Significance
Human T lymphotropic virus type 1 (HTLV-1) is a retrovirus that mainly transforms CD4+ T cells, ultimately leading to the development of adult T cell leukemia/lymphoma or HTLV-1-associated myelopathy/tropical spastic paraparesis, both of which have limited treatment options. A deeper understanding of how the virus hijacks host factors for replication is critical for the development of new therapies. In this study, we discovered that a host transcription factor, Yin Yang 1 (YY1) is a potent activator of HTLV-1 gene expression. Surprisingly, it does so by binding to viral RNA, instead of the classical DNA–transcription factor interaction. Our results provide insights into host–viral interactions, as well as the biology of cellular transcription factors.
Keywords: transcription factor Yin Yang 1 (YY1), human T cell leukemia virus type 1 (HTLV-1), RNA binding, transcriptional activation
Abstract
Yin Yang 1 (YY1) is a DNA-binding transcription factor that either activates or represses gene expression. YY1 has previously been implicated in the transcriptional silencing of many retroviruses by binding to DNA sequences in the U3 region of the viral long terminal repeat (LTR). We here show that YY1 overexpression leads to profound activation, rather than repression, of human T lymphotropic virus type 1 (HTLV-1) expression, while YY1 down-regulation reduces HTLV-1 expression. The YY1 responsive element mapped not to YY1 DNA-binding sites in the HTLV-1 LTR but to the R region. The HTLV-1 R sequence alone is sufficient to provide YY1 responsiveness to a nonresponsive promoter, but only in the sense orientation and only when included as part of the mRNA. YY1 binds to the R region of HTLV-1 RNA in vitro and in vivo, leading to increased transcription initiation and elongation. The findings indicate that YY1 is a potent transactivator of HTLV-1 gene expression acting via binding viral RNA, rather than DNA.
Human T lymphotropic virus type 1 (HTLV-1) is a retrovirus that mainly transforms CD4+ T cells in vivo. For most infected individuals, de novo infection is followed by a long and clinically latent period before the development of adult T cell leukemia/lymphoma (ATL) (1, 2), or the neurological disorder HTLV-1-associated myelopathy/tropical spastic paraparesis (3, 4). Both conditions have limited treatment options and ATL carries a poor prognosis.
Infection of susceptible cells by HTLV-1, as with all retroviruses, results in the establishment of an integrated proviral copy of the viral genome, flanked by directly repeated sequences termed the long terminal repeats (LTRs). Following integration of HTLV-1 viral DNA into the host genome, the 5′ LTR, with the help of host factors, functions as the viral promoter and directs transcription of viral genes by the RNA polymerase II enzyme. Retroviral LTR sequences are divided into three parts, named U3 (for unique 3′), R (for repeated), and U5 (for unique 5′). The U3 region contains regulatory sequences, the binding sites for numerous transcription factors (5), as well as the TATA box, binding site for the TATA-binding protein (TBP) factor. Transcription starts at the U3–R boundary, and the resulting transcripts begin with the R and U5 sequences as part of the long 5′ untranslated region (UTR) of the viral transcripts.
HTLV-1 encodes a transactivator protein Tax, which activates viral transcription through three 21 base pair repeats in the U3 region of the LTR, termed the Tax-responsive element (TRE) (6, 7). Tax does not bind the TRE directly. Instead, each TRE contains a cAMP response element (CRE) that is recognized by members of the CRE-binding protein/activating transcription factor (CREB/ATF) family of proteins. CREB recruits Tax to the TRE (8–10) and allows for subsequent recruitment of downstream transcriptional activators such as CREB-binding protein (CBP), p300, and p300/CBP-associated factor (P/CAF), leading to potent activation of viral transcription (11–13). In addition, Tax itself also interacts with many cellular transcription factors, including Ets1 (14), NF-kappa B (15), and others, resulting in their recruitment to the LTR and further activation of viral gene expression. Tax also activates expression of numerous cellular genes.
Yin Yang 1 (YY1) is a member of the GLI-Krüppel class of transcription factors capable of binding either DNA or RNA, leading to the activation or repression of gene expression during embryogenesis, cell growth, and differentiation (16). Mice lacking YY1 show peri-implantation lethality, highlighting its essential role in development (17). YY1 binds to DNA through a consensus sequence VDCCATNWY (18). Many cellular genes are regulated by YY1, including c-Myc, c-Fos, β-actin, IFN-γ, CREB, Sp1, and others (19, 20). In addition, YY1 regulates transcription from retroviral promoters. In T cells, YY1 binds to DNA sequences in the HIV-1 LTR, leading to transcriptional repression and establishment of viral latency (21, 22). In mouse embryonic cells, YY1 binds to a short DNA sequence located in the U3 region of the murine leukemia virus (MLV) LTR (23), leading to transcriptional silencing (24). Apart from DNA binding, there is emerging evidence supporting the role of YY1 as an RNA-binding protein. In Xenopus oocytes, YY1 associates with structurally divergent RNAs and contributes to the assembly of messenger ribonucleoprotein particles (mRNPs) (25, 26). Both DNA and RNA binding by YY1 are important for tethering Xist, a long noncoding RNA (lncRNA), to DNA regions on the X chromosome, leading to X chromosome inactivation in maternal cells (27). Moreover, a recent study showed the ability of YY1 to bind nascent mRNA transcripts, leading to local retention of YY1 at actively transcribed promoters (28).
The N-terminal domain of YY1 contains features required for transcriptional activation, which includes stretches of acidic residues (amino acids 16 to 29 and amino acids 43 to 53), a glycine-rich region (amino acids 54 to 69), a histidine-rich cluster (amino acids 70 to 80), and sequences rich in proline and glutamine (amino acids 80 to 100) (29–32). The C-terminal domain of YY1 contains four C2H2-type zinc finger motifs (amino acids 298 to 414) required for nucleic acid binding (33). In addition, portions of the zinc finger motifs (amino acids 333 to 397), together with a region rich in alanine and glycine (amino acids 157 to 201) contribute to transcriptional repression (30, 34). Whether YY1 acts as a repressor or an activator is likely dictated by its interaction with various cofactors.
In this study, we report the unexpected observation that YY1 is a potent transactivator, rather than repressor, of expression of the HTLV-1 LTR. Both N- and C-terminal domains of YY1 are required for activation. The YY1 responsive element in the HTLV-1 LTR maps to the R region and functions independently of YY1 binding to DNA. Using chimeric promoter constructs, we show that HTLV-1 R could provide YY1 responsiveness to a nonresponsive SV40 early promoter, but only when R is present as part of the mRNA and not when it is outside the transcriptional unit. Furthermore, R only functioned in the sense orientation and not when inverted, a behavior that is consistent with it acting as an RNA, rather than a DNA element. We found that YY1 associates with HTLV-1 R RNA in vitro and in vivo, leading to increased rates of transcription initiation and elongation. Taken together, our findings indicate that YY1 acts as a potent transactivator of the HTLV-1 LTR via interacting with RNA, rather than DNA.
Results
YY1 Activates Reporter Expression from HTLV-1 LTR.
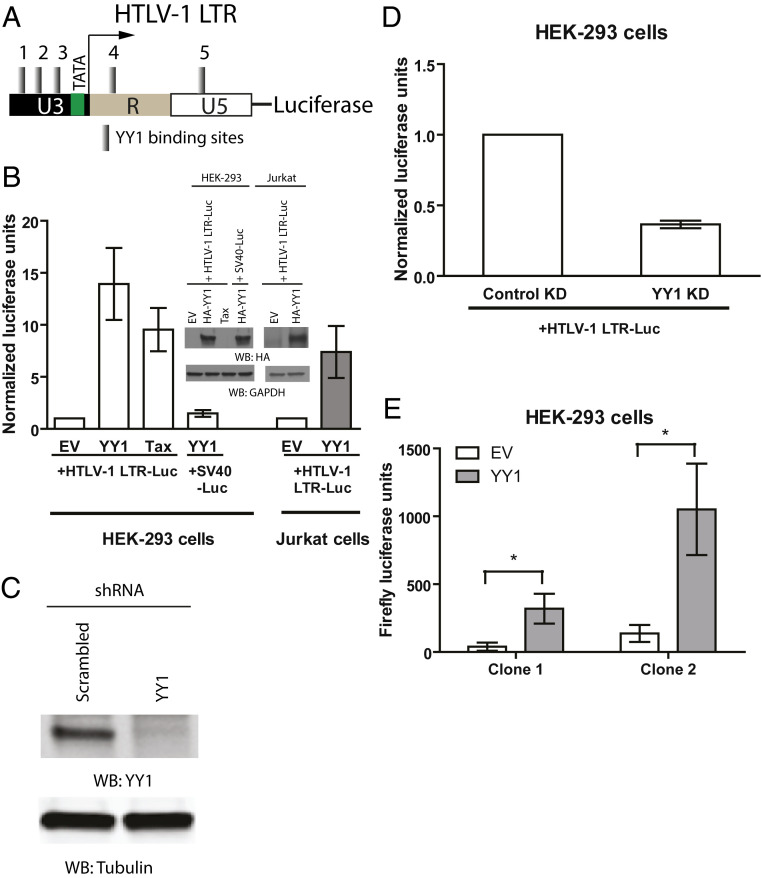
YY1 is a GLI-Krüppel transcription factor known to bind specifically to a short consensus DNA sequence (VDCCATNWY) (18). Analysis of the HTLV-1 LTR sequence revealed the presence of five potential YY1 DNA-binding sites (Fig. 1A and SI Appendix, Table S1), which could mediate regulation of HTLV-1 expression. To test the effects of YY1 overexpression on HTLV-1 LTR activity, HEK-293 cells were cotransfected with empty vector (EV) or a plasmid encoding HA-tagged YY1, together with two reporter plasmid DNAs: one containing the HTLV-1 LTR-driving expression of firefly luciferase, and one containing the thymidine kinase (TK) promoter-driving expression of Renilla luciferase for normalization of transfection efficiency. At 24 to 48 h after transfection, cells were harvested, lysed, and subjected to dual-luciferase assays, and the ratio of expression of the two reporters was determined. A plasmid expressing HTLV-1 Tax was used as a positive control in lieu of YY1 (35). To our surprise, rather than repressing transcription, YY1 overexpression resulted in robust activation of the HTLV-1 LTR (14-fold), which was even higher than cells expressing Tax (10-fold). Similar activation was observed in Jurkat T cells (Fig. 1B). The ability of YY1 to activate HTLV-1 LTR occurs in a dose-dependent manner (SI Appendix, Fig. S1A). As a negative control, a construct expressing firefly luciferase from the SV40 promoter, which lacks potential YY1 DNA-binding sites, was used in lieu of the HTLV-1 LTR luciferase construct. YY1 overexpression had no effect on the SV40 promoter (Fig. 1B). YY1 overexpression in HEK-293 cells also had no effect on MLV LTR activity, consistent with the notion that YY1-mediated transcriptional silencing of MLV is specific to mouse embryonic cells (SI Appendix, Fig. S1C) (24).
Fig. 1.
YY1 transactivates reporter expression from HTLV-1 LTR. (A) Schematic of predicted YY1 DNA-binding sites in HTLV-1 LTR. YY1-binding sites are numbered as indicated. (B) HEK-293 or Jurkat cells were transfected with empty vector (EV) or DNAs encoding YY1 or Tax, together with HTLV-1 LTR firefly luciferase or SV40 early promoter firefly luciferase, and HSV-TK Renilla luciferase control plasmid. Relative luciferase expression was calculated by first dividing the firefly luciferase signal by the Renilla luciferase signal in a given cell line, and the resulting ratio was then normalized to the ratio obtained from EV-transfected cells (set to 1). Results shown are means ± SEs from three independent experiments performed in duplicate. HA-YY1 overexpression is confirmed with Western blotting (WB) using an anti-HA antibody (Upper). (C) Lysate prepared from HEK-293 cells stably transduced with either control shRNA or YY1-specific shRNA were subjected to WB as indicated. (D) Similar assay in B in YY1 knockdown cells. Results shown are means ± SEs from three independent experiments performed in duplicate. (E) Cells with stably integrated HTLV-1 LTR luciferase reporter were transfected with EV or plasmid expressing YY1. Forty-eight hours later the activity of HTLV-1 LTR was determined by firefly luciferase assay. Data shown are mean firefly luciferase signal ± SEs from four independent experiments performed in duplicate. Student’s t test was used for statistical analysis. *P < 0.05.
To eliminate the possibility that the effect of YY1 overexpression is limited to transiently transfected reporter plasmids, HEK-293 cells containing stably integrated HTLV-1 LTR luciferase was generated by cotransfection of HTLV-1 LTR luciferase with a plasmid encoding neomycin resistance, followed by selection with G418. Individual G418-resistant clones were isolated and genomic DNA qPCR using primers specific for the HTLV-1 LTR confirmed the presence of stable integrants, with each clone containing approximately six copies of LTR-luciferase (SI Appendix, Fig. S1D). Consistent with our transient transfection experiments, overexpression of YY1 caused robust up-regulation of luciferase expression in two independent HTLV-1 LTR-luciferase clones (Fig. 1E). These results suggest that YY1-mediated transactivation of the HTLV-1 LTR occurs in the context of both plasmid-based and chromosomal-based reporter genes.
To examine the effect of YY1 knockdown on HTLV-1 LTR activity, HEK-293 cells were transduced with lentivirus vectors expressing shRNAs either targeting YY1 mRNA or a scrambled sequence, together with a puromycin resistance gene. After puromycin selection, efficient YY1 knockdown in the cell population was confirmed with Western blotting (Fig. 1C). These YY1 knockdown cells and control knockdown cells were then transfected with HTLV-1 LTR firefly luciferase DNA and subjected to luciferase assay. In the absence of Tax, HTLV-1 LTR basal transcription was very low in both cell pools. Consistent with our overexpression experiments, YY1 knockdown resulted in a threefold decrease in the basal HTLV-1 LTR activity (Fig. 1D).
Both N-Terminal Activation and C-Terminal Zinc Finger Domains of YY1 Are Required for Transactivation of HTLV-1 LTR.
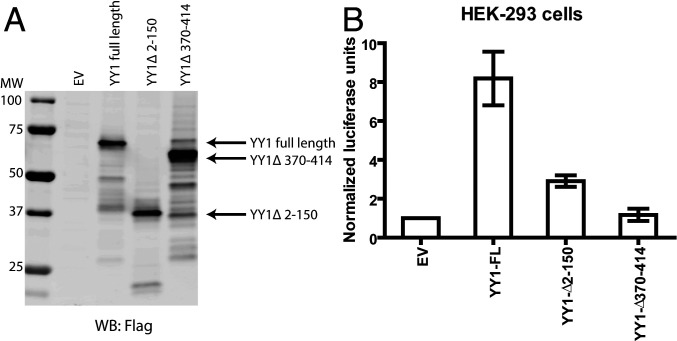
YY1 contains four C2H2-type zinc finger motifs at its C terminus responsible for DNA binding and transcriptional repression, while the N-terminal region is involved in transcriptional activation (30). To define regions of YY1 required for transactivation of the HTLV-1 LTR, we carried out luciferase reporter assays outlined above in the presence of either N-terminal (YY1Δ2-150) or C-terminal (YY1Δ370-414) truncation mutants. Both mutants were expressed at levels comparable to that of full-length YY1 (Fig. 2A). However, selective elimination of either the zinc finger region or activation domain severely compromised YY1’s ability to induce expression from the HTLV-1 LTR (Fig. 2B).
Fig. 2.
Both N-terminal activation and C-terminal zinc finger domains of YY1 are required for transactivation of HTLV-1 LTR. (A) Lysate prepared from HEK-293 cells transfected with EV or DNAs encoding various Flag-tagged YY1 truncation mutants were subjected to WB as indicated. (B) Similar assay as Fig. 1B using Flag-tagged YY1 truncation mutants. Results shown are means ± SEs from three independent experiments performed in duplicate.
The YY1 Responsive Element in HTLV-1 LTR Maps to the R Region.
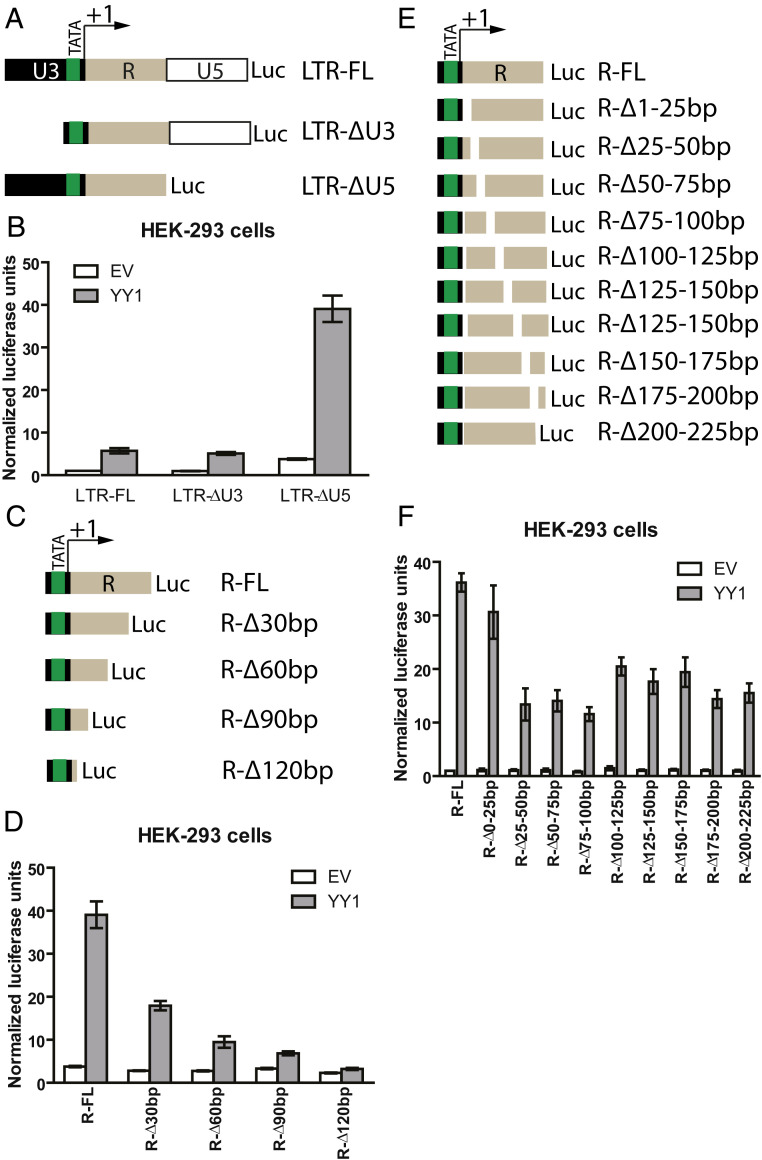
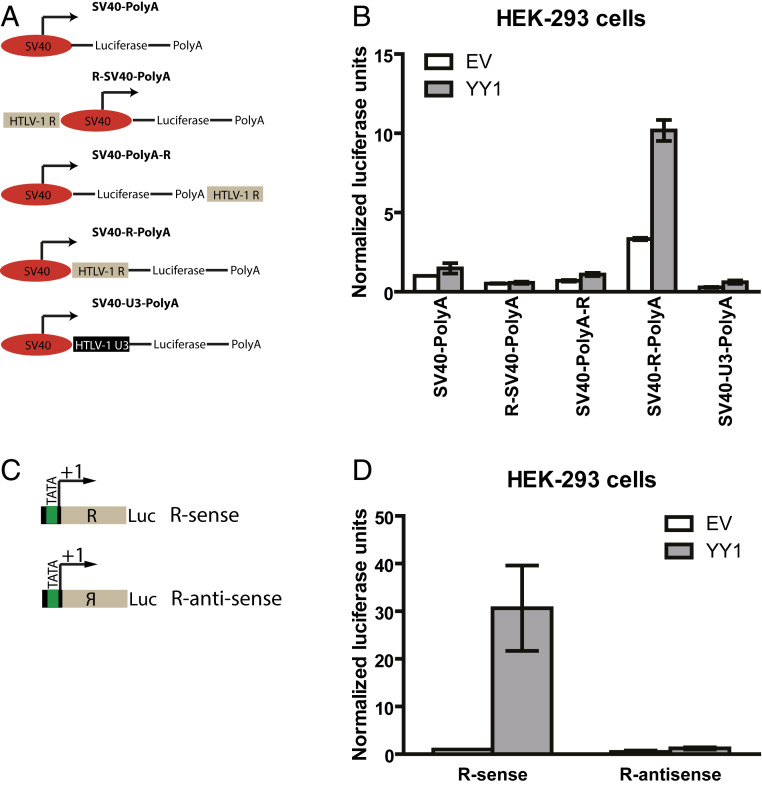
To map the DNA sequences in HTLV-1 LTR required for YY1 activity, we constructed a series of LTR truncation mutants (Fig. 3A) and tested their responsiveness to YY1 overexpression. YY1 transactivation was not affected by deleting large portions of U3, U5 (Fig. 3B), or the TRE (SI Appendix, Fig. S2), suggesting YY1-mediated activation occurs independently of Tax or other regulatory sequences in the U3 or U5 regions of the LTR. Of note, a mutant LTR lacking U5 showed significantly higher basal, as well as YY1-stimulated reporter expression, compared to the full-length LTR. This observation is consistent with prior reports showing the presence of a cis-repressive element causing nuclear retention of viral RNAs (36). For our subsequent studies, we chose to use an LTR construct lacking the U5 region to bypass this additional layer of regulation. Mutagenesis of this construct indicated that the R region contained the major YY1 response elements, as successive truncation of the R region from the 3′ end (Fig. 3C) resulted in progressive loss of YY1-mediated transactivation (Fig. 3D). To further map the YY1 responsive element in R, we also created a series of 25 base pair deletions across the entire R region (Fig. 3E). With the exception of the first 25 base pairs of R, partial loss of YY1-mediated transactivation was observed for almost all mutants, while no single mutation completely abolished transactivation in vivo (Fig. 3F). These findings suggest the presence of multiple YY1 responsive elements, or of a large structured element, in the R region of the HTLV-1 LTR.
Fig. 3.
YY1 responsive element in HTLV-1 LTR maps to the R region. (A) Schematic of LTR truncation mutants. (B) Similar assay as Fig. 1B using LTR truncation mutants. Relative luciferase expression was calculated by first dividing the firefly luciferase (FL) signal by the Renilla luciferase signal in a given cell, and the resulting ratio was then normalized to the ratio obtained from cells transfected with LTR-FL (set to 1). Results shown are means ± SEs from three independent experiments performed in duplicate. (C) Schematic of HTLV-1 R region 3′ truncation mutants. (D) Similar assay as Fig. 1B using HTLV-1 R region 3′ truncation mutants. Results shown are means ± SEs from three independent experiments performed in duplicate. (E) Schematic of HTLV-1 R region serial deletion mutants. (F) Similar assay as Fig. 1B using HTLV-1 R region serial deletion mutants. Results shown are means ± SEs from three independent experiments performed in duplicate.
Putative YY1 DNA-Binding Sites in the HTLV-1 LTR Are Not Required for YY1-Mediated Transactivation.
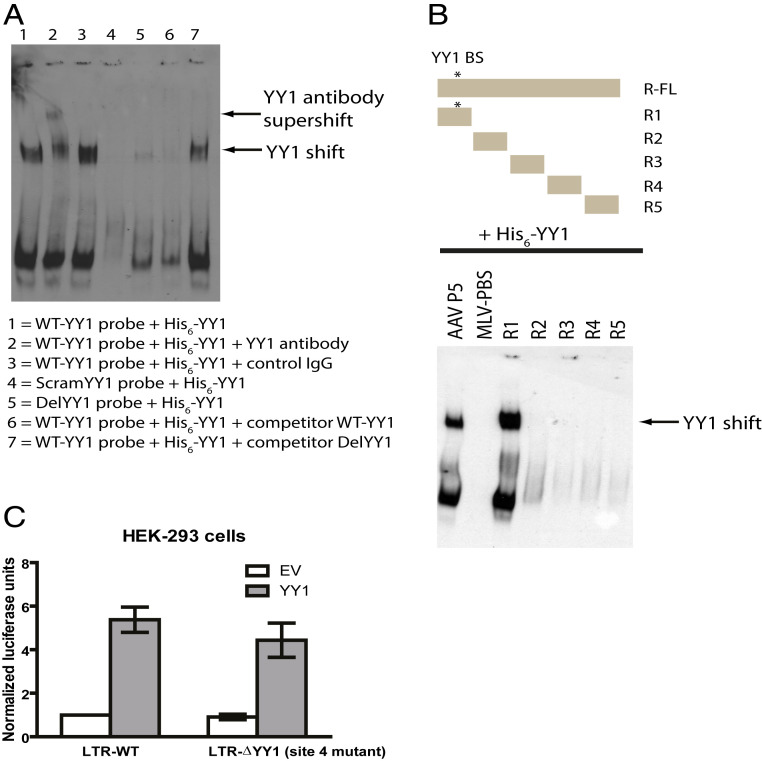
The R region contains a putative YY1 DNA-binding site near its 5′ end. We first carried out gel-shift assays to detect the binding of recombinant YY1 to this sequence in vitro. Biotinylated oligonucleotides containing the YY1-binding site were incubated with His-tagged YY1. DNA–protein complexes were resolved on a native polyacrylamide gel, transferred to membrane and probed with a streptavidin-horseradish peroxidase (HRP) conjugate to detect the migration of DNA–protein complexes. Recombinant YY1 specifically bound the oligonucleotide containing the putative DNA-binding site, and this complex could be supershifted by incubation with an anti-YY1 antibody (Fig. 4A). Binding was not observed when the binding site was mutated or deleted from the sequence, and YY1 binding to the probe could be competed by excess unmodified probe (Fig. 4A). To address whether purified YY1 can bind to other regions of R DNA, we used a series of biotinylated oligonucleotides spanning the R region (Fig. 4 B, Upper). Biotinylated DNA derived from adeno-associated virus (AAV-P5) promoter was used as a positive control (37), and MLV-PBS (primer-binding site) was used as a negative control. Apart from the R fragment containing the YY1 consensus-binding site, no binding was observed between YY1 and any other regions of R (Fig. 4 B, Lower). We next deleted the YY1 DNA-binding site in the context of our luciferase reporter construct. YY1-mediated transactivation was unaffected by this deletion (Fig. 4C), suggesting that the YY1 DNA-binding site is dispensable for transactivation. In addition, we ruled out the possible involvement of the other four YY1 DNA-binding sites located in the U3 and U5 regions of the LTR by creating binding site mutants individually or in tandem; none of these mutants significantly reduced YY1-mediated transactivation (SI Appendix, Fig. S3A). Together, these results are consistent with our above truncation analysis (Fig. 3 D and F), demonstrating the presence of multiple YY1 responsive elements, or of a large structured element, in the R region.
Fig. 4.
YY1 binds to the YY1 binding site in the HTLV-1 R region DNA in vitro, but the binding site is not required for transactivation in vivo. (A) Electrophoretic mobility shift assays with recombinant His-tagged YY1 were performed using biotin-labeled HTLV-1 R YY1-binding site (WT-YY1 probe), or Scrambled (ScramYY1 probe), or deleted (DelYY1 probe) YY1-binding site DNAs. Anti-YY1 (lane 2) or nonspecific IgG (lane 3) antibodies were used for supershift assay. Unmodified probes (WT-YY1 or Del-YY1) were used for competition experiments. Data shown are representative of two independent experiments. (B) Similar experiments in A using HTLV-1 R fragments (Upper). Location of YY1 binding site (YY1 BS) is indicated by the "*". AAV-P5 denotes adeno-associated virus P5 promoter DNA. MLV-PBS denotes Moloney murine leukemia virus primer-binding site DNA. Data shown are representative of two independent experiments. (C) Similar experiments in Fig. 1B using HTLV-1 LTR YY1-binding site 4 deletion mutant. Results shown are means ± SEs from three independent experiments performed in duplicate.
Characterizing the Orientation and Position Dependence of the YY1 Responsive Element.
The R region is present at the 5′ UTR of viral transcripts, and YY1 is known to bind RNA (25, 26, 28). This prompted us to investigate the possibility that the HTLV-1 R region may function as an RNA, rather than a DNA element. Since the SV40 early promoter is by itself unaffected by YY1 overexpression (Fig. 1B), we reasoned that adding the HTLV-1 R region in cis might render the SV40 promoter YY1 responsive. Various fusion promoter constructs were made to test the activity and position dependence of the HTLV-1 R element (Fig. 5A). Insertion of the R region did indeed confer YY1 responsiveness to the promoter. Interestingly, YY1-mediated transactivation of the SV40-R fusion promoter was observed only when R was inserted downstream of the SV40 promoter transcriptional start site (TSS), while no transactivation was seen when R was inserted either upstream of the TSS or downstream of the site of addition of the poly-A tail (Fig. 5B). As a control, insertion of the HTLV-1 U3 sequence downstream of the SV40 TSS did not induce YY1 responsiveness. These observations suggest that R must be part of the RNA transcript for efficient activation. Consistent with this, YY1-mediated transactivation of R was also found to be orientation dependent, as inversion of R (Fig. 5C) completely abolished YY1-mediated transactivation (Fig. 5D). Together, these observations suggest that YY1-mediated activation of HTLV-1 may occur via an RNA, rather than a DNA element.
Fig. 5.
Position and orientation dependence of HTLV-1 YY1 responsive element. (A) Schematic of HTLV-1 R-SV40 fusion promoter constructs. (B) Similar experiment as Fig. 1B using HTLV-1 R-SV40 fusion promoter constructs. Results shown are means ± SEs from three independent experiments performed in duplicate. (C) Schematic of HTLV-1 R sense and antisense orientation constructs. (D) Similar experiments in Fig. 1B using HTLV-1 R sense and antisense orientation constructs. Results shown are means ± SEs from three independent experiments performed in duplicate.
YY1 Associates with RNA Derived from HTLV-1 R Region In Vitro and In Vivo.
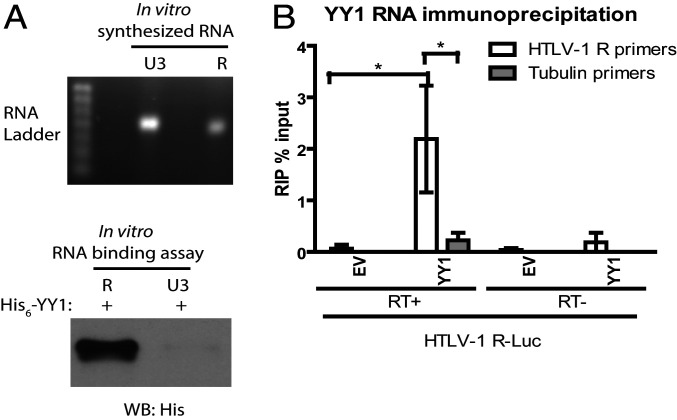
To address the possibility that YY1 may bind specifically to HTLV-1 R RNA, we first carried out in vitro RNA pulldown assays. Biotin-labeled RNAs containing the HTLV-1 R region, or the HTLV-1 U3 region as control, were synthesized in vitro (Fig. 6 A, Upper) and immobilized on streptavidin beads. Beads were then incubated with recombinant His-tagged YY1 and washed, and the bound proteins were eluted, resolved by sodium dodecyl sulfate/polyacrylamide gel electrophoresis (SDS/PAGE), and subjected to Western blotting with an anti-His antibody (Fig. 6 A, Lower). YY1 associated specifically with HTLV-1 R region, but not U3 region RNA under stringent high salt conditions (Fig. 6A). To detect YY1 HTLV-1 RNA interaction in vivo, HEK-293 cells were transfected with plasmids expressing HA-tagged YY1 and an HTLV-1 R luciferase reporter, followed by UV cross-linking and immunoprecipitation of YY1–RNA complexes using an anti-HA antibody. Bound RNA was eluted from beads and subjected to qRT-PCR with primers targeting the HTLV-1 R region. YY1 associated specifically with HTLV-1 R RNA, and much less so with tubulin mRNA (Fig. 6B). Very little qRT-PCR signal was observed in samples analyzed without RT, arguing against DNA contamination. Taken together, these results suggest a specific and direct interaction between YY1 and the HTLV-1 R RNA sequence in vivo.
Fig. 6.
YY1 binds to HTLV-1 R in vitro and in vivo. (A) Bacterially expressed His-tagged YY1 was subjected to an in vitro RNA-binding assay using biotin-labeled HTLV-1 R RNA or control RNA. (Upper) In vitro synthesized RNA used for pulldown was visualized using agarose electrophoresis. (Lower) Formation of YY1–RNA complex monitored using WB with an anti-His antibody. (B) UV-crosslink RNA immunoprecipitation (RIP) of HEK-293 cells transfected with HA-YY1 together with HTLV-1 R luciferase followed by qRT-PCR with primers specific for HTLV-1 R or tubulin. RIP data are presented as percent of input RNA (means ± SEs from three independent experiments). Student’s t test was used for statistical analysis. *P < 0.05.
YY1 Overexpression Causes Increased Transcription Initiation and Elongation from the HTLV-1 LTR.
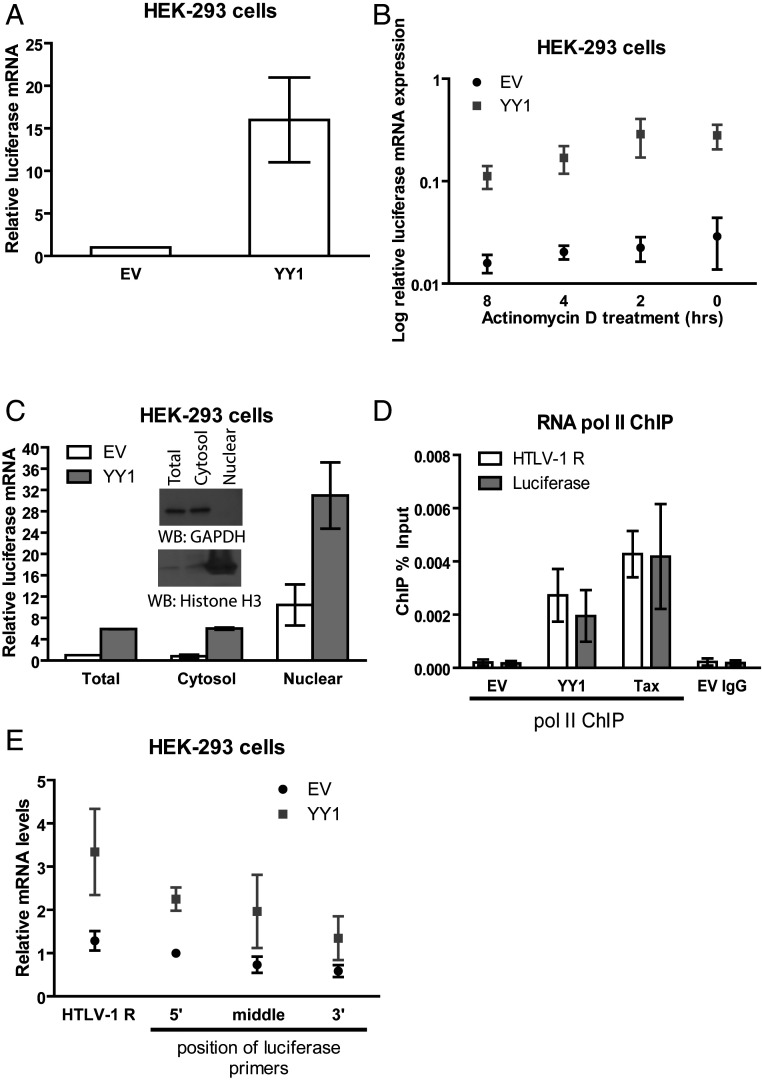
Since YY1 binds HTLV-1 R RNA and causes robust increase in HTLV-1 LTR-driven reporter expression, we hypothesized that the induced expression is a consequence of increased mRNA levels and unlikely to be an effect on reporter protein stability. To address this, we measured reporter mRNA levels by qRT-PCR and found that, indeed, total luciferase mRNA levels were markedly up-regulated in YY1-overexpressing cells (Fig. 7A). This increase in principle could be due to either increased transcription initiation, transcription elongation, nuclear export, mRNA stability, or a combination of these. To assess mRNA stability, control or YY1-overexpressing cells were treated with actinomycin D for different times to inhibit transcription and then subjected to qRT-PCR to measure mRNA abundance. Compared to cells transfected with empty vector, we detected significantly higher luciferase mRNA levels in YY1-overexpressing cells at all time points tested (Fig. 7B). However, the rate of mRNA decay as determined by the slope of the mRNA abundance versus time curve was similar between control and YY1-overexpressing cells (Fig. 7B). Thus, increased mRNA stability is unlikely to account for YY1’s mechanism of action. To assess whether YY1 overexpression enhances nuclear export of HTLV-1 RNA, we carried out nuclear/cytoplasmic fractionation of HEK-293 cells, followed by RNA extraction and quantification of luciferase mRNA levels using qRT-PCR. The efficiency of our fractionation of nuclear and cytoplasmic extracts was confirmed using anti-histone H3 and GAPDH antibodies, respectively (Fig. 7C). If YY1 enhanced nuclear export of HTLV-1 RNA, one would expect increased luciferase mRNA levels in the cytoplasm with a corresponding decrease in levels in the nuclear fraction. Though YY1 overexpression resulted in the increased levels of luciferase mRNA in the cytoplasmic fraction, increased levels were also observed in the nuclear fraction (Fig. 7C), making increased mRNA export unlikely. To assess the rate of transcription of the reporter DNA, we used chromatin IP (ChIP) assays to measure the abundance of RNA Pol II localization on the viral DNA with and without overexpression of YY1. This assay revealed a dramatic increase in the recovery of viral reporter DNA precipitated with antibodies to Pol II with YY1 overexpression (Fig. 7D). A similar effect was observed in positive control cells overexpressing the viral-encoded transactivator Tax (Fig. 7D). Finally, to provide another direct readout of the rate of transcription of the reporter genome, nuclear run-on assays were carried out. Nuclei isolated from HEK-293 cells were supplied with biotin-uridine triphosphate (UTP), and incubated at 30 °C for 30 min to allow for run-on transcription (38). Newly synthesized transcripts labeled with biotin-UTP were then isolated using streptavidin-coated magnetic beads, followed by qRT-PCR using primers spanning the entire luciferase gene transcript to monitor transcript abundance. YY1 overexpression resulted in a 2.5- to 3-fold increase in the rates of nascent transcript synthesis in the levels of both the R and luciferase RNAs (Fig. 7E). This suggests that increased transcription per se is responsible for the activity of YY1. The increase in RNAs from both regions of the genome further suggests that increased initiation of transcription at the TSS, rather than an increased release from a block to elongation occurring after synthesis of the R region, is the mechanism of action of YY1. Taken together, these results suggest YY1 binding to HTLV-1 R RNA results in enhanced transcription initiation and elongation, increased mRNA levels, and increased reporter gene expression.
Fig. 7.
YY1 overexpression causes increased transcriptional initiation and elongation from the HTLV-1 LTR. (A) Total RNA prepared from HEK-293 cells was subjected to qRT-PCR. Levels of luciferase transcripts were first normalized using the 2-ΔΔCT method to the value obtained for the alpha-tubulin gene; the obtained values were then normalized to the values obtained from HEK-293 cells transfected with EV (set to 1). Results shown are means ± SEs from three independent experiments. (B) Total RNA prepared from HEK-293 cells treated with 1 μM actinomycin D for the indicated time points was subjected to qRT-PCR as in A. Results shown are means ± SDs from two independent experiments. (C) Total RNA prepared from nuclear/cytoplasmic fractionation was subjected to qRT-PCR as in A. Results shown are means ± SDs from two independent experiments. Effectiveness of nuclear/cytoplasmic fractionation was confirmed by WB using anti-GAPDH (cytosolic control) or anti-histone H3 (nuclear control) antibodies. (D) RNA polymerase II ChIP analysis of chromatin harvested from HEK-293 cells transfected with empty vector, HA-YY1, or Tax plasmid together with HTLV-1 LTR luciferase plasmid. ChIP data are presented as percent of input DNA (means ± SDs from two independent experiments). (E) Biotin-labeled RNA prepared from nuclear run-on assay in HEK-293 cells was isolated using streptavidin-coated magnetic beads, followed by qRT-PCR to monitor transcript abundance using primers spanning the entire luciferase transcript, including the 5′ UTR. Levels of luciferase transcripts were first normalized using the 2-ΔΔCT method to the value obtained for the alpha-tubulin gene; the obtained values were then normalized to the values obtained from 5′ luciferase primer of HEK-293 cells transfected with EV (set to 1). Results shown are means ± SDs from two independent experiments.
Discussion
Here we report the finding that YY1 acts as a potent transactivator of the HTLV-1 LTR, in sharp contrast to its repression of other retroviral genomes. The HTLV-1 LTR sequence required for YY1 responsiveness lies in the R region, present in the 5′ UTR of the viral mRNA. Based on YY1’s previous roles in mediating transcriptional repression of HIV-1 (21, 22) and MLV (23, 24) via DNA binding, we initially expected that YY1’s effect on HTLV-1 would also occur through DNA binding. Recombinant YY1 specifically associates with DNAs containing a YY1-binding site at the 5′ end of R (Fig. 4A). However, deletion of this site (Fig. 4C), or any other YY1 DNA-binding site in the HTLV-1 LTR had no effect on YY1-mediated transactivation in vivo (SI Appendix, Fig. S3A). Instead, deletion of R from the 3′ end resulted in progressive loss of YY1 responsiveness (Fig. 3D), and sequential 25 base pair deletion across the entire region of R resulted in partial loss of YY1 responsiveness in almost all mutants (Fig. 3F), suggesting the presence of multiple YY1 responsive elements, or binding to a large structure. Since YY1 does not bind R DNA lacking the YY1-binding site (Fig. 4B), our data raised the possibility that the HTLV-1 R may be acting as an RNA element. Consistent with this, insertion of the R region sequence provided YY1 responsiveness to a nonresponsive SV40 early promoter, but only when R was included as part of the transcription unit (Fig. 5B). In addition, the R region lost YY1 responsiveness when it was inverted (Fig. 5D). Together, the position and orientation dependence of the HTLV-1 R region argues for its functioning as an RNA element.
Recombinant YY1 specifically associated in vitro with HTLV-1 R RNA, and not U3 RNA, under stringent conditions (Fig. 6A). Furthermore, RNA immunoprecipitation experiments in HEK-293 cells revealed YY1 binding to HTLV-1 R RNA, but not to tubulin mRNA in vivo (Fig. 6B). Previous studies examining the RNA-binding activity of YY1 showed that YY1 does not bind to the RNA equivalent of its consensus DNA sequence (26), and in vitro binding assays show a preference for AU-rich and poly-U containing duplexes (26). HTLV-1 R does not contain AU-rich or poly-U stretches. It is possible that HTLV-1 R region RNA adopts a secondary structure recognized by YY1, in a manner similar to the Tar element of HIV-1 recognized by the HIV-1 Tat protein. In preliminary experiments, structures of the R region predicted by RNA folding programs (MFold and RNAfold) resulted in several nearly isoenergetic folds with distinct paired regions. We have made several deletions aimed at selectively removing predicted stem loops based on these predicted structures and tested the mutant sequences for YY1 responsiveness. Some deletions had no effect, while others resulted in modest loss of YY1 responsiveness. The mutagenesis has not yet strongly supported any of the in silico predicted RNA structures, suggesting that they do not fully reflect the native fold in vivo. It is also possible that the HTLV-1 R RNA exhibits conformational flexibility in vivo, capable of adopting multiple secondary structures that associate with YY1 with similar affinities. More work will be required to determine the folded structure of this RNA.
Both the N-terminal activation and C-terminal zinc finger domains of YY1 are required for full activation of the HTLV-1 LTR (Fig. 2B). Little is known about the nature of YY1–RNA interactions. Previously, the N-terminal domain of YY1 was suggested to mediate RNA binding (28), while another study showed the C-terminal zinc fingers are sufficient (39). It is possible that the N-terminal domain of YY1 may regulate RNA binding either on its own or via the recruitment of protein partners. It has also been previously shown that YY1 forms an oligomer, which allows it to bind to DNA sequences lacking the consensus recognition site (40). In the case of HIV-1, the virus encodes the Rev protein capable of binding to and oligomerizing on the highly structured Rev response element (RRE) RNA, creating a large RNP that directs the nuclear export of fully and partially unspliced viral RNAs (41–43). It is possible that YY1 oligomerization also affects the avidity and specificity of RNA binding.
YY1 promotes MLV silencing in mouse embryonic cells (24) and HIV-1 latency in T cells (44). In the case of MLV, YY1 binding to a short consensus DNA-binding motif in the U3 region of the viral LTR led to the recruitment of the universal transcriptional repressor TRIM28, as well as downstream chromatin modifiers such as histone methyltransferase SETDB1, histone deacetylases, and DNA methyltransferases to repress viral gene expression (24). In the case of HIV-1, YY1 also binds directly to the LTR, and its affinity for DNA is enhanced via its interaction with its cofactor LSF (late SV40 factor), which in turn recruits histone deacetylase 1 (HDAC1) to promote viral latency (44, 45). In the context of this history of viral repression, YY1’s functioning to transactivate the HTLV-1 LTR is surprising. It is further surprising that it seems to act via RNA binding rather than DNA binding. YY1’s activation of expression is unlikely to be a result of increased mRNA stability (Fig. 7B) or nuclear export (Fig. 7C). but rather most likely involves directly increasing transcription from the HTLV-1 LTR (Fig. 7 D and E). YY1 appears to function independently of the known viral transactivator Tax, as deletion of the Tax response element, which is located in the U3 region of the LTR, had no effect on YY1-mediated transactivation (SI Appendix, Fig. S2). In ATL cells, the 5′ LTR is frequently deleted, resulting in a defective provirus (46, 47). YY1 likely only functions on intact 5′ LTRs to drive initial viral transcription during early infection, prior to and after the production of Tax. In addition, YY1 could contribute to the initial, transient burst of sense transcript production upon reactivation from latency (48). We do not expect YY1 to play a major role in the antisense expression of the HBZ protein in transformed cells, though this possibility remains to be tested. Future studies should examine the effect of manipulating YY1 levels on early viral spread in acutely infected primary T cells. YY1’s ability to associate with HTLV-1 R RNA may be analogous to the association of HIV-1 Tat with TAR RNA, which also maps to the R region of the LTR. However, their mechanism of action is likely different. YY1 overexpression resulted in 1) increased Pol II occupancy on R and luciferase DNA (Fig. 7D), and 2) increased rates of nascent transcript synthesis in the sequences of both the R region and luciferase RNA (Fig. 7E). These observations suggest YY1 functions to increase the initiation of transcription rather than increasing the release from a block to elongation occurring after synthesis of the R region, as it is the case for HIV-1 Tat.
Very few examples of activation of expression by YY1 have been studied in detail. YY1 was shown to bind the AAV P5 DNA at the +1 region and to initiate efficient basal transcription by polymerase II in the absence of TATA-box binding protein (49, 50). Another study showed that YY1 binds to a consensus DNA-binding site in the GDAP1 promoter to activate transcription (51). Our observations that YY1 promotes transcription from the HTLV-1 LTR via RNA binding may be similar to findings in a recent study showing widespread YY1 trapping at gene regulatory elements via binding to nascent transcripts (28). In preliminary experiments, knockdown of several known YY1 coactivators of transcription (19) using RNAi did not abolish YY1-mediated HTLV-1 transactivation (SI Appendix, Fig. S4). It is possible that these cofactors are functionally redundant, or that the set of coactivators required for YY1-mediated transcription via RNA binding differs from those mediated through DNA binding. Another possible player is CTCF, the CCCTC-binding factor important in chromosome topology. YY1 interacts with CTCF and shows extensive colocalization at CTCF-bound regions with active histone marks (52). The HTLV-1 genome contains a CTCF-binding site located in the pX region (53), but the site is far away from the YY1 responsive element, and absent in our reporter constructs, making it unlikely that CTCF is involved in YY1-mediated transactivation of the HTLV-1 LTR. In other contexts, YY1 is able to exert its effect on transcription through microRNAs (54), but we have no evidence that YY1 is utilizing microRNAs for the activation of HTLV-1. Recently, RNA binding motif protein 25 (RBM25), an RNA-binding protein previously implicated in splicing regulation, was shown to colocalize with YY1 on gene promoters, and RBM25 depletion results in attenuation of YY1-dependent activities, including chromatin binding, DNA looping, and transcription (55). In preliminary experiments, RBM25 knockdown only resulted in a modest (twofold) decrease in YY1-mediated HTLV-1 transactivation, and no decrease in YY1 binding to HTLV-1 R RNA was observed. More work is required to determine the possible involvement of RBM25 in YY1-mediated transactivation of the HTLV-1 LTR.
In sum, we here demonstrated that the HTLV-1 LTR is a YY1 responsive promoter whose activation involves RNA binding. Our observations thus suggest a significant expansion of the potential activities and targets of the YY1 factor.
Materials and Methods
Cells and Transfections.
HEK-293 cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% heat-inactivated fetal bovine serum (FBS). Jurkat cells were cultured in RPMI medium supplemented with 10% heat-inactivated FBS. All cells were maintained in a 37 °C incubator with 5% CO2. Transient transfections of HEK-293 cells and Jurkat cells were carried out using Lipofectamine LTX reagent (Invitrogen) according to the manufacturer’s instructions.
Dual-Luciferase Assays.
Dual-luciferase assays were performed as previously described (56). Briefly, HEK-293 or Jurkat cells cultured in 24-well plates were transfected with 400 ng of pCMV vector, pCMV-HA-YY1 or pCMV-Tax DNA, 100 ng of HTLV-1 LTR firefly luciferase construct and 10 ng of HSV-TK Renilla luciferase control plasmid using Lipofectamine LTX (Life Sciences). One to 2 d later, cells were lysed in 1× reporter lysis buffer (Promega) and levels of luciferase were quantified using a POLARstar Omega multimode plate reader (BMG Labtech) according to the manufacturer’s instructions. Relative luciferase expression was calculated by first dividing the firefly luciferase signal by the Renilla luciferase signal in the transfected cells, and the resulting ratio was then normalized to the ratio obtained from cells transfected with EV, set to 1.
In Vitro RNA-Binding Assay.
In vitro RNA-binding assays were performed as previously described (56). Briefly, RNA fragments were transcribed in vitro from linearized plasmid DNA using T7 polymerase in the presence of biotin-16-UTP (Roche Applied Science), treated with DNase, purified, and renatured by heating to 95 °C for 10 min followed by slow cooling to room temperature. Each pulldown assay consists of incubating 1 μg of renatured RNA immobilized on Dynabeads MyOne streptavidin (Invitrogen) with 1 μg of recombinant His-YY1, purified from Escherichia coli, for 2 h at room temperature in high salt phosphate buffered saline (PBS) supplemented with 500 mM NaCl, 2 mM MgCl2, 0.2 mM ZnCl2, 15 mM β-mercaptoethanol, 100 units/mL RNase inhibitor, 0.1 mg/mL yeast tRNA (Ambion), 0.05% bovine serum albumin, and 0.2% Nonidet P-40. Beads were then washed four times using the same binding buffer and twice with regular PBS and subjected to SDS/PAGE and immunoblotting with anti-His antibodies.
RNA Immunoprecipitation.
Ten million HEK-293 cells cotransfected with plasmids encoding HA-YY1 and LTR luciferase reporter plasmids were UV cross-linked at 254 nm (4,000 J/m2) and lysed in RIPA buffer (10 mM Tris-Cl pH 8.0, 1 mM ethylenediaminetetraacetic acid (EDTA), 1% Triton X-100, 140 mM NaCl, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, 1 mM dithiothreitol, 10 U RNaseOUT/mL) for 30 min at 4 °C. After centrifugation for 20 min at 13,000 rpm, the supernatant was incubated with 10 μL of anti-HA magnetic beads (Pierce) for 8 h at 4 °C. Beads were washed three times in RIPA buffer, once in PBS, and DNase treated (10 U) for 30 min at 37 °C. Bound RNA was eluted by incubating in elution buffer (100 mM Tris⋅HCl pH 7.5, 50 mM NaCl, 10 mM EDTA, 30 μg of Proteinase K, 0.5% SDS) for 30 min at 55 °C. RNA was recovered by TRIzol (Invitrogen) extraction and reverse transcribed with random primers using the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). qRT-PCR was performed using SYBR Green PCR Master Mix (Roche) containing 15 pmol of indicated primers (SI Appendix, Table S1) in 25-μL reactions. PCRs were performed in 96-well plates using the 7900 Fast Real-Time PCR System (Applied Biosystems) with the following reaction conditions: 10 min at 95 °C, followed by 40 cycles of 30 s at 95 °C, 30 s at 60 °C, and 30 s at 72 °C. Relative enrichment was calculated by determining the bound/input value using the 2-ΔΔCT method (57).
Cell Fractionation and RT-PCR.
HEK-293 cells from a confluent 10-cm2 plate were washed twice with ice-cold PBS and lysed on ice for 10 min with buffer A (10 mM Tris⋅HCl, pH 7.5, 140 mM NaCl, 1.5 mM MgCl2, 10 mM EDTA, 0.5% Nonidet P-40, 100 U/mL of RNaseOUT [Invitrogen]). Following centrifugation at 3,000 rpm for 5 min, the resulting supernatant was taken as the cytosolic fraction. The pellet was washed twice in buffer A and taken as the nuclear fraction. Total RNAs from cytosolic and nuclear fractions were isolated using TRIzol and subjected to reverse transcription and qRT-PCR as described above.
Supplementary Material
Acknowledgments
This study was supported in part by a grant from the NIH (R01 CA 030488). S.P.G. is a Howard Hughes Medical Institute investigator.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2005726117/-/DCSupplemental.
Data Availability.
Data supporting the findings of this study are included in the main manuscript and SI Appendix. All reagents made (e.g., plasmids) are available upon request.
References
- 1.Blattner W. A. et al., The human type-C retrovirus, HTLV, in Blacks from the Caribbean region, and relationship to adult T-cell leukemia/lymphoma. Int. J. Cancer 30, 257–264 (1982). [DOI] [PubMed] [Google Scholar]
- 2.Hinuma Y. et al., Adult T-cell leukemia: Antigen in an ATL cell line and detection of antibodies to the antigen in human sera. Proc. Natl. Acad. Sci. U.S.A. 78, 6476–6480 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gessain A. et al., Antibodies to human T-lymphotropic virus type-I in patients with tropical spastic paraparesis. Lancet 2, 407–410 (1985). [DOI] [PubMed] [Google Scholar]
- 4.Osame M. et al., HTLV-I associated myelopathy, a new clinical entity. Lancet 1, 1031–1032 (1986). [DOI] [PubMed] [Google Scholar]
- 5.Speck N. A., Baltimore D., Six distinct nuclear factors interact with the 75-base-pair repeat of the Moloney murine leukemia virus enhancer. Mol. Cell. Biol. 7, 1101–1110 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brady J., Jeang K. T., Duvall J., Khoury G., Identification of p40x-responsive regulatory sequences within the human T-cell leukemia virus type I long terminal repeat. J. Virol. 61, 2175–2181 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jeang K. T., Boros I., Brady J., Radonovich M., Khoury G., Characterization of cellular factors that interact with the human T-cell leukemia virus type I p40x-responsive 21-base-pair sequence. J. Virol. 62, 4499–4509 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adya N., Giam C. Z., Distinct regions in human T-cell lymphotropic virus type I tax mediate interactions with activator protein CREB and basal transcription factors. J. Virol. 69, 1834–1841 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franklin A. A. et al., Transactivation by the human T-cell leukemia virus Tax protein is mediated through enhanced binding of activating transcription factor-2 (ATF-2) ATF-2 response and cAMP element-binding protein (CREB). J. Biol. Chem. 268, 21225–21231 (1993). [PubMed] [Google Scholar]
- 10.Yoshimura T., Fujisawa J., Yoshida M., Multiple cDNA clones encoding nuclear proteins that bind to the tax-dependent enhancer of HTLV-1: All contain a leucine zipper structure and basic amino acid domain. EMBO J. 9, 2537–2542 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harrod R. et al., p300 and p300/cAMP-responsive element-binding protein associated factor interact with human T-cell lymphotropic virus type-1 Tax in a multi-histone acetyltransferase/activator-enhancer complex. J. Biol. Chem. 275, 11852–11857 (2000). [DOI] [PubMed] [Google Scholar]
- 12.Jiang H. et al., PCAF interacts with tax and stimulates tax transactivation in a histone acetyltransferase-independent manner. Mol. Cell. Biol. 19, 8136–8145 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwok R. P. et al., Control of cAMP-regulated enhancers by the viral transactivator Tax through CREB and the co-activator CBP. Nature 380, 642–646 (1996). [DOI] [PubMed] [Google Scholar]
- 14.Gitlin S. D., Dittmer J., Shin R. C., Brady J. N., Transcriptional activation of the human T-lymphotropic virus type I long terminal repeat by functional interaction of Tax1 and Ets1. J. Virol. 67, 7307–7316 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suzuki T., Hirai H., Fujisawa J., Fujita T., Yoshida M., A trans-activator Tax of human T-cell leukemia virus type 1 binds to NF-kappa B p50 and serum response factor (SRF) and associates with enhancer DNAs of the NF-kappa B site and CArG box. Oncogene 8, 2391–2397 (1993). [PubMed] [Google Scholar]
- 16.Chen F., Sun H., Zhao Y., Wang H., YY1 in cell differentiation and tissue development. Crit. Rev. Oncog. 22, 131–141 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Donohoe M. E. et al., Targeted disruption of mouse Yin Yang 1 transcription factor results in peri-implantation lethality. Mol. Cell. Biol. 19, 7237–7244 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yant S. R. et al., High affinity YY1 binding motifs: Identification of two core types (ACAT and CCAT) and distribution of potential binding sites within the human beta globin cluster. Nucleic Acids Res. 23, 4353–4362 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gordon S., Akopyan G., Garban H., Bonavida B., Transcription factor YY1: Structure, function, and therapeutic implications in cancer biology. Oncogene 25, 1125–1142 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Shi Y., Lee J. S., Galvin K. M., Everything you have ever wanted to know about Yin Yang 1. Biochim. Biophys. Acta 1332, F49–F66 (1997). [DOI] [PubMed] [Google Scholar]
- 21.Bernhard W., Barreto K., Raithatha S., Sadowski I., An upstream YY1 binding site on the HIV-1 LTR contributes to latent infection. PLoS One 8, e77052 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Margolis D. M., Somasundaran M., Green M. R., Human transcription factor YY1 represses human immunodeficiency virus type 1 transcription and virion production. J. Virol. 68, 905–910 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flanagan J. R. et al., Cloning of a negative transcription factor that binds to the upstream conserved region of Moloney murine leukemia virus. Mol. Cell. Biol. 12, 38–44 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schlesinger S., Lee A. H., Wang G. Z., Green L., Goff S. P., Proviral silencing in embryonic cells is regulated by Yin Yang 1. Cell Rep. 4, 50–58 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Belak Z. R., Ficzycz A., Ovsenek N., Biochemical characterization of Yin Yang 1-RNA complexes. Biochem. Cell Biol. 86, 31–36 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Belak Z. R., Ovsenek N., Assembly of the Yin Yang 1 transcription factor into messenger ribonucleoprotein particles requires direct RNA binding activity. J. Biol. Chem. 282, 37913–37920 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Jeon Y., Lee J. T., YY1 tethers Xist RNA to the inactive X nucleation center. Cell 146, 119–133 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sigova A. A. et al., Transcription factor trapping by RNA in gene regulatory elements. Science 350, 978–981 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee T. C., Zhang Y., Schwartz R. J., Bifunctional transcriptional properties of YY1 in regulating muscle actin and c-myc gene expression during myogenesis. Oncogene 9, 1047–1052 (1994). [PubMed] [Google Scholar]
- 30.Bushmeyer S., Park K., Atchison M. L., Characterization of functional domains within the multifunctional transcription factor, YY1. J. Biol. Chem. 270, 30213–30220 (1995). [DOI] [PubMed] [Google Scholar]
- 31.Lee J. S., See R. H., Galvin K. M., Wang J., Shi Y., Functional interactions between YY1 and adenovirus E1A. Nucleic Acids Res. 23, 925–931 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Austen M., Lüscher B., Lüscher-Firzlaff J. M., Characterization of the transcriptional regulator YY1. The bipartite transactivation domain is independent of interaction with the TATA box-binding protein, transcription factor IIB, TAFII55, or cAMP-responsive element-binding protein (CPB)-binding protein. J. Biol. Chem. 272, 1709–1717 (1997). [DOI] [PubMed] [Google Scholar]
- 33.Hariharan N., Kelley D. E., Perry R. P., Delta, a transcription factor that binds to downstream elements in several polymerase II promoters, is a functionally versatile zinc finger protein. Proc. Natl. Acad. Sci. U.S.A. 88, 9799–9803 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Galvin K. M., Shi Y., Multiple mechanisms of transcriptional repression by YY1. Mol. Cell. Biol. 17, 3723–3732 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park R. E., Haseltine W. A., Rosen C. A., A nuclear factor is required for transactivation of HTLV-I gene expression. Oncogene 3, 275–279 (1988). [PubMed] [Google Scholar]
- 36.Seiki M., Hikikoshi A., Yoshida M., The U5 sequence is a cis-acting repressive element for genomic RNA expression of human T cell leukemia virus type I. Virology 176, 81–86 (1990). [DOI] [PubMed] [Google Scholar]
- 37.Shi Y., Seto E., Chang L. S., Shenk T., Transcriptional repression by YY1, a human GLI-Krüppel-related protein, and relief of repression by adenovirus E1A protein. Cell 67, 377–388 (1991). [DOI] [PubMed] [Google Scholar]
- 38.Patrone G. et al., Nuclear run-on assay using biotin labeling, magnetic bead capture and analysis by fluorescence-based RT-PCR. Biotechniques 29, 1012–1014–1016–1017 (2000). [DOI] [PubMed] [Google Scholar]
- 39.Wai D. C., Shihab M., Low J. K., Mackay J. P., The zinc fingers of YY1 bind single-stranded RNA with low sequence specificity. Nucleic Acids Res. 44, 9153–9165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.López-Perrote A. et al., Structure of Yin Yang 1 oligomers that cooperate with RuvBL1-RuvBL2 ATPases. J. Biol. Chem. 289, 22614–22629 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Daugherty M. D., Liu B., Frankel A. D., Structural basis for cooperative RNA binding and export complex assembly by HIV Rev. Nat. Struct. Mol. Biol. 17, 1337–1342 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Malim M. H., Cullen B. R., HIV-1 structural gene expression requires the binding of multiple Rev monomers to the viral RRE: Implications for HIV-1 latency. Cell 65, 241–248 (1991). [DOI] [PubMed] [Google Scholar]
- 43.Daugherty M. D., Booth D. S., Jayaraman B., Cheng Y., Frankel A. D., HIV Rev response element (RRE) directs assembly of the Rev homooligomer into discrete asymmetric complexes. Proc. Natl. Acad. Sci. U.S.A. 107, 12481–12486 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coull J. J. et al., The human factors YY1 and LSF repress the human immunodeficiency virus type 1 long terminal repeat via recruitment of histone deacetylase 1. J. Virol. 74, 6790–6799 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He G., Margolis D. M., Counterregulation of chromatin deacetylation and histone deacetylase occupancy at the integrated promoter of human immunodeficiency virus type 1 (HIV-1) by the HIV-1 repressor YY1 and HIV-1 activator Tat. Mol. Cell. Biol. 22, 2965–2973 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miyazaki M. et al., Preferential selection of human T-cell leukemia virus type 1 provirus lacking the 5′ long terminal repeat during oncogenesis. J. Virol. 81, 5714–5723 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tamiya S. et al., Two types of defective human T-lymphotropic virus type I provirus in adult T-cell leukemia. Blood 88, 3065–3073 (1996). [PubMed] [Google Scholar]
- 48.Miura M. et al., Kinetics of HTLV-1 reactivation from latency quantified by single-molecule RNA FISH and stochastic modelling. PLoS Pathog. 15, e1008164 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seto E., Shi Y., Shenk T., YY1 is an initiator sequence-binding protein that directs and activates transcription in vitro. Nature 354, 241–245 (1991). [DOI] [PubMed] [Google Scholar]
- 50.Usheva A., Shenk T., TATA-binding protein-independent initiation: YY1, TFIIB, and RNA polymerase II direct basal transcription on supercoiled template DNA. Cell 76, 1115–1121 (1994). [DOI] [PubMed] [Google Scholar]
- 51.Ratajewski M., Pulaski L., YY1-dependent transcriptional regulation of the human GDAP1 gene. Genomics 94, 407–413 (2009). [DOI] [PubMed] [Google Scholar]
- 52.Schwalie P. C. et al., Co-binding by YY1 identifies the transcriptionally active, highly conserved set of CTCF-bound regions in primate genomes. Genome Biol. 14, R148 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Satou Y. et al., The retrovirus HTLV-1 inserts an ectopic CTCF-binding site into the human genome. Proc. Natl. Acad. Sci. U.S.A. 113, 3054–3059 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Khachigian L. M., The Yin and Yang of YY1 in tumor growth and suppression. Int. J. Cancer 143, 460–465 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Xiao R. et al., Pervasive chromatin-RNA binding protein interactions enable RNA-based regulation of transcription. Cell 178, 107–121.e18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang G. Z., Goff S. P., Regulation of Yin Yang 1 by tyrosine phosphorylation. J. Biol. Chem. 290, 21890–21900 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Livak K. J., Schmittgen T. D., Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting the findings of this study are included in the main manuscript and SI Appendix. All reagents made (e.g., plasmids) are available upon request.