Significance
The hallmark of neurodegenerative disorders (e.g., Alzheimer’s, Parkinson’s, and Huntington’s disease) is the generation of aggregated proteins in different brain regions. These large aggregates originate from smaller, invisible, and soluble oligomers/aggregates and are believed to be sequestered from essential cellular machineries and, therefore, to attenuate/postpone the pathologic process. That, in contrast to the smaller ones that are thought to inhibit basic cellular pathways by trapping their components. The role played in this process by ubiquitin—which normally targets proteins for degradation—is unknown. Here, we show that ubiquitination of pathogenic Htt is important for its efficient aggregation and probably sequestration. Abolishing its ubiquitination results in generation of numerous smaller and less visible aggregates that are particularly harmful to cells.
Keywords: huntingtin, ubiquitin, aggregation, cell death
Abstract
Huntington’s disease (HD) is a progressive incurable neurodegenerative disorder characterized by motor and neuropsychiatric symptoms. It is caused by expansion of a cytosine–adenine–guanine triplet in the N-terminal domain of exon 1 in the huntingtin (HTT) gene that codes for an expanded polyglutamine stretch in the protein product which becomes aggregation prone. The mutant Htt (mHtt) aggregates are associated with components of the ubiquitin–proteasome system, suggesting that mHtt is marked for proteasomal degradation and that, for reasons still debated, are not properly degraded. We used a novel HD rat model, proteomic analysis, and long-term live neuronal imaging to characterize the effects of ubiquitination on aggregation of mHtt and subsequent cellular responses. We identified two lysine residues, 6 and 9, in the first exon of mHtt that are specifically ubiquitinated in striatal and cortical brain tissues of mHtt-transgenic animals. Expression of mHtt exon 1 lacking these ubiquitination sites in cortical neurons and cultured cells was found to slow aggregate appearance rates and reduce their size but at the same time increase the number of much smaller and less visible ones. Importantly, expression of this form of mHtt was associated with elevated death rates. Proteomic analysis indicated that cellular reactions to mHtt expression were weaker in cells expressing the lysineless protein, possibly implying a reduced capacity to cope with the proteotoxic stress. Taken together, the findings suggest a novel role for ubiquitination—attenuation of the pathogenic effect of mHtt.
Huntington’s disease (HD) is an autosomal dominant progressive and fatal neurological disorder caused by genomic expansion of the cytosine–adenine–guanine (CAG) repeat in the domain that codes for the N-terminal segment of Htt. The expansion results in mHtt in which consecutive polyglutamine (polyQ) stretches in this region are much longer than those occurring normally (15–20 repeats on average). HD is most known for the severe motor dysfunction it causes; yet the earliest symptoms, often manifested years before severe motor symptoms develop, are cognitive deficits and neuropsychiatric abnormalities, which gradually deteriorate into dementia and serious behavioral anomalies. HD symptoms are attributed to the degeneration of neurons in the striatum, yet neurons in the neocortex and other brain regions are also affected, explaining, perhaps, the extensive range of both cognitive and motor symptoms (1–3).
Htt is a large (∼350 kDa) protein, whose physiological roles are still not entirely understood (see ref. 4 for a recent comprehensive review). It is now clear that the disease etiology is tightly linked to a gain of function exerted by the polyQ expansion as a single copy of mHtt or even of exon 1 containing this expansion is sufficient to induce HD symptoms (5). mHtt cytotoxicity seems to be related to its propensity to form relatively stable aggregates, that accumulate in the cytosol and as nuclear inclusion bodies (2). The mechanisms by which such aggregates induce cell damage and ultimately cell death are not entirely clear; it has been suggested, for example, that cytotoxicity is related to the tendency of aggregates to sequester crucial proteins, such as transcription factors (6–9), proteasomes, or other ubiquitin–proteasome system (UPS) components (10–13). Paradoxically, the formation of aggregates was also suggested to fulfill a protective role against the polyQ-expanded protein by reducing levels of toxic, soluble, and smaller mHtt oligomers that are more harmful (9, 14, 15).
mHtt aggregates are commonly found in tight association with ubiquitin, different conjugating and deubiquitinating enzymes, proteasomes, and chaperones (2, 13, 16–19), which has been interpreted as a cellular attempt to cope with stress induced by the misfolded and/or proteotoxic variants of this protein (reviewed in refs. 20 and 21). The association of mHtt aggregates with UPS components would seem to indicate that mHtt is targeted for ubiquitin- and proteasome-mediated degradations, but for yet unknown reasons, fails to be degraded (2, 16). This might be due to direct effects of mHtt on proteasome function (20, 22) but also by overwhelming the cellular protein quality control systems by the excess of the misfolded protein (23, 24). However, none of these hypotheses has been confirmed rigorously (25). Thus, the relationships among mHtt ubiquitination, aggregation, disposal, and cell fate are still elusive.
Here, we used a novel HD model (bacterial artificial chromosome HD [BACHD] rats, ref. 26), long-term imaging of neurons expressing different mHtts, viability and biochemical assays, and proteomic analyses to characterize the effects of ubiquitination of mHtt on its aggregation and the subsequent cell’s response. We found that two lysine residues, 6 and 9, in the first exon of mHtt (upstream of the polyQ stretch) are specifically ubiquitinated in the striatal and cortical brain regions of BACHD but not wild-type (WT) rats. Expression of exon 1 of mHtt (fused to enhanced green fluorescent protein [EGFP]) that contains 134 repeats of glutamine showed that, when these ubiquitination sites are altered, the numbers and sizes of visible aggregates are significantly reduced. However, biochemical analysis revealed that the lysine-mutated aggregates are less soluble, their number is significantly higher than that of the ubiquitinated protein, and that many of them are probably small and, therefore, optically invisible but biochemically detectable. Importantly, these small aggregates appear to be more harmful to the cell than their nonmutated, yet, polyQ-expanded counterparts as they induce more cell death. In correlation with these findings, proteomic analysis demonstrated that the ubiquitinated mHtt induces more effectively multiple cellular responses probably as an attempt to cope with the cellular stress imposed by mHtt expression. Conversely, the mHtt variant that lacked these ubiquitination sites evoked a weaker response.
Taken together, these findings suggest a novel and rather unexpected role for ubiquitin modification in the formation of a lower number but larger-size aggregates, which are less harmful to the cell. It appears that mHtt recruits the ubiquitin system proactively as in WT Htt, the two lysines are not ubiquitinated.
Results
mHtt but Not WT Htt Is Specifically Ubiquitinated on Lysine 6 and 9 in a HD Model Rat.
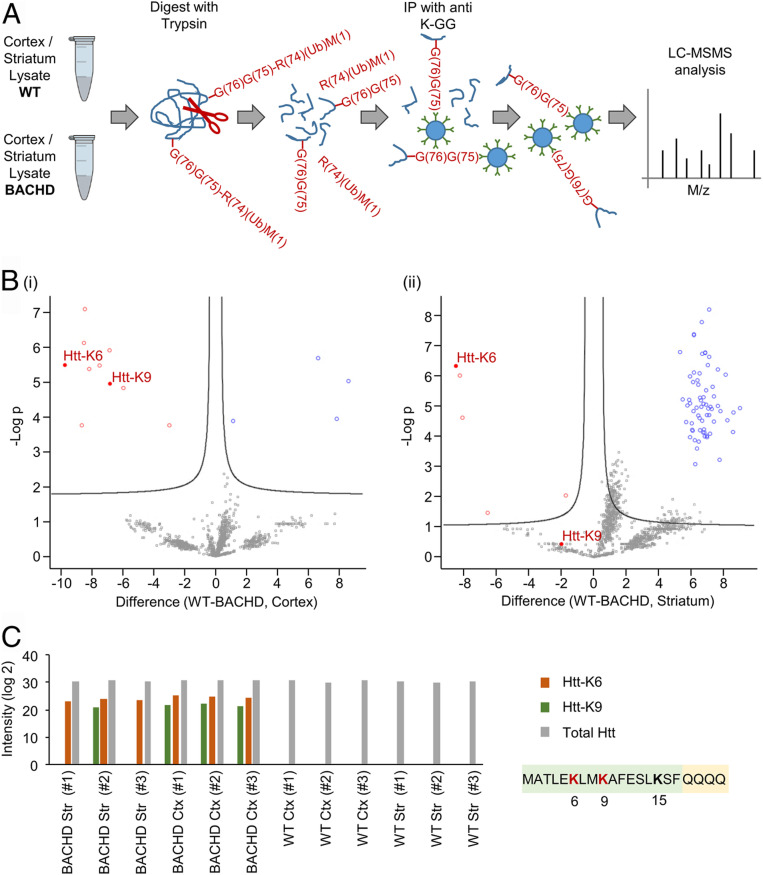
Animal models of HD are invaluable to elucidate important pathways of this disease. A unique HD model—the BACHD rat (26) was created to overcome many of the shortcomings of prior mouse models, such as overexpression of only a fragment of the protein or of a nonhuman Htt. BACHD rats express the full length human Htt protein with a stretch of 97 glutamine residues. The expressing gene is under control of the human HTT promoter and all of its regulatory elements. These rats exhibit a robust and early onset of HD-like deficits, including accumulation of mHtt aggregates in neurons as early as at 3 mo of age. To measure the ubiquitination profile of mHtt in BACHD animals which fully display HD-like deficits, brain sections (striatal and cortical) of 15-mo-old BACHD rats were subjected to proteomic analysis and compared to tissue samples collected in age-matched WT rats (samples were taken from each brain region from three different BACHD and three different WT rats). The ubiquitination profile was identified following enrichment of trypsin-treated lysates for ubiquitinated proteins (Fig. 1A) using a specific antibody that recognizes the two ubiquitin C-terminal glycines ligated in an isopeptide bond to an internal lysine residue in the target protein (K-GG; see also refs. 27–29). We found that lysine residues 6 and 9 in the N-terminal domain of mHtt are highly ubiquitinated in both the cortical and the striatal tissues, in striking contrast to the homologous two lysine residues in WT Htt that were not (Fig. 1 B and C). Importantly, we did not find any evidence for ubiquitination of lysine residue 15 in the N-terminal domain of mHtt in both BACHD and WT rats, even though overall Htt expression levels were identical in both populations (Fig. 1C).
Fig. 1.
mHtt exon1 is highly and specifically ubiquitinated in HD. (A) Workflow for identifying ubiquitination targets and sites in cortical and striatal neurons from brains of BACHD and WT rats, following enrichment of ubiquitin-modified peptides by bead-conjugated anti–K-GG antibodies. R(74)(Ub)M(1) denotes the ubiquitin fragment (residues 1–74) released by trypsin. —G(76)G(75) denotes the two C-terminal Gly residues of ubiquitin remained bound in an isopeptide bond to an internal Lys residue in the target substrate. Details of the methods are described in the Ubiquitinome analysis of brain samples of the BACHD and WT rats section in Materials and Methods. (B) Volcano plots of differentially expressed ubiquitinated peptides from brain samples of WT and BACHD rats. The plots display significant results of a t test with permutation-based false discovery rate (FDR) calculation of fold change (difference) and significance (−Log p) using the Perseus software. (i) Visualization of ubiquitinated peptides from cortical brain samples. (ii) Visualization of ubiquitinated peptides from striatal brain samples. (C) Histogram showing intensities of Htt N-terminal peptides ubiquitinated on lysines 6 and 9 as well as total Htt measured in the same samples. Each sample is from the cortex or striatum of one WT or BACHD rat brain.
Ubiquitination of mHtt Results in the Formation of a Higher Number of Larger Visible Aggregates.
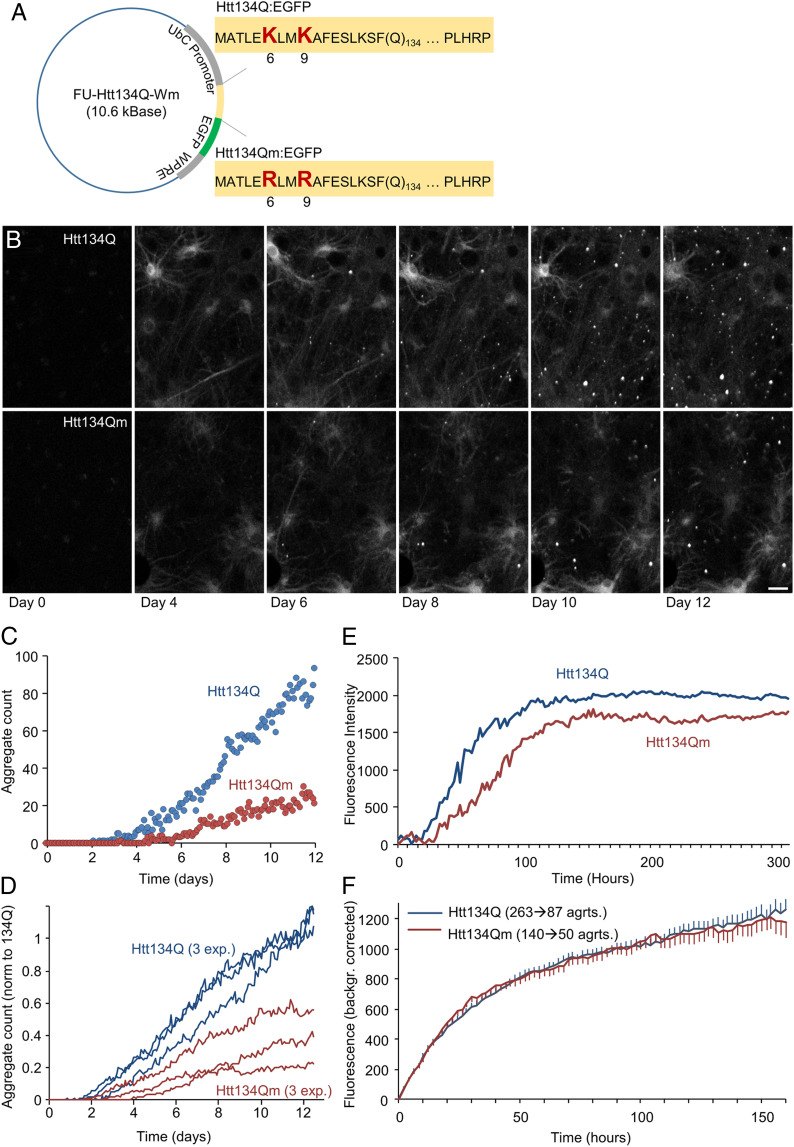
To test the potential role of the specific ubiquitination on aggregate formation dynamics, we replaced the two lysine residues with arginine residues that cannot be ubiquitinated and constructed two plasmids coding for exon 1 of mHtt that contain 134 residues of glutamine (Fig. 2A). One expresses the protein with the WT lysines (designated as “Htt134Q”) and the other in which lysines 6 and 9 were mutated to arginine (designated as “Htt134Qm”). EGFP was fused to the C terminus of both variants to allow visualization of aggregate formation. We used long-term imaging in primary cultures of rat cortical neurons to follow the fate of the two polyQ-expanded proteins. We found that both the numbers and the sizes of visible aggregates were significantly larger in cells expressing the protein that contains the two lysine residues, compared to cells that express the mHtt form that cannot be ubiquitinated (Fig. 2 B–E and two video clips Movie S1 A and B). No effect on formation rates of individual aggregates was observed (Fig. 2F and SI Appendix, Fig. S1 A and B) indicating, perhaps, that (large) aggregate nucleation rates are slower in the absence of lysine 6 and 9.
Fig. 2.
Site-specific ubiquitination of Htt134Q:EGFP N-terminal domain affects aggregates’ numbers and sizes but not their growth kinetics. (A) Schematic of FU-Htt134Q:EGFP-Wm and FU-Htt134Qm:EGFP-Wm coding vectors. The first 17 amino acids are followed by 134 glutamine residues and a proline rich domain (PLHRP) which is fused to EGFP. Upper sequence is from FU-Htt134Q:EGFP-Wm; native lysine residues at positions 6 and 9 shown in red large letters. Lower sequence is from FU-Htt134Qm:EGFP-Wm; lysine-to-arginine–mutated residues at positions 6 and 9 shown in red large letters. (B) Time-lapse images of cortical neurons following lentiviral transduction with FU-Htt134Q:EGFP-Wm or FU-Htt134Qm:EGFP-Wm. Times posttransduction indicated at the bottom. (Scale bar: 20 µm.) (C–F) Quantitative long-term imaging of cortical neurons expressing FU-Htt134Q:EGFP-Wm or FU-Htt134Qm:EGFP-Wm. (C) Changes in aggregate numbers with time from transduction (one region of interest in each condition in a single experiment). (D) Changes in aggregate numbers with time from transduction, three different experiments. Aggregate numbers were normalized in each experiment to the average Htt134Q:EGFP aggregate count at days 11 and 12. (E) Changes in aggregate fluorescence with time from transduction. Averages for three independent experiments. (F) Formation rates of individual aggregates. Individual aggregates were tracked backward in time up until the moment of their appearance. Fluorescence values over time were then obtained for each aggregate and corrected for background fluorescence values measured just before aggregate appearance. All fluorescence trajectories were then aligned to the moment of aggregate appearance, and average trajectories were then calculated (see SI Appendix, Fig. S1 for further details). Based on aggregates tracked in three experiments. Numbers of aggregates indicated above curves. Error bars: SEM.
Ubiquitinated Htt’s Aggregates Are Fewer and More Soluble than Their Lysine-Mutated Counterparts.
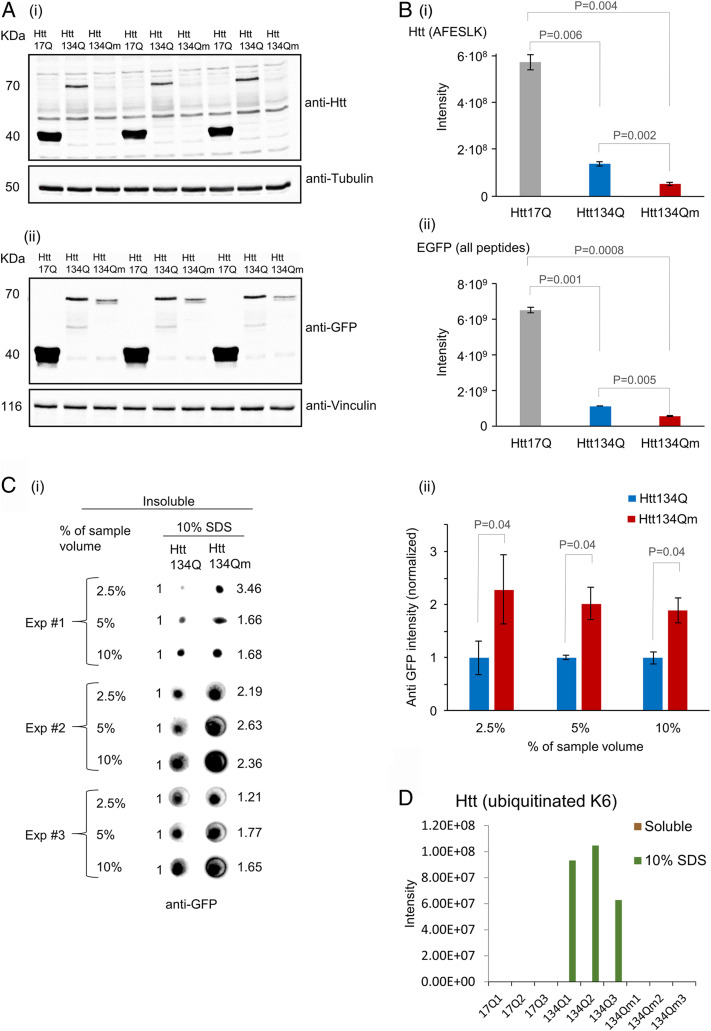
To study the effect of ubiquitination on the characteristics of the aggregates, we solubilized HEK293 cells expressing Htt134Q:EGFP, Htt134Qm:EGFP, as well as Htt17:EGFP (exon 1 with 17 glutamine repeats) initially under mild conditions using NP-40, followed by harsher solubilization conditions (10% sodium dodecyl sulfate [SDS]). As can be seen in the experiment depicted in Fig. 3 A, i and ii where we resolved the NP-40-soluble fraction, Htt134Q:EGFP appears to be more prominent than its mutant lysineless counterpart. Proteomic analyses of the soluble fractions for an Htt-specific peptide and peptides derived from EGFP (Fig. 3 B, i and ii) resulted in findings similar to those observed in the electrophoretic separation: We identified a high number of peptides derived from Htt17Q:EGFP which was completely soluble, a smaller number derived from the Htt134Q:EGFP protein, and an even smaller number of those derived from the Htt134Qm:EGFP protein. Dot blot analysis of the 10% SDS-soluble fraction revealed that most Htt134Qm:EGFP resided in this fraction (Fig. 3C). Although the numbers and sizes of visible Htt134Qm:EGFP aggregates were significantly smaller than those generated by Htt134Q:EGFP (Fig. 2 C–E), the amount of the pathologic protein in Htt134Qm:EGFP expressing cells was consistently higher as evident from the blot’s intensities (Fig. 3 C, i and ii). The most plausible interpretation for these two independent findings is that the number of Htt134Qm:EGFP aggregates is much larger than those of Htt134Q:EGFP, but most of them are invisible when examined by fluorescence microscopy due to the limited sensitivity and resolution of this method (Fig. 2 C and D). An interesting finding relates to identification of ubiquitination on K6 that was found only in the Htt134Q:EGFP variant and only in its fraction that is insoluble (Fig. 3D). It appears, therefore, that ubiquitination is unique only to insoluble aggregates, which probably recruit the ubiquitination machinery. It might well be that ubiquitination affects aggregate forms, determining their sizes and numbers.
Fig. 3.
Htt134Qm:EGFP generates a larger number of less soluble aggregates than Htt134Q:EGFP. HEK293 cells were transfected with FU-Htt17Q:EGFP-Wm, FU-Htt134Q:EGFP-Wm, or FU-Htt134Qm:EGFP-Wm. Four days later, cells were collected and pelleted. Two sequential fractions were prepared (as described in SI Appendix, Materials and Methods). NP-40-soluble fraction and insoluble fraction (that were dissolved in 10% SDS). Samples were analyzed by Western blots, dot blots, and mass spectrometry (MS). (A) Western blot analysis of mHtt variants (17Q:EGFP, 134Q:EGFP, and 134Qm:EGFP) in the NP-40-soluble fraction using either anti-Htt (i) or anti-EGFP (ii) antibodies. (B) MS analysis of the NP-40-soluble fraction: (i) MS based quantification of the peptide AFESLK—a region in the N terminus of mHtt that does not contain K/R6 or K/R9. (ii) MS-based quantification of all EGFP peptides. Quantification was carried out using MaxQuant 1.5.2.8 against the Human Uniprot database with a FDR <0.01 (three replicates). (C) NP-40-insoluble and 10% SDS-soluble fraction analysis. (i) Dot blot analysis of the insoluble fraction from three independent experiments. Numbers on the right hand side indicate intensity fold change in Htt134Qm:EGFP relative to Htt134Q:EGFP (Htt134Q:EGFP intensity defined as 1.0, left hand side). Dot blot intensities quantified using ImageJ. (ii) Average intensities of Htt134Q:EGFP and Htt134Qm:EGFP shown in C, i. (D) MS-based quantification of Htt-K6 ubiquitinated peptides from NP-40-soluble and insoluble (but 10% SDS-soluble) fractions. P values from two-sample t tests assuming unequal variances are shown (two tailed for B, i and ii; one tailed for C, ii).
Site-Specific Ubiquitination of Htt Reduces Cell Death.
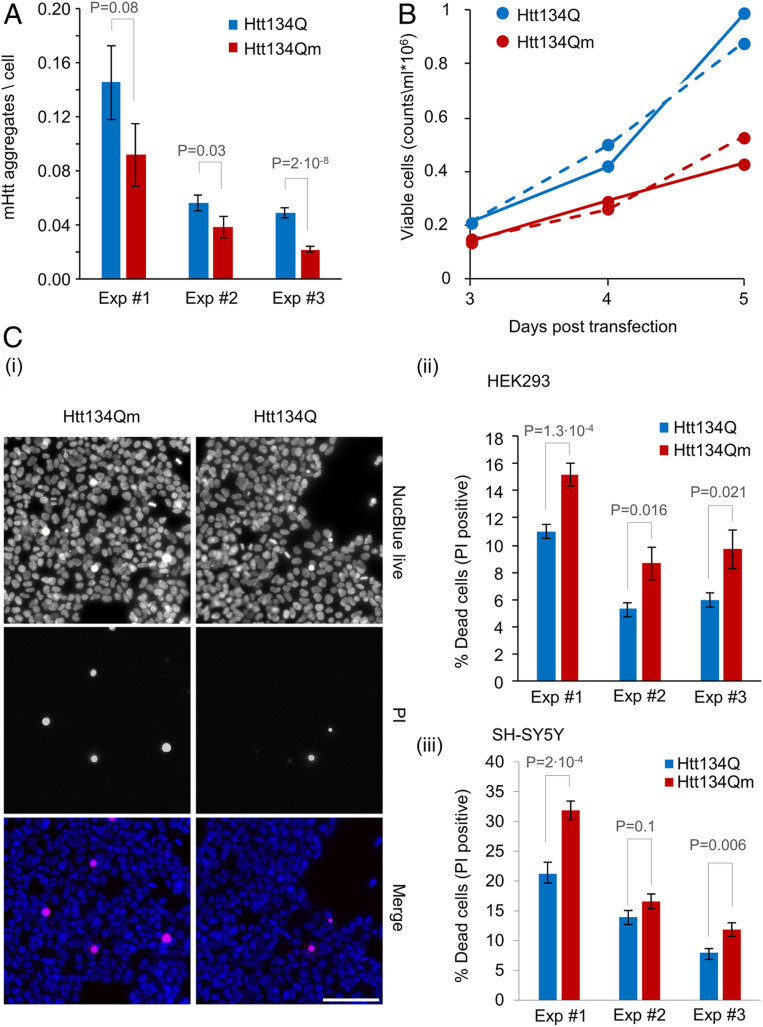
To study the potential pathophysiological effect of ubiquitination of lysine residues 6 and 9, we first reproduced, in HEK293 cells, the results obtained in cultured neurons. To that end, we used a high content imaging system to analyze large numbers of HEK293 cells expressing Htt134Q:EGFP and Htt134Qm:EGFP (as well as HttQ17:EGFP). As shown in Fig. 4A, the number of visible aggregates following transfection with Htt134Qm:EGFP was lower than that generated by Htt134Q:EGFP, basically reproducing the phenomenon observed in primary neurons (Fig. 2 C and D). Cell counts (Fig. 4B) and live/dead labeling (Fig. 4C, SI Appendix, Fig. S2, and see SI Appendix, Materials and Methods) revealed that expression of Htt134Qm:EGFP was associated with significant reductions in numbers of viable cells (which was not due to a decrease in cell division rates; SI Appendix, Fig. S3 A, i and ii) and higher levels of cell death as compared to cells expressing Htt134Q:EGFP (and HttQ17:GFP, SI Appendix, Fig. S2). A similar phenomenon was observed also when Htt134Q:EGFP and Htt134Qm:EGFP were expressed in SH-SY5Y neuroblastoma cells (Fig. 4 C, iii).
Fig. 4.
Htt134Q:EGFP ubiquitination alleviates the proteotoxic effects of mHtt. (A) HEK293 cells overexpressing FU-Htt134Q:EGFP-Wm or FU-Htt134Qm:EGFP-Wm were stained with the DNA-binding dye NucBlue, and visualized using an ImageXpress Micro Confocal system. Numbers of mHtt aggregates per cell were calculated as the total number of EGFP-positive aggregates divided by the number of NucBlue-positive cells as described in the SI Appendix, Materials and Methods. Aggregate counts per cell in cells expressing Htt134Q:EGFP or Htt134Qm:EGFP (25 fields of view per well and 49 wells from three independent experiments). (B) HEK293 cells were transfected with FU-Htt134Q:EGFP-Wm or FU-Htt134Qm:EGFP-Wm. The cells were harvested at indicated times following transfection, labeled with Tryptan blue, and live cells were counted by a Vi Cell XR cell counter. The continuous and dotted lines represent two independent experiments. (C) HEK293 and SH-SY5Y cells were transfected with FU-Htt134Q:EGFP-Wm or FU-Htt134Qm:EGFP-Wm. Four days following transfection, the cells were treated with propidium Iodide (PI) and NucBlue reagents and visualized using ImageXpress Micro Confocal system. The percentage of dead cells was calculated as the number of PI-positive cells relative to NucBlue-positive cells. (i) Representative images of PI-positive (red), and NucBlue-positive (blue) HEK293 cells. (ii and iii) Percentage of dead cells in HEK293 (ii) and SH-SY5Y (iii) cells (three independent experiments). Error bars: SEM; P values from two-sample t tests assuming unequal variances are shown (one tailed for A; two tailed for C, ii and iii). (Scale bar in C, i: 100 µm.)
Changes in Cellular Proteomes Associated with Elimination of N-Terminal Ubiquitination Sites.
Substitution of lysines 6 and 9 with arginines in Htt134Q:EGFP was associated with the delayed appearance, smaller and less soluble aggregates, and with higher levels of cell death. To obtain clues as to potential mechanisms by which these mutations impair cell viability, we carried out proteomic analyses of HEK293 cells expressing Htt134Q:EGFP, Htt134Qm:EGFP, or Htt17Q:EGFP. Two types of experiments were carried out: In the first, cells were solubilized in 10% SDS and resolved via SDS/polyacrylamide gel electrophoresis. We subjected both the material accumulated in the stacking gel and in the resolving gel to MS analysis. In the second type, cells were extracted under milder conditions (NP-40), followed by solublization of the pellets in 10% SDS (as described above). Each fraction was then subjected to proteomic analysis. Proteomic analyses of insoluble fractions revealed as expected strong differences between the Htt134Q:EGFP /Htt134Qm:EGFP and the Htt17Q:EGFP; differences between Htt134Q:EGFP and Htt134Qm:EGFP, however, did not reveal significant differences between the cells expressing the different proteins and across the two types of experiments. It, thus, seems that numerous proteins associate with mHtt aggregates, but this association is relatively nonspecific and not strongly dependent on the presence or absence of the N-terminal ubiquitination sites.
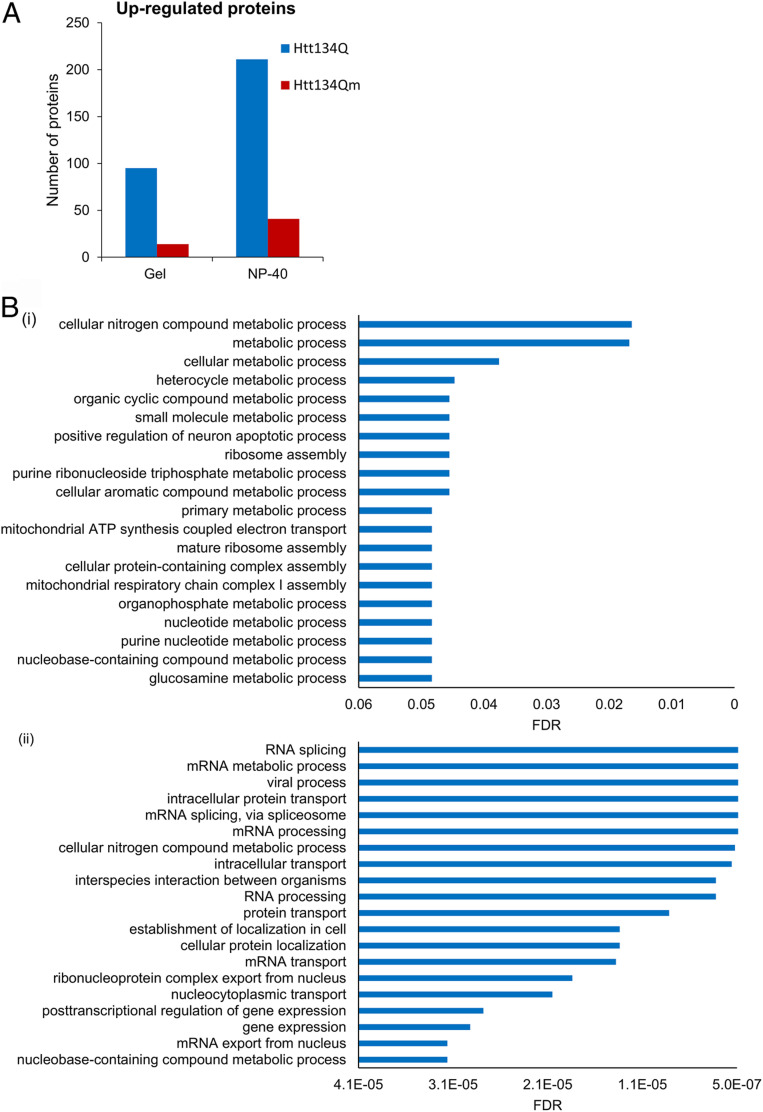
In contrast, analysis of the soluble fractions provided interesting clues. In all experiments, we noted a significant number of proteins that were more abundant in cells expressing Htt134Q:EGFP as compared to those expressing Htt134Qm:EGFP (Fig. 5A and see Materials and Methods for inclusion criteria; lists of affected proteins and fold changes are provided in SI Appendix, Tables S1 and S2). Functional analysis of these data pointed to significant changes in multiple biological processes (Fig. 5B). The overlap in affected pathways in cells solubilized with 10% SDS and NP-40 was partial, but this might be expected given the different populations of proteins (and organelles) extracted under harsh and mild conditions. The most strongly affected pathways seemed to involve RNA metabolism, intracellular transport, mitochondrial-related metabolism, and gene regulation. In contrast, carrying out similar analyses for proteins whose abundance was increased in cells expressing Htt134Qm suggested much more moderate changes in these and other pathways (see Dataset S1, for all gene ontology analyses, see also SI Appendix, Fig. S4 for protein abundance changes associated with HD-related pathways). These analyses suggest that cellular reactions to mHtt expression were weaker in cells expressing the lysineless protein, possibly implying a reduced induction of mechanisms required for coping with proteotoxic stress.
Fig. 5.
Htt134Q:EGFP ubiquitination affects the cellular proteome. Proteomic profile of soluble fractions from HEK293 cells expressing FU-Htt17Q:EGFP-Wm, FU-Htt134Q:EGFP-Wm, or FU-Htt134Qm:EGFP-Wm extracted with 10% SDS (Gel, three replicates from one experiment) or NP-40 (six replicates from two experiments). Both populations were analyzed by MS as described in Materials and Methods. (A) Numbers of proteins up-regulated in Htt134Q:EGFP- relative to Htt134Qm:EGFP-expressing cells and vice versa. (B) Biological process enrichment of the differentially expressed proteins shown in A analyzed using the STRING database version 11.0. (i) Data derived from analysis of gel-resolved proteins; (ii) data derived from NP-40 dissolved proteins.
In addition, and in order deepen the analysis, we performed a search aimed at identifying specific proteins, the abundance of which changed significantly and consistently in cells expressing Htt134Q:EGFP or Htt134Qm:EGFP relative to cells expressing Htt17Q:EGFP. To that end, we reanalyzed the soluble NP-40 fractions (six replicates from two experiments). We applied rather stringent inclusion criteria: 1) comparisons were made only for proteins identified in at least four replicates in each condition; 2) the minimal acceptable change was a factor of ∼1.75; 3) a P value ≤0.05 (t test assuming unequal variance) in comparisons between Htt134Qm:EGFP and Htt134Qm:EGFP. Twelve differentially expressed proteins that fulfilled these criteria were identified (SI Appendix, Table S3; the full data set is provided in Dataset S2). The observed changes in some of these differentially expressed proteins provide some interesting clues regarding the poorer survival of cells expressing mHtt exon 1 lacking ubiquitination sites (see Discussion).
Discussion
An important, yet unresolved, problem in the field of neurodegenerative diseases is whether protein aggregation reflects a failure of cellular protein degradation systems. Since ubiquitination is essential for targeting proteins for both proteasomal and lysosome-autophagy mediated degradation, we decided to characterize the mode and function of mHtt ubiquitination. Proteomic analysis of brain samples from rats that express the complete human mHTT gene with 97 glutamine repeats revealed that lysines 6 and 9, upstream to the glutamine repeat, are specifically ubiquitinated in mutant but not WT rats (Fig. 1). The mHtt-specific ubiquitination of these sites indicated that these ubiquitination events may be related to the aggregation process. To address this possibility, we used long-term imaging to measure aggregate formation dynamics in rat cortical neurons expressing fluorescently tagged mHtt exon 1 variants in which lysines 6 and 9 were (or were not) mutated to arginine. Elimination of these ubiquitination sites was found to significantly slow aggregate appearance rates and reduce mean aggregate size (Fig. 2 and Movie S1). Conversely, biochemical analysis revealed that elimination of lysines 6 and 9 significantly increased the amount of insoluble mHtt, most likely explained by enhanced formation of very small aggregates, undetectable by conventional light microscopy (Fig. 3). Expression of this exon 1 variant was associated with more cell death in comparison to the variant containing lysines 6 and 9 (Fig. 4). Finally, proteomic analysis suggested that cells expressing lysine-containing variants up-regulate more strongly the synthesis of a significant numbers of proteins associated, among other pathways, with RNA metabolism, intracellular transport, mitochondrial-related metabolism, and gene regulation (Fig. 5).
The importance of posttranslational modifications in regulating mHtt pathology is well known (reviewed in ref. 30). In particular, posttranslational modification of lysines 6, 9 (and 15) through small ubiquitin like modifier (SUMO)lation (31) or acetylation (32) have been suggested to exacerbate or reduce mHtt toxicity, respectively. Ubiquitination of these sites was suggested to modestly abrogate neurodegeneration (31), although experiments in Drosophila led the authors to conclude that SUMOlation, rather than ubiquitination, is more physiologically relevant. Yet, our proteomic analysis (Fig. 1) shows that lysines 6 and 9 are, in fact, selectively ubiquitinated in brain sections from a well-characterized HD model. Moreover, a nearly identical observation was very recently reported in a different HD animal model (Q175 HD mice, ref. 33). In line with these observations, ubiquitination of lysine 6 was observed only in aggregated Htt134Q and not in its soluble forms (Fig. 3D). Thus, ubiquitination is likely to represent a physiologically relevant posttranslational modification of these sites, although this does not preclude roles for other modifications (see also ref. 34).
The manners by which N-terminal ubiquitination affects cell viability is not known. Prior studies have suggested that mHtt aggregates sequester essential proteins and that the loss of these proteins negatively affects cellular physiology (10, 12). In our proteomic analyses, however, we found no evidence that levels of proteins associated with mHtt aggregates of either form were significantly reduced in soluble fractions of the same biochemical preparations. On the other hand, these analyses indicated that mHtt expression, in particular, Htt134Q:EGFP, increased the abundance of proteins that might enhance the cells capacity to cope with proteotoxic stress. Furthermore, the mutation of lysines 6 and 9 to arginine resulted in the formation of more, yet smaller mHtt aggregates, which are likely to consist of mHtt oligomers or prefibrillar aggregates now believed to be the most toxic forms of mHtt (9, 35, 36). Interestingly, the 17-amino acid N terminus of mHtt has been shown repeatedly to play crucial roles in mHtt oligomerization and fibril nucleation in manners highly sensitive to posttranslational modifications (reviewed in ref. 35). Such effects on nucleation rates might possibly explain our observations that the appearance of large aggregates—but not their growth—was slower in neurons expressing mHtt exon 1 lacking the aforementioned ubiquitination sites (Fig. 2, SI Appendix, Fig. S1, and Movie S1).
Our detailed proteomic analysis pointed to 12 proteins whose abundance was selectively altered in cells expressing either Htt134Q:EGFP or Htt134Qm:EGFP. Within this group, two interesting proteins were GLTSCR2 and WDR48. Glioma tumor suppressor candidate region gene 2 (GLTSCR2) is a protein previously shown to promote apoptosis through multiple pathways (37). Interestingly, abundance of this protein was elevated more than twofold in cells expressing Htt134Qm:EGFP, possibly explaining elevated death rates in these cell populations. WDR48 (WD repeat-containing protein 48) is a potent activator of multiple deubiquitinating enzymes (38), whose expression was reduced nearly twofold in cells expressing Htt134Q:EGFP. This change might be expected to increase the overall level of ubiquitinated proteins. Consequently, the extent of ubiquitination of mHtt may increase as well, assisting the cell to cope with the proteotoxic stress. Increasing the extent of ubiquitination of other proteins might stimulate their degradation, which is probably also important to mitigate the sequelae of the stress.
To summarize, our findings shed light on a novel role of ubiquitination in shaping the size, number, possibly the morphology of mHtt aggregates, and on manners by which it may attenuate mHtt pathogenicity.
Materials and Methods
Ethics
All experiments were approved by the local ethics committee at Regierungspraesidium Tuebingen (approval ID: HG11/12), and by the "Technion-Israel Institute of Technology Committee for the Supervision of Animal Experiments" (ethics approval number IL-151-00-32). All animal-related experiments were carried out in accordance with the German Animal Welfare Act and the guidelines of the Federation of European Laboratory Animal Science Associations, based on European Union legislation (Directive 2010/63/EU).
Primary Cortical Neuronal Culture.
Primary cultures of rat cortical neurons for live imaging were prepared as described previously (39). Briefly, cortices of 1- to 2-d-old rats were dissected, dissociated by trypsin which was followed by trituration using a siliconized Pasteur pipette. Cells were then seeded on a 6-well plate (2 × 106 cells/well) or on thin glass multielectrode array (MEA) dishes (Multichannel Systems), divided into two chambers (0.6–1.2 × 106/chamber), which were pretreated with polyethylenimine (Sigma-Aldrich) to facilitate cell attachment. Cells were initially grown in a medium containing Eagle’s minimal essential medium (Sigma-Aldrich), 25-µg/mL insulin (Sigma-Aldrich), 20-mM glucose (Sigma-Aldrich), 2-mM l-glutamine (Sigma-Aldrich), 11.16-µg/mL gentamicin sulfate (Sigma-Aldrich), 10% NuSerum (Becton Dickinson Labware), and 0.5% fetal bovine serum (HyClone). The preparation was then transferred to a humidified tissue culture incubator and maintained at 37 °C in a 95% air and 5% CO2 mixture. Half the volume of the culture medium was replaced three times a week with a feeding medium similar to the medium described above, but devoid of NuSerum and serum, and contains a lower concentration of l-glutamine (0.5 mM, Sigma-Aldrich) and a 2% B-27 supplement (Gibco).
BACHD Rats.
The BACHD rat is a transgene expressing the full length human HTT gene with 97 CAG repeats under the human HTT promoter and all its regulatory elements (26). The rats exhibit an early onset HD-like behavioral phenotype, progressive aggregation of mHtt, and cell death. Briefly, rats were sacrificed by CO2 inhalation in a carbon dioxide chamber. Brains were immediately dissected on ice, and brain regions were sampled. Tissue was shock frozen in liquid nitrogen and stored at −80 °C.
Ubiquitinome Analysis of Brain Samples of the BACHD and WT Rats.
Lysates were prepared from 60-mg cortical and striatal samples of 15-mo-old BACHD and WT rats using mechanical disruption in 2 mL of urea buffer (9-M urea, 400-mM ammonium bicarbonate [ABC], and 10-mM dithiothreitol [DTT]), followed by two cycles of sonication. Amounts of 3.02-mg proteins from each sample were reduced with 10-mM DTT (60 °C for 30 min), that was neutralized with 4-mM iodoacetamide in 100-mM ABC (in the dark at room temperature for 30 min). The proteins were then digested (overnight at 37 °C) with modified trypsin (Promega) at a 1:50 enzyme to substrate ratio in 2-M urea and 100-mM ABC. A second digestion with trypsin at a 1:100 enzyme-to-substrate ratio was carried out for 4 h at 37 °C. The tryptic peptides from each sample were subjected to Ub enrichment in order to obtain quantitative profiles of nonredundant ubiquitinated proteins. The trypsin cleaves also ubiquitin C terminally to Arg74, leaving the two last amino acids in the ubiquitin molecule, Gly75 and Gly76, attached in an isopeptide bond to an internal lysine residue in the peptides released from the intact substrate by the same tryptic digestion. These Gly-Gly–modified peptides were isolated using the UbiScan ubiquitin remnant motif (K-GG) antibody conjugated to beads (cell signaling) following overnight incubation at 4 °C. The beads were washed once with the Kit’s immunoaffinity purification buffer and twice in phosphate-buffered saline, 0.1% acetic glycoside, and 500-mM NaCl. The ubiquitinated peptides were then eluted twice at room temperature with 0.2% trifluoroacetic buffer and centrifuged for 10 min at 850 rpm. Both eluates were pooled, and the enriched peptides from each sample were desalted using an UltraMicro tip columan C18 (Harvard), dried, and resuspended in 0.1% formic acid. MS was performed by a Q Exactive plus mass spectrometer (ThermoFisher) in a positive mode using repetitively full MS scans, followed by collision-induced dissociation (HCD) of the 10 most dominant ions selected from the first MS scan. The MS data from all of the biological repeats were analyzed using the MaxQuant software 1.5.2.8 (Mathias Mann’s group) vs. the Rattus norvegicus proteome from the Uniprot database (with 1% FDR). The data were quantified by label free analysis using the same software based on extracted ion currents of peptides enabling quantitation from each liquid chromatography/MS run for each peptide identified in any of the experiments. Statistical analyses of the identification and quantization were performed using the Perseus software.
Long-Term Imaging.
Cortical neurons were plated on thin glass MEA dishes divided into two chambers as described in Primary Cortical Neuronal Culture. Approximately 0.6–1.2 × 106 cells were plated per chamber and allowed to develop for 14 d. At day in vitro (DIV) 14, viral particles of FU-Htt134Q:EGFP-Wm or FU-Htt134Qm:EGFP-Wm were added to each chamber. The imaging, initiated at DIV 15, was carried out by an automated multisite time-lapse confocal microscope at 2-h intervals (three sections at each location) for a period of 2 wk. MEAs were fabricated on thin glass (180 µm), which allowed for the use of a high numerical aperture (N.A.), oil immersion objectives suited for high resolution imaging (39–41). Scanning fluorescence and bright field images were acquired using a custom designed confocal laser scanning microscope based on a Zeiss Axio Observer Z1 using a ×40, 1.3 N.A. Fluar objective. The system was controlled by custom written software and included provisions for automated multisite time-lapse microscopy. MEA dishes containing networks of cortical neurons were mounted on headstage/amplifier which was attached to the microscope’s motorized stage. FU-Htt134Q:EGFP-Wm and FU-Htt134Qm:EGFP-Wm were excited using a 488-nm solid state laser (Coherent). Fluorescence emissions were read through a 500–550-nm band-pass filter (Chroma Technology). All data were collected at a resolution of 640 × 480 pixels at 12 bits/pixel with the confocal aperture fully open. Data were collected sequentially from predefined sites using the confocal microscope robotic XYZ stage to cycle automatically through these sites. Focal drift during the experiment was corrected automatically by using the microscopes’ “autofocus” feature. MEA dishes were covered with a custom designed cap containing two inlet and two outlet ports for separate perfusion of media, a fifth inlet for air mixture, and two platinum wire reference ground electrodes. MEA dishes were continuously perfused with feeding media (described above) at a rate of 2 mL/d by means of a custom built perfusion system based on two ultraslow flow peristaltic pumps (Instech Laboratories, Inc.) and silicone tubing. The tubes were connected to the dish through the appropriate ports in the custom designed cap. A 95% air/5% CO2 sterile mixture was continuously streamed into the dish at very low rates through a port with flow rates regulated by a high precision flow meter (Gilmont Instruments). The bases of the headstage/amplifier and the objective were heated to 37 °C using resistive elements and separate temperature sensors and controllers, stabilizing a temperature of 36–37 °C in the culture media.
All other routine methods are described in the SI Appendix.
Supplementary Material
Acknowledgments
A.C. was supported by grants from the Adelson Medical Research Foundation (AMRF), the Israel Science Foundation (ISF), and the Technion-University of Michigan at Ann Arbor Collaborative Program. A.C. is an Israel Cancer Research Fund (ICRF) recipient. N.E.Z. and A.C. were supported by grants from the European Community’s Seventh Framework Programme FP7/2012 (TreatPolyQ, Grant Agreement 264508), the Rappaport Institute, and the Allen and Jewel Prince Center for Neurodegenerative Disorders of the Brain. A.C., N.E.Z., H.H.P.N., and O.R. were supported by a grant from the Germany-Israeli Foundation for Scientific Research and Development (GIF; I-1437-418.13/2017).
Footnotes
Competing interest statement: A.C. and G.M. were coauthors on a 2018 nomenclature proposal with an additional 170 authors; they did not collaborate directly on the article.
Data deposition: The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (accession nos. PXD019552 and PXD019553).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2007667117/-/DCSupplemental.
Data Availability.
Underlying data for Figs. 1–5 as well as SI Appendix figures are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0) at Figshare (42).
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (43) partner repository (accession nos. PXD019552 and PXD019553) (44, 45).
References
- 1.Rosas H. D. et al., Cerebral cortex and the clinical expression of Huntington’s disease: Complexity and heterogeneity. Brain 131, 1057–1068 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zuccato C., Valenza M., Cattaneo E., Molecular mechanisms and potential therapeutical targets in Huntington’s disease. Physiol. Rev. 90, 905–981 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Aylward E. H. et al., Author manuscript; Available in PMC. J. Neurol. Neurosurg. Psychiatry 82, 405–410 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saudou F., Humbert S., The biology of huntingtin. Neuron 89, 910–926 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Ross C. A., Tabrizi S. J., Huntington’s disease: From molecular pathogenesis to clinical treatment. Lancet Neurol. 10, 83–98 (2011). [DOI] [PubMed] [Google Scholar]
- 6.McCampbell A. et al., CREB-binding protein sequestration by expanded polyglutamine. Hum. Mol. Genet. 9, 2197–2202 (2000). [DOI] [PubMed] [Google Scholar]
- 7.Nucifora J. et al., Interference by huntingtin and atrophin-1 with CBP-mediated transcription leading to cellular toxicity. Science 291, 2423–2428 (2001). [DOI] [PubMed] [Google Scholar]
- 8.Steffan J. S. et al., The Huntington’s disease protein interacts with p53 and CREB-binding protein and represses transcription. Proc. Natl. Acad. Sci. U.S.A. 97, 6763–6768 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim Y. E. et al., Soluble oligomers of PolyQ-expanded huntingtin target a multiplicity of key cellular factors. Mol. Cell 63, 951–964 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Donaldson K. M. et al., Ubiquitin-mediated sequestration of normal cellular proteins into polyglutamine aggregates. Proc. Natl. Acad. Sci. U.S.A. 100, 8892–8897 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arrasate M., Finkbeiner S., Protein aggregates in Huntington’s disease. Exp. Neurol. 238, 1–11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hosp F. et al., Spatiotemporal proteomic profiling of huntington’s disease inclusions reveals widespread loss of protein function. Cell Rep. 21, 2291–2303 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cummings C. J. et al., Chaperone suppression of aggregation and altered subcellular proteasome localization imply protein misfolding in SCA1. Nat. Genet. 19, 148–154 (1998). [DOI] [PubMed] [Google Scholar]
- 14.Miller J. et al., Quantitative relationships between huntingtin levels, polyglutamine length, inclusion body formation, and neuronal death provide novel insight into huntington’s disease molecular pathogenesis. J. Neurosci. 30, 10541–10550 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bodner R. A. et al., Pharmacological promotion of inclusion formation: A therapeutic approach for Huntington’s and Parkinson’s diseases. Proc. Natl. Acad. Sci. U.S.A. 103, 4246–4251 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ciechanover A., Brundin P., The ubiquitin proteasome system in neurodegenerative diseases: Sometimes the chicken, sometimes the egg. Neuron 40, 427–446 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Doi H. et al., Identification of ubiquitin-interacting proteins in purified polyglutamine aggregates. FEBS Lett. 571, 171–176 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Zeng L. et al., Differential recruitment of UBQLN2 to nuclear inclusions in the polyglutamine diseases HD and SCA3. Neurobiol. Dis. 82, 281–288 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang H. et al., PolyQ-expanded huntingtin and ataxin-3 sequester ubiquitin adaptors hHR23B and UBQLN2 into aggregates via conjugated ubiquitin. FASEB J. 32, 2923–2933 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Schipper-Krom S., Juenemann K., Reits E. A. J., The ubiquitin-proteasome system in huntington’s disease: Are proteasomes impaired, initiators of disease, or coming to the rescue? Biochem. Res. Int. 2012, 837015 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKinnon C., Tabrizi S. J., The ubiquitin-proteasome system in neurodegeneration. Antioxid. Redox Signal. 21, 2302–2321 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Bence N. F., Sampat R. M., Kopito R. R., Impairment of the ubiquitin-proteasome system by protein aggregation. Science 292, 1552–1555 (2001). [DOI] [PubMed] [Google Scholar]
- 23.Labbadia J., Morimoto R. I., Huntington’s disease: Underlying molecular mechanisms and emerging concepts. Trends Biochem. Sci. 38, 378–385 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ciechanover A., Kwon Y. T., Degradation of misfolded proteins in neurodegenerative diseases: Therapeutic targets and strategies. Exp. Mol. Med. 47, e147 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bowman A. B., Yoo S. Y., Dantuma N. P., Zoghbi H. Y., Neuronal dysfunction in a polyglutamine disease model occurs in the absence of ubiquitin-proteasome system impairment and inversely correlates with the degree of nuclear inclusion formation. Hum. Mol. Genet. 14, 679–691 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Yu-Taeger L. et al., A novel BACHD transgenic rat exhibits characteristic neuropathological features of Huntington disease. J. Neurosci. 32, 15426–15438 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim W. et al., Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44, 325–340 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bustos D., Bakalarski C. E., Yang Y., Peng J., Kirkpatrick D. S., Characterizing ubiquitination sites by peptide-based immunoaffinity enrichment. Mol. Cell. Proteomics 11, 1529–1540 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Williamson A., Werner A., Rape M., The Colossus of ubiquitylation: Decrypting a cellular code. Mol. Cell 49, 591–600 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambataro F., Pennuto M., Post-translational modifications and protein quality control in motor neuron and polyglutamine diseases. Front. Mol. Neurosci. 10, 82 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steffan J. S. et al., SUMO modification of Huntingtin and Huntington’s disease pathology. Science 304, 100–104 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Chaibva M. et al., Acetylation within the first 17 residues of Huntingtin exon 1 alters aggregation and lipid binding. Biophys. J. 111, 349–362 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sap K. A. et al., Global proteome and ubiquitinome changes in the soluble and insoluble fractions of Q175 huntington mice brains. Mol. Cell. Proteomics 18, 1705–1720 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cong X. et al., Mass spectrometric identification of novel lysine acetylation sites in Huntingtin. Mol. Cell. Proteomics 10, M111.009829 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arndt J. R., Chaibva M., Legleiter J., The emerging role of the first 17 amino acids of huntingtin in Huntington’s disease. Biomol. Concepts 6, 33–46 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leitman J., Ulrich Hartl F., Lederkremer G. Z., Soluble forms of polyQ-expanded huntingtin rather than large aggregates cause endoplasmic reticulum stress. Nat. Commun. 4, 2753 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Chen H. et al., Moesin-ezrin-radixin-like protein (merlin) mediates protein interacting with the carboxyl terminus-1 (PICT-1)-induced growth inhibition of glioblastoma cells in the nucleus. Int. J. Biochem. Cell Biol. 43, 545–555 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Cohn M. A., Kee Y., Haas W., Gygi S. P., D’Andrea A. D., UAF1 is a subunit of multiple deubiquitinating enzyme complexes. J. Biol. Chem. 284, 5343–5351 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Minerbi A. et al., Long-term relationships between synaptic tenacity, synaptic remodeling, and network activity. PLoS Biol. 7, e1000136 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rubinski A., Ziv N. E., Remodeling and tenacity of inhibitory synapses: Relationships with network activity and neighboring excitatory synapses. PLoS Comput. Biol. 11, e1004632 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaufman M., Corner M. A., Ziv N. E., Long-term relationships between cholinergic tone, synchronous bursting and synaptic remodeling. PLoS One 7, e40980 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hakim-Eshed V., et al. , Site-specific ubiquitination of pathogenic huntingtin attenuates its deleterious effects. FigShare. 10.6084/m9.figshare.12417278. Deposited 3 June 2020. [DOI] [PMC free article] [PubMed]
- 43.Vizcaíno J. A. et al., The PRoteomics IDEntifications (PRIDE) database and associated tools: Status in 2013. Nucleic Acids Res. 41, D1063–D1069 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hakim-Eshed V., et al. , Site-specific ubiquitination of pathogenic huntingtin attenuates its deleterious effects. PRIDE. https://www.ebi.ac.uk/pride/archive/projects/PXD019553. Deposited 4 June 2020. [DOI] [PMC free article] [PubMed]
- 45.Hakim-Eshed V., et al. , Site-specific ubiquitination of pathogenic huntingtin attenuates its deleterious effects. PRIDE. https://www.ebi.ac.uk/pride/archive/projects/PXD019552. Deposited 4 June 2020. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Underlying data for Figs. 1–5 as well as SI Appendix figures are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0) at Figshare (42).
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (43) partner repository (accession nos. PXD019552 and PXD019553) (44, 45).