Abstract
The vacuolar-type H+-ATPases (V-ATPase) hydrolyze ATP to pump protons across the plasma or intracellular membrane, secreting acids to the lumen or acidifying intracellular compartments. It has been implicated in tumor metastasis, renal tubular acidosis, and osteoporosis. Here, we report two cryo-EM structures of the intact V-ATPase from bovine brain with all the subunits including the subunit H, which is essential for ATPase activity. Two type-I transmembrane proteins, Ac45 and (pro)renin receptor, along with subunit c”, constitute the core of the c-ring. Three different conformations of A/B heterodimers suggest a mechanism for ATP hydrolysis that triggers a rotation of subunits DF, inducing spinning of subunit d with respect to the entire c-ring. Moreover, many lipid molecules have been observed in the Vo domain to mediate the interactions between subunit c, c”, (pro)renin receptor, and Ac45. These two structures reveal unique features of mammalian V-ATPase and suggest a mechanism of V1-Vo torque transmission.
Subject terms: Cryoelectron microscopy, Membrane structure and assembly, Permeation and transport
The vacuolar-type H+ -ATPases (V-ATPase) hydrolyze ATP to pump protons across the plasma or intracellular membrane. Here authors report two cryo-EM structures of the intact V-ATPase from bovine brain with all the subunits including the subunit H, which is essential for ATPase activity.
Introduction
The pH homeostasis is essential for the physiological function of cells and cellular organelles. An acidic pH is important for many physiological processes, such as renal acidification, bone resorption, and sperm maturation. V-ATPases localize to the plasma membrane, endosomes, and membrane vesicles and play an important role in proton homeostasis1–4. V-ATPase is involved in autophagy5, metabolism6,7, and neurotransmitter secretion3. Recycling of cell receptors is facilitated by V-ATPase through endocytosis8,9. For example, the low pH generated by V-ATPase promotes the dissociation of ligands, such as mannose 6-phosphate receptor10, which recognizes lysosomal enzymes in Golgi and releases them in prelysosomes by its multiple repeats, facilitating the receptor recycling11. Moreover, V-ATPase activity is involved in Wnt, mTOR, and Notch signaling12–14. The aberrant activity of V-ATPases in humans is associated with many pathological processes and diseases, such as tumor metastasis, cutis laxa, distal renal tubular acidosis, autosomal recessive osteopetrosis, infectious diseases, and neurodegenerative diseases3.
Mammalian V-ATPase is a rotary machine made up of two domains: the ATP-hydrolytic V1 domain and the proton-translocation Vo domain. The V1 domain consists of three catalytic AB heterodimers that form a heterohexamer with threefold rotational pseudosymmetry, three peripheral stalks each consisting of the subunits EG, one central rotor including subunits D and F, and the regulatory subunits C and H; the proton-translocation domain Vo consists of the proton transport subunit a, a ring of proteolipid subunits c9c”, rotary subunit d, and subunits e and Ac45. The subunits B, C, E, G, H, a, d, and e of mammalian V-ATPase have multiple isoforms in various tissues and cells15 causing a technical barrier to study its structural and biochemical features due to heterogeneity.
The V-ATPase is structurally and evolutionarily related to the F1F0-ATP synthases and the archaebacterial ATP synthases termed V/A-ATPases2. The composition of the mammalian V1 domain is similar to that of the yeast domain, which is a well-characterized model for V-ATPase studies. The c-ring of yeast’s Vo domain contains eight subunit c, one subunit c’, and one subunit c”. In the V/A-ATPase, there are two subunit E and two subunit G in the V1 domain, and twelve subunits c that form a homogeneous c-ring in the Vo domain. Structural insights into V-ATPase function has been gleaned from structures of the intact V-ATPase from Saccharomyces cerevisiae (scV-ATPase) at 7-Å resolution16, structures of the intact V/A-ATPase from Thermus thermophilus (ttV/A-ATPase)17, cryo-EM structures of the scVo domain at atomic resolution18,19, and most recently published, the structure of rat V-ATPase with its bacterial inhibitor SidK20. However, the intact structure of a mammalian V-ATPase with the ATPase activity is still unknown.
In this paper, we purified the intact V-ATPase from bovine brain with the specific ATP hydrolysis activity at about 1.4 μmols of Pi·mg of protein−1·min−1 and report two cryo-EM structures in the distinct rotational states at overall 3.4-Å and 3.8-Å resolution, respectively. The structures show two unique mammalian V-ATPase components, Ac45 and (pro)renin receptor (PRR) in the Vo domain. This structural work reveals a high-resolution map of the V-type ATPase of bovine brain showing the interaction details between PRR and Ac45. It will also aid in the design of small molecules for the treatment of related human diseases.
Results
Overall structure of the Bos taurus V-ATPase
The Bos taurus V-ATPase (btV-ATPase) was purified from bovine brain as previously reported21. The resulting complex was further purified by gel filtration in the presence of 0.1% CHAPS and 0.004% glyco-diosgenin (GDN) (Supplementary Fig. 1a). The purified protein exhibits high ATPase activity and can be inhibited by bafilomycin A1 in vitro (Supplementary Fig. 1b). The identity of each component of the btV-ATPase was confirmed by mass spectrometry following SDS-PAGE (Supplementary Table 1). This complex was prepared on grids and subject to cryo-EM. The btV-ATPase particles are homogenous showing clear features in the cryo-EM images, making them suitable for high-resolution structure determination (Supplementary Fig. 2). The 3D classification enabled us to distinguish two different states. The 3D refinement of individual classes yielded a resolution of 3.4 Å (state 1) and 3.8 Å (state 2) (Fig. 1a–d). The homology models for subunits A-H of V1 and subunits a, c, c”, d, e of Vo were generated by MODELLER22 based on the multiple sequence alignments between the bovine subunits and the corresponding subunits of previously determined structures (Supplementary Fig. 3).
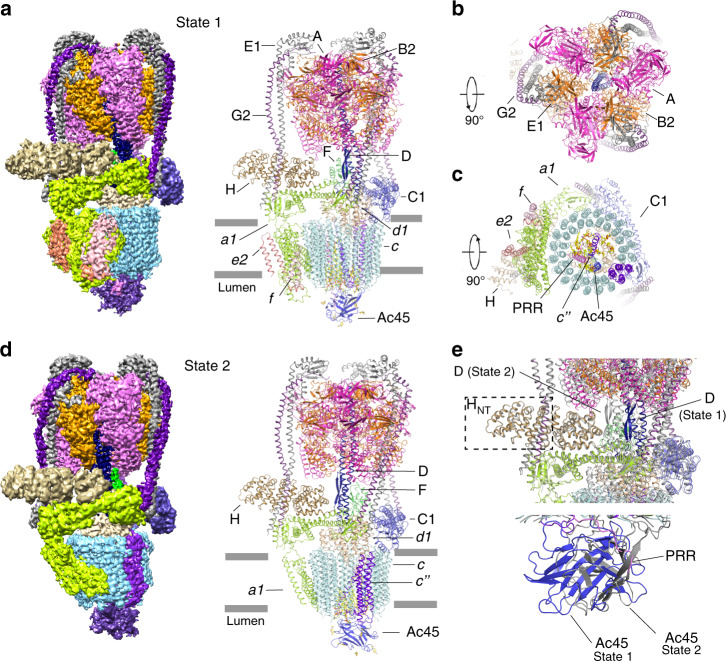
Fig. 1. Overall structure of V-ATPase from bovine brain.
a Overall cryo-EM map and structure showing V-ATPase in state 1 viewed from the side of the membrane. b The cytosolic view of the V-ATPase. c The luminal view of the V-ATPase. d Overall cryo-EM map and structure showing V-ATPase in state 2. Each subunit is colored and indicated. e The structural comparison of the two states.
The local resolution of the V1 domain is better than that of the Vo domain in both states 1 and 2 (Supplementary Fig. 4). To improve the quality of the Vo and V1 density, we performed a local refinement by specific Vo and V1 masks, respectively, which resulted in a density that shows clear features of the side chains, helping the unambiguous assignment of most residues (Supplementary Figs. 5 and 6). We built B2, C1, E1, G2, a1, d1, and e2 into the final model based on mass spectrometry results and the expression distribution in brain tissues3. The maps after local refinement were merged by “phenix.combine_focused _maps” for model building (Supplementary Fig. 6 and Supplementary Table 2). The overall dimensions of btV-ATPase are 160 Å*100 Å*270 Å. Due to the higher resolution of state 1, we discuss the structure of state 1 in the following section, except for the structural comparison of state 1 and state 2.
The subunit H is crucial for the V-ATPase activity23,24. A structural study showed that without the Vo domain, subunit H can bind to the A3B3 hexamer to prevent the rotation of V1 domain, thereby blocking ATPase activity25. In both states of our study, subunit H is well determined in the cryo-EM map (Fig. 1a, d). Structural comparison did not reveal notable conformational changes in subunit H with a root-mean-square deviation (RMSD) of 0.656 Å for 365 Cα atoms. The N-terminal helices of subunit H (HNT) bind to the subunits E1 and G2 (dash box in Fig. 1e) and—along with the C-terminal helices of subunit H (HCT)—interact with the cytosolic domain of subunit a1 (Fig. 1e). This contact may keep the subunit a1 in a certain conformation conducive for proton translocation.
A3B3 hexamer for ATP hydrolysis
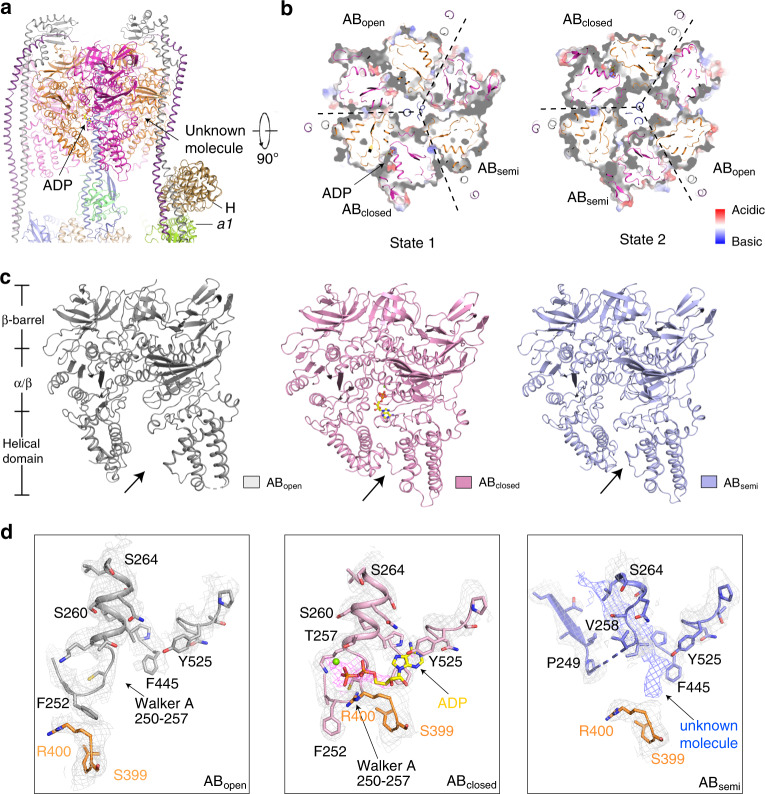
The A3B3 hexamer hydrolyzes ATP to provide energy for the rotation of the Vo domain (Fig. 2a, b). Both the A and B subunits have three domains: an N-terminal β-barrel domain, a middle nucleotide-binding α/β domain, and a C-terminal α-helical domain (Fig. 2c). ATP hydrolysis occurs at the interface between the nucleotide-binding α/β domains of subunits A and B26. In state 1, three AB heterodimers exhibit distinct conformations (ABsemi, ABclosed, and ABopen) (Fig. 2c; Supplementary Fig. 7). The structural comparison of the AB heterodimers with those of ttV/A-ATPase shows that the three AB heterodimers share a similar conformation with ABsemi, ABclosed (ADP-bound), and ABopen of ttV/A-ATPase with RMSD at 2.0 Å, 0.8 Å, and 0.9 Å, respectively (Supplementary Fig. 8). ABopen has a pocket open to the cytosol showing a high affinity to accommodate an ATP molecule26. Density representing the di-phosphate of ADP with magnesium was found in ABclosed (Fig. 2d) that binds ADP in ttV/A-ATPase17. Therefore, we built an ADP with a magnesium ion into ABclosed (Fig. 2c, d).
Fig. 2. Structure of the V1 domain showing three AB heterodimers with distinct conformations.
a Overall structure showing the V1 domain. b The cytosolic view of the V1 domain in state 1 (left) and state 2 (right). c, d The structural comparison of ABopen, ABclosed, and ABsemi in state 1. The domains of the AB heterodimer have been indicated in the left side of panel c. Cryo-EM maps in state 1 are shown at the 4σ level.
Notably, an unknown density was observed in ABsemi (Fig. 2d). This unknown molecule binds to a hydrophobic area of subunit A of ABsemi, and the Walker A motif (G250AFGCGKT257) of this subunit in the cryo-EM map is not clear (Fig. 2d). The residues 258–264 in the ABsemi form a loop to accommodate this unknown molecule; by contrast, these residues form an α-helix in ABclosed and ABopen (Fig. 2d). It is possible that it represents a nucleotide, since the ABsemi exhibits an intermediate conformation for releasing ADP. This small molecule also may be a CHAPS detergent or another unknown molecule. Further investigation is required for identification of this molecule. The C-terminal α-helical domains of ABsemi and ABclosed are shifted by ~10 Å compared with those in ABopen (Fig. 2c), demonstrating substantial conformational plasticity in the A3B3 hexamer. The structural analysis shows that the ABsemi, ABclosed, and ABopen heterodimers share similar conformations in states 1 and 2 (Supplementary Fig. 7).
Overall structure of Vo domain
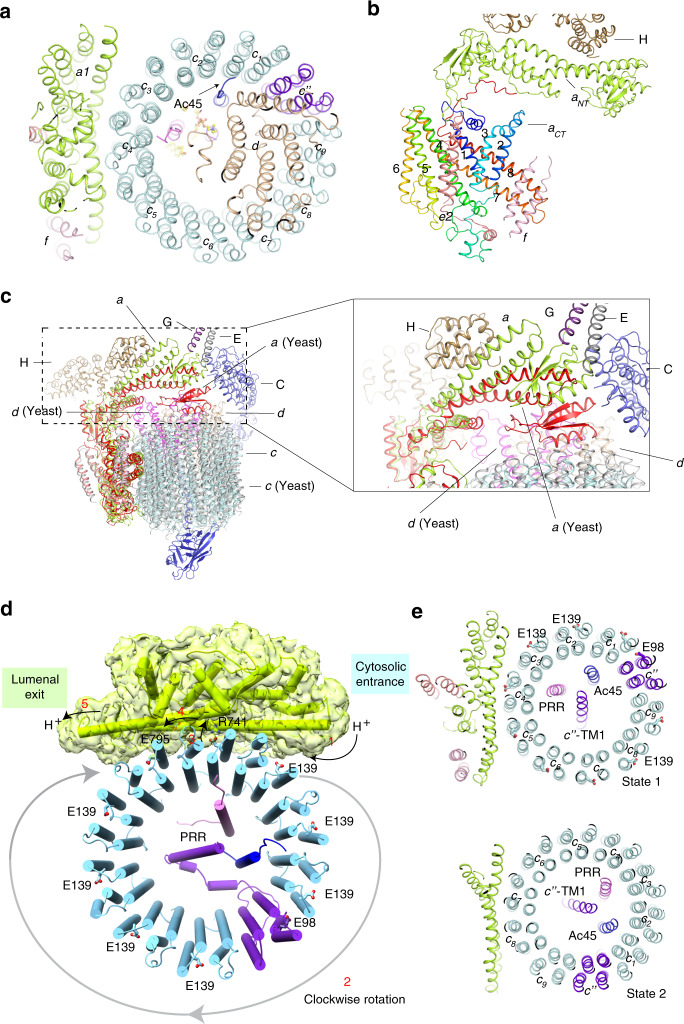
The subunit composition of the Vo domain considerably differs among species. The Vo domain of ttV/A-ATPase includes subunits adc12, whereas the yeast V-ATPase contains subunits ac8c’ c”def. However, the Vo domain of bovine V-ATPase possesses subunits ac9c”deAc45, as previously reported17,27. Three extra TMs (transmembrane helices) in the center of btVo domain have been observed in the cryo-EM map (Fig. 1c). In order to unambiguously identify these TMs, we combined the results of mass spectrometry, previously reported scVo domain at atomic resolution and of our density map. This analysis revealed that one of these helices is TM1 of subunit c”, while TMs 2–5 of subunit c” form the c-ring with the nine copies of subunit c (designated as c1-c9 counterclockwise from cytosolic view) (Fig. 3a). The second helix in the center of the c-ring belongs to Ac45, which is essential for the activity of V-ATPase28; remarkably, the last TM is part of the (pro)renin receptor (PRR) (Figs. 3a, b and 4a), a 39-kD type-I membrane protein, which is involved in various physiological processes like the cell cycle, water homeostasis, and blood pressure regulation29. Knocking out the PRR leads to a dysfunctional V-ATPase, interfering with vesicular acidification in organs30,31.
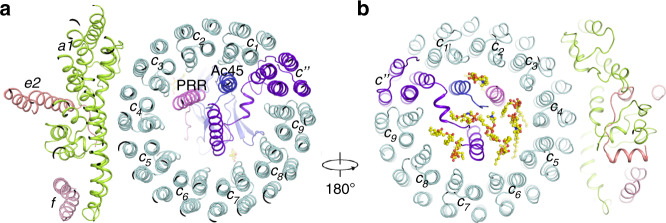
Fig. 3. Structure of the Vo domain with its associated lipids.
a Overall structure showing the Vo domain. b The luminal view of the Vo domain in state 1 showing the lipids that are involved in the Vo assembly. The putative PCs are shown in yellow sticks.
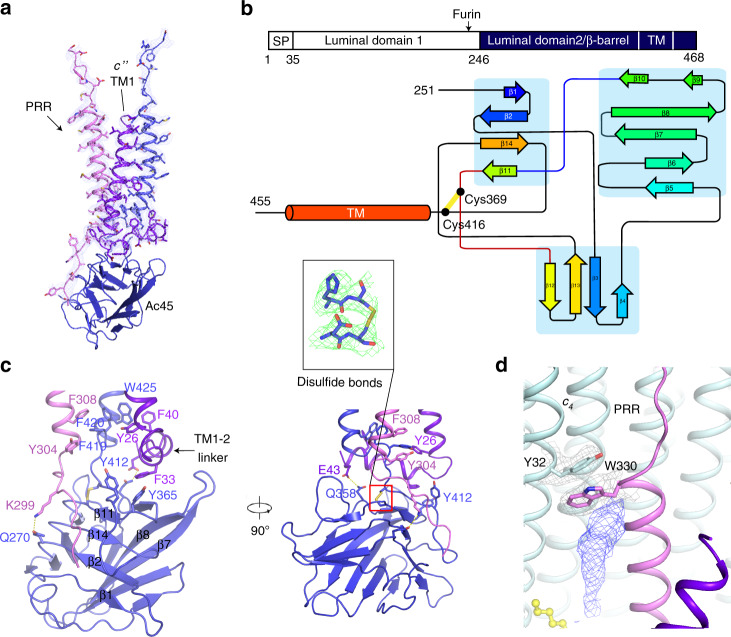
Fig. 4. Structure of the core of Vo domain.
a The core of c-ring. Cryo-EM map of TMs of PRR, Ac45, and c” in state 1 is shown at the 5σ level. b The topology diagram of Ac45. The schematic diagram of full-length Ac45 is shown on the top. c The interaction details of luminal peptide of PRR with Ac45 and TM1-2 linker of c”. The individual β-strand of Ac45 is labeled. The cryo-EM map of the disulfide bonds is shown at the 5σ level. d The interaction details of unknown lipid-mediated PRR and c4 interaction. Cryo-EM maps are shown at the 5σ level.
In addition to subunit e2, two α-helices associated with subunit a1 have been observed at the edge of the c-ring (Figs. 1c and 3a). Because two analogous α-helices have been identified as subunit f in the scVo structure, we followed this nomenclature and named these helices as subunit f. To date, the function of subunit f is unknown, and knockout of this subunit does not affect the function of V-ATPase in yeast. Mass spectrometry results and our cryo-EM data of the btV-ATPase indicate that subunit f may be the RNAseK that was shown to associate with V-ATPase32. The specific function of RNAseK in V-ATPase still requires further characterization.
Interaction details of Vo domain
The N-terminus of Ac45 contains two luminal domains (Fig. 4b). A previous study showed that furin protease can cleave the linker between the two domains33. Consistent with this observation, only the second luminal domain and TM domain (residues 251–455) are present in our cryo-EM map. Glycosylation sites in this domain facilitated unambiguous residue assignment (Supplementary Fig. 6c). The second luminal domain of Ac45 was also found in a low-resolution cryo-EM map of bovine V-ATPase, supporting our observation34. Similarly, the luminal domain of PRR is not observed in the map, since it has been cleaved by furin protease35. The residues 293–313 of renin receptor binds to the luminal domain of Ac45 that contains 14 β-strands and a disulfide bridge between Cys369 and Cys416 (Fig. 4b, c). The structural analysis shows that PRR engages in several hydrophobic interactions with the TM of Ac45 and TM1 of c” (Fig. 4c). Besides the hydrophobic contacts between PRR, Ac45, and subunit c”, Lys299 of PRR interacts with Gln270 of Ac45 and Glu43 of subunit c” have hydrophilic interactions with Gln358 of Ac45 to further stabilize the core of the c-ring (Fig. 4c).
Many phospholipid molecules were observed in the Vo domain, particularly in the lumenal leaflet, indicating that they may play a role in stabilizing the core of the Vo domain (Fig. 3b; Supplementary Fig. 6c). From the density, we can distinguish that these densities belong to the phospholipids, including phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylserine (PS). The previous studies showed that purified endogenous btV-ATPase could be associated with a mixture for these lipids36. Reconstitution of the V-ATPase complex with proton-pumping activity requires the liposomes containing a mixture of PC, PE, PS, and cholesterol36. Since we could not identify these phospholipids in each map, we tentatively docked PC as a lipid representative into our cryo-EM map. These lipids may provide a hydrophobic environment to maintain the conformation of the c-ring core (Fig. 3). Interestingly, a sterol-like density was observed at Trp330 of PRR in the cryo-EM map (Fig. 4d), suggesting that a lipid molecule may stabilize the interaction of PRR with subunit c4 in Vo.
Proton translocation by Vo
In the state 1, one pair of subunit EG interacts with subunit a, while the other pair engages with subunit C, which binds to subunit d; moreover, subunit d binds to subunits DF to connect the V1 and Vo domains (Fig. 1a). The interaction between subunits d and C will be released during the rotation (Fig. 1e). The C-terminal TM of Ac45 interacts with subunit d and subunit c1 (Fig. 5a). The subunit d also makes several contacts with c”, c1, c7, c8, and c9 (Fig. 5a). These interactions are retained in the two states, suggesting that the fixed conformations of the peripheral subunits can facilitate the rotation of subunits DFd and of the c-ring during ATP hydrolysis and proton translocation. These observations imply that the subunit d is the key component to connect the c-ring with subunits DF of the V1 domain.
Fig. 5. Proton translocation and the rotation of Vo domain.
a Overall structure showing the connection between subunit d and c-ring. b The structural elements of subunit a1 with subunit H, e2, and f. The transmembrane helices of a1 have been labeled. c Structural comparison with auto-inhibitory yeast Vo domain (pdb: 6O7T). d The putative proton-translocation mechanism. The residues for the proton transfer are shown in sticks. The cryo-EM map of subunit a is shown. e The cytosolic view of Vo showing the rotation of c-ring in two states. The Glu residues for protonation in state 1 are shown in stick.
The subunit a contains two domains: an N-terminal cytoplasmic region (aNT) and eight C-terminal TMs (aCT) (Fig. 5b). Comparing the structure to yeast Vo, the yeast Vo domain is in an autoinhibit state after dissociation with V1 with subunit a N-terminal domain (residues186–258) bound to subunits c and d, the conformational change on the N-terminus of subunit a in non-inhibitory state induces it interacting with subunits H, C, and two peripheral stalks which facilitate the V1 and Vo assembly and ATP hydrolysis coupled proton translocation37,38 (Fig. 5c).
There are two aqueous half-channels in subunit a near the interface to the c-ring providing access for protons from the cytoplasm to lumen or extracellular space (Fig. 5d). We hypothesize that a proton accesses the lipid bilayer through the cytoplasmic half-channel to neutralize the negative charge of a conserved residue Glu139 of subunit c that would directly transfer the proton to Arg741 of subunit a; then, the protonated Arg741 would deliver the proton into the luminal/extracellular half-channel18,20,39–41. Proton release may be facilitated by the residue Glu795 of subunit a, which is conserved in eukaryotic V-ATPases (Fig. 5d).
The structures of the A3B3 hexamer in the two states showed that ATP hydrolysis triggers a conformational change in the subunit DF, which further induces a shift of subunit d. The c-ring is subsequently rotated and results in a continuous proton translocation across the membrane. Notably, Glu98 of the subunit c” disrupts the pattern that is created by the key Glu residues of the nine c subunits in the c-ring, causing an asymmetric distribution of these Glu residues (Fig. 5e). The structural analysis shows that the TM1 of subunit c”, Ac45, and PRR do not change their relative positions in these two states. However, they have rotated ~120o with the c-ring (Fig. 5e), suggesting that the entire c-ring including subunits c, c”, Ac45, and PRR function as a rigid domain in proton translocation.
Discussion
We compared the structures of state 1 and state 2, with the previously reported model of V/A-ATPase17 to elucidate the rotational mechanism. We found that the switch of AB heterodimers (Fig. 2b) provides the energy necessary to trigger a rotation of subunits DF resulting in the spinning of subunit d coupled with the entire c-ring region, and each state will rotate ~120° (Fig. 5e). Remarkably, the complex interaction network between subunit d, TM1 of subunit c”, Ac45, PRR, and lipids in our cryo-EM data, provides multiple fulcrums to facilitate the rotation of the entire c-ring region between different states.
Three different rotational states of yeast V-ATPase16 and ttV/A-ATPase17 had been found in the cryo-EM maps; in contrast, our cryo-EM maps reveal only two rotational states of btV-ATPase (Figs. 1a, d and 5e). The previous structural studies revealed that the endogenous ATPase exhibits the unequal population for three different classes, such as the yeast V-ATPase16 with state 1 (47%, 6.9 Å), state 2 (36%, 7.6 Å), state 3 (17%, 8.3 Å), and the ttV/A-ATPase42 with state 1 (66%, 5.0 Å), state 2 (16%, 6.7 Å), state 3 (7%, 7.5 Å), and 11% bad class. These findings imply that state 3 shows the least population and the worst resolution after 3D refinement. We speculate that the third state, where the subunit c” faces subunit a, may not be stable in our bovine tissue preparation or exists in very few populations that could not be captured, as the similar results observed in some studies42,43.
The structures of rat V-ATPase with SidK, a bacterial inhibitor of V-ATPase, has been reported recently20. This remarkable work showed that Ac45, PRR, and RNAseK are essential components of Vo, consistent with our analysis. In the rat V-ATPase structures, three conformationally different AB heterodimers have been reported: one is bound to ADP and the other two represent nucleotide-free states. When comparing our structure with the rat V-ATPase structure, we find that their conformations of ABopen and ABoccluded share similar conformations with our AB heterodimers (Supplementary Fig. 9a, b); however, the C-terminal α-helical domain of subunit A in the ABclosed state of SidK bound V-ATPase has a ~3 Å shift to make the ABclosed state more compact than the active form (Supplementary Fig. 9c). The structural comparison reveals that the subunits EG have a ~4 Å shift between the two structures; this difference may be caused by the presence of subunit H that has contact with the subunits EG directly (Supplementary Fig. 9d). The V1 and Vo structures of rat V-ATPase adopt a similar conformation to those in bovine V-ATPase structures state 1 and state 2, respectively. Due to the limitation of the resolution of the rat V-ATPase structure, the luminal domain of Ac45 could not be modeled; in contrast, we built the entire second luminal domain of Ac45 showing the interaction details of the Ac45 and PRR (Fig. 4c; Supplementary Fig. 9e). In summary, we report the mammalian V-ATPase structure including all the essential subunits, revealing a more complete map of this important rotary machine.
Methods
Gain clathrin-coated vesicles
About three defatted bovine brains were rinsed and blended with 0.1 M Na-MES, pH 6.5, 1 mM EGTA, 0.5 mM MgCl2, and 3 mM NaN3 (buffer A). Usually, one kilogram of the cleaned tissue is homogenized in 900 ml of buffer A. The homogenate is centrifuged in a GSA (Sorvall) rotor for 50 min at 20,000 g, and the supernatant is centrifuged at 140,000 g for 1 h to sediment the membrane vesicles. The pellet containing enriched coated vesicles is resuspended in buffer A at a protein concentration of 15 mg/ml, frozen in liquid nitrogen, and stored at −80 °C.
Purification of V-ATPase
The V-ATPase was solubilized and purified using an established protocol21. Briefly, the membrane vesicles incubated with 0.75 M Tris-HCl pH 8.0 on ice for 30 min to strip clathrin. After centrifugation at 150,000 g for 35 min, the pellet was resuspended in 0.5% Na-cholate and incubated at 0 °C for 30 min. After centrifugation at 150,000 g for 35 min, the pellet was resuspended in 0.75% C12E9, 10 mM Tris-MES pH 6.75, incubated on ice for 60 min and centrifuged at 150,000 g for 90 min.
The supernatant was applied to a hydroxylapatite column which had been equilibrated with 0.1% C12E9, 10% glycerol, 0.5 mM DTT, 10 mM Tris-MES pH 7.0 (buffer B). The column was washed with 2CV of buffer B and eluted with 0–0.3 M Na-phosphate in buffer B. Saturated ammonium sulfate solution was added dropwise to the active hydroxylapatite fractions (selected by either SDS-PAGE or ATPase assay), to a final concentration of 1.65 M. The mixture was centrifuged at 100,000 g for 30 min, the pellet was harvested and dissolved in buffer B without glycerol.
Then the sample was loaded on to a glycerol gradient (12 ml, 10–30%) prepared in buffer B. The gradient was centrifuged at 170,000 g for 20 h, and fractions were collected. The fractions containing V-ATPase were concentrated and further purified by gel filtration using a Superose 6 10/300 column (GE Healthcare) pre-equilibrated with buffer C (20 mM HEPES pH 7.5, 150 mM NaCl, 0.1% CHAPS, 0.004% glycodiosgenin (Anatrace)). Mass spectrometry confirmed the identity of the subunits of V-ATPase (Supplementary Table 1). The peak fractions were collected and concentrated to 2.5–3.5 mg/ml for cryo-EM grid preparation.
V-ATPase activity assay
ATPase activity was measured as the liberation of 32Pi from [γ−32P]ATP21. The assay was carried out in a total volume of 200 μl under the following conditions: 4 μl phosphatidylserine (26 μM concentration), 5 μl V-ATPase in buffer C (at 10 nM concentration), and 189 μl ATPase assay solution A (30 mM KCl, 50 mM Tris-MES, pH 7.0, 3 mM MgCl2, and 3 mM [γ−32P]ATP (400 cpm/nmol)) and 2 μl of ethanol or ethanol dissolved bafilomycin A1 (Sigma-Aldrich, 1 μM concentration), The V-ATPase was first incubated with phosphatidylserine for 2 min, and then the reaction was started by addition ATPase assay solution A and ethanol/bafilomycin A1, and continued for 10 min at 37 °C. The ATP hydrolysis reaction was terminated by adding 1.0 ml of 1.25 N perchloric acid, and the released 32Pi was extracted and counted in a Beckman scintillation counter44. The results were expressed as specific activity (μmol of Pi/min per mg of protein), The experiment has been repeated twice, and the results were performed using GraphPad Prism8.
EM sample preparation and imaging for 200 kV Cryo-TEM
Freshly purified 2.5–3.5 mg/ml V-ATPase in buffer C was applied to Quantifoil R1.2/1.3 300 mesh Au holey carbon grids (Quantifoil). The grids were then blotted and plunged into liquid ethane for flash freezing using a Vitrobot Mark IV (FEI). The grids were imaged in a 200 keV Talos Arctica (FEI) with a Gatan K3 Summit direct electron detector (Gatan) in super-resolution mode using the data-collection software Serial EM. Data were collected at 0.735 Å/pixel with a counted rate of 42 electrons per physical pixel per second. Images were recorded for 4 s exposures in 50 subframes with a total dose of 80 electrons per Å2 and a defocus range of −1.0 to −2.0 μm.
Imaging processing for 200 kV Cryo-TEM
Dark-subtracted images were first normalized by gain reference and then binned twofold that resulted in a pixel size of 1.47 Å. Motion correction and gain reference were performed using the program MotionCor245. The contrast transfer function (CTF) was estimated using CTFFIND446. To generate V-ATPase templates for automatic picking, around 2000 particles were manually picked and classified by 2D classification in RELION-347. After auto-picking, the low-quality images and false-positive particles were removed manually. The remaining 420,602 particles were extracted for subsequent 2D. A low-resolution cryo-EM map of V-ATPase, which was generated from 3200 particles by RELION-3, was used as the initial model for 3D classification. The best two classes, containing 75,386 particles, provided a 8.53 Å map after 3D auto-refinement with a mask and postprocess in RELION-3, then CTF refinement and Bayesian polishing of particles were performed using RELION-3 for once followed by 3D refinement using a soft mask. Next, the second 3D classification was performed, two rotational states were separated by the obvious position of subunits D and F. One class of state 1, including 61,520 particles, provided a 7.61 Å map after 3D auto-refinement with a mask and postprocess in RELION-3. After the second CTF refinement with beam tilt correction, the resulting particles of state 1 were used for the final 3D refinement with a soft mask and solvent-flattened FSCs yielded a reconstruction at 7.22 Å revealing clear secondary structural elements. The resolution was estimated using “post-processing” with the FSC criteria of 0.143.
EM sample preparation and imaging for 300 kV Cryo-TEM
Freshly purified 2.5–3.5 mg/ml V-ATPase in buffer C was applied to Quantifoil R1.2/1.3 300 or 400 mesh Au holey carbon grids (Quantifoil). The grids were then blotted and plunged into liquid ethane for flash freezing using a Vitrobot Mark IV (FEI). The grids were imaged in a 300 keV Titan Krios (FEI) with a Gatan K3 Summit direct electron detector (Gatan) in super-resolution mode using the data-collection software Serial EM.
Dark-subtracted images collected at super-resolution mode were first normalized by gain reference and binned twofold, which resulted in a pixel size of 0.833 Å with a counted rate of 23 electrons per physical pixel per second48. Images were recorded for 1.8-s exposures in 60 subframes with a total dose of 60 electrons per Å2 and a defocus range of −1.0 to −2.0 μm.
Imaging processing and 3D reconstruction for 300 kV Cryo-TEM
The images were collected in two sessions (images from 400 mesh, images from 300 mesh Au holey carbon grids). The images used for the second session were divided into two parts for data processing. Dark-subtracted images were first normalized by gain reference and then binned twofold that resulted in a pixel size of 0.833 Å. Motion correction and gain reference was performed using the program MotionCor245. The contrast transfer function (CTF) was estimated using CTFFIND446. To generate V-ATPase templates for automatic picking, about 2000 particles were manually picked and classified by 2D classification in RELION-347.
After auto-picking, low-quality images and false-positive particles were removed manually. About 112 K/174 K/210 K particles of V-ATPase were extracted. We used the cryo-EM structure of V-ATPase, which was determined by us with the data collected from a 200 keV Arctica (FEI) low-pass filtered to 40 Å as the initial model and applied a mask for 3D classification. The best class, containing 28,125/45,034/56,149 particles, provided a 4.29 Å /5.00 Å/4.88 Å map after 3D auto-refinement with a mask and postprocess in RELION-3.
For the particles in each dataset, CTF refinement and Bayesian polishing of particles were performed using RELION-3 for once followed by 3D refinement using a soft mask. Then, we combined the particles from CTF refinement of dataset 1, the particles from CTF refinement of dataset 2, and particles from Bayesian polishing of dataset 3 with a total of 129,308 particles and performed a 3D classification with six classes. Two rotational states were separated by the obvious position of subunits D and F. Two classes of state 1 including 84,345 particles refined using a soft mask and solvent-flattened Fourier shell correlations (FSCs) yielded a reconstruction at 3.45 Å. Applying a soft mask in RELION-3 post processing yielded a final cryo-EM map of 3.37 Å. One class of state 2 including 41,821 particles refined using a soft mask and solvent-flattened Fourier shell correlations (FSCs) yielded a reconstruction at 3.86 Å. Applying a soft mask in RELION-3 post processing yielded a final cryo-EM map of 3.79 Å. Resolution was estimated using the Fourier shell correlation (FSC) 0.143 criterion.
For rotational state 1, after 3D refinement, applying a full mask in RELION-3 post processing yielded a 3.37 Å overall resolution based on the Fourier shell correlation (FSC) 0.143 criterion. But due to the flexibility of V1 and Vo, Vo was not well resolved. Several focused refinements with different masks were attempted to get a high-quality local map. These focused refinements included subunits A3B3DE3FG3, subunits A3B3CDE3FG3H, subunits adCFH, subunits CFHVo provided some good local maps for this state, especially for the Vo part (Supplementary Fig. 5). After post processing, these refinements gave a resolution of 3.22 Å, 3.26 Å, 3.89 Å, and 3.54 Å, respectively. For rotational state 2, after 3D refinement, applying a full mask in RELION-3 post processing yielded a 3.79 Å overall resolution based on the Fourier shell correlation (FSC) 0.143 criterion. The same strategy had been applied to improve the local maps of state 2. These focused refinements included subunits A3B3DE3FG3, subunits A3B3CDE3FG3H, subunits CFHVo, subunits c9c”defAc45 PRR, provided good local maps for this state. After post processing, these refinements gave a resolution of 3.60 Å, 3.79 Å, 4.47 Å, and 3.73 Å, respectively. All these focused maps of rotational state 1 and state 2 can be used to generate a composite map for refinement.
Model construction
To obtain better side-chain densities for model building, we sharpened the map of rotational state 1 of V-ATPase using post processing in RELION-3 with a B-factor value of −79.3 Å2. Based on the previously reported structures including subunit AB (PDB code 5ZE9), subunit C (PDB code 1U7L), subunit D (PDB code 4RND), subunits EG (PDB code 4EFA), subunit F (PDB code 4IX9), subunit H (PDB code 1HO8), and subunits a, c, c”, d, e (PDB code 6O7U), the homology models for subunits A-H of V1 and subunits a, c, c”, d, e, of Vo were generated by MODELLER22, Ac45 and subunit f were predicted by Phyre249. Each subunit was docked into the cryo-EM map, respectively, using Chimera50 to generate the initial model, and then manually adjusted using COOT51. The transmembrane domain of PRR was built de novo. The glycosylation sites of Ac45 luminal domain were assigned to facilitate the register of Ac45. The residues of bovine V-ATPase that were not resolved or built are shown in Supplementary Table 1. For rotational state 2, we sharpened the map of the complex using post processing in RELION-3 with a B-factor value of −85.8 Å2. Atomic models of each subunit were based on the structure of rotational state 1. Each subunit was docked into the cryo-EM map, respectively, using Chimera50 to generate the initial model, and then manually adjusted by COOT51. Besides the unmodelled residues in state 1, the densities of subunit a (residues 4–8, 167–169, 382–529, 564–658, 830–834), subunits e and f were not resolved nor built. Side chains were removed to generate a model for subunit C (residues 42–310, 325–343), subunit H and subunit a (residues 530–563) due to the limited local resolution.
Model refinement and validation
For state 1, to generate a composite map for refinement, several focused maps that have been described above were combined and aligned using phenix.combine_focused _maps which would coalescence the best parts of several maps together. The model was refined in real space using PHENIX52 with secondary-structure restraints and stereochemical restraints53. The same strategy has also been used for state 2. For cross-validations, the final model was refined against one of the half maps generated by 3D auto-refine, and the model vs. map FSC curves were generated in the comprehensive validation module in PHENIX. MolProbity54 and PHENIX were used to validate the final model. Local resolutions were estimated using RELION-3. Structure Figures were generated using PyMOL and Chimera50.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
This research was done in the memory of our colleague Dennis K. Stone. The data were collected at the UT Southwestern Medical Center Cryo-EM Facility (funded in part by the CPRIT Core Facility Support Award RP170644); we thank D. Stoddard for assistance in data collection. We thank E. Debler for discussion during paper preparation and A. Lemoff at the UT Southwestern Proteomics Core for mass spectrometry identification. This work was supported by NIH grant R01 HL072304 (to X.X.), P01 HL020948, the Endowed Scholars Program in Medical Science of UT Southwestern Medical Center, O’Donnell Junior Faculty Funds, Welch Foundation (I-1957) (to X.L.). X.L. is a Damon Runyon-Rachleff Innovator supported by the Damon Runyon Cancer Research Foundation (DRR-53-19) and Rita C. and William P. Clements Jr. Scholar in Biomedical Research at UT Southwestern Medical Center.
Source data
Author contributions
X.L. and X.X. conceived the project and designed the research. R.W., A.H., and J.W. performed the biochemical study. R.W., T.L., and Y.S. carried out cryo-EM work, built the model, and refined the structure. R.W., T.L., X.X., and X.L. analyzed the data, and all the authors contributed to paper preparation. X.L. wrote the paper.
Data availability
Data supporting the findings of this paper are available from the corresponding authors upon reasonable request. A reporting summary for this Article is available as a Supplementary Information file. The 3D cryo-EM density maps of rotational state 1 and state 2 of V-ATPase have been deposited in the Electron Microscopy Data Bank under the accession numbers EMD-22121 and EMD-22122. Atomic coordinates for the atomic model of rotational state 1 and state 2 of V-ATPase have been deposited in the Protein Data Bank under the accession numbers PDB 6XBW and PDB 6XBY. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review informationNature Communications thanks the anonymous, reviewers for their contributions to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xiao-Song Xie, Email: xiao-song.xie@utsouthwestern.edu.
Xiaochun Li, Email: xiaochun.li@utsouthwestern.edu.
Supplementary information
Supplementary information is available for this paper at 10.1038/s41467-020-17762-9.
References
- 1.Futai M, Sun-Wada GH, Wada Y, Matsumoto N, Nakanishi-Matsui M. Vacuolar-type ATPase: a proton pump to lysosomal trafficking. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2019;95:261–277. doi: 10.2183/pjab.95.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vasanthakumar T, Rubinstein JL. Structure and roles of V-type ATPases. Trends Biochem. Sci. 2020;45:295–307. doi: 10.1016/j.tibs.2019.12.007. [DOI] [PubMed] [Google Scholar]
- 3.Forgac M. Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat. Rev. Mol. Cell Biol. 2007;8:917–929. doi: 10.1038/nrm2272. [DOI] [PubMed] [Google Scholar]
- 4.Marshansky V, Futai M. The V-type H+-ATPase in vesicular trafficking: targeting, regulation and function. Curr. Opin. Cell Biol. 2008;20:415–426. doi: 10.1016/j.ceb.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mauvezin C, Neufeld TP. Bafilomycin A1 disrupts autophagic flux by inhibiting both V-ATPase-dependent acidification and Ca-P60A/SERCA-dependent autophagosome-lysosome fusion. Autophagy. 2015;11:1437–1438. doi: 10.1080/15548627.2015.1066957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang CS, et al. The lysosomal v-ATPase-ragulator complex is a common activator for AMPK and mTORC1, acting as a switch between catabolism and anabolism. Cell Metab. 2014;20:526–540. doi: 10.1016/j.cmet.2014.06.014. [DOI] [PubMed] [Google Scholar]
- 7.Karet FE. Physiological and metabolic implications of V-ATPase isoforms in the kidney. J. Bioenerg. Biomembr. 2005;37:425–429. doi: 10.1007/s10863-005-9484-x. [DOI] [PubMed] [Google Scholar]
- 8.Perzov N, Padler-Karavani V, Nelson H, Nelson N. Characterization of yeast V-ATPase mutants lacking Vph1p or Stv1p and the effect on endocytosis. J. Exp. Biol. 2002;205:1209–1219. doi: 10.1242/jeb.205.9.1209. [DOI] [PubMed] [Google Scholar]
- 9.Dettmer J, Hong-Hermesdorf A, Stierhof YD, Schumacher K. Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell. 2006;18:715–730. doi: 10.1105/tpc.105.037978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishi T, Forgac M. The vacuolar (H+)-ATPases–nature’s most versatile proton pumps. Nat. Rev. Mol. Cell Biol. 2002;3:94–103. doi: 10.1038/nrm729. [DOI] [PubMed] [Google Scholar]
- 11.Wang R, Qi X, Schmiege P, Coutavas E, Li X. Marked structural rearrangement of mannose 6-phosphate/IGF2 receptor at different pH environments. Sci. Adv. 2020;6:eaaz1466. doi: 10.1126/sciadv.aaz1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cruciat CM, et al. Requirement of prorenin receptor and vacuolar H+-ATPase-mediated acidification for Wnt signaling. Science. 2010;327:459–463. doi: 10.1126/science.1179802. [DOI] [PubMed] [Google Scholar]
- 13.Abu-Remaileh M, et al. Lysosomal metabolomics reveals V-ATPase- and mTOR-dependent regulation of amino acid efflux from lysosomes. Science. 2017;358:807–813. doi: 10.1126/science.aan6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vaccari T, Duchi S, Cortese K, Tacchetti C, Bilder D. The vacuolar ATPase is required for physiological as well as pathological activation of the Notch receptor. Development. 2010;137:1825–1832. doi: 10.1242/dev.045484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gogarten JP, Starke T, Kibak H, Fishman J, Taiz L. Evolution and isoforms of V-ATPase subunits. J. Exp. Biol. 1992;172:137–147. doi: 10.1242/jeb.172.1.137. [DOI] [PubMed] [Google Scholar]
- 16.Zhao J, Benlekbir S, Rubinstein JL. Electron cryomicroscopy observation of rotational states in a eukaryotic V-ATPase. Nature. 2015;521:241–245. doi: 10.1038/nature14365. [DOI] [PubMed] [Google Scholar]
- 17.Zhou, L. & Sazanov, L. A. Structure and conformational plasticity of the intact Thermus thermophilus V/A-type ATPase. Science365, 10.1126/science.aaw9144 (2019). [DOI] [PubMed]
- 18.Mazhab-Jafari MT, et al. Atomic model for the membrane-embedded VO motor of a eukaryotic V-ATPase. Nature. 2016;539:118–122. doi: 10.1038/nature19828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roh SH, et al. The 3.5-A CryoEM structure of nanodisc-reconstituted yeast vacuolar ATPase Vo proton channel. Mol. Cell. 2018;69:993–1004 e1003. doi: 10.1016/j.molcel.2018.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abbas YM, Wu D, Bueler SA, Robinson CV, Rubinstein JL. Structure of V-ATPase from the mammalian brain. Science. 2020;367:1240–1246. doi: 10.1126/science.aaz2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie XS, Stone DK. Isolation and reconstitution of the clathrin-coated vesicle proton translocating complex. J. Biol. Chem. 1986;261:2492–2495. [PubMed] [Google Scholar]
- 22.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 23.Geyer M, et al. Subunit H of the V-ATPase binds to the medium chain of adaptor protein complex 2 and connects Nef to the endocytic machinery. J. Biol. Chem. 2002;277:28521–28529. doi: 10.1074/jbc.M200522200. [DOI] [PubMed] [Google Scholar]
- 24.Crider BP, Xie XS. Characterization of the functional coupling of bovine brain vacuolar-type H(+)-translocating ATPase. Effect of divalent cations, phospholipids, and subunit H (SFD) J. Biol. Chem. 2003;278:44281–44288. doi: 10.1074/jbc.M307372200. [DOI] [PubMed] [Google Scholar]
- 25.Oot RA, Kane PM, Berry EA, Wilkens S. Crystal structure of yeast V1-ATPase in the autoinhibited state. EMBO J. 2016;35:1694–1706. doi: 10.15252/embj.201593447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arai S, et al. Rotation mechanism of Enterococcus hirae V1-ATPase based on asymmetric crystal structures. Nature. 2013;493:703–707. doi: 10.1038/nature11778. [DOI] [PubMed] [Google Scholar]
- 27.Stransky L, Cotter K, Forgac M. The function of V-ATPases in cancer. Physiol. Rev. 2016;96:1071–1091. doi: 10.1152/physrev.00035.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Supek F, et al. A novel accessory subunit for vacuolar H(+)-ATPase from chromaffin granules. J. Biol. Chem. 1994;269:24102–24106. [PubMed] [Google Scholar]
- 29.Ichihara A, Yatabe MS. The (pro)renin receptor in health and disease. Nat. Rev. Nephrol. 2019;15:693–712. doi: 10.1038/s41581-019-0160-5. [DOI] [PubMed] [Google Scholar]
- 30.Kinouchi K, et al. The (pro)renin receptor/ATP6AP2 is essential for vacuolar H+-ATPase assembly in murine cardiomyocytes. Circ. Res. 2010;107:30–34. doi: 10.1161/CIRCRESAHA.110.224667. [DOI] [PubMed] [Google Scholar]
- 31.Trepiccione F, et al. Renal Atp6ap2/(Pro)renin receptor is required for normal vacuolar H+-ATPase function but not for the renin-angiotensin system. J. Am. Soc. Nephrol. 2016;27:3320–3330. doi: 10.1681/ASN.2015080915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perreira JM, et al. RNASEK is a V-ATPase-associated factor required for endocytosis and the replication of rhinovirus, influenza A virus, and dengue virus. Cell Rep. 2015;12:850–863. doi: 10.1016/j.celrep.2015.06.076. [DOI] [PubMed] [Google Scholar]
- 33.Louagie E, et al. Role of furin in granular acidification in the endocrine pancreas: identification of the V-ATPase subunit Ac45 as a candidate substrate. Proc. Natl Acad. Sci. USA. 2008;105:12319–12324. doi: 10.1073/pnas.0800340105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gregorini M, Wang J, Xie XS, Milligan RA, Engel A. Three-dimensional reconstruction of bovine brain V-ATPase by cryo-electron microscopy and single particle analysis. J. Struct. Biol. 2007;158:445–454. doi: 10.1016/j.jsb.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 35.Cousin C, et al. Soluble form of the (pro)renin receptor generated by intracellular cleavage by furin is secreted in plasma. Hypertension. 2009;53:1077–1082. doi: 10.1161/HYPERTENSIONAHA.108.127258. [DOI] [PubMed] [Google Scholar]
- 36.Xie XS, Tsai SJ, Stone DK. Lipid requirements for reconstitution of the proton-translocating complex of clathrin-coated vesicles. Proc. Natl Acad. Sci. USA. 1986;83:8913–8917. doi: 10.1073/pnas.83.23.8913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qi J, Forgac M. Function and subunit interactions of the N-terminal domain of subunit a (Vph1p) of the yeast V-ATPase. J. Biol. Chem. 2008;283:19274–19282. doi: 10.1074/jbc.M802442200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kane PM. Disassembly and reassembly of the yeast vacuolar H(+)-ATPase in vivo. J. Biol. Chem. 1995;270:17025–17032. [PubMed] [Google Scholar]
- 39.Junge W, Lill H, Engelbrecht S. ATP synthase: an electrochemical transducer with rotatory mechanics. Trends Biochem. Sci. 1997;22:420–423. doi: 10.1016/s0968-0004(97)01129-8. [DOI] [PubMed] [Google Scholar]
- 40.Vik SB, Antonio BJ. A mechanism of proton translocation by F1F0 ATP synthases suggested by double mutants of the a subunit. J. Biol. Chem. 1994;269:30364–30369. [PubMed] [Google Scholar]
- 41.Hahn, A., Vonck, J., Mills, D. J., Meier, T. & Kuhlbrandt, W. Structure, mechanism, and regulation of the chloroplast ATP synthase. Science360, 10.1126/science.aat4318 (2018). [DOI] [PMC free article] [PubMed]
- 42.Nakanishi A, et al. structure of intact rotary H(+)-ATPase/synthase from Thermus thermophilus. Nat. Commun. 2018;9:89. doi: 10.1038/s41467-017-02553-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schep DG, Zhao J, Rubinstein JL. Models for the a subunits of the Thermus thermophilus V/A-ATPase and Saccharomyces cerevisiae V-ATPase enzymes by cryo-EM and evolutionary covariance. Proc. Natl Acad. Sci. USA. 2016;113:3245–3250. doi: 10.1073/pnas.1521990113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stone DK, Xie XS, Racker E. Inhibition of clathrin-coated vesicle acidification by duramycin. J. Biol. Chem. 1984;259:2701–2703. [PubMed] [Google Scholar]
- 45.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rohou A, Grigorieff N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife7, 10.7554/eLife.42166 (2018). [DOI] [PMC free article] [PubMed]
- 48.Long T, Sun Y, Hassan A, Qi X, Li X. Structure of nevanimibe-bound tetrameric human ACAT1. Nature. 2020;581:339–343. doi: 10.1038/s41586-020-2295-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pettersen EF, et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 51.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 52.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D. Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 54.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting the findings of this paper are available from the corresponding authors upon reasonable request. A reporting summary for this Article is available as a Supplementary Information file. The 3D cryo-EM density maps of rotational state 1 and state 2 of V-ATPase have been deposited in the Electron Microscopy Data Bank under the accession numbers EMD-22121 and EMD-22122. Atomic coordinates for the atomic model of rotational state 1 and state 2 of V-ATPase have been deposited in the Protein Data Bank under the accession numbers PDB 6XBW and PDB 6XBY. Source data are provided with this paper.