed published high-resolution ChIP-exonuclease (ChIP-exo) data of the yeast TFIIH subunit Ssl2 (also known as Rad25) (16). Ssl2 is a 3′ to 5′ DNA helicase (homolog of human XPB) and is directly involved in NER (15). Consistent with the high Rad26-independent TC-NER activity on the TSS-proximal half of +1 nucleosome (Fig. 3A), ChIP-exo data revealed high occupancy of Ssl2 in the +1 nucleosome, particularly on the TSS-proximal side (Fig. 3B). Furthermore, Ssl2 occupancy decreased to the basal level in the +2 and the following nucleosomes (Fig. 3B), which was correlated with severely impaired TC-NER from the +2 nucleosome in rad26∆ (Fig. 3A). The TFIIH distribution was further confirmed by analyzing published ChIP-seq data of another two TFIIH subunits, Rad3 and Kin28 (34). Rad3 is a 5′ to 3′ DNA helicase (homolog of human XPD), and Kin28 is a protein kinase that phosphorylates the Ser5 residue of Pol II C-terminal domain (CTD) (15). ChIP-seq analyses showed high occupancy of both TFIIH subunits in the +1 nucleosome but significantly reduced occupancy downstream of the +1 nucleosome (SI Appendix, Fig. S4A), which was similar to the Ssl2 ChIP-exo data, suggesting that the TFIIH complex is enriched in the +1 nucleosome.
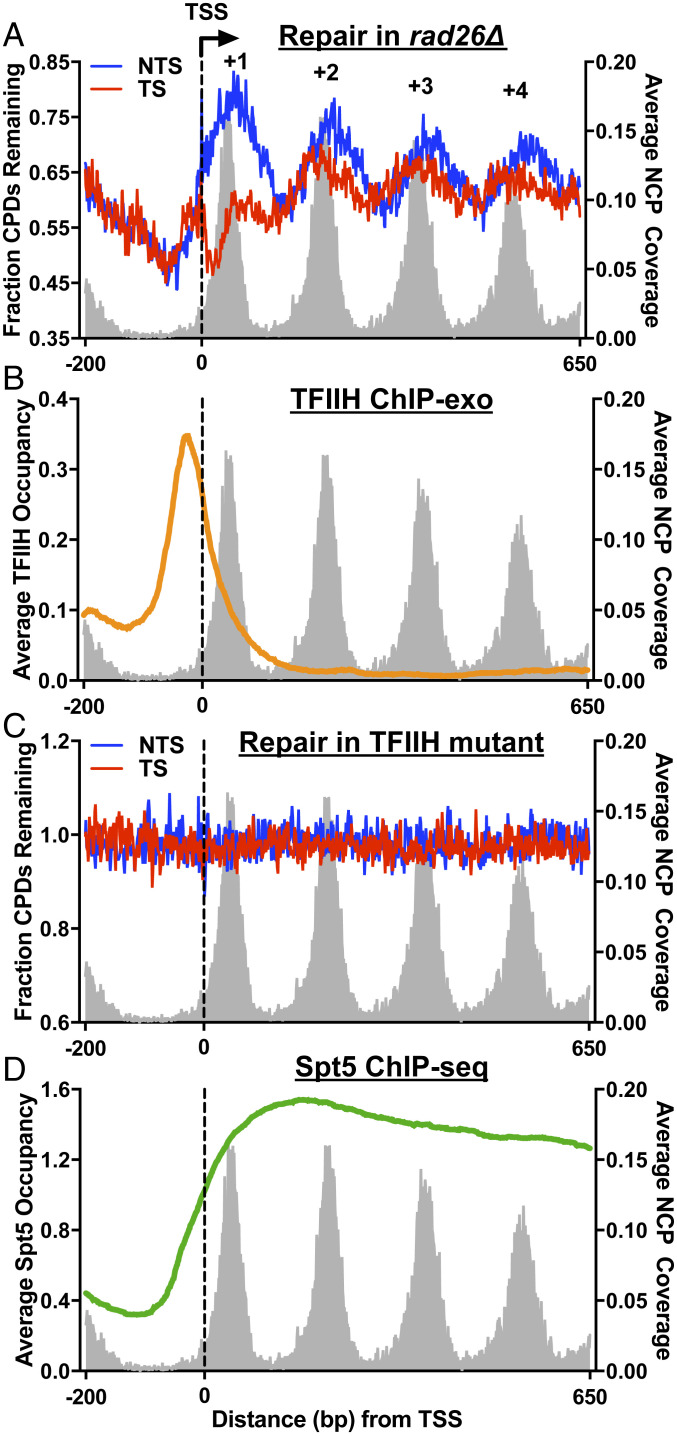
Fig. 3.
The requirement for Rad26 in TC-NER is correlated with TFIIH and Spt5 distribution. (A) Plot of remaining CPDs in the +1 and the downstream nucleosomes in rad26∆. This plot is the same as Fig. 2B and is presented to demonstrate the correlation between repair and TFIIH and Spt5 distribution. (B) ChIP-exo analysis shows high TFIIH occupancy in the promoter and the +1 nucleosome. TFIIH ChIP-exo data were obtained from the published study (16). Peaks of the TFIIH subunit Ssl2 were called, and the peak density was plotted. (C) The DNA helicase activity of TFIIH is essential for DNA repair. CPD-seq data from the helicase-dead mutant (Rad3-K48R) was analyzed around the TSS. (D) ChIP-seq analysis showing Spt5 occupancy. The Spt5 ChIP-seq data were derived from the published study (20). The normalized Spt5 occupancy was plotted around the TSS.
The high TFIIH occupancy in the +1 nucleosome implies that the helicases (Rad3 and Ssl2) are readily available for DNA unwinding upon Pol II stalling, which may allow TC-NER to occur in the absence of Rad26. To test this hypothesis, we generated a yeast strain harboring a point mutation in the conserved helicase motif of Rad3 with CRISPR-Cas9 (25). This point mutation, Lys48Arg, has been shown to abolish Rad3 ATPase and DNA helicase activities but retain Rad3 protein stability in yeast cells (35). We confirmed that this Rad3 helicase-dead (i.e., Rad3-K48R) yeast mutant was highly sensitive to UV killing, even more sensitive than the rad26∆rad16∆ double mutant (SI Appendix, Fig. S4B). Next, we analyzed CPD repair with CPD-seq. Our repair data showed that the K48R mutation in Rad3 abolished both GG-NER and TC-NER in yeast genes, including the +1 nucleosome (Fig. 3C), which explains the extremely high UV sensitivity in this mutant (SI Appendix, Fig. S4B). These findings are consistent with our hypothesis and indicate that the helicase activity of TFIIH is essential for TC-NER in the +1 nucleosome.
Analysis of the published Spt5 ChIP-seq data (20) showed low occupancy of Spt5 in the TSS-proximal region within the +1 nucleosome but high occupancy in the downstream region (Fig. 3D). Considering the suppressive role of Spt4/Spt5 in TC-NER (18, 19), the high occupancy of Spt5 likely inhibits TC-NER in the downstream region in the absence of Rad26. Indeed, our repair data showed significantly reduced TC-NER in this region in rad26∆ (Fig. 3A), but not in WT cells (Fig. 2A). Taken together, these data revealed a correlation between Rad26 requirement in TC-NER and the combined distribution of TFIIH and Spt4/Spt5.
Rad26-Independent TC-NER on the TSS-Proximal Side of the +1 Nucleosome Is Associated with High TFIIH Occupancy in Active Genes.
We next sought to understand why the TSS-proximal half of the +1 nucleosome exhibits the most prominent Rad26-independent TC-NER (e.g., see SI Appendix, Fig. S2F). Genome-wide studies with Native Elongating Transcript (NET)-seq have shown that Pol II density peaks near the TSS in yeast (36) and human genes (37) and decreases toward the 3′ end. The uneven Pol II distribution suggests that damage located near the 5′ end of transcribed genes is likely scanned by Pol II more frequently than damage near the 3′ end, which may contribute to the more robust Rad26-independent TC-NER.
To test if high-Pol II density is responsible for high Rad26-independent TC-NER, we sorted yeast genes with their Pol II density. The Pol II density for each gene was determined by NET-seq counts (38) normalized by the gene length. We then compared DNA repair between high-Pol II (i.e., top one-third of genes; n = 1,653) and low-Pol II groups (i.e., bottom 1,653 genes). We found that the Pol II density for high-Pol II genes was significantly higher than low-Pol II genes in the coding region (P < 0.0001; paired t test) (SI Appendix, Fig. S5 A and B). More importantly, we found that the robust Rad26-independent TC-NER on the TSS-proximal side of the +1 nucleosome was mainly associated with high-Pol II genes (Fig. 4A). In low-Pol II genes, the TS repair in the +1 nucleosome was similar between the TSS-proximal and TSS-distal halves (Fig. 4B). This finding was further confirmed by a closer comparison of TS repair within the +1 nucleosome between the two groups, which showed that the repair difference between high-Pol II and low-Pol II genes mainly occurred on the TSS-proximal side, not the TSS-distal side (Fig. 4C). As the high-Pol II genes had much higher Pol II density than low-Pol II genes, including the TSS-distal side (Fig. 4D), our data suggest that the high-Pol II density alone is insufficient to activate Rad26-independent TC-NER.
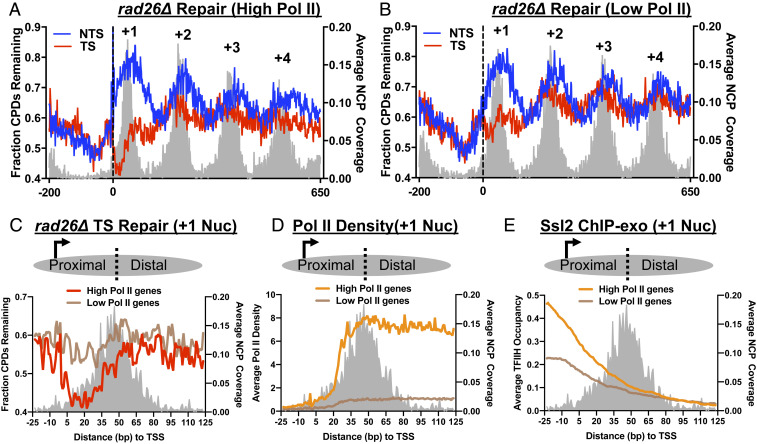
Fig. 4.
High TFIIH occupancy on the TSS-proximal side of the +1 nucleosome promotes Rad26-independent TC-NER. (A) High-Pol II genes exhibit prominent Rad26-independent TC-NER on the TSS-proximal side of the +1 nucleosome. Yeast genes were sorted by their Pol II density, using NET-seq data derived from the published study (38). High-Pol II genes (top 1,653 genes) were aligned at the TSS, and the fraction of remaining CPDs following 2 h repair was analyzed using rad26∆ CPD-seq data. (B) CPD-seq data in the rad26∆ mutant was analyzed for low-Pol II genes. (C) High-Pol II genes have higher Rad26-independent TC-NER on the TSS-proximal side of the +1 nucleosome than low-Pol II genes. The +1 nucleosome was shown as the region from −23 bp to +123 bp relative to the TSS. Fraction of remaining CPDs in the TS at 2 h in the rad26∆ mutant was plotted for high-Pol II and low-Pol II genes. (D) High-Pol II genes have significantly higher Pol II density in the +1 nucleosome. NET-seq data were analyzed in the +1 nucleosome (−23 bp to +123 bp relative to the TSS). (E) High-Pol II genes have higher TFIIH occupancy on the TSS-proximal side of the +1 nucleosome relative to low-Pol II genes. Ssl2 ChIP-exo data were analyzed in the +1 nucleosome in high-Pol II and low-Pol II genes.
These observations imply that Pol II on the TSS-distal side of the +1 nucleosome may not be competent for TC-NER unless Rad26 is available. One likely explanation is that the repair factor TFIIH is released from Pol II on the TSS-distal side. To test this hypothesis, we analyzed the TFIIH ChIP-exo data (16) and compared Ssl2 occupancy between high-Pol II and low-Pol II genes. Indeed, high-Pol II genes had significantly higher Ssl2 occupancy than low-Pol II genes on the TSS-proximal side, but not the TSS-distal side (Fig. 4E). Consistent with our analysis, in vitro data have shown that TFIIH disassociates from Pol II downstream of the TSS, starting from ∼30 bp after transcription synthesis (39). Thus, in highly transcribed genes, high TFIIH occupancy allows active Rad26-independent TC-NER to occur on the TSS-proximal side of the +1 nucleosome. On the TSS-distal side of the +1 nucleosome, low TFIIH availability hinders Rad26-independent TC-NER, although Pol II density is high.
Deletion of SPT4 Significantly Rescues TC-NER in the rad26∆ Mutant.
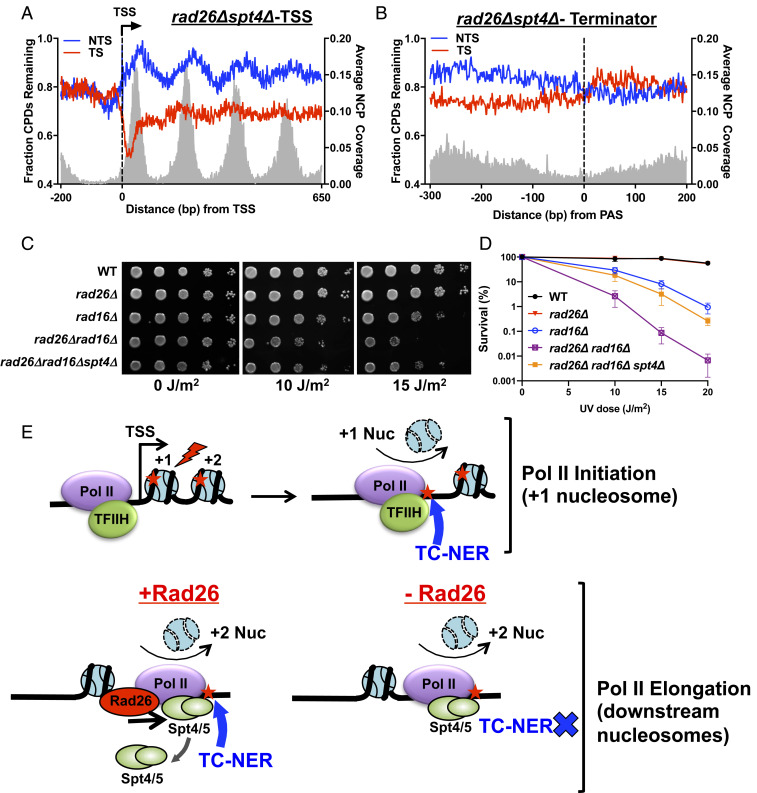
Lastly, we investigated the role of Rad26 in the downstream nucleosomes. Recent structural data indicates that Rad26 and Spt4/Spt5 may compete for the binding site on Pol II (12), suggesting a potential role for Rad26 in evicting Spt4/Spt5 from Pol II to initiate repair. To test this model, we knocked out SPT4, which encodes the nonessential subunit of the Spt4/Spt5 complex, and mapped remaining CPDs in a rad26∆spt4∆ double mutant.
Analysis of CPD-seq data revealed significantly restored TC-NER in the rad26∆spt4∆ strain (Fig. 5A and SI Appendix, Fig. S6A). Compared to the rad26∆ single mutant (Fig. 2B), we found apparent TC-NER in the downstream nucleosomes (e.g., +2, +3, and +4 nucleosomes) in the rad26∆spt4∆ double mutant (Fig. 5A and SI Appendix, Fig. S6A), as shown by less remaining CPDs in the TS relative to the NTS. Furthermore, TC-NER on the TSS-distal side of the +1 nucleosome was also enhanced by SPT4 deletion (Fig. 5A). These data suggest that TC-NER on the TSS-distal side of the +1 nucleosome as well as in the downstream nucleosomes was suppressed by Spt4/Spt5 in rad26∆, which is consistent with the distribution of Spt5 in the coding region (Fig. 3D). However, TC-NER in the downstream nucleosomes was not as active as on the TSS-proximal side of the +1 nucleosome in rad26∆spt4∆ (Fig. 5A and SI Appendix, Fig. S6A), suggesting that TC-NER was not fully recovered by deletion of SPT4. Additionally, TC-NER restoration was also seen in the termination region in the rad26∆spt4∆ strain (Fig. 5B), consistent with the Spt5 ChIP-seq data showing that there were considerable levels of Spt5 upstream of the PAS (SI Appendix, Fig. S6B).
Fig. 5.
Deletion of SPT4 significantly restores TC-NER in the rad26∆ mutant. (A) Yeast genes (n = 5,205) were aligned at the TSS and the number of CPD-seq reads in the double mutant at 2 h (normalized by 0 h) was plotted at base resolution. (B) TC-NER is recovered in the termination region by SPT4 deletion. Remaining CPDs at 2 h in the rad26Δspt4Δ mutant were plotted around the PAS. (C) Yeast strains were spotted on yeast extract-peptone-dextrose plates and exposed to different UV doses. (D) Quantitative UV survival data shows significant but not full recovery of UV resistance by deletion of SPT4. (E) Model depicting the function of Rad26 in promoting TC-NER. After the +1 nucleosome, Pol II releases TFIIH and binds Spt4/Spt5 for productive elongation. When Pol II is stalled by UV damage (red star) in the +1 nucleosome, TFIIH bound to Pol II is able to activate TC-NER, without the need of Rad26 (Upper). When Pol II stalls in the +2 or more downstream nucleosomes, Rad26 is required to remove Spt4/Spt5, the TC-NER suppressor (Lower).
The restoration of TC-NER by SPT4 deletion was further confirmed by UV survival data. We found that SPT4 deletion significantly increased UV resistance in the rad26∆rad16∆ double mutant (Fig. 5C), which is in agreement with the published data (18). Furthermore, quantitative analysis of UV survival showed that the rad26∆rad16∆spt4∆ triple mutant was less resistant than the rad16∆ single mutant (Fig. 5D), consistent with our data showing partial but not full restoration of TC-NER by SPT4 deletion.
Discussion
Here, we utilized CPD-seq and analyzed repair of CPDs at single nucleotide resolution in both WT and Rad26-deficient yeast strains. Our genomic data revealed that Rad26 is generally required for TC-NER in most yeast genes; however, TC-NER in a small number of genes and in specific chromatin regions can occur without Rad26. We showed that the ATP-dependent DNA translocase activity of Rad26 is essential for TC-NER, likely by countering the TC-NER suppressor, Spt4/Spt5. We further showed that the Rad26-independent TC-NER in the +1 nucleosome is correlated with high occupancy of TFIIH.
Our genome-wide data indicates that a small number of yeast genes can activate TC-NER in a Rad26-independent manner (SI Appendix, Table S1). These genes tend to have very high transcription frequency, and the Rad26-independent TC-NER appears to govern their entire transcribed strand, from the TSS to the transcription termination site (TTS) (SI Appendix, Fig. S7A). Although Rad26 is dispensable, we found that these highly transcribed genes are dependent on the TFIIH helicase activity to repair their TS (SI Appendix, Fig. S7B). Furthermore, these genes have low TFIIH occupancy downstream of the +1 nucleosome (SI Appendix, Fig. S7C), suggesting TFIIH recruitment is required following Pol II stalling. It is unclear how TFIIH is recruited to stalled Pol II in the absence of Rad26 in these genes; however, previous studies have shown that Rpb9, a nonessential subunit of Pol II complex, can mediate TC-NER for highly active genes (27). Therefore, it is likely that TC-NER in these highly transcribed genes is dependent on Rpb9, which may assist the recruitment of TFIIH to stalled Pol II.
Our analysis revealed a generic Rad26-independent TC-NER near the TSS for almost all yeast genes. This Rad26-independent TC-NER is associated with the stereotypic nucleosome organization in the coding region and appears to be strictly limited within the TSS-proximal half of the +1 nucleosome. Mechanistically, the Rad26-independent TC-NER in the +1 nucleosome is correlated with the early elongation status of Pol II, characterized by its association with the transcription initiation factor TFIIH (39). Indeed, analysis of high-resolution TFIIH ChIP-exo data revealed high TFIIH occupancy in the +1 nucleosome, particularly on the TSS-proximal side. The TFIIH occupancy data suggests that TFIIH is bound to Pol II during initiation and travels with Pol II into the +1 nucleosome and reaches the TSS-proximal half of this nucleosome. At this point, TFIIH is readily available to initiate repair when Pol II stalls at a lesion, presumably by utilizing its DNA helicases to create an open intermediate for dual incision (15), thus bypassing the need of Rad26 (Fig. 5 E, Upper). Consistent with this notion, no TC-NER was observed in the +1 nucleosome when the TFIIH helicase activity is deficient. On the TSS-distal side of the +1 nucleosome, TFIIH gradually disassociates from Pol II. TFIIH dissociation is likely due to the exchange between Pol II initiation and elongation factors (40) within the +1 nucleosome, which facilitates productive elongation in the downstream chromatin. This exchange results in reduced TFIIH occupancy on the TSS-distal side of the +1 nucleosome and lack of TFIIH in the downstream nucleosomes (Fig. 3B), and increased occupancy of the elongation factor Spt4/Spt5 (Fig. 3D).
The requirement for Rad26 in TC-NER in the downstream nucleosomes may be in part due to low occupancy of TFIIH, and Rad26 may be needed to recruit TFIIH. In line with this notion, human CSB has been shown to physically interact with TFIIH in vitro (14). However, Rad26 does not appear to play a direct role in recruiting TFIIH in yeast, as TC-NER can function actively in the rad26∆spt4∆ mutant. Hence, the more important function of Rad26 appears to counter Spt4/Spt5 in the downstream nucleosomes. Yeast Spt4/Spt5 is a known TC-NER suppressor (18, 19). It suppresses repair by holding the elongation complex in a closed confirmation, which enhances elongation but is intrinsically suppressive to TC-NER (41). The significantly recovered TC-NER in the rad26∆spt4∆ double mutant suggests that Rad26 likely evicts Spt4/Spt5 from Pol II to switch Pol II from transcription elongation to DNA repair (Fig. 5 E, Lower). Failing to evict Spt4/Spt5 upon Pol II stalling is associated with poor TC-NER, as observed in the downstream nucleosomes in Rad26-deficient cells. Furthermore, our repair data in the Rad26 ATPase-dead mutant suggests that the DNA translocase activity of Rad26 is important for evicting Spt4/Spt5. A likely mechanism is that the ATP-dependent translocase activity allows Rad26 to move closer to the stalled Pol II in order to evict Spt4/Spt5. Consistent with this model, structural data indicates that Rad26 binds to DNA upstream of transcribing Pol II and utilizes its ATPase activity to move toward Pol II. The overlapping binding sites for Rad26 and Spt4/Spt5 on Pol II imply eviction of Spt4/Spt5 by Rad26 during the initiation of TC-NER (12). Intriguingly, TC-NER deficiency in Rad26-deficient cells is compensated by GG-NER to repair the TS. In this case, the prolonged stalling may trigger degradation of Pol II from the TS, as previous studies have shown increased Pol II degradation mediated by Def1 when Rad26 is mutated (42). After Pol II degradation or displacement, the damage in the TS is reassembled into nucleosomes and repaired by GG-NER, which can explain the periodic CPD pattern in the downstream nucleosomes of the TS in rad26∆ (Fig. 2B).
In summary, we have shown that Rad26-dependent and Rad26-independent TC-NER occur in different chromatin regions. We further showed that the differential requirement of Rad26 is correlated with the distribution of TFIIH and Spt4/Spt5 along transcribed genes. Considering the functional similarity between Rad26 and human CSB, our results provide insights into the mechanism of CSB in TC-NER in the human genome. For example, CSB-independent TC-NER is also found in the TSS-proximal coding region in human cells (33), suggesting that the TFIIH occupancy may be high in the coding region near the TSS in human genes so that it can bypass CSB to initiate repair. Additionally, the elongation factor Spt4/Spt5 is highly conserved among eukaryotes. Future studies in Spt4-deficient or Spt5-deficient human cells will uncover if human Spt4/Spt5 also functions as a TC-NER suppressor and if CSB is important for antagonizing Spt4/Spt5 in human TC-NER.
Materials and Methods
Yeast cells (BY4741; WT and mutant strains) were grown to midlog phase and irradiated with 125 J/m2 or 100 J/m2 UVC light (254 nm). After UV treatment, cells were incubated in prewarmed fresh yeast extract-peptone-dextrose medium for repair. Detailed CPD-seq library preparation and data analysis are given in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Mark Wildung and Wei Wei Du for technical assistance with Ion Proton sequencing. We also thank Drs. Amelia Hodges and Hua-ying Fan for reading the manuscript. This work was supported by National Institute of Environmental Health Sciences Grants R01ES028698 (to J.J.W.), R21ES029655 (to J.J.W.), R21ES027937 (to J.J.W.), R21ES029302 (to P.M. and J.J.W.), and R03ES027945 (to P.M.). This research was partially supported by the University of New Mexico Comprehensive Cancer Center Support Grant NCI P30CA118100.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The data have been deposited with the Gene Expression Omnibus, https://www.ncbi.nlm.nih.gov/geo/ (accession code GSE145911).
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2003868117/-/DCSupplemental.
References
- 1.Friedberg E. C. et al., DNA Repair and Mutagenesis, (ASM Press, 2006). [Google Scholar]
- 2.Hanawalt P. C., Spivak G., Transcription-coupled DNA repair: Two decades of progress and surprises. Nat. Rev. Mol. Cell Biol. 9, 958–970 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Hu J., Adar S., Selby C. P., Lieb J. D., Sancar A., Genome-wide analysis of human global and transcription-coupled excision repair of UV damage at single-nucleotide resolution. Genes Dev. 29, 948–960 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haradhvala N. J. et al., Mutational strand asymmetries in cancer genomes reveal mechanisms of DNA damage and repair. Cell 164, 538–549 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gregersen L. H., Svejstrup J. Q., The cellular response to transcription-blocking DNA damage. Trends Biochem. Sci. 43, 327–341 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Selby C. P., Witkin E. M., Sancar A., Escherichia coli mfd mutant deficient in “mutation frequency decline” lacks strand-specific repair: In vitro complementation with purified coupling factor. Proc. Natl. Acad. Sci. U.S.A. 88, 11574–11578 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Gool A. J. et al., RAD26, the functional S. cerevisiae homolog of the Cockayne syndrome B gene ERCC6. EMBO J. 13, 5361–5369 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Venema J., Mullenders L. H., Natarajan A. T., van Zeeland A. A., Mayne L. V., The genetic defect in Cockayne syndrome is associated with a defect in repair of UV-induced DNA damage in transcriptionally active DNA. Proc. Natl. Acad. Sci. U.S.A. 87, 4707–4711 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park J.-S., Marr M. T., Roberts J. W., E. coli Transcription repair coupling factor (Mfd protein) rescues arrested complexes by promoting forward translocation. Cell 109, 757–767 (2002). [DOI] [PubMed] [Google Scholar]
- 10.Selby C. P., Sancar A., Molecular mechanism of transcription-repair coupling. Science 260, 53–58 (1993). [DOI] [PubMed] [Google Scholar]
- 11.Fan J., Leroux-Coyau M., Savery N. J., Strick T. R., Reconstruction of bacterial transcription-coupled repair at single-molecule resolution. Nature 536, 234–237 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Xu J. et al., Structural basis for the initiation of eukaryotic transcription-coupled DNA repair. Nature 551, 653–657 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Selby C. P., Sancar A., Cockayne syndrome group B protein enhances elongation by RNA polymerase II. Proc. Natl. Acad. Sci. U.S.A. 94, 11205–11209 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Selby C. P., Sancar A., Human transcription-repair coupling factor CSB/ERCC6 is a DNA-stimulated ATPase but is not a helicase and does not disrupt the ternary transcription complex of stalled RNA polymerase II. J. Biol. Chem. 272, 1885–1890 (1997). [DOI] [PubMed] [Google Scholar]
- 15.Compe E., Egly J.-M., TFIIH: When transcription met DNA repair. Nat. Rev. Mol. Cell Biol. 13, 343–354 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Vinayachandran V. et al., Widespread and precise reprogramming of yeast protein-genome interactions in response to heat shock. Genome Res. 28, 357–366 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rhee H. S., Pugh B. F., Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature 483, 295–301 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jansen L. E. T. et al., Spt4 modulates Rad26 requirement in transcription-coupled nucleotide excision repair. EMBO J. 19, 6498–6507 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding B., LeJeune D., Li S., The C-terminal repeat domain of Spt5 plays an important role in suppression of Rad26-independent transcription coupled repair. J. Biol. Chem. 285, 5317–5326 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baejen C. et al., Genome-wide analysis of RNA polymerase II termination at protein-coding genes. Mol. Cell 66, 38–49.e6 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Mao P., Smerdon M. J., Roberts S. A., Wyrick J. J., Chromosomal landscape of UV damage formation and repair at single-nucleotide resolution. Proc. Natl. Acad. Sci. U.S.A. 113, 9057–9062 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park D., Morris A. R., Battenhouse A., Iyer V. R., Simultaneous mapping of transcript ends at single-nucleotide resolution and identification of widespread promoter-associated non-coding RNA governed by TATA elements. Nucleic Acids Res. 42, 3736–3749 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mao P. et al., ETS transcription factors induce a unique UV damage signature that drives recurrent mutagenesis in melanoma. Nat. Commun. 9, 2626 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li S., Transcription coupled nucleotide excision repair in the yeast Saccharomyces cerevisiae: The ambiguous role of Rad26. DNA Repair (Amst.) 36, 43–48 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Laughery M. F. et al., New vectors for simple and streamlined CRISPR-Cas9 genome editing in Saccharomyces cerevisiae. Yeast 32, 711–720 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Selvam K., Ding B., Sharma R., Li S., Evidence that moderate eviction of Spt5 and promotion of error-free transcriptional bypass by Rad26 facilitates transcription coupled nucleotide excision repair. J. Mol. Biol. 431, 1322–1338 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Li S., Smerdon M. J., Rpb4 and Rpb9 mediate subpathways of transcription-coupled DNA repair in Saccharomyces cerevisiae. EMBO J. 21, 5921–5929 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holstege F. C. et al., Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95, 717–728 (1998). [DOI] [PubMed] [Google Scholar]
- 29.Jiang C., Pugh B. F., Nucleosome positioning and gene regulation: Advances through genomics. Nat. Rev. Genet. 10, 161–172 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mao P., Smerdon M. J., Roberts S. A., Wyrick J. J., Asymmetric repair of UV damage in nucleosomes imposes a DNA strand polarity on somatic mutations in skin cancer. Genome Res. 30, 12–21 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Verhage R. et al., The RAD7 and RAD16 genes, which are essential for pyrimidine dimer removal from the silent mating type loci, are also required for repair of the nontranscribed strand of an active gene in Saccharomyces cerevisiae. Mol. Cell. Biol. 14, 6135–6142 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tijsterman M., Verhage R. A., van de Putte P., Tasseron-de Jong J. G., Brouwer J., Transitions in the coupling of transcription and nucleotide excision repair within RNA polymerase II-transcribed genes of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 94, 8027–8032 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tu Y., Bates S., Pfeifer G. P., Sequence-specific and domain-specific DNA repair in xeroderma pigmentosum and Cockayne syndrome cells. J. Biol. Chem. 272, 20747–20755 (1997). [DOI] [PubMed] [Google Scholar]
- 34.Eyboulet F. et al., Mediator links transcription and DNA repair by facilitating Rad2/XPG recruitment. Genes Dev. 27, 2549–2562 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sung P., Higgins D., Prakash L., Prakash S., Mutation of lysine-48 to arginine in the yeast RAD3 protein abolishes its ATPase and DNA helicase activities but not the ability to bind ATP. EMBO J. 7, 3263–3269 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Churchman L. S., Weissman J. S., Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature 469, 368–373 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nojima T. et al., Mammalian NET-seq reveals genome-wide nascent transcription coupled to RNA processing. Cell 161, 526–540 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harlen K. M. et al., Comprehensive RNA polymerase II interactomes reveal distinct and varied roles for each phospho-CTD residue. Cell Rep. 15, 2147–2158 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zawel L., Kumar K. P., Reinberg D., Recycling of the general transcription factors during RNA polymerase II transcription. Genes Dev. 9, 1479–1490 (1995). [DOI] [PubMed] [Google Scholar]
- 40.Pokholok D. K., Hannett N. M., Young R. A., Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol. Cell 9, 799–809 (2002). [DOI] [PubMed] [Google Scholar]
- 41.Li W., Giles C., Li S., Insights into how Spt5 functions in transcription elongation and repressing transcription coupled DNA repair. Nucleic Acids Res. 42, 7069–7083 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Woudstra E. C. et al., A Rad26-Def1 complex coordinates repair and RNA pol II proteolysis in response to DNA damage. Nature 415, 929–933 (2002). [DOI] [PubMed] [Google Scholar]
- 43.Weiner A. et al., High-resolution chromatin dynamics during a yeast stress response. Mol. Cell 58, 371–386 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.