Significance
Green coloration of vertebrates is normally attributed to pigments and structural components inside skin chromatophores cells. However, these components do not account for the vivid blue-green colors of hundreds of species of frogs with sparse chromatophores. Our study shows that green coloration originates in proteins of the serpin superfamily that bind the pigment biliverdin, modulating its absorbance properties. Using a South American treefrog, we demonstrated that these serpins have a clear ecological role in modulating the reflectance properties and rendering animals cryptic in the foliage even in the near-infrared portion of the spectrum. These findings open up exciting research perspectives both in biochemistry and evolution of serpins, as well as in the study of extracellular protein-mediated coloration in vertebrates.
Abstract
Many vertebrates have distinctive blue-green bones and other tissues due to unusually high biliverdin concentrations—a phenomenon called chlorosis. Despite its prevalence, the biochemical basis, biology, and evolution of chlorosis are poorly understood. In this study, we show that the occurrence of high biliverdin in anurans (frogs and toads) has evolved multiple times during their evolutionary history, and relies on the same mechanism—the presence of a class of serpin family proteins that bind biliverdin. Using a diverse combination of techniques, we purified these serpins from several species of nonmodel treefrogs and developed a pipeline that allowed us to assemble their complete amino acid and nucleotide sequences. The described proteins, hereafter named biliverdin-binding serpins (BBS), have absorption spectra that mimic those of phytochromes and bacteriophytochromes. Our models showed that physiological concentration of BBSs fine-tune the color of the animals, providing the physiological basis for crypsis in green foliage even under near-infrared light. Additionally, we found that these BBSs are most similar to human glycoprotein alpha-1-antitrypsin, but with a remarkable functional diversification. Our results present molecular and functional evidence of recurrent evolution of chlorosis, describe a biliverdin-binding protein in vertebrates, and introduce a function for a member of the serpin superfamily, the largest and most ubiquitous group of protease inhibitors.
The green coloration of vertebrates is normally attributed to a particular spatial arrangement of chromatophore cells in their skins (1). Using a complex multilayered system of structural and pigmentary components inside of the cells, animals can attain a diverse array of hues. Interestingly, hundreds of frog species have translucent skins, partially or completely devoid of melanophores and other pigmentary cells (2, 3). In these species, it is expected that other extracellular pigments and structures may account for the vivid colors they can display. In fact, recent studies have demonstrated the importance of noncellular, subcutaneous (s.c.), and glandular chromophores and fluorophores in the development of amphibian blue-green coloration (2). Among the extracellular pigments, it is known that many amphibian species have distinctive blue-green pigments in blood, lymph, other soft tissues, and bones, a phenomenon known as physiological chlorosis (4), that is prevalent in numerous arboreal species and renders them a characteristic blue-green hue (2, 3) (Figs. 1 and 2A). Previous studies have shown that this coloration is caused by high concentrations of the pigment biliverdin (BV) (4, 5), which is the first intermediate of heme catabolism from senescent red blood cells. In birds (6, 7) and at least some amphibians (8), fish (9), and reptiles (10), BV is the end product of heme catabolism and is excreted directly without further reduction to bilirubin (BR). In most mammals though, BV is rapidly reduced to BR and promptly excreted, thus making BV normally undetectable in their bile or blood even under extreme hemolytic conditions (11). In fact, BV concentrations reach detectable values in humans only when some conditions are met: severe cirrhotic pathologies, bile duct obstructions, and impairment of metabolic function (12–14). Remarkably, chlorotic frog species show plasma BV concentrations at least four times larger than in the described pathologies (4, 12–14), and at least 200 times larger than in nonchlorotic species (4). Interestingly, attempts to induce chlorosis in nonchlorotic species by hemolysis or direct injection of BV have been unsuccessful, with the animals immediately excreting the excess BV (5, 15). These observations suggest an unusual physiology of chlorotic frogs.
Fig. 1.
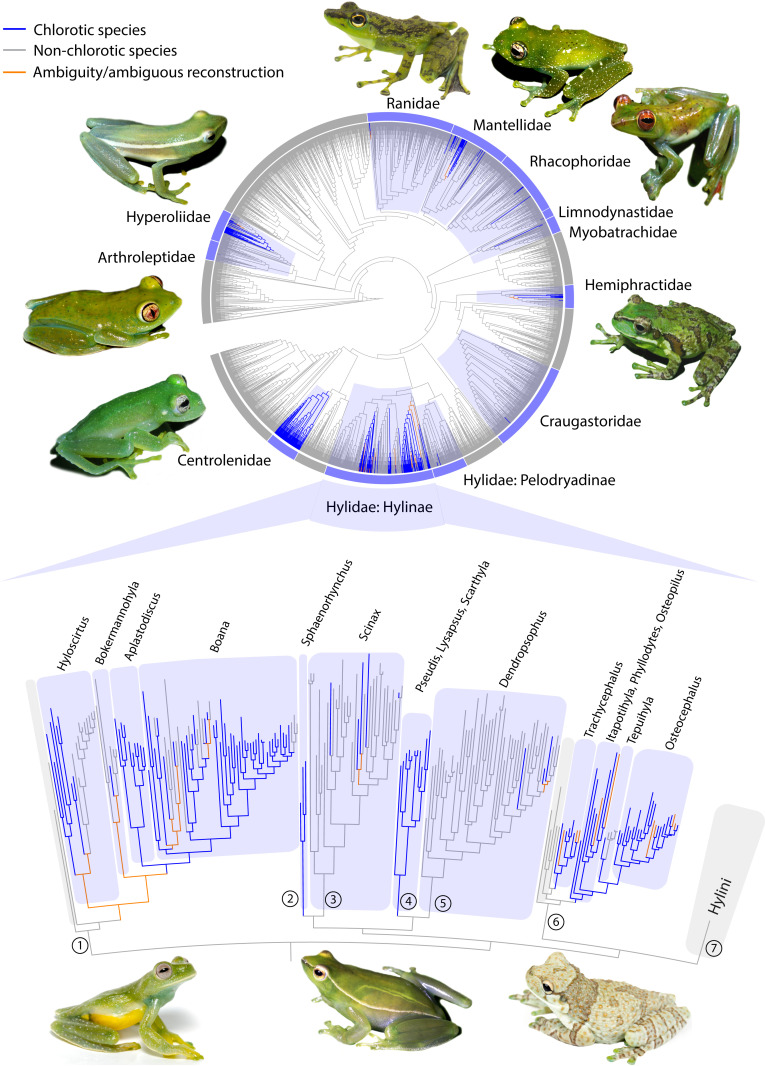
Physiological chlorosis distribution in anurans evolved multiple times. Phylogenetic tree of anurans (16) showing ancestral character state reconstructions for physiological chlorosis (circular display). Chlorotic species are found in 11 anuran families in which it evolved independently. Shaded in blue are those families where chlorosis evolved at least once during their evolutionary history. In the lower section, a detailed topology shows that chlorosis evolved at least 11 times within the hylid subfamily Hylinae. Numbers refer to the tribes: (1) Cophomantini (2) Sphaenorhynchini (3) Scinaxini (4) Pseudini (5) Dendropsophini (6) Lophyohylini (7) Hylini.
Fig. 2.
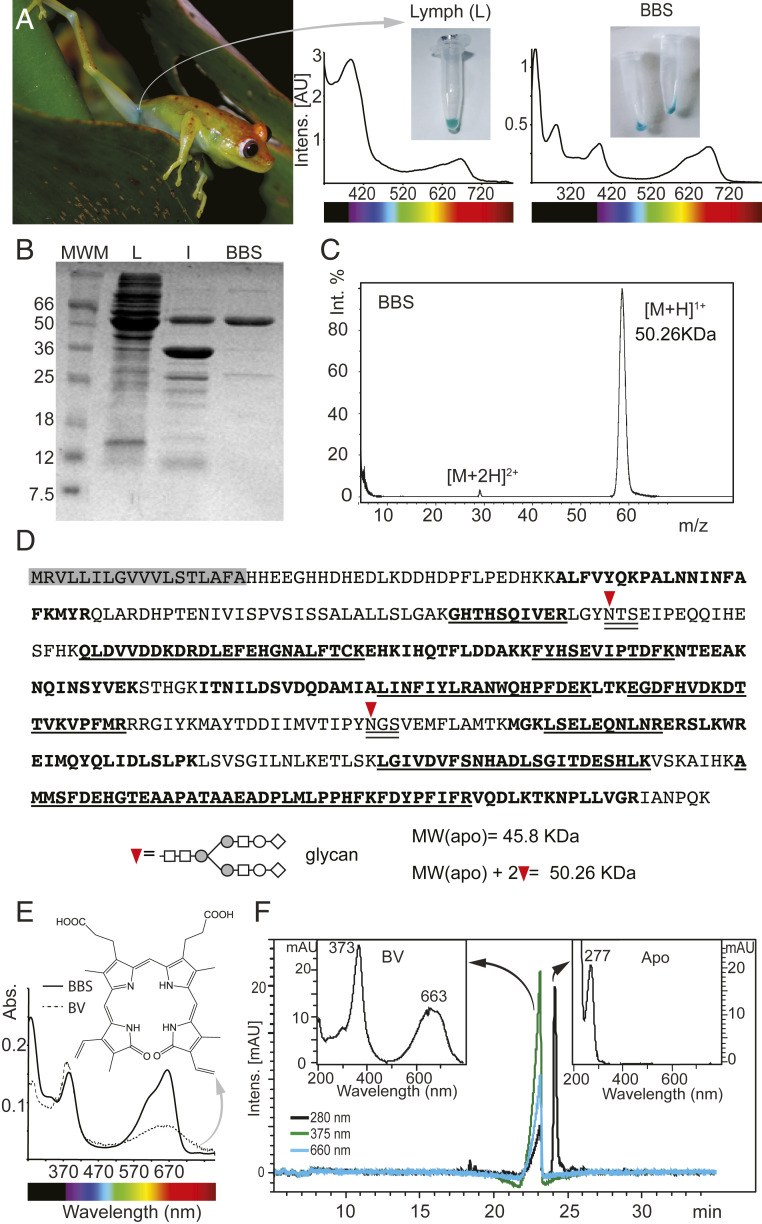
Purification of BBS from B. punctata green lymph. S.c. green lymph is clearly observable through the skin (A) and shows strong absorbance bands in the 400 nm and 660 nm regions. After removal of hyloins (2) and other yellow pigments, purified BBS is cyan with Soret band maximum absorbance at 390 nm and Q band at 667 nm. (B) PAGE of Lymph (L), lymph 80% ammonium sulfate precipitate (I) and purified BBS (B). MWM: Molecular weight marker (C) MALDI-TOF of purified BBS. Molecular weight is ∼50 kDa, which is consistent with (B). (D) Sequence of BBS. Shaded in gray is the predicted signal peptide. Bold fonts: Peptides obtained by MALDI-TOF that map to the sequence. Underlined fonts: Sequences that were verified by MSMS. Double underlined fonts: N-glycosylation consensus sequences. Inverted red triangle: N-glycosylation site. The predicted MW of the apoprotein obtained from the sequence is smaller than the empirical value obtained by MALDI-TOF (Fig. 2C). Two glycans at N-109 and N-269 account for the difference (see also SI Appendix, Fig. S5 and Table S3). Further analyses using Orbitrap showed 100% coverage of the sequence (SI Appendix, Table S3). (E) Organic solvent extraction of biliverdin (BV) from BBS. Ratio Soret/Q changes from 3.2 for the isolated BV to 1.05 in BBS. (F) HPLC of BBS. BV is separated from the apoprotein, which has no absorbance at visible wavelengths.
The function of BV in animals is controversial (11), and it was initially only regarded as an intermediate product of heme catabolism in vertebrates. However, some evidence has shown that it may be a potent hydrophilic antioxidant (17, 18). Besides frogs, large BV concentrations have been documented in fish (19) and lizards (20, 21), but its function has remained a mystery. Indeed, little is known about the physiology and evolution of chlorosis, and its occurrence has remained an enigma in amphibian biology for more than a century.
In this report, we combined protein purifications, RNA-seq, ancestral character reconstruction, and optical modeling to better understand chlorosis. We show that chlorosis is highly prevalent within amphibians, and that it evolved multiple times along the evolutionary history of frogs. We isolated a serpin protein that binds BV, identified as the IXα isomer, and is responsible for its observed high concentration in chlorotic animals. The described serpin, called biliverdin-binding serpin (BBS), is a highly expressed glycoprotein belonging to the most diversified clade of the serpin superfamily, the largest group of protease inhibitors in nature (22–26). We also show that the binding of BV to the serpin changes its coloration, modulating the shape and intensity of absorbance in the visible region. Using Boana punctata as an animal model, we demonstrate that BBSs play a crucial role in the fine-tuning of their coloration, explaining not only the green hue of the animals, but also the existence of a red-edge change of their reflectance profile, which makes them unnoticeable against the foliage where they live. This report thus provides insights into serpin function diversification, showing different serpins that independently evolved the ability to bind BV. We also show that biliverdin has a strong function in vertebrate camouflage, which demonstrate a clear function for BV and upending the common belief that BV is only a waste product of heme catabolism.
Results and Discussion
Multiple Evolutionary Origins of Chlorosis in Frogs.
To understand the evolution of physiological chlorosis, we first undertook a comprehensive survey of its occurrence across anurans. The existence of blue-green pigmentation in the bones and soft tissues of frogs has been documented in hundreds of species from different clades (3–5). Our survey showed that it is present in more than 430 anuran species (SI Appendix, Table S1), distributed among 11 families (Arthroleptidae, Centrolenidae, Craugastoridae, Hemiphractidae, Hylidae, Hyperoliidae, Limnodynastidae, Mantellidae, Myobatrachidae, Ranidae, Rhacophoridae). We inferred that chlorosis evolved at least 41 times during anuran evolutionary history (Fig. 1 and SI Appendix, Discussion). Indeed, with the exception of glassfrogs (Centrolenidae), we documented multiple origins of physiological chlorosis within most other families (Fig. 1 and SI Appendix, Figs. S1 and S2). Of the species known to have chlorosis, 99% are treefrogs or are nested within treefrog clades (e.g., the aquatic Pseudini hylids).
Purification and Identification of Biliverdin-Binding Serpin from Boana punctata.
To study the biochemical origin of the blue-green coloration of frogs, we used B. punctata as a model. We chose this species because 1) its plasma BV concentration was reported in a previous study (205 μM) (4), and 2) some of the optical properties of its skin, muscle, and lymph were thoroughly studied in a previous work (2). B. punctata s.c. lymph is blue-green, which is easily observable through its translucent skin (2) (Fig. 2A). We extracted lymph samples and interstitial fluid from skin and muscle tissue, and performed chemical extractions followed by size exclusion separation to isolate the blue-green phase. After removal of the yellow components [e.g., hyloins (2) and carotenoids], we traced the remaining blue coloration to a protein that we termed “biliverdin-binding serpin” (BBS) (Fig. 2 A and B). The purified protein (∼95% purity) had a molecular weight of ∼50 kDa (Fig. 2 B and C) and was conformationally homogeneous with a folded core as indicated by circular dichroism, fluorescence spectroscopy, and size exclusion chromatography (SI Appendix, Fig. S3). Due to the lack of genomic sequence data in available databases, we identified the BBS via a combination of peptide mass fingerprint analysis (PMF), manual de novo amino acid sequencing of some of selected fragments using matrix-assisted laser desorption/ionization–time of flight tandem mass spectrometry (MALDI-TOF/TOF), and Edman sequencing of the N terminus (SI Appendix, Fig. S4). The resulting peptide dataset (SI Appendix, Fig. S4C) was subjected to MSBlast (27), which identified the biliprotein as a member of the glycoprotein serpin superfamily, particularly of the alpha-1-antitrypsin-like clade (α1-AT). Since serpins of this group are normally expressed in the liver (28), to obtain the complete nucleotide sequence, we extracted total RNA from this tissue and converted it to a complementary DNA (cDNA) library. Then we designed degenerate primers based on de novo and EDMAN amino acid sequences and proceeded with 3′ rapid amplification of cDNA ends and thermal asymmetric interlaced polymerase chain reaction (SI Appendix, Fig. S4D). Based on the complete amino acid sequence, we found that the BBS of B. punctata is a glycoprotein (Fig. 2D and SI Appendix, Fig. S5 and Table S3) from the Clade A serpin superfamily, showing a 45% identity to human α1-AT and ∼17–33% identity to other human serpins of the same clade (Table 1).
Table 1.
Sequence identity between B. punctata BBS and human Clade A serpins
| UniProt Accession Number| Protein Name | Identity, % | Identity Protein Core, % |
| P01009| SerpinA1 alpha-1-antitrypsin | 41.7 | 45.47 |
| P20848| SerpinA2 putative alpha-1-antitrypsin-related protein | 32.6 | 36.16 |
| P01011| SerpinA3 alpha-1-antichymotrypsin | 32.1 | 35.61 |
| P29622| SerpinA4 kallistatin | 31.9 | 35.61 |
| P05154I| SerpinA5 plasma serine protease inhibitor | 32.3 | 35.06 |
| P08185| SerpinA6 corticosteroid-binding globulin | 32.9 | 35.89 |
| P05543| SerpinA7 thyroxine-binding globulin | 32.0 | 35.06 |
| P01019| SerpinA8 angiotensinogen | 17.3 | 19.45 |
| Q86WD7| SerpinA9 | 30.9 | 32.23 |
| Q9UK55| SerpinA10 protein Z-dependent protease inhibitor | 25.2 | 29.04 |
| Q86U17| SerpinA11 | 31.2 | 34.8 |
| Q8IW75| SerpinA12 vaspin | 33.3 | 37.8 |
Sequence identities are shown considering the complete alignments (Identity, %) or excluding the poorly conserved leader sequences and signal peptides in the N-terminal region of the protein (Identity Core, %) (23). Protein core was defined from the N terminus of the first alpha helix to the C terminus of the protein (see SI Appendix, Fig. S5 for an interpretation of the secondary structure composition of the serpins). The position of the first alpha helix was based on the alignment to human α1-AT (23).
Identification of BV Isomer from B. punctata BBS.
Considering the existence of multiple BV isomers in nature, with different occurrences across animal taxa as well as ontogenetic changes during their development (29–31), we first characterized the isomery of BV from BBS in B. punctata. BV was extracted from BBS with organic solvents in mild acidic conditions (Fig. 2E), and it also dissociated form the apoprotein during liquid chromatography (Fig. 2F), suggesting a noncovalent association. High-resolution molecular-weight determination showed a mass-to-charge ratio (m/z) compatible with that of BV (observed m/z 583.2536, theoretical m/z for C33H34N4O6 plus one proton [M + H]+ is 583.2551, Δm/z = 2.5 ppm). We used ultraviolet-visible (UV-VIS) spectrophotometry (SI Appendix, Fig. S6A), one-dimensional (1D) and two-dimensional (2D) nuclear magnetic resonance (NMR) of the derivatized pigment (SI Appendix, Fig. S6B and Table S2) and liquid chromatography coupled to diode array detection and electrospray ionization tandem mass spectrometry (LC-DAD-MS/MS) (SI Appendix, Figs. S7A and S8). BV in B. punctata was identified as the IXα isomer, the same isomer detected in eggs from nonchlorotic species (32, 33) (SI Appendix, Fig. S6). Using the complete amino acid sequence and the BV absorbance, we determined the stoichiometry of apoprotein/BV to be 1:1. Absorbance spectra of BBS showed a Soret (390 nm)/Q(667 nm) ratio of 1.05 ± 0.05, which is compatible with a partially extended BV conformation inside the protein core (34) (SI Appendix, Discussion). This ratio is about three times smaller than the expected value for free helical BVs at physiological pH (35) (Soret/Q = 3.2).
BBSs Identification and Gene Expression across Evolutionary Origin of Chlorotic Frogs.
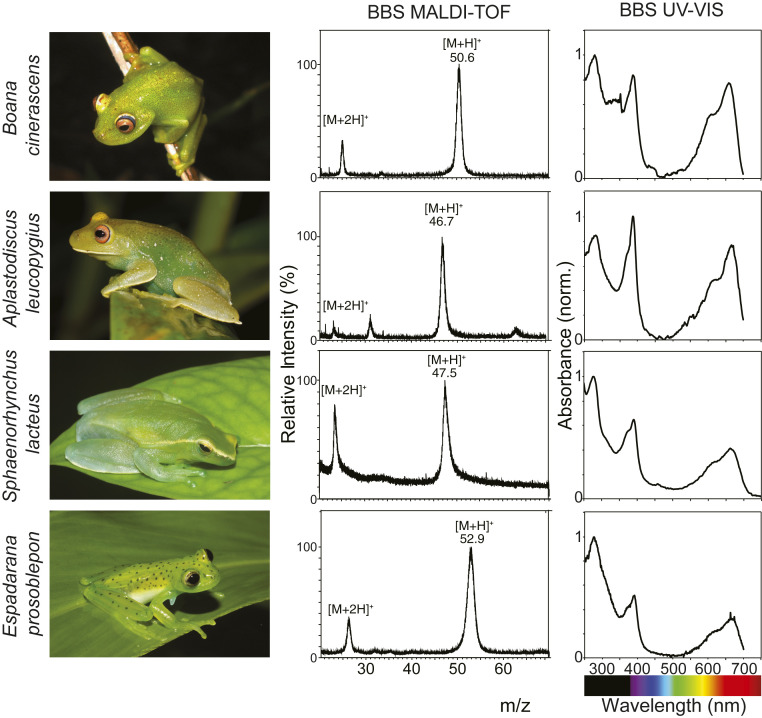
To evaluate the generality of the results obtained for B. punctata, we studied eight other species from different clades of chlorotic frogs. We purified the BBSs from lymph and bones and extracted liver tissue for RNA-seq analyses. BV was identified as the IXα isomer in all of the studied species and was noncovalently bound to the apoproteins (SI Appendix, Fig. S7 B and C). The ratio of absorbance Soret/Q ranged from 1.0 to 1.5, which was consistent with the partially extended chromophore conformation predicted in B. punctata inside the protein matrixes (Fig. 3 and SI Appendix, Fig. S9), suggesting a highly conserved mode of BV binding. The BBSs had molecular weights in the range of 46 to 54 kDa (Fig. 3 and SI Appendix, Fig. S10). In some of the species, BBS from bones was the same as that from lymph, while in others they were different isoforms or paralogs (SI Appendix, Fig. S11 and Discussion). The intrinsic multiplicity of clade A serpins (13 paralogs in humans, 9 paralogs in Xenopus tropicalis) (24, 36) implies several difficulties for the accurate de novo assembly of the sequences. Indeed, if the divergence among the sequences is low, considerable uncertainty is normally present in the assemblies (37) and the sequences are not reliable (e.g., many of them are chimeras). To overcome this, we implemented an iterative assembly approach based on RNA-seq read frequency, followed by PMF of the purified BBSs and tandem mass spectrometry to corroborate the sequences (SI Appendix, Fig. S12 and Tables S3–S12). In all of the species, lymphatic BBSs were identified as clade A serpins, with maximum identity to human α1-AT (SI Appendix, Fig. S13 and Table S13). Expression values ranged between 104–105 fragments per kilobase per million (SI Appendix, Table S14) indicating that BBSs are highly expressed in chlorotic frogs. Altogether, these findings introduce a class of carrier serpins, which broadens the repertoire of the already known ligand-binding proteins of the serpin superfamily, such as corticosteroid binding globulin (CBG) in several vertebrates and thyroxine-binding globulin (TBG) present in some mammals (38, 39).
Fig. 3.
MALDI-TOF and UV-VIS spectra of BBS from selected chlorotic species. BBSs are highly concentrated in soft tissues of chlorotic animals and can normally be seen through the skins, which are partially devoid of chromatophore cells. Purified BBSs of these selected species were analyzed by MALDI-TOF. Molecular weights range from 46 to 54 kDa (SI Appendix, Fig. S9 and Table S3). Absorbance spectra of the same BBSs (phosphate buffer 0.01 M, pH 7) show similar features to those in B. punctata (Fig. 2E). Absorbance peaks are shifted compared to those of isolated BV (Fig. 2E) and have maxima at 390 nm (Soret band) and 667 nm (Q band). Ratio of absorbance Soret/Q ranges from 1 to 1.5, which suggests a partially extended chromophore conformation inside the protein matrix (SI Appendix, Fig. S10 and Discussion) and a putative conserved mode of BV binding to the serpin.
Our discovery that in all of the studied species BV is bound to at least one member of the clade A serpins raises the question about the homology of the different BBSs (see SI Appendix for details). The scarcity of highly curated amphibian sequences, in addition to the large number of clade A paralogs in vertebrate genomes (36), precludes any conclusion on the evolutionary relationships of BBSs based on amino acid or nucleotide sequences. Indeed, broad phylogenomic studies have shown that it is not possible to do one-to-one orthology comparisons for many of the serpins of the group (36).
However, analyses of BBSs sequences assembled in this study provide some evidence to hypothesize about their homology. Our results revealed that BBSs sequence identities are ∼80 to 85% between different genera of glass frogs (Centrolenidae) (SI Appendix, Fig. S13 and Table S13) whereas they range from 44 to 54% between different hylid species in which chlorosis evolved independently. These results are consistent with a common origin of chlorosis in centrolenid frogs and may also support the common origin—and thus the orthology—of their BBSs (Fig. 1). On the other hand, the results obtained for hylids suggest that it is likely that different serpin paralogs are responsible for physiological chlorosis in the different clades where it evolved independently (SI Appendix, Fig. S13, Table S13, and Discussion). The only exception might be the case of the closely related B. punctata and Boana cinerascens, whose BBSs sequences share 75% identity (SI Appendix, Discussion). These results suggest that different serpin paralogs are likely responsible for physiological chlorosis in the different groups where it evolved independently. The fact that in at least some of the species there are two different BBSs, one in lymph and one in bones, supports the observation that multiple, different serpin paralogs may be responsible for chlorosis (SI Appendix, Fig. S11).
Functional Diversification of Serpins and the Mechanistic Basis of Green Coloration in Frogs.
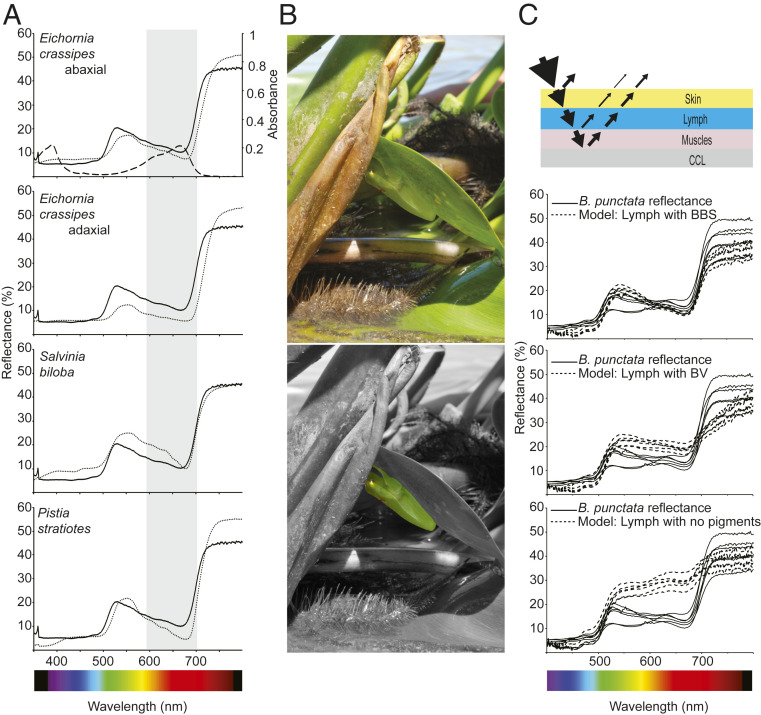
Pathological hyperbiliverdinemia in humans has only been described in a limited number of conditions (12–14), with bile duct obstruction. In chlorotic anuran species, however, anatomical observations cannot account for any morphological cause of the accumulation (5) (SI Appendix, Fig. S14), and there are no signs of increased hemolysis revealed by up-regulation of heme oxygenase (SI Appendix, Table S15 and Discussion). The fact that BV is bound to a serpin could change the normal BV excretion rates, which is supported by the fact that BV injected into nonchlorotic species can be easily excreted without increase in plasma concentration (5, 15). The high expression of BBSs, combined with their recurrent evolution, raise the question of their potential biological functions. Indeed, the fact that they modulate the spectral absorbance of isolated BV (Fig. 2E) could have an effect on animal coloration. Within amphibians, several chlorotic species have translucent skins, completely or partially devoid of melanophores and other chromatophore cells (2, 3) (Fig. 2A and 3). Thus, BBSs as well as other skin interstitial fluid and s.c. lymph pigments may be important in the overall coloration of anurans, complementing the role of dermal chromatophores (2). To test this, we evaluated the influence of BBSs in crypsis in B. punctata. We first measured reflectance spectra of different native plants from where they rest during the day or perch at night (2) and found that these plant spectra closely matched those of the frog (Fig. 4 A and B). The reflectance match extends to the red and near-infrared regions of the spectrum, showing a clear, red-edge sharp change in reflectance with an inflection point at 695–700 nm, comparable to that in plants (40). The absorbance spectrum of the visible band of BBS overlaps with the region of lower reflectance (600–670 nm) of the animals (Fig. 4 A and B), which suggests a putative role of BBS in color modulation. To test this, we employed an optical model based on the same anatomical structural organization described in previous studies (2) (Fig. 4C). This consisted of four layers of tissue: an outer skin layer (S), a s.c. lymphatic sac containing lymph (L), dorsal muscle (M), and a broadband reflector composed from an underlying layer of connective tissue with guanine crystals (CCL) (2). We simulated the expected reflectance under three scenarios: 1) 0.7 mm path length of s.c. lymph + skin with BBS at a physiological concentration of 104 μM (n = 12, range 88–204 μM), 2) the same path length with the same concentration of free helical BV at neutral physiological pH, and 3) with neither BV nor BBS. Only the occurrence of s.c. BBS in combination with an underlying broadband reflector (CCL) can account for the fine-tuning of the frog’s reflectance spectra and thus provide a mechanism to explain the red edge effect and the saturated green coloration on chlorotic frogs. Thus, it provides the anatomical and biochemical basis to understand how the animal can naturally camouflage in the surrounding green vegetation. The repeated evolution of BBSs with similar absorption profiles in arboreal frogs (SI Appendix, Fig. S9), in combination with the scarcity of chromatophore cells, suggest that this mechanism is the outcome of ecological and evolutionary processes and provide the mechanistic basis to understand the evolution of cryptic leaf coloration in chlorotic frogs (41).
Fig. 4.
Influence of BBS in B. punctata coloration. (A) Reflectance spectra of frogs obtained from (2) (solid line) and different plants (dotted line) from their environment. BBS absorbance spectrum is shown in the Upper graph (dashed line), and both the shape and position of the Q band (shaded in gray) are consistent with the reduced reflectance of the animals in the red portion of the spectrum. Red-edge reflectance corresponds to a region of sharp change in reflectance at ∼695–700 nm, a phenomenon widely known in plants. The blue region is normally dominated by other pigments and can be modulated by fluorescence of hyloins (2). (B, Upper) Specimen of B. punctata in a normal resting position in the abaxial surface of a leaf of the water hyacinth Eichhornia crassipes during daytime. (B, Lower) Background is displayed in grayscale to highlight the frog position. (C) A simplified model of reflectance for the frogs to evaluate the influence of BBS in coloration. Light is reflected at the air skin interphase, transmitted through the translucent skins, and red and blue light are highly absorbed by lymphatic BBS. Under the lymphatic sacs, light is transmitted through the muscles and is reflected back in a broadband guanine-based reflector in the fascia (CCL) in the retroperitoneal cavity. Lymph can modulate the spectral properties of light emerging from the animals and it depends on its pigments’ composition. Physiological concentration of BBS exerts an important role in B. punctata coloration, and it outperforms the modeled effect in control experiments with isolated biliverdin (Middle graph) or with lymph devoid of BBS (Bottom graph). Solid lines: reflectance of six individuals. Dashed line: modeled reflectance for six independent skin samples.
Other BBSs Functions.
Our results highlight the fact that the functional study of chlorosis goes beyond the study of elevated BV concentrations, to understanding the influence of biliproteins on ecologically relevant traits. Our work presents a comprehensive report on the identity, sequence, and function of an amphibian serpin (SI Appendix, Discussion) and describes an entire group of previously unknown BV carriers in animals. Even though we showed a clear role of these serpins in anuran color tuning, there may be other functions for BBSs, especially in the few chlorotic species without translucent skin that have normal chromatophores distribution (e.g., functions mediated by their serpin nature). The serpins are a superfamily of proteins (350–500 amino acids in size), normally serine proteases (in humans 27/36 serpins) (24) with a conserved fold and a unique mechanism of action (42). This involves a drastic conformational change and is highly dependent on proteases recognition of the reactive center loop (RCL) sequence and length (43) (SI Appendix, Fig. S14). The sequence identity of BBSs to human α1-AT, a common neutrophil elastase inhibitor during inflammatory processes, suggests that BBSs may have an inhibitory function too. However, a detailed analysis of BBS sequences showed that there are atypical amino acid substitutions in the RCL for B. punctata, B. cinerascens and Hyloscirtus phyllognathus, which are known to preclude the inhibitory mechanism (44–46), making uncertain their role as protease inhibitors. Other roles of BBSs may be related to the function of the noninhibitory hormone carriers CBG and TBG, which includes the transport of their ligands to target organs. For instance, it is known that CBG is involved in the proteinase-triggered release of steroids to sites of inflammation (39). An analogous function of BBSs can include a delivery system of BV to sites of inflammation, where it could act as an antioxidant (17, 18) or free radical scavenger (47).
Materials and Methods
Full experimental details including citations are provided in the SI Appendix.
To study the prevalence and distribution of chlorosis in amphibians, we identified those species that show clear accumulations of green lymph and/or green tissues (many times evident in the oral mucosa and inguinal region) or green bones (often evident by transparent phalanges, humerus, femur, tibiofibula, maxilla, and premaxilla). We surveyed the taxonomic literature of all extant anurans and utilized colleagues with expertise on the relevant taxonomic groups.
To infer the number of independent origins of chlorosis during the evolutionary history of anurans, we performed ancestral character reconstruction using the phylogenetic results of Jetz & Pyron (16) (Fig. 1) to have a general perspective and recent, densely sampled phylogenetic hypotheses for several families. The ancestral character reconstruction was done using Fitch optimization (48) as implemented in TNT (49, 50). Phylogenetic tree graphical editing was done using the iTOL webserver (51).
For the study of the origin of green coloration on soft tissues and bones, we selected 11 chlorotic species. We purified BBSs and performed RNA-seq from nine of them: Family Hylidae: Aplastodiscus leucopygius, B. puncata, B. cinerascens, H. phyllognathus, Pseudis minuta, and Sphaenorhynchus lacteus; family Centrolenidae: Chimerella mariaelenae, Espadarana prosoblepon and Nymphargus posadae. Additionally, we purified BBSs from Boana atlantica and Aplastodiscus flumineus.
BBSs purification was assessed by PAGE, high-performance liquid chromatography (HPLC), MALDI-TOF. For B. punctata BBS, we also performed circular dichroism, fluorescence spectroscopy, and size exclusion chromatography.
Structural characterization of the isolated BV was performed by 1D and 2D NMR and LC-DAD-MS/MS. To characterize the BBSs, we performed trypsinization of the isolated proteins followed by peptide mass fingerprinting, de novo sequencing of MALDI TOF-TOF spectra of selected peptides, and MSBLAST searches. For B. punctata, we also performed Edman sequencing of the N terminus. To obtain the full sequence of the proteins, we assembled hepatic transcriptomes using a custom pipeline, mapped the peptide masses to the assembled sequences and verified the matches by MSMS. With the complete protein sequences, we used BLASTP against different databases for identification of the apoproteins.
To assess the influence of BBS in coloration, we considered the same anatomical model published elsewhere (2). Since chlorosis was normally assumed to be a product of biliverdin accumulation, we compared the influence of isolated biliverdin with that of BBSs in the overall animal coloration. Diffuse reflectance and transmittance of plant species were obtained by means of a spectrophotometer (UV3101PC; Shimadzu) equipped with an integrating sphere (ISR-3100; Shimadzu).
All procedures involving animals were carried out according to the regulations specified by the Institutional Animal Care and Use Committee of the Facultad de Ciencias Exactas y Naturales, Universidad de Buenos Aires (Res C/D 140/00), and by Conselho Nacional de Controle de Experimentação Animal, Ministério da Ciência, Tecnologia e Inovação, Brazil. Collection permits were issued by Secretaria de Medio Ambiente, Ministerio de Aguas, Servicios Públicos y Medio Ambiente, Province of Santa Fe, Argentina (021-2011 and 063–2013), Ministerio de Ecología, Province of Misiones, Argentina (010-2015), and Instituto Chico Mendes de Conservação da Biodiversidade/SISBIO (Permits 41508–8, 50071–1, 50071–2, A1FC113).
Supplementary Material
Acknowledgments
C.T. was supported by a Human Frontier Science Program (LT 000660/2018-L) postdoctoral fellowship and a Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) doctoral fellowship. A.E.B. and M.L.L. were supported by São Paulo Research Foundation (FAPESP) postdoctoral fellowships (#2014/20915-6, # 2017/ 23725-1, #2017/26162-8). A.E.B., M.G.L., J.F., L.B.C., S.E.B. are CONICET researchers. This research was supported by grants of Agencia Nacional de Promoción Científica y Tecnológica, Proyecto de Investigación Científica y Tecnológica 2011-1895, 2013-404, 2013-1895 (to L.B.C.), 2014-1022, and 2015-820, CONICET grant 11220150100394CO, the Universidad de Buenos Aires, Universidad de Buenos Aires Ciencia y Técnica 20020170100037BA, FAPESP, and FAPESP/Fundação Grupo Boticário de Proteção à Natureza (grants #2013/50741-7, #2012/10000-5, #2013/50741-7, #2014/50342-8, and #2018/15425-0), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) (306623/2018-8). Fieldwork in Ecuador was funded by Secretaría Nacional de Educación Superior, Ciencia, Tecnología e Innovación del Ecuador (SENESCYT) (Arca de Noé initiative; S.R.R. and Omar Torres, principal investigators), and a grant from Dirección General Académica Pontificia Universidad Católica del Ecuador. We thank the Duke Computer Cluster for providing computational resources and Jesse Delia, Vaclav Gvozdik, Alan Channing, Alexander Devin Edmonds, Alex Haas, and Taran Grant, for kindly authorizing the use of their photographs. We thank Vivian Trevine, Délio Baeta, Diego Baldo, Edgar Lehr, Victor G. D. Orrico, Tiago L. Pezutti, Paulo D. Pinheiro, Marco Rada, and Pablo Suárez for their input to Supp. Table 1, and Laura Andrade for assistance in proteins extraction and purification. We also thank Andrés Merino for providing access to live frogs from Balsa de los Sapos at PUCE, Josefina Awruch for suggestions and providing reagents, and Jesse Delia for critical comments and suggestions.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The RNA-Seq data generated and analyzed in this study are available in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) (BioProject ID number PRJNA625657; https://www.ncbi.nlm.nih.gov/bioproject/625657). Biliverdin-binding serpin nucleotide sequences reported in this paper are available in the GenBank repository (accession nos. MT358317–MT358325).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2006771117/-/DCSupplemental.
Data Availability.
The RNA-Seq data generated and analyzed in this study are available in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA), BioProject ID number PRJNA625657 (https://www.ncbi.nlm.nih.gov/bioproject/625657). Biliverdin-binding serpin nucleotide sequences reported in this paper were deposited into GenBank under accession numbers MT358317-358325.
References
- 1.Bagnara J. T., Taylor J. D., Hadley M. E., The dermal chromatophore unit. J. Cell Biol. 38, 67–79 (1968). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taboada C. et al., Naturally occurring fluorescence in frogs. Proc. Natl. Acad. Sci. U.S.A. 114, 3672–3677 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taboada C., Brunetti A. E., Alexandre C., Lagorio M. G., Faivovich J., Fluorescent frogs: A herpetological perspective. South Am. J. Herpetol. 12, 1–13 (2017). [Google Scholar]
- 4.Barrio A., Cloricia fisiológica en batracios anuros. Physis 25, 137–142 (1965). [Google Scholar]
- 5.Jones D. A., “Green pigmentation in neotropical frogs,” Ph.D. dissertation, University of Florida, Gainesville, FL (1967).
- 6.Himes J. A., Cornelius C. E., Biliverdin and bilirubin excretion in the Turkey. Cornell Vet. 65, 374–379 (1975). [PubMed] [Google Scholar]
- 7.Lin G. L., Himes J. A., Cornelius C. E., Bilirubin and biliverdin excretion by the chicken. Am. J. Physiol. 226, 881–885 (1974). [DOI] [PubMed] [Google Scholar]
- 8.Colleran E., O’Carra P., “Enzymology and comparative physiology of biliverdin reduction” in Chemistry and Physiology of Bile Pigments, Berk P. D., Berlin N. I., Eds. (U.S Department of Health, Education, and Welfare, 1977), pp. 69–80. [Google Scholar]
- 9.Cornelius C. E., Bile pigments in fishes: A review. Vet. Clin. Pathol. 20, 106–115 (1991). [DOI] [PubMed] [Google Scholar]
- 10.Noonan N. E., Olsen G. A., Cornelius C. E., A new animal model with hyperbilirubinemia: The indigo snake. Dig. Dis. Sci. 24, 521–524 (1979). [DOI] [PubMed] [Google Scholar]
- 11.McDonagh A. F., Turning green to gold. Nat. Struct. Biol. 8, 198–200 (2001). [DOI] [PubMed] [Google Scholar]
- 12.Greenberg A. J., Bossenmaier I., Schwartz S., Green jaundice. A study of serum biliverdin, mesobiliverdin and other green pigments. Am. J. Dig. Dis. 16, 873–880 (1971). [DOI] [PubMed] [Google Scholar]
- 13.Gåfvels M. et al., A novel mutation in the biliverdin reductase-A gene combined with liver cirrhosis results in hyperbiliverdinaemia (green jaundice). Liver Int. 29, 1116–1124 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Nytofte N. S. et al., A homozygous nonsense mutation (c.214C->A) in the biliverdin reductase alpha gene (BLVRA) results in accumulation of biliverdin during episodes of cholestasis. J. Med. Genet. 48, 219–225 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Cabello Ruz J., Fijación de bilirrubina y biliverdina por el hígado de sapo. Rev. Soc. Argent. Biol. 19, 71–76 (1943). [Google Scholar]
- 16.Jetz W., Pyron R. A., The interplay of past diversification and evolutionary isolation with present imperilment across the amphibian tree of life. Nat. Ecol. Evol. 2, 850–858 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Stocker R., Yamamoto Y., McDonagh A. F., Glazer A. N., Ames B. N., Bilirubin is an antioxidant of possible physiological importance. Science 235, 1043–1046 (1987). [DOI] [PubMed] [Google Scholar]
- 18.Stocker R., Antioxidant activities of bile pigments. Antioxid. Redox Signal. 6, 841–849 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Fang L. S., Bada J. L., The blue-green blood plasma of marine fish. Comp. Biochem. Physiol. B 97, 37–45 (1990). [DOI] [PubMed] [Google Scholar]
- 20.Austin C. C., Jessing K. W., Green-blood pigmentation in lizards. Comp. Biochem. Physiol. A Physiol. 109, 619–626 (1994). [Google Scholar]
- 21.Rodriguez Z. B., Perkins S. L., Austin C. C., Multiple origins of green blood in New Guinea lizards. Sci. Adv. 4, eaao5017 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huntington J. A., Serpin structure, function and dysfunction. J. Thromb. Haemost. 9 (suppl. 1), 26–34 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Irving J. A., Pike R. N., Lesk A. M., Whisstock J. C., Phylogeny of the serpin superfamily: Implications of patterns of amino acid conservation for structure and function. Genome Res. 10, 1845–1864 (2000). [DOI] [PubMed] [Google Scholar]
- 24.Law R. H. et al., An overview of the serpin superfamily. Genome Biol. 7, 216 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Silverman G. A. et al., The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Evolution, mechanism of inhibition, novel functions, and a revised nomenclature. J. Biol. Chem. 276, 33293–33296 (2001). [DOI] [PubMed] [Google Scholar]
- 26.Gettins P. G., Serpin structure, mechanism, and function. Chem. Rev. 102, 4751–4804 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Shevchenko A. et al., Charting the proteomes of organisms with unsequenced genomes by MALDI-quadrupole time-of-flight mass spectrometry and BLAST homology searching. Anal. Chem. 73, 1917–1926 (2001). [DOI] [PubMed] [Google Scholar]
- 28.Badola S. et al., Correlation of serpin-protease expression by comparative analysis of real-time PCR profiling data. Genomics 88, 173–184 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Yamaguchi T., Nakajima H., Changes in the composition of bilirubin-IX isomers during human prenatal development. Eur. J. Biochem. 233, 467–472 (1995). [DOI] [PubMed] [Google Scholar]
- 30.Cole K. D., Little G. H., UDP-glucuronosyltransferase activity and bilirubin conjugation in the bullfrog. Biochem. J. 212, 265–269 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rüdiger W., Klose W., Vuillaume M., Barbier M., On the structure of pterobilin, the blue pigment of Pieris brassicae. Experientia 24, 1000 (1968). [DOI] [PubMed] [Google Scholar]
- 32.Marinetti G. V., Bagnara J. T., Yolk pigments of the Mexican leaf frog. Science 219, 985–987 (1983). [DOI] [PubMed] [Google Scholar]
- 33.Falchuk K. H. et al., A role for biliverdin IXα in dorsal axis development of Xenopus laevis embryos. Proc. Natl. Acad. Sci. U.S.A. 99, 251–256 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iturraspe J. B., Bari S. E., Frydman B., Total synthesis of “extended” biliverdins. The relation between their conformation and their spectroscopic properties. J. Am. Chem. Soc. 111, 1525–1527 (1989). [Google Scholar]
- 35.Heirwegh K. P., Blanckaert N., Van Hees G., Synthesis, chromatographic purification, and analysis of isomers of biliverdin IX and bilirubin IX. Anal. Biochem. 195, 273–278 (1991). [DOI] [PubMed] [Google Scholar]
- 36.Kumar A., Bayesian phylogeny analysis of vertebrate serpins illustrates evolutionary conservation of the intron and indels based six groups classification system from lampreys for ∼500 MY. PeerJ 3, e1026 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ranwez V. et al., Disentangling homeologous contigs in allo-tetraploid assembly: Application to durum wheat. BMC Bioinformatics 14 (suppl. 15), S15 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou A., Wei Z., Read R. J., Carrell R. W., Structural mechanism for the carriage and release of thyroxine in the blood. Proc. Natl. Acad. Sci. U.S.A. 103, 13321–13326 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klieber M. A., Underhill C., Hammond G. L., Muller Y. A., Corticosteroid-binding globulin, a structural basis for steroid transport and proteinase-triggered release. J. Biol. Chem. 282, 29594–29603 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Horler D. N. H., Dockray M., Barber J., The red edge of plant leaf reflectance. Int. J. Remote Sens. 4, 273–288 (1983). [Google Scholar]
- 41.Blount C. C., “Near infrared refletance in Anura,” Ph.D. dissertation, The University of Manchester, Manchester, UK (2018).
- 42.Huntington J. A., Read R. J., Carrell R. W., Structure of a serpin-protease complex shows inhibition by deformation. Nature 407, 923–926 (2000). [DOI] [PubMed] [Google Scholar]
- 43.Zhou A., Carrell R. W., Huntington J. A., The serpin inhibitory mechanism is critically dependent on the length of the reactive center loop. J. Biol. Chem. 276, 27541–27547 (2001). [DOI] [PubMed] [Google Scholar]
- 44.Hopkins P. C., Carrell R. W., Stone S. R., Effects of mutations in the hinge region of serpins. Biochemistry 32, 7650–7657 (1993). [DOI] [PubMed] [Google Scholar]
- 45.Lawrence D. A. et al., Partitioning of serpin-proteinase reactions between stable inhibition and substrate cleavage is regulated by the rate of serpin reactive center loop insertion into beta-sheet A. J. Biol. Chem. 275, 5839–5844 (2000). [DOI] [PubMed] [Google Scholar]
- 46.Simonovic M., Gettins P. G., Volz K., Crystal structure of human PEDF, a potent anti-angiogenic and neurite growth-promoting factor. Proc. Natl. Acad. Sci. U.S.A. 98, 11131–11135 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Farrera J.-A. et al., The antioxidant role of bile pigments evaluated by chemical tests. Bioorg. Med. Chem. 2, 181–185 (1994). [DOI] [PubMed] [Google Scholar]
- 48.Fitch W. M., Toward defining the course of evolution: Minimum change for a specific tree topology. Syst. Zool. 20, 406–416 (1971). [Google Scholar]
- 49.Goloboff P. A., Farris J. S., Nixon K. C., TNT, a free program for phylogenetic analysis. Cladistics 24, 774–786 (2008). [Google Scholar]
- 50.Goloboff P. A., Catalano S. A., TNT version 1.5, including a full implementation of phylogenetic morphometrics. Cladistics 32, 221–238 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-Seq data generated and analyzed in this study are available in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA), BioProject ID number PRJNA625657 (https://www.ncbi.nlm.nih.gov/bioproject/625657). Biliverdin-binding serpin nucleotide sequences reported in this paper were deposited into GenBank under accession numbers MT358317-358325.