Significance
Animal NLRs form wheel-like structures called inflammasomes upon perception of pathogen-associated molecules. The induced proximity of the signaling domains at the center of the wheel is hypothesized to recruit caspases for the first step of immune signal transduction. We expressed a plant-animal NLR fusion to demonstrate that induced proximity of TIR signaling domains from plant NLRs is sufficient to activate plant immune signaling. This demonstrates that a signaling-competent inflammasome can be formed from known, minimal components. The intrinsic NADase activity of plant TIRs is necessary for immune signaling, but fusions to a bacterial or a mammalian TIR domain with NADase activity, which also lead to accumulation of NAD+ hydrolysis products (e.g. cyclic ADP-ribose), were unable to activate immune signaling.
Keywords: NLR immune receptors, plant immunity, inflammasome, effector-triggered immunity
Abstract
Plant and animal intracellular nucleotide-binding, leucine-rich repeat (NLR) immune receptors detect pathogen-derived molecules and activate defense. Plant NLRs can be divided into several classes based upon their N-terminal signaling domains, including TIR (Toll-like, Interleukin-1 receptor, Resistance protein)- and CC (coiled-coil)-NLRs. Upon ligand detection, mammalian NAIP and NLRC4 NLRs oligomerize, forming an inflammasome that induces proximity of its N-terminal signaling domains. Recently, a plant CC-NLR was revealed to form an inflammasome-like hetero-oligomer. To further investigate plant NLR signaling mechanisms, we fused the N-terminal TIR domain of several plant NLRs to the N terminus of NLRC4. Inflammasome-dependent induced proximity of the TIR domain in planta initiated defense signaling. Thus, induced proximity of a plant TIR domain imposed by oligomerization of a mammalian inflammasome is sufficient to activate authentic plant defense. Ligand detection and inflammasome formation is maintained when the known components of the NLRC4 inflammasome is transferred across kingdoms, indicating that NLRC4 complex can robustly function without any additional mammalian proteins. Additionally, we found NADase activity of a plant TIR domain is necessary for plant defense activation, but NADase activity of a mammalian or a bacterial TIR is not sufficient to activate defense in plants.
Plants lack an adaptive immune system and rely on innate immunity to defend against pathogens (1, 2). Upon perception of pathogen effector proteins by plant nucleotide-binding, leucine-rich repeat (NLR) intracellular receptors, immune signaling is initiated that often culminates in a programmed cell death called the hypersensitive response (HR) (3). NLRs carry an N-terminal signaling domain, a nucleotide-binding (NB) domain, and a C-terminal leucine-rich repeat (LRR) domain (2). The P-loop (or Walker A) motif contained in the NB domain binds ATP or ADP. NLRs typically bind ADP in their resting state and exchange it for ATP when activated (1, 2, 4). The two major classes of plant NLRs are defined by the N-terminal signaling domain they contain: either a TIR (Toll-like, Interleukin-1 receptor, Resistance protein) or a CC (coiled-coil) domain (2). Ectopic expression of TIR and CC domains can activate immune signaling (1, 5–7). Self-association interfaces are required for TIR-mediated immune signaling (8–10), but the mechanism of signaling activation is unknown.
Animal NLR domain architecture resembles plant NLRs (1, 2, 11). NLRC4, a mammalian NLR, contains an N-terminal caspase activation and recruitment domain (CARD), a NACHT NB domain, and a C-terminal LRR. NLRC4 cooperates with NAIP NLRs to detect bacterial PAMPs (pathogen-associated molecular patterns); in mice, NAIP1 and NAIP2 detect type III secretion system (T3SS) needle and rod components, respectively, and NAIP5 detects flagellin (12, 13). The PAMP binds to the NAIP protein, altering its conformation and provoking recruitment of an NLRC4 molecule, which initiates stepwise recruitment of additional NLRC4 molecules, forming a wheel-like oligomer called an inflammasome (14–16). This complex brings the N-terminal CARDs into close proximity, allowing recruitment and activation of caspases (1, 17). Hence, activation of NLRC4-mediated immune signaling occurs via induced proximity of N-terminal signaling domains.
Recently, the structure of Arabidopsis ZAR1, an NLR with a CC N-terminal signaling domain, was resolved in complex with the pseudokinase RKS1 and the decoy kinase PBL2 (18, 19). ZAR1 associates with RKS1, and the effector AvrAC uridylylates PBL2 and induces its recruitment to the ZAR1-RKS1 heterodimer (20). Subsequently, a wheel-like structure forms, termed a “resistosome,” consisting of five heterotrimeric ZAR1-RKS1-PBL2 protomers, and activates an immune response (18, 19, 21). Similar to the NLRC4-inflammasome, induced proximity is imposed on the N-terminal CC signaling domains leading to a significant structural change in this domain. This suggests that induced proximity of N-terminal signaling domains may be a conserved mechanism of signaling activation in NLRs, although it has not yet been observed in TIR-domain containing NLRs. Here, we fused the TIR domain from RPS4, a well-characterized Arabidopsis NLR (22–24), to NLRC4 to investigate whether induced proximity imposed by an animal NLR is sufficient to activate an N-terminal TIR signaling domain of a plant NLR in planta.
Some but not all TIR domains can hydrolyze NAD+ to nicotinamide and various forms of ADP ribose (ADPR) (25–28). A conserved catalytic glutamate is required for NAD+ hydrolysis (25, 26). This catalytic glutamate is also required for defense activation for plant TIRs (22, 27). Plant and bacterial TIRs, in contrast to the TIR domain of mammalian SARM1, can make a variant cyclic ADPR (v-cADPR) (25, 27, 28). Here, we use the TIR-NLRC4 platform to demonstrate that while NADase activity of plant TIRs is necessary for their activation of cell death, the in vivo generation of v-cADPR or cADPR is not sufficient to induce cell death.
Results and Discussion
We examined whether NLRC4-imposed induced proximity is sufficient to activate defense mediated by the RPS4 TIR domain (TIRRPS4). We generated a TIRRPS4-NLRC4 chimaera under control of the CaMV 35S promoter for expression in plant leaves (Fig. 1A). To determine if this construct could form a TIRRPS4-NLRC4/NAIP/PAMP inflammasome in plants, we transiently coexpressed epitope-tagged TIRRPS4-NLRC4, NAIP2, NAIP5, and either Legionella pneumophila flagellin (FlaA) or Salmonella typhimurium T3SS rod protein PrgJ in Nicotiana benthamiana. To detect inflammasomes, we separated proteins extracted from infiltrated leaves by blue native polyacrylamide gel electrophoresis (BN-PAGE) (SI Appendix, Fig. S1). We additionally purified the complex by immunoprecipitating NAIPs to remove background immunoblot signal (Fig. 1 B and C). Upon coexpression in leaves of TIRRPS4-NLRC4, NAIPs, and PAMPs, all inflammasome proteins interacted when in cognate combinations: NAIP5 and FlaA, or NAIP2 and PrgJ, but not in other NAIP-ligand combinations (Fig. 1B).
Fig. 1.
Induced proximity of TIRRPS4 triggers HR-like cell death when fused to NLRC4. (A) Schematic diagram of domain architecture of TIRRPS4-NLRC4 drawn to scale. TIR domain, Toll-like, interleukin-1 receptor, Resistance protein domain; CARD, caspase activation and recruitment domain; NACHT, NAIP, CIITA, HET-E, TP1; LRR, leucine rich repeat. TIRRPS4 contains amino acids residues 1–236 of RPS4. (B and C) Oligomerization assay to test the formation of inflammasome-like complexes in plants. N. benthamiana leaves were transiently cotransformed with combinations of TIRRPS4-NLRC4, NAIP, PAMP (as indicated by a + or – symbol), and silencing suppressor p19 by A. tumefaciens infiltration. After 3 d, leaves were harvested, proteins tagged with a FLAG epitope were immunoprecipitated and subjected to SDS-PAGE (B) and BN-PAGE (C), and immunoblotted for V5, FLAG or Myc. Results shown are representative of at least three independent replicates. See also SI Appendix, Fig. S1. Arrowhead indicates predicted inflammasome complex. HF, (His)6-(FLAG)3 tag. SDS-PAGE (D) and BN-PAGE (E) of mutant versions of TIRRPS4-NLRC4 or NAIP5. TIRRPS4-NLRC4, NAIP, and FlaA were immunoprecipitated from extracts from coinfiltrated N. benthamiana leaves. Results shown are representative of at least three independent replicates. D/A and L/D indicate NLRC4 mutations D125A and L435D, respectively; R/A indicates TIRRPS4 mutation R116A; ΔP indicates NAIP5 with a deleted P-loop (amino acids 464–487; NAIP5ΔPloop). Arrowhead indicates predicted inflammasome complex.
An oligomeric complex of >1,200 kDa, consistent with an inflammasome, also appeared upon coexpression in leaves of TIRRPS4-NLRC4 and cognate NAIP-PAMP pairs (Fig. 1C).
NLRC4 has two interaction surfaces that mediate oligomerization: the “donor” surface, which recruits NLRC4 monomers to the complex, and the “acceptor” surface, which interacts with the donor surface of other NLRC4s and NAIPs (14–16). We transiently coexpressed either an acceptor surface mutant (D125A) or a donor surface mutant (L435D) of TIRRPS4-NLRC4 with NAIP5 and FlaA. As expected, the acceptor surface mutant TIRRPS4-NLRC4(D125A) did not immunoprecipitate with NAIP5 (Fig. 1D), nor was it capable of forming an inflammasome, because the acceptor surface was prevented from associating with the donor surface of NAIP5 (Fig. 1E). The donor surface mutant TIRRPS4-NLRC4(L435D) did immunoprecipitate with NAIP5 but did not form a >1,200 kDa inflammasome (Fig. 1E). NAIP5 and FlaA immunoprecipitated with TIRRPS4-NLRC4(L435D) at a lower apparent affinity than with TIRRPS4-NLRC4 (Fig. 1D). This may be due to an avidity defect: A fully assembled inflammasome ring likely increases the apparent affinity of all members of the complex. Therefore, the TIRRPS4-NLRC4(L435D)/NAIP5/FlaA trimer may dissociate more readily than the full ∼13-mer inflammasome. Consistent with previous results (12), a P-loop motif deletion in NAIP5 prevented in planta inflammasome assembly (Fig. 1 D and E). Taken together, these results indicate that TIRRPS4-NLRC4/NAIP/PAMP can form an authentic inflammasome complex in plants.
We next asked whether a mammalian inflammasome signaling platform could activate TIR-dependent plant responses, like HR. Transient coexpression of the following inflammasome-forming combination triggered HR in N. tabacum leaves: TIRRPS4-NLRC4, NAIP1, and the T3SS needle protein from Yersinia pestis (YscF); TIRRPS4-NLRC4/NAIP2/PrgJ; and TIRRPS4-NLRC4/NAIP5/FlaA (Fig. 2 A and B; HR index scale shown in SI Appendix, Fig. S2F). No cell death was observed when combinations that do not form inflammasomes were coexpressed (Fig. 2 A and B). TIRRPS4-NLRC4 donor and acceptor surface mutants, which are incapable of forming inflammasomes, coexpressed with NAIP5 and FlaA did not trigger HR (Fig. 2C); this indicates that a higher-order complex containing multiple TIR domains is required, and not just a conformational change in TIRRPS4-NLRC4 induced by interaction with NAIP5/FlaA. Similarly, NAIP5 P-loop mutants did not trigger HR when expressed with TIRRPS4-NLRC4 and FlaA (SI Appendix, Fig. S2A). These data suggest that induced proximity of the RPS4 TIR domain is sufficient to activate immune signaling and HR.
Fig. 2.
TIRRPS4-NLRC4 inflammasome oligomerization triggers HR-like cell death. (A) HR-like cell death is elicited by coexpression of RPS4TIR-NLRC4 and inflammasome components that oligomerize. N. tabacum leaf sections were coinfiltrated with A. tumefaciens strains (each at OD600 = 0.5) carrying RPS4TIR-NLRC4, a NAIP, and a PAMP. HR was visually assessed and scored after 3 days postinfiltration (dpi). (B) Mean HR index of visually assessed tobacco infiltrations. Each graph represents coinfiltrations of a different NAIP with TIRRPS4-NLRC4 and three ligands (indicated on the x axis), with a sample size of n = 6. Error bars show SD. HR index scale shown in SI Appendix, Fig. S2F. (C) HR cell death assay of NLRC4 interface mutants. N. tabacum leaf sections were coinfiltrated with NAIP5-HA, 4xMyc-FlaA, and TIRRPS4-NLRC4-HF, or donor or acceptor surface mutants of TIRRPS4-NLRC4-HF. HR was visually assessed and photographed after 3 dpi. The numbers in parentheses are the number of leaves displaying HR equivalent to the image shown out of the total number of leaves infiltrated.
Structure-function analyses of TIR domains revealed two surfaces of the RPS4 TIR essential for RPS4-mediated immunity: The AE interface surface, required for heterodimerization and homodimerization, and the DE surface, predicted to be a self-interaction surface (8, 9). To test whether inflammasome-mediated signaling acts by promoting self-association through these interfaces, we introduced an AE interface mutation (S33A, H34A; SH/AA) and a DE interface mutation (R116A) into separate TIRRPS4-NLRC4 constructs. NAIP5 and FlaA formed inflammasomes with TIRRPS4(SH/AA)-NLRC4 and TIRRPS4(R116A)-NLRC4 (Fig. 1 D and E and SI Appendix, Fig. S2 B and C). Inflammasomes containing TIRRPS4(SH/AA)-NLRC4 exhibited no HR and TIRRPS4(R116A)-NLRC4 exhibited occasional, weak HR (SI Appendix, Fig. S2 D and E). Taken together, these data demonstrate that oligomerization via the NLRC4-NACHT was not sufficient for TIRRPS4 HR and that both TIR self-association interfaces are required. Wan et al. and Horsefield et al. reached similar conclusions by fusing TIRRPS4 to the SAM oligomerization domain from SARM1 (27, 28). Hence, induced proximity promotes formation of a TIR domain oligomer that is sufficient for signaling through the AE and DE interfaces.
Induced proximity of N-terminal signaling domains may be a general mechanism of TIR-NLR signaling activation. To test this, we fused to NLRC4 the TIRs of the plant NLRs SNC1 from Arabidopsis; N from tobacco; L6, M, and P from flax; and RRS1, an NLR that acts with RPS4 to confer effector perception in Arabidopsis (refs. 23, 24, and 29; alignment of TIRs shown in SI Appendix, Fig. S3). Two fragments of TIRSNC1 activated HR when fused to NLRC4 coexpressed with NAIP5 and FlaA: TIRSNC1(1-179), the minimal TIR domain containing amino acid residues 1–179, and TIRSNC1(1-226), an autoactive fragment (Fig. 3A) (9). TIRL6 activated HR when fused to NLRC4 coexpressed with NAIP5 and FlaA (Fig. 3A) but was also active in the absence of the appropriate ligand, suggesting autoactivity of this construct. TIRP2/L6, an L6 construct with its N-terminal Golgi membrane anchor replaced with the N terminus of the flax NLR P2 (30), showed HR only in the presence of the correct ligand (Fig. 3A). AE and DE interface mutations in these TIRs also abolished HR (Fig. 3B), except for TIRSNC1(K112E) (Fig. 3B). Although this mutation reduces the visible HR induced by TIRSNC1, it does not affect self-association in solution and has only a minor effect on ion leakage, suggesting that it only partially impairs function (9). TIRRRS1-NLRC4 did not trigger inflammasome-dependent HR (SI Appendix, Fig. S2E), consistent with RRS1’s “sensor” function in the RRS1/RPS4 pair: RPS4 monitors RRS1 and initiates immune signaling when RRS1’s WRKY domain interacts with effectors (24).
Fig. 3.
HR phenotype of transiently expressed plant TIR-NLRC4 fusions. (A) HR phenotype of NLRC4 fusions of L6, SNC1, or RPS4 TIR domains. N. tabacum leaves were coinfiltrated with A. tumefaciens strains carrying TIR-NLRC4 fusions, NAIP5, and PAMPs (FlaA or PrgJ). HR was photographed 5 dpi. TIRP2/L6 represents a TIRL6 construct in which the signal anchor is replaced by the N-terminal sequence from flax NLR P2. Two fragments of TIRSNC1 were tested, the minimal TIR domain (residue 1–179) and the autoactive fragment (residue 1–226). The numbers in parentheses are the number of leaves displaying HR equivalent to the image shown out of the total number of leaves infiltrated. (Lower) Expression analysis of TIR-NLRC4 fusions. Total proteins from N. tabacum leaves with expression of HF tag-fused TIR-NLRC4 proteins were immunoblotted with anti-FLAG antibodies. The dash indicates noninfiltrated N. tabacum leaf tissue. Staining of RuBisCO with Ponceau S was used as a loading control. (B and C) Mutations in self-association interfaces of TIRP2/L6 or TIRSNC1 (residue 1–226) affect cell death triggered by TIR-NLRC4 oligomerization. N. tabacum leaves were coinfiltrated with A. tumefaciens strains carrying TIR-NLRC4 or interface mutants, with NAIP5 and FlaA. HR was photographed 5 dpi. Mutations of the AE interface are shown in orange and DE interface in blue. (Lower) Expression analysis of TIR-NLRC4 and interface mutants.
In contrast to TIRSNC1 and TIRL6, the TIRs from N, M, and P did not trigger HR when fused to NLRC4 and coexpressed with FlaA and NAIP5 (SI Appendix, Fig. S4). Tobacco leaf sections infiltrated with TIRN-NLRC4, NAIP5, and FlaA displayed yellowing, but TIRM-NLRC4 and TIRP-NLRC4 fusions were green and healthy (SI Appendix, Fig. S4). Therefore, other factors likely influence TIR domain capacity to trigger HR upon induced proximity.
TIR-mediated immunity is dependent on proteins that act as hubs for signaling, including the lipase-like nucleocytoplasmic protein, EDS1, and the RPW8-NLR, NRG1 (2, 31, 32). TIRRPS4-NLRC4/NAIP5/FlaA did not trigger HR in an EDS1-silenced tobacco RNA interference (RNAi) line but did trigger HR in nonsilenced tobacco (SI Appendix, Fig. S5A). We also tested the activation of HR by TIRRPS4-NLRC4/NAIP5/FlaA in N. benthamiana nrg1 mutants. TIRRPS4-NLRC4/NAIP5/FlaA triggered HR in WT N. benthamiana but not in the nrg1 mutant (SI Appendix, Fig. S5B). Furthermore, we demonstrated the requirement of EDS1 for HR triggered by other TIR-NLRC4 constructs by coinfiltrating NAIP5, FlaA, and NLRC4 fusions to either TIRL6, TIRP2/L6, or TIRSNC1 in N. benthamiana eds1 mutants (SI Appendix, Fig. S5C). Complementing these mutants transiently with N. benthamiana EDS1 rescued HR triggered by these TIR-NLRC4 constructs (SI Appendix, Fig. S5C) (33). The genetic requirement for the immune signaling hubs EDS1 and NRG1, as well as the requirement for intact TIR interaction interfaces, demonstrates that HR triggered by TIRRPS4-NLRC4/NAIP5/FlaA mimics RPS4/RRS1-mediated HR.
The TIRs used in this study (except for that of RRS1) contain a conserved glutamate, which is at position 88 in RPS4 (E88) (SI Appendix, Fig. S3). The glutamate at this position is required for TIR NADase activity (26–28). Mutating this residue in full-length RPS4 or TIRRPS4 abolishes HR (22, 27, 34). An E88A substitution to TIRRPS4-NLRC4 abolished HR upon inflammasome formation (SI Appendix, Fig. S2 B, C, and E). We hypothesized that the induced inflammasome would activate plant TIR NADase activity, which is required for HR in our system. To test this hypothesis, we coexpressed the inflammasome components in N. benthamiana and purified the TIR-NLRC4 proteins or complexes by immunoprecipitation. Beads with the purified TIR-NLRC4 proteins were then incubated with NAD+ (5 µM), and metabolites were extracted and analyzed using liquid chromatography with tandem mass spectrometry (LC-MS/MS). NAD+ can be cleaved into nicotinamide (Nam), ADPR, and cADPR (25). A TIRSARM1-NLRC4/NAIP5/FlaA inflammasome complex showed strong production of Nam as well as a reduction in NAD+ levels in the reaction mix (SI Appendix, Fig. S6 A and B). However, when purified from leaves expressing TIRSARM1-NLRC4/NAIP2/FlaA, the monomeric TIRSARM1-NLRC4 did not show NADase activity (SI Appendix, Fig. S6 A and B), consistent with a requirement for oligomerization for SARM1-TIR enzymatic activity (26). In contrast, we detected no significant loss of NAD+ and no detectable production of Nam with TIRRPS4-NLRC4/NAIP5/FlaA or TIRP2/L6-NLRC4/NAIP5/FlaA (SI Appendix, Fig. S6 A and B). TIRL6 has been reported to have higher enzymatic activity than TIRRPS4, but both proteins have much lower activity than detected for SARM1 (28).
Recent reports revealed that plant TIR domains, like certain prokaryotic TIR domains, cleave NAD+ into a variant cADPR (v-cADPR), and v-cADPR may serve as a signaling molecule to execute cell death (25, 27). The bacterial A. baumannii TIR (TIRAbTir) domain, which produces v-cADPR (25), was fused to NLRC4. We first tested its NADase activity after expressing TIRAbTir-NLRC4 with NAIPs and FlaA in N. benthamiana and immunoprecipitating TIRAbTir-NLRC4 and associated proteins. Production of v-cADPR was detected, but no significant depletion of NAD+ or production of Nam was detected, from in vitro reaction mixes containing immunoprecipitated protein from extracts from leaves coexpressing either the inflammasome-forming combination of TIRAbTir-NLRC4/NAIP5/FlaA, but also in reaction mixes containing TIRAbTir-NLRC4/NAIP2/FlaA, a combination that does not form an inflammasome (SI Appendix, Fig. S7 A–C). This suggests that oligomerization is not required for NADase activity in the TIRAbTir-NLRC4 fusion. To test whether v-cADPR can signal cell death in planta, we measured v-cADPR production in leaves expressing TIR-NLRC4 fusions. v-cADPR was detected upon transient expression of TIRAbTir-NLRC4/NAIP5/FlaA and TIRAbTir-NLRC4/NAIP2/FlaA in N. bethamiana but not for other TIR-NLRC4 combinations, again demonstrating that TIRAbTir-NLRC4 NADase activity is oligomerization-independent (SI Appendix, Fig. S7D). However, TIRAbTir-NLRC4/NAIP5/FlaA did not cause HR in tobacco (SI Appendix, Fig. S7F). In addition, cADPR accumulated in leaves coexpressing TIRSARM1-NLRC4/NAIP5/FlaA, and also in threefold lower levels in leaves coexpressing TIRSARM1-NLRC4/NAIP2/FlaA in planta (SI Appendix, Fig. S7E), but TIRSARM1-NLRC4/NAIP5/FlaA also did not induce HR (SI Appendix, Fig. S6C). Both the results from TIRAbTir and TIRSARM1 suggest that neither v-cADPR nor cADPR are sufficient for cell death activation. Our data are consistent with previous results suggesting that plant TIRs have much lower NADase activity than TIRAbTIR and TIRSARM1 (27, 28). We infer that plant TIR NADase activity, while essential, is not sufficient for cell death and defense activation in plant cells.
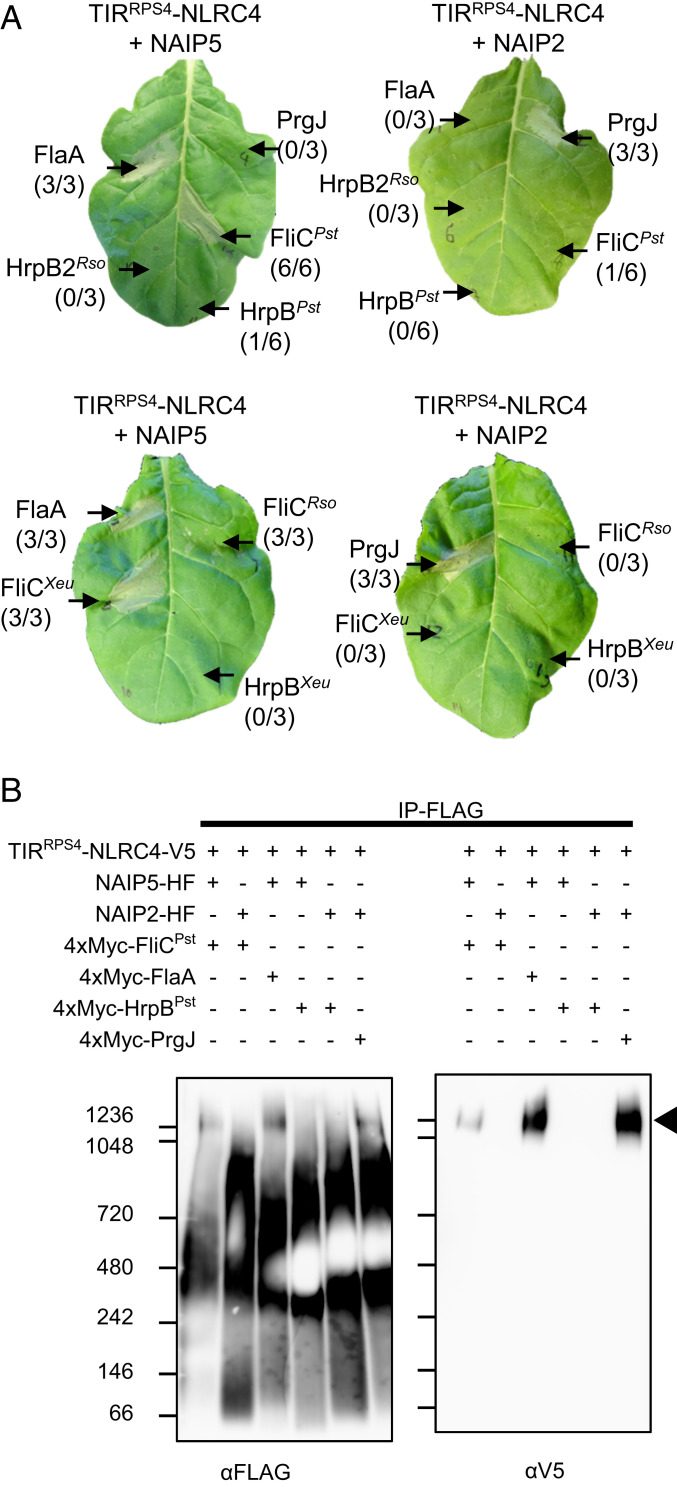
Plants recognize bacterial PAMPs, including flagellin and EF-Tu, via cell-surface receptors. Although bacterial PAMPs enter the plant cytosol during infection, plants lack cytosolic receptors that can detect them (35). We sought to test if TIRRPS4-NLRC4/NAIP would function as an intracellular surveillance system in plants for PAMPs secreted by plant-pathogenic bacteria. We generated expression vectors carrying FlaA or PrgJ homologs from the plant-pathogenic bacteria Pseudomonas syringae pv. tomato (Pst) DC3000 (FliCPst and HrpBPst), Ralstonia solanacearum (FliCRso and HrpB2Rso), and Xanthomonas euvesicatoria (FliCXeu and HrpBXeu). FliCPst triggered HR specifically when coexpressed in tobacco with TIRRPS4-NLRC4 and NAIP5, while FliCRso and FliCXeu triggered a weaker cell death; the PrgJ homologs HrpBPst, HrpB2Rso, and HrpBXeu did not trigger HR when coexpressed with TIRRPS4-NLRC4 and NAIP2 (Fig. 4A). Consistent with these HR phenotypes, FliCPst induced TIRRPS4-NLRC4/NAIP5 inflammasome formation but HrpBPst did not induce TIRRPS4-NLRC4/NAIP2 inflammasome formation (Fig. 4B and SI Appendix, Fig. S8). Therefore, the TIRRPS4-NLRC4/NAIP inflammasome can assemble and trigger HR in plants in response to flagellin from plant pathogenic bacteria, but not in response to rod components. However, despite reports that bacterial flagellin from Pst DC3000 can enter plant cells (35), we were unable to detect enhanced bacterial resistance in transgenic Arabidopsis lines carrying TIRRPS4-NLRC4 and NAIP5 (SI Appendix, Fig. S9). The discrepancy between the observation of HR in transient assays and the lack of an immune response in stably transformed Arabidopsis may be due to the different amounts of flagellin present in plant cells in each assay: Overexpression of flagellin in transient assays is likely to result in much more intracellular flagellin accumulation than occurs during Pst DC3000 infection.
Fig. 4.
TIRRPS4-NLRC4 and NAIP5 confer perception of flagellin from plant-pathogenic bacteria in plants. (A) Transient coexpression of TIRRPS4-NLRC4 and plant-pathogenic bacterial flagellin and NAIP5, but not NAIP2 and T3SS rod components, triggers HR-like cell death when coexpressed with TIRRPS4-NLRC4. N. tabacum leaf sections were coinfiltrated with A. tumefaciens strains (each at OD600 = 0.5) carrying TIRRPS4-NLRC4, a NAIP, and a PAMP. HR was visually assessed and photographed after 3 dpi. The numbers in parentheses are the number of leaves displaying HR equivalent to the image shown out of the total number of leaves infiltrated. Superscript abbreviations indicate plant-pathogenic bacterial origin of genes encoding each PAMP: Pst, Pst DC3000; Xeu, X. euvesicatoria; Rso, Ralstonia solanacearum. (B) BN-PAGE of immunoprecipitated combinations of TIRRPS4-NLRC4, NAIP, and Pst DC3000 PAMPs. Arrowheads indicate inflammasome oligomer. See also SI Appendix, Fig. S8.
By fusing plant NLR TIR domains to the N terminus of the mammalian NLR NLRC4, we show here that induced proximity of plant NLR TIRs is sufficient for immune activation, resembling the activation mechanism of mammalian NLR CARDs. These data emphasize the modular nature of NLRs, where the NB domain provides a regulated mechanism for oligomerization and inducing proximity of N-terminal domains, and N-terminal domains can be swapped between NLRs and retain functionality. Similarly, our data demonstrate that the known components of the NAIP/NLRC4 inflammasome can detect ligands and functionally assemble when transplanted across kingdoms, suggesting that there are no additional unidentified mammalian-specific components required for NAIP/NLRC4 activation.
TIR activation required authentic oligomerization at the center of the assembled inflammasome. The linear helix hypothesized to form in an active TIR oligomer (9), or another signaling active conformation, must assemble in the TIR-NLRC4/NAIP5/FlaA inflammasome. Although the structure of the NLRC4 CARDs in an inflammasome has not been resolved, cryoelectron tomography predicts a helix at the center of an inflammasome (36), while purified NLRC4 CARDs in vitro form a tetramer that comprises the base of a helical filament (37, 38). Steric constraints imposed by CARD oligomerization may have prevented the assembly into a signaling-active conformation of some of the plant TIRs we tested.
The assembly of the ZAR1/RKS1/PBL2 resistosome coordinates the ZAR1-CCs into a pore-like structure. This structure resembles a pore-forming toxin, leading to the hypothesis that the ZAR1 resistosome triggers HR cell death through integration and pore formation in the plant plasma membrane (18). Although ZAR1 similarly requires induced proximity of N-terminal signaling domains, the TIR domains of RPS4 and other TNLs are not predicted to form pores.
We were unable to detect plant TIR domain NADase activity in immunopurified plant TIR-NLRC4 complex, perhaps due to its low enzymatic activity and abundance. However, NADase enzymatic activity is a conserved function across prokaryotic and eukaryotic TIR domain proteins (25–28), and the key catalytic glutamate residue is widely present in plant TIR domains (SI Appendix, Fig. S3). We infer that NADase activity of plant TIRs and the production of v-cADPR may be necessary but not sufficient for cell death and defense activation. Our data (SI Appendix, Fig. S5) and other reports show that plant TIR domain-induced HR requires EDS1, SAG101, and NRG1 (31, 32, 39–42). EDS1-SAG101-NRG1 have coevolved as a module to mediate cell death signaling by TIR-domain immune receptors within plant species (42, 43). Future work will focus on the additional components required for v-cADPR activation of immune signaling.
Materials and Methods
Plant Material and Growth Conditions.
Nicotiana tabacum cv. “Petite Gerard” or “Samsun” and N. benthamiana were grown on soil at 24 °C, 55% relative humidity with a 16/8-h light/dark photoperiod. Arabidopsis thaliana accession “Col-0” was grown at 21 °C, 70% humidity with 10/14-h light/dark photoperiod. The N. tabacum cv. “Samsun” EDS1 RNAi line was provided by Barbara Baker, Department of Plant and Microbial Biology, University of California, Berkeley, CA, the N. benthamiana eds1 line was provided by Brian Staskawicz, Department of Plant and Microbial Biology, University of California, Berkeley, CA (33), and we previously generated the N. benthamina nrg1 line (32).
Plasmid Construction.
Plasmids were constructed using Golden Gate cloning, as described in refs. 24 and 44. The TIR domain of RPS4 was PCR amplified with primers containing BpiI recognition sites and a specific 4-bp overhang, then cloned into the N-terminal tag module pICSL01002. All other TIR domains were similarly cloned. The TIRSARM1 was PCR amplified from pGW1-Myc-Sarm1 (45), a gift from Yi-Ping Hsueh, Institute of Molecular Biology, Academia Sinica, Taipei, Taiwan (Addgene plasmid no. 50707; http://www.addgene.org/50707/; RRID:Addgene_50707). NLRC4, NAIPs, and PAMPs were similarly cloned into the coding sequence module pICSL01005. Modules in pICSL01005 and pICSL01002 were released with BsaI digestion and assembled in pICSL86900 with the CaMV35S promoter, Ocs terminator, and N-terminal and/or C-terminal epitope tags.
Leaf Infiltration.
Transient transformation by agroinfiltration of N. tabacum lamina sections between veins for HR and whole N. benthamiana leaves for protein analyses was performed on 4- to 5-wk-old plants. Agrobacterium tumefaciens strains were mixed in infiltration medium (10 mM MgCl2, 10 mM 2-(N-morpholino) ethanesulfonic acid [MES], pH 5.6) each at an OD600 of 0.5, and hand-infiltrated with a 1-mL needle-less syringe.
Protein Assays.
Protein was extracted from transiently transformed N. benthamiana leaves 72 h postinfiltration (hpi) with A. tumefaciens as previously described (23). Briefly, leaves were harvested and ground in liquid nitrogen, and extracted in GTEN buffer (10% glycerol, 100 mM Tris⋅HCl, pH 7.5, 1 mM EDTA, 150 mM NaCl, 5 mM 1,4-dithiothreitol (DTT), 1× cOmplete protease inhibitor mixture [Roche] and 0.2% [vol/vol] Nonidet P-40). Cleared samples were separated into subsamples for input for BN-PAGE, input for SDS-PAGE and samples for immunoprecipitation. Immunoprecipitations were performed for 4 h at 4 °C with gentle agitation, in the presence of 10 μL per 1 mL of protein extract of anti-FLAG M2 affinity Gel (A2220 Sigma-Aldrich), anti-V5 (A7345 Sigma-Aldrich) or anti-Myc (9E10 ThermoFisher). Beads were washed four times in GTEN buffer. FLAG beads were incubated with 150 ng/μL 3XFLAG peptide (Sigma-Aldrich) for 30 min at 4 °C. Other samples were eluted by boiling in SDS-PAGE loading buffer. For BN-PAGE, IP-FLAG eluate and input samples were mixed with 10× BN-PAGE loading buffer, loaded onto Invitrogen NativePAGE 3–12% Bis-Tris Protein Gels, and electrophoresed according to the manufacturer’s instructions. For SDS-PAGE, anti-V5 and anti-Myc beads, as well as IP-FLAG eluate and input samples, were mixed with 3× SDS-PAGE loading buffer and heated for 20 min at 80 °C. After electrophoresis, separated proteins were transferred to Immunobilon-P PVDF (Merck Millipore) membranes for immunoblotting. Membranes were blocked for 2 h in 5% nonfat milk, probed with horseradish peroxidase (HRP)-conjugated antibodies overnight and imaged.
NADase Assay and LC-MS/MS Metabolite Measurement.
Transiently expressed proteins were extracted from N. benthamiana as described above. NADase activity was measured as described in Essuman et al. (26). To be brief, 20 μL anti-FLAG beads with bound protein was mixed with 5 μL of reaction buffer (924 mM NaCl and 6.4× phosphate buffered saline [PBS]), 5 μL of 100 μM NAD+ and 20 μL of water. Reactions were either terminated immediately, or after 30 min at 20 °C with periodic vortexing, by the addition of 50 μL of 1 M perchloric acid and incubation on ice for 10 min. Terminated reactions were neutralized by the addition of 16.7 μL of 3 M K2CO3. Samples were then centrifuged, and 90 μL of supernatant was frozen in liquid nitrogen and stored at −80 °C. NAD+ and nicotinamide (Nam) were measured in reaction supernatants by LC-MS/MS. Samples were separated on 100 × 2.1 mm 2.6 μ Kinetex EVO C18 column with guard, whose outflow was attached to a 50 × 2.1 mm 2.6 μ Kinetex F5 column. The high performance liquid chromatography (HPLC) was run with an aqueous solvent of 0.1% formic acid adjusted to pH 6.02 by addition of ammonium hydroxide, with a gradient to 60% methanol. Target compounds were detected by electrospray MS using the 2020 single quadrupole’s dual ion source in ESI mode, with spray chamber conditions of 200 °C heat block, 250 °C desorbation line, 1.5 L·min−1 nebulizer gas, and 15 L·min−1 drying gas. The instrument collected positive mode scan data from m/z 100–800 and single ion monitoring data for masses 123.1, 664, 560, and 542 (positive, total even time 0.1 s).
Plant extracts were prepared and analyzed according to Wan et al. (27). Briefly, after transiently transforming N. benthamiana leaves 45 hpi with A. tumefaciences, leaves were harvested and ground in liquid nitrogen. Samples were extracted with 50% methanol in water and deproteinized with chloroform. The aqueous phase was lyophilized and reconstituted in 5 mM ammonium formate, centrifuged at 13,000 rpm for 10 min, and the supernatant analyzed by LC-MS/MS. Analysis of v-cADPR was on an Acquity ultra performance liquid chromatography (UPLC) attached to a Xevo TQS tandem quadrupole mass spectrometer. Chromatography was exactly the same as for the single quadrupole. E. coli lysate of AbTIR was used as a reference for v-cADPR. Spray chamber conditions were 500 °C desolvation temperature, 900 L·hr−1 desolvation gas, 150 L·hr−1 cone gas, 7 bar nebulizer pressure.
Bacterial Growth Assay.
Pst DC3000 strains were grown on selective King’s B (KB) medium agar plates for 48 h at 28 °C. Bacteria were harvested from the plates and resuspended in infiltration buffer (10 mM MgCl2, pH 5.6), and the OD600 of the resuspended cell was adjusted to 0.001. Leaves were hand-infiltrated by a needleless syringe. Leaf discs were harvested using a 6-mm cork borer. Two leaf discs per seedling were used as a single treatment, with four replicates sampled after infiltration and eight replicates after 3 days postinfiltration (dpi). Samples were ground and the lysate was diluted in infiltration buffer, and then spotted on selective KB medium plates.
Supplementary Material
Acknowledgments
We thank Barbara Baker for the N. tabacum cv “Samsun” EDS1 RNAi line, Brian Staskawicz for N. benthamiana eds1, and Matthew Moscou for providing an MLA7 construct. We thank Jeffrey Milbrandt for providing the AbTIR construct. We thank Matthew Smoker and Jodie Taylor for help with Arabidopsis transformation. We thank the reviewers for their constructive suggestions for improving the manuscript. Z.D., S.U.H., S.W., H.G., and L.Hu were supported on European Research Council (ERC) Grant “Immunitybypairdesign” Project ID 669926 (to J.D.G.J.). Y.M. was supported on Biotechnology and Biological Sciences Research Council (BBSRC) Grant BB/M008193/1 (to J.D.G.J.). S.U.H. was supported on Next-Generation BioGreen 21 Program (Project No. PJ01365301), Rural Development Administration, Republic of Korea. J.C. was supported by a Chinese Scholarship Council Postgraduate Fellowship. P.D. was supported by the European Union’s Horizon 2020 Research and Innovation Program under Marie Skłodowska-Curie Individual Fellowship (Project ID 656243) and a Future Leader Fellowship from BBSRC (Grant Agreement BB/R012172/1). P.N.M. was supported by a Marie Skłodowska-Curie Action Individual Fellowship (Project ID 656011).
Footnotes
The authors declare no competing interest.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2001185117/-/DCSupplemental.
Data Availability.
All data are contained in the manuscript, supplemental figures, or Dataset S1.
References
- 1.Duxbury Z. et al., Pathogen perception by NLRs in plants and animals: Parallel worlds. BioEssays 38, 769–781 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Jones J. D. G., Vance R. E., Dangl J. L., Intracellular innate immune surveillance devices in plants and animals. Science 354, aaf6395 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Jones J. D. G., Dangl J. L., The plant immune system. Nature 444, 323–329 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Bernoux M. et al., Comparative analysis of the flax immune receptors L6 and L7 suggests an equilibrium-based switch activation model. Plant Cell 28, 146–159 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frost D. et al., Tobacco transgenic for the flax rust resistance gene L expresses allele-specific activation of defense responses. Mol. Plant Microbe Interact. 17, 224–232 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Cesari S. et al., Cytosolic activation of cell death and stem rust resistance by cereal MLA-family CC-NLR proteins. Proc. Natl. Acad. Sci. U.S.A. 113, 10204–10209 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maekawa T. et al., Coiled-coil domain-dependent homodimerization of intracellular barley immune receptors defines a minimal functional module for triggering cell death. Cell Host Microbe 9, 187–199 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Williams S. J. et al., Structural basis for assembly and function of a heterodimeric plant immune receptor. Science 344, 299–303 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Zhang X. et al., Multiple functional self-association interfaces in plant TIR domains. Proc. Natl. Acad. Sci. U.S.A. 114, E2046–E2052 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishimura M. T. et al., TIR-only protein RBA1 recognizes a pathogen effector to regulate cell death in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 114, E2053–E2062 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bentham A., Burdett H., Anderson P. A., Williams S. J., Kobe B., Animal NLRs provide structural insights into plant NLR function. Ann. Bot. 119, 827–702 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kofoed E. M., Vance R. E., Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature 477, 592–595 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao Y. et al., The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature 477, 596–600 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Zhang L. et al., Cryo-EM structure of the activated NAIP2-NLRC4 inflammasome reveals nucleated polymerization. Science 350, 404–409 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu Z. et al., Structural and biochemical basis for induced self-propagation of NLRC4. Science 350, 399–404 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Tenthorey J. L. et al., The structural basis of flagellin detection by NAIP5: A strategy to limit pathogen immune evasion. Science 358, 888–893 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salvesen G. S., Dixit V. M., Caspase activation: the induced-proximity model. Proc. Natl. Acad. Sci. U.S.A. 96, 10964–10967 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang J. et al., Reconstitution and structure of a plant NLR resistosome conferring immunity. Science 364, eaav5870 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Wang J. et al., Ligand-triggered allosteric ADP release primes a plant NLR complex. Science 364, eaav5868 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Wang G. et al., The decoy substrate of a pathogen effector and a pseudokinase specify pathogen-induced modified-self recognition and immunity in plants. Cell Host Microbe 18, 285–295 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Hu M., Qi J., Bi G., Zhou J.-M., Bacterial effectors induce oligomerization of immune receptor ZAR1 in vivo. Mol. Plant 13, 793–801 (2020). [DOI] [PubMed] [Google Scholar]
- 22.Swiderski M. R., Birker D., Jones J. D. G., The TIR domain of TIR-NB-LRR resistance proteins is a signaling domain involved in cell death induction. Mol. Plant Microbe Interact. 22, 157–165 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Sarris P. F. et al., A plant immune receptor detects pathogen effectors that target WRKY transcription factors. Cell 161, 1089–1100 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Ma Y. et al., Distinct modes of derepression of an Arabidopsis immune receptor complex by two different bacterial effectors. Proc. Natl. Acad. Sci. U.S.A. 115, 10218–10227 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Essuman K. et al., TIR domain proteins are an ancient family of NAD+-consuming enzymes. Curr. Biol. 28, 421–430.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Essuman K. et al., The SARM1 Toll/Interleukin-1 Receptor domain possesses intrinsic NAD+ cleavage activity that promotes pathological axonal degeneration. Neuron 93, 1334–1343.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wan L. et al., TIR domains of plant immune receptors are NAD+-cleaving enzymes that promote cell death. Science 365, 799–803 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horsefield S. et al., NAD+ cleavage activity by animal and plant TIR domains in cell death pathways. Science 365, 793–799 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Guo H. et al., Phosphorylation-regulated activation of the Arabidopsis RRS1-R/RPS4 immune receptor complex reveals two distinct effector recognition mechanisms. Cell Host Microbe 27, 769–781.e6 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Takemoto D. et al., N-terminal motifs in some plant disease resistance proteins function in membrane attachment and contribute to disease resistance. Mol. Plant Microbe Interact. 25, 379–392 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Qi T. et al., NRG1 functions downstream of EDS1 to regulate TIR-NLR-mediated plant immunity in Nicotiana benthamiana. Proc. Natl. Acad. Sci. U.S.A. 115, E10979–E10987 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Castel B. et al., Diverse NLR immune receptors activate defence via the RPW8-NLR NRG1. New Phytol. 222, 966–980 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Schultink A., Qi T., Lee A., Steinbrenner A. D., Staskawicz B., Roq1 mediates recognition of the Xanthomonas and Pseudomonas effector proteins XopQ and HopQ1. Plant J. 92, 787–795 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Sohn K. H. et al., The nuclear immune receptor RPS4 is required for RRS1SLH1-dependent constitutive defense activation in Arabidopsis thaliana. PLoS Genet. 10, e1004655 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei H.-L., Chakravarthy S., Worley J. N., Collmer A., Consequences of flagellin export through the type III secretion system of Pseudomonas syringae reveal a major difference in the innate immune systems of mammals and the model plant Nicotiana benthamiana. Cell. Microbiol. 15, 601–618 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Diebolder C. A., Halff E. F., Koster A. J., Huizinga E. G., Koning R. I., Cryoelectron tomography of the NAIP5/NLRC4 inflammasome: Implications for NLR activation. Structure 23, 2349–2357 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Matyszewski M. et al., Cryo-EM structure of the NLRC4CARD filament provides insights into how symmetric and asymmetric supramolecular structures drive inflammasome assembly. J. Biol. Chem. 293, 20240–20248 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Y. et al., Cryo-EM structures of ASC and NLRC4 CARD filaments reveal a unified mechanism of nucleation and activation of caspase-1. Proc. Natl. Acad. Sci. U.S.A. 115, 10845–10852 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aarts N. et al., Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 95, 10306–10311 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feys B. J. et al., Arabidopsis SENESCENCE-ASSOCIATED GENE101 stabilizes and signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. Plant Cell 17, 2601–2613 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.García A. V. et al., Balanced nuclear and cytoplasmic activities of EDS1 are required for a complete plant innate immune response. PLoS Pathog. 6, e1000970 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lapin D. et al., A coevolved EDS1-SAG101-NRG1 module mediates cell death signaling by TIR-domain immune receptors. Plant Cell 31, 2430–2455 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gantner J., Ordon J., Kretschmer C., Guerois R., Stuttmann J., An EDS1-SAG101 complex is essential for TNL-mediated immunity in Nicotiana benthamiana. Plant Cell 31, 2456–2474 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weber E., Engler C., Gruetzner R., Werner S., Marillonnet S., A modular cloning system for standardized assembly of multigene constructs. PLoS One 6, e16765 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen C. Y., Lin C. W., Chang C. Y., Jiang S. T., Hsueh Y. P., Sarm1, a negative regulator of innate immunity, interacts with syndecan-2 and regulates neuronal morphology. J. Cell Biol. 193, 769–784 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained in the manuscript, supplemental figures, or Dataset S1.