Significance
Genome-wide association studies have identified common noncoding variants associated with ECG parameters in the TBX3 locus, suggesting that a modest change in TBX3/Tbx3 expression level affects heart function. We characterized the morphology, physiology, and gene expression profile of the atrioventricular conduction system in a Tbx3 haploinsufficient mouse model. We identified Tbx3-dependent regulatory DNA elements active in the atrioventricular conduction system and validated the functionality of these elements in vivo. Using genome editing, we deleted a regulatory DNA element from the mouse genome and show it drives expression of Cacna1g in the cardiac conduction system. Our study unveils a Tbx3-dependent gene regulatory network in the AV conduction system that contributes to heart electrophysiology.
Keywords: Tbx3, Cacna1g, Ryr2, atrioventricular conduction system, electrical patterning
Abstract
Genome-wide association studies have identified noncoding variants near TBX3 that are associated with PR interval and QRS duration, suggesting that subtle changes in TBX3 expression affect atrioventricular conduction system function. To explore whether and to what extent the atrioventricular conduction system is affected by Tbx3 dose reduction, we first characterized electrophysiological properties and morphology of heterozygous Tbx3 mutant (Tbx3+/−) mouse hearts. We found PR interval shortening and prolonged QRS duration, as well as atrioventricular bundle hypoplasia after birth in heterozygous mice. The atrioventricular node size was unaffected. Transcriptomic analysis of atrioventricular nodes isolated by laser capture microdissection revealed hundreds of deregulated genes in Tbx3+/− mutants. Notably, Tbx3+/− atrioventricular nodes showed increased expression of working myocardial gene programs (mitochondrial and metabolic processes, muscle contractility) and reduced expression of pacemaker gene programs (neuronal, Wnt signaling, calcium/ion channel activity). By integrating chromatin accessibility profiles (ATAC sequencing) of atrioventricular tissue and other epigenetic data, we identified Tbx3-dependent atrioventricular regulatory DNA elements (REs) on a genome-wide scale. We used transgenic reporter assays to determine the functionality of candidate REs near Ryr2, an up-regulated chamber-enriched gene, and in Cacna1g, a down-regulated conduction system-specific gene. Using genome editing to delete candidate REs, we showed that a strong intronic bipartite RE selectively governs Cacna1g expression in the conduction system in vivo. Our data provide insights into the multifactorial Tbx3-dependent transcriptional network that regulates the structure and function of the cardiac conduction system, which may underlie the differences in PR duration and QRS interval between individuals carrying variants in the TBX3 locus.
The atrioventricular (AV) conduction system facilitates electrical impulse conduction from the atria to the ventricles and coordinates synchronized ventricular activation. T-box transcription factor 3-encoding gene Tbx3 is specifically expressed in the AV conduction system, and loss-of-function and gain-of-function experiments have demonstrated that Tbx3 is required for cardiac conduction system development and homeostasis (1). Common variation in the gene desert upstream of TBX3 has been associated with PR interval and QRS duration (2–6) (SI Appendix, Fig. S1). Since the activity of regulatory DNA elements (REs) is largely confined to a topologically associating domain (TAD) (7–12) we hypothesize that the PR and QRS interval-associated variants in the TAD cause subtle differences in TBX3 expression resulting in altered AV conduction system function. The mechanistic relation between modest TBX3 expression differences and AV conduction system function has not been investigated yet.
Insight into the transcriptional regulatory network controlling the development and function of the AV conduction system has been largely derived from animal knockout studies on individual transcription factors. These studies uncovered crucial roles for transcription factors including Tbx5, Tbx3, Nkx2-5, and Etv1 (1, 13, 14). Nevertheless, full molecular characterization of the AV conduction system has been hampered by the small size and heterogeneous composition of the AV node and bundle (15, 16). In addition, relevant cell lines modeling this tissue and human AV conduction system samples are insufficiently available. To investigate the repercussions of lower Tbx3 expression on the AV conduction system, we characterized the AV conduction system morphology and function of Tbx3 haploinsufficient (Tbx3+/−) mice, isolated AV nodes and defined their transcriptome, generated a genome-wide chromatin accessibility map of AV conduction system cells, and identified putative Tbx3-dependent AV conduction system REs. To gain insight into the in vivo function of such REs, we deleted an example bipartite RE with a strong epigenetic signature in Cacna1g (conduction system-enriched voltage-dependent T-type calcium channel Cav3.1; ref. 17) using genome editing.
Results
Heterozygous Loss of Tbx3 Causes Postnatal AV Bundle-Bundle Branch Hypoplasia and Affects AV Conduction.
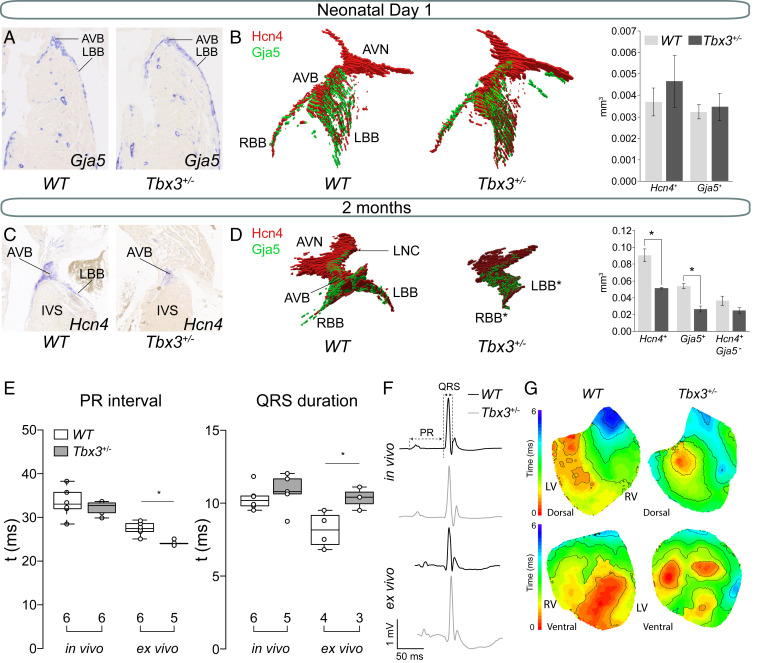
We studied the morphology and volume of the AV conduction system of neonatal Tbx3+/− and WT littermates by in situ hybridization on serial sections of hearts. The AV node was defined by the presence of Hcn4 (Hyperpolarization activated cyclic nucleotide gated potassium channel 4) and absence of Gja5 (Gap junction protein, alpha 5) expression in the AV junction. The AV bundle and bundle branches (BBs) were identified by coexpression of Hcn4 and Gja5 in the AV junction and ventricular septum (16, 18–20). Reconstructing and measuring the volumes of the Hcn4+Gja5− AV node and Hcn4+Gja5+ AV bundle-BBs using three-dimensional (3D) quantitative morphometric analysis showed that AV node, AV bundle, and BBs were not different between neonatal WT and Tbx3+/− hearts (Fig. 1 A and B). In contrast, AV bundle-BBs of 2-mo-old Tbx3+/− mice were hypoplastic, while the AV node remained similar in size to WT littermates (Fig. 1 C and D), thus indicating that AV bundle-BBs hypoplasia in Tbx3+/− mice develops after birth. We found no evidence of apoptosis (cleaved caspase-3) in hypoplastic AV bundles of Tbx3+/− mice at postnatal day (P) 28 (SI Appendix, Fig. S2).
Fig. 1.
Hypoplasia of the AV bundle-bundle branches and conduction disturbances in adult Tbx3+/− mice. (A) Representative sections of newborn WT and Tbx3+/− AVB-BBs. (B) Examples of a reconstruction of newborn WT and Tbx3+/− AV conduction system showing expression of Hcn4 (red) and Gja5 (green). Average volumes of Hcn4 (AV conduction system) and Gja5 (AV bundle and bundle branches) and Hcn4+-Gja5+ (AV node) expression domains in the AV conduction system of three WT (dark gray) and three Tbx3+/− (light gray) mice. Error bars represent SEM. *P < 0.05 was considered significant using a two-tailed Student’s t test. (C) Representative sections of 2-mo-old WT and Tbx3+/− AVB-BBs. (D) Example of a reconstruction of WT AV conduction system showing expression of Hcn4 (red) and Gja5 (green). Average volumes of Hcn4+, Gja5+, and Hcn4+-Gja5− expression domains in the AV conduction system of three WT (dark gray) and three Tbx3+/− (light gray) mice. Error bars represent SEM. (E) Boxplots visualizing the individually measured and average PR interval and QRS duration in vivo and ex vivo. Numbers below boxplot reflect the number of animals measured. (E) Boxplots visualizing the individually measured and average PR interval and QRS duration in vivo and ex vivo. (F) In vivo and ex vivo ECG traces from WT and Tbx3+/− mice. (G) Dorsal and ventral activation maps from WT and Tbx3+/− mice based on optical mapping. *P < 0.05 was considered significant using two-way ANOVA. AVB, atrioventricular bundle; AVN, atrioventricular node; IVS, interventricular septum; LBB, left bundle branch; LNC, lower nodal cells; RBB, right bundle branch.
Next, we recorded electrocardiograms (ECGs) in vivo from 2-mo-old mice and ex vivo from hearts and found significantly shorter PR interval (WT 27.57 ± 0.62 ms and Tbx3 ± 24.33 ± 0.22 ms) and prolonged QRS duration (WT 8.15 ± 0.62 ms and Tbx3 ± 10.33 ± 0.44 ms) in Tbx3 ± mutant hearts ex vivo. In contrast, there were no differences in vivo (PR: WT 33.34 ± 1.27 ms and Tbx3 ± 32.27 ± 1.05 ms; QRS: 10.35 ± 0.41 ms and Tbx3 ± 10.70 ± 0.51 ms) (Fig. 1E). Furthermore, we did not observe delta waves in ECG traces (Fig. 1F). To test whether the autonomic nervous system concealed a possible phenotype in vivo, we recorded ECGs in the presence of propranolol and atropine. QRS duration in WTs did not change after autonomic blockade (before, 9.07 ± 0.30 ms; after, 8.95 ± 0.40 ms), whereas Tbx3+/− mutants had showed a trend toward QRS prolongation from 9.07 ± 0.25 ms to 9.59 ± 0.38 ms (P = 0.08) (SI Appendix, Fig. S3). Optical mapping of isolated Langendorff-perfused hearts showed delayed apical activation at the dorsal side in Tbx3+/− mutants compared to controls (Fig. 1G). The anterior side showed two near simultaneous epicardial breakthroughs in WT hearts. In mutant hearts, multiple small breakthroughs separated by areas of late activation were present (Fig. 1G). The epicardial activation patterns indicate a slower and less homogeneous activation of the ventricular chambers possibly as a consequence of the AV bundle/BB hypoplasia. Together, these findings reveal previously underappreciated changes in AV conduction system morphology and function in a Tbx3 haploinsufficient mouse model.
Gene Expression Profiles of WT and Tbx3+/− AV Nodes.
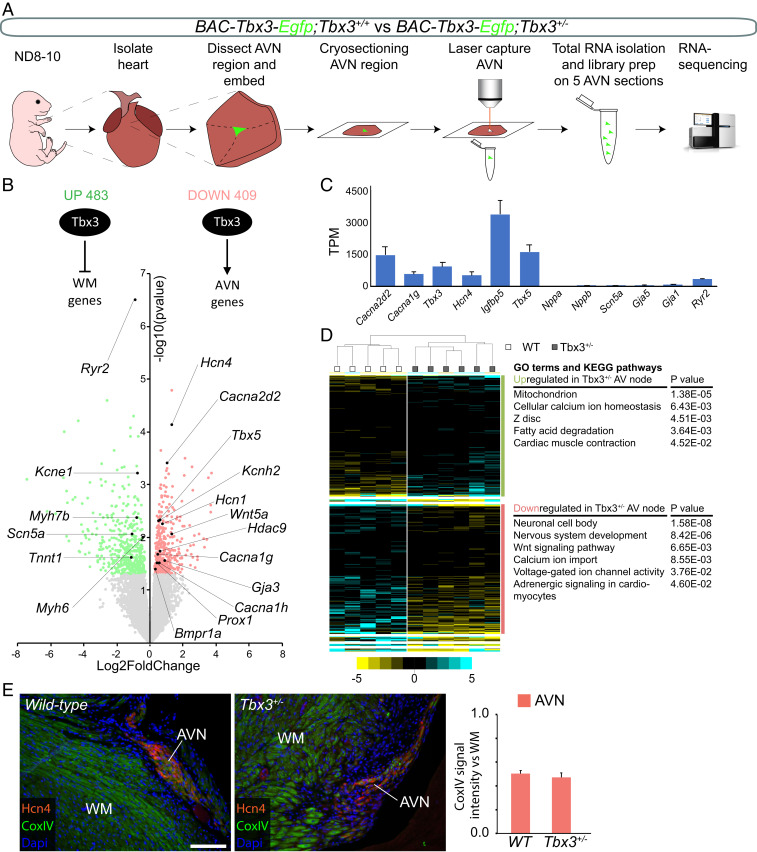
Deregulation of the AV nodal gene program is likely to contribute to PR interval shortening. Therefore, we characterized the transcriptomes of Tbx3+/− and WT AV nodes. We performed fluorescence-guided laser capture microdissection to isolate labeled AV node tissue from P8–10 unfixed cryosections (Fig. 2A). To specifically identify AV nodes of WT and Tbx3 heterozygotes, we generated Tbx3+/+;BAC-Tbx3-Egfp and Tbx3+/CreERT2;BAC-Tbx3-Egfp mice (21). BAC-Tbx3-Egfp mice express Egfp in the compact AV node, but not in the other tissues surrounding the AV node (22).
Fig. 2.
Tbx3 maintains pacemaker and conduction properties in the AV node. (A) Approach to obtain and define transcriptomes of WT and Tbx3+/− AV nodes. (B) Volcano plot of normalized expression levels in WT AV nodes over Tbx3+/− AV nodes. All significantly up-regulated genes are labeled green, and all significantly down-regulated genes are labeled red. P < 0.05 (red and green), P < 0.1 (light red and light green). (C) Average WT expression levels (in TPM) of conduction system and chamber genes. (D) Unsupervised hierarchical clustering of genes differentially expressed between Tbx3+/− AV nodes and WT AV nodes. GO terms and KEGG pathways associated with up-regulated (green) and down-regulated (red) genes in Tbx3+/− AV nodes. (E) Representative immunostaining for Hcn4 (red), CoxIV (green), and nuclei (blue). Graph shows quantification of CoxIV signal intensity in AVN normalized to WM in WT and Tbx3+/− sections (n = 3). AVN, AV node; WM, working myocardium. (Scale bar: 10 µm.)
As a first validation, we examined the expression of established AV nodal, chamber, and ventricular conduction system marker genes (15, 16, 18, 22–24). Hcn4, Cacna1g (Calcium channel, voltage-dependent, T-type, alpha 1G subunit), Cacna2d2 (Calcium voltage-gated channel auxiliary subunit α2-δ2), and Igfbp5 (Insulin like growth factor binding protein 5) were highly enriched in our AV nodal samples. In contrast, genes specific for atrial and ventricular chamber myocardium, and ventricular conduction system, Nppa (Natriuretic peptide type A), Gja1 (Gap junction protein, alpha 1), and Gja5, were virtually absent in our samples (Fig. 2C) (16, 24, 25).
Transcriptomic analysis revealed that 892 transcripts were differentially expressed in Tbx3+/− AV nodes (12,033 total transcripts detected, P < 0.05). Of these, 409 genes were down-regulated, (e.g., Hcn1, Hcn4, Cacna2d2, Kcnh2, and Cacna1g) and 483 genes were up-regulated (e.g., Myh6, Scn5a, Kcne1, and Ryr2; Fig. 2B and see SI Appendix, Table S1). Unsupervised hierarchical clustering of the differentially expressed genes resulted in two primary groups based on genotype (Fig. 2D). Gene ontology (GO) term and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis identified genes involved in Wnt signaling, neuronal, and voltage-gated ion channel activity to be overrepresented in the down-regulated gene set of Tbx3+/− AV nodes. Neuronal gene profiles are found in the AV conduction system (22), and Wnt signaling plays crucial roles in its development (26–28). Genes important for mitochondria, metabolism, and cardiac muscle contraction were found to be overrepresented in the up-regulated genes (Fig. 2D). As chamber (working) cardiomyocytes have more mitochondria and better developed sarcomeric structures and sarcoplasmic reticula compared to nodal cells (29–31), it stands to reason that genes associated with working myocardial gene programs are up-regulated in AV nodal cells of Tbx3+/− mice. However, the overall changes in expression are small and are therefore unlikely to affect detectable AV nodal properties. Indeed, we found no difference in mitochondrial abundance (CoxIV immunolabeling) between WT and Tbx3+/− AV nodes (Fig. 2E and see SI Appendix, Fig. S4). Interestingly, genes involved in calcium homeostasis were found in both the down-regulated and up-regulated gene sets. This could reflect the existence of complex Tbx3-dependent transcriptional modulation that controls AV node function.
To gain further insight into the transcriptional regulatory hierarchy in the AV node, we cross-referenced our dataset with a catalog of genes encoding transcription factors (32). In total, transcripts of 710 transcription factor-encoding genes were expressed in the AV node samples. The top 50 highest expressed transcription factor genes include Tbx5, Tbx3, Tbx20, Mef2a, Gata4, Gata6, and Prox1. Interestingly, Tbx5 and Prox1 were down-regulated in Tbx3+/− AV nodes (Fig. 2B) (33, 34). Together, our data indicate Tbx3+/− AV nodes acquired a modest working myocardial gene program while maintaining pacemaker properties.
Identification of Tbx3-Dependent AV Conduction System Regulatory Elements.
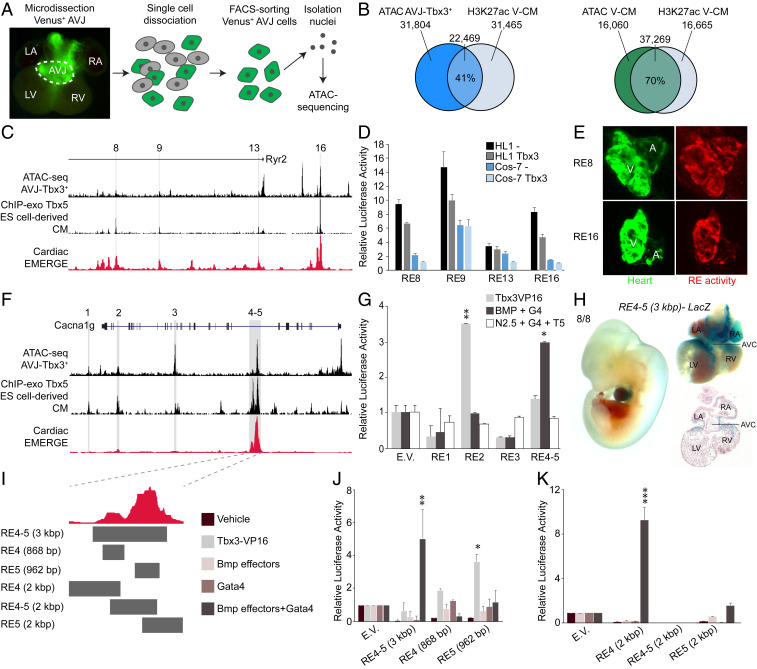
To identify regulatory elements active in the AV conduction system cardiomyocytes, we used Assay for Transposase-Accessible Chromatin (ATAC)-sequencing (seq) (35) on fluorescence-activated cell sorting (FACS)-purified Venus+ cells from microdissected AV junctions of Tbx3+/Venus fetuses (Fig. 3A). Peak calling (SI Appendix, Fig. S5A) identified 54,273 accessible regions, of which 41% overlapped with an available histone H3K27ac ventricular cardiomyocyte chromatin immunoprecipitation (ChIP)-sequencing dataset (36) (Fig. 3B and see SI Appendix, Fig. S5A). As expected, the overlap between ATAC-seq data from ventricular cardiomyocytes (37) and this H3K27ac ChIP-seq dataset was much broader (70%) (Fig. 3B), suggesting that the AV conduction ATAC-seq dataset could be useful to pinpoint AV conduction system-specific REs. Indeed, three previously established AV conduction system-specific REs (12, 38, 39) were readily identified in this dataset (SI Appendix, Fig. S5 B and C).
Fig. 3.
Identification and functional testing of Tbx3-dependent REs. (A) Strategy to assay the accessible chromatin profiles in E12.5–E14.5 AV conduction system cardiomyocytes using ATAC-seq. (B) Venn diagrams showing overlap between Venus+ AV conduction system ATAC-seq signals (Left) or ventricular cardiomyocyte (V-CM) ATAC-seq signals (Right) and ventricular cardiomyocyte H3K27ac enrichment. (C) Genomic region around transcription start site of Ryr2 showing relevant ATAC-seq and ChIP-seq signals and four potential REs. (D) Basal activity (−) and Tbx3 repressed activity of the four regions in transiently transfected HL1 (black and gray) and Cos-7 (dark and light blue) cells. Relative luciferase activity was normalized to the empty vector. (E) In vivo RE8 and RE16 activity pattern in 5 d postfertilization AttP2P zebrafish embryos. A, atrium; CM, cardiomyocytes; V, ventricle. (F) A large region including Canca1g locus showing relevant ATAC-seq and ChIP-seq signals and five potential REs. (G) Basal activity of REs and activity in the presence of several forms of stimuli included by cotransfection. (H) E11.5 RE4-5-LacZ transient transgenic embryo showing expression in the heart and specifically in the AV canal. Eight of eight transient transgenic embryos had expression in the heart. (I) Tested smaller fragments of RE4-5. (J and K) Basal activity of REs and activity in the presence of several forms of stimuli included by cotransfection. For all luciferase assays, relative luciferase activity was normalized to the empty vector. *P < 0.05 was considered significant using a two-tailed Student’s t test. **P < 0.01, ***P < 0.001. AVB, AV bundle; AVC, AV canal; IVS, interventricular septum; LA, left atrium; LBB, left bundle branch; LV, left ventricle; RA, right atrium; R-AVRB, right AV ring bundle; RBB, right bundle branch; RV, right ventricle.
Next, we intersected our AV conduction system ATAC-seq with available cardiac Tbx3 and Tbx5 ChIP-seq datasets (Tbx3 and Tbx5 share DNA binding sites), as well as the recently improved EMERGE cardiac enhancer prediction tool (40–43) to identify candidate Tbx3-dependent, AV conduction system specific REs. We chose to focus on two loci harboring genes representative of up-regulated and down-regulated transcripts in Tbx3+/− AV nodes: Ryr2 (up), and Cacna1g (down). Both genes are found in loci containing variants associated with PR-interval (44) and had straightforward epigenomic signatures.
Ryr2 encodes the Ryanodine receptor 2, which plays a role in calcium homeostasis (45) and is enriched in chamber myocardium (15). In this locus, we selected 19 regions as candidate REs based solely on chromatin accessibility in the AV conduction system cardiomyocytes and scored these regions for Tbx3 and Tbx5 occupancy derived from cardiomyocyte-specific datasets, and whether they were predicted to be a cardiac enhancer by EMERGE (41–43) (SI Appendix, Fig. S6). Four of these regions (Fig. 3C) were selected for transfection luciferase assays in HL-1, an atrial cardiomyocyte-like cell line, and Cos-7, a noncardiomyocyte cell line. HL-1 cells express Tbx5 and Ryr2, but not Tbx3, and based on transcriptome analysis, are similar to fetal right atrial cardiomyocytes (SI Appendix, Fig. S7). All regions were more active in HL-1 than in Cos-7 cells (Fig. 3D), thus implying the candidate REs are cell-type specific. Additionally, the activity of three of the REs was repressed by TBX3 (Fig. 3D). We determined the spatial activity of RE8 and RE16 in a zebrafish reporter assay and confirmed that both regions show specific activity in the atrium and ventricles of the fish heart and not in other tissues (Fig. 3E). Our data indicate that these Ryr2 REs are active in chamber myocardium and are repressed by Tbx3.
The second Tbx3 target we chose to study is Cacna1g. This gene encodes the T-type calcium channel Cav3.1, is important for calcium homeostasis, is specifically expressed throughout development and adulthood in the conduction system in an expression pattern similar to Tbx3 (SI Appendix, Fig. S8) (46), and was down-regulated in Tbx3+/− AV nodes (Fig. 2B). Furthermore, previous studies have demonstrated that Cacna1g is induced by Tbx3 (47) and knockout mice present with PR interval phenotypes (17). We selected eight chromatin regions accessible in the Tbx3+ AV junction and then scored each region for Tbx3 and Tbx5 occupancy and EMERGE cardiac enhancer prediction signal (SI Appendix, Fig. S9). A previous study showed that many AV canal-specific REs are GATA-dependent switches; synergistic activity of Gata4 and Bmp2/Smad signaling results in H3K27 acetylation and transcription of AV-specific target genes (48). Therefore, we tested the responsiveness of five of the candidate regions to Tbx3, Gata4, and/or Bmp signaling effectors using transfection luciferase assays in HEK 293 cells (Fig. 3F). We found that RE2 was responsive to Tbx3 and RE4-5 to Bmp signaling effectors (constitutively active Alk3, Smad1, Smad4, and Gata4) (Fig. 3G), whereas none of the REs were responsive to other cardiogenic transcription factors (Nkx2.5, Gata4, and Tbx5). This result suggests that RE2 and RE4-5 are specifically active in the AV conduction system (48). Next, we examined the spatial activity of RE4-5 in transient transgenic embryos and found it was active in the E11.5 AV canal (precursor of the AV conduction system) in 8/8 transgenic embryos, with 3/8 showing no activity outside the heart (Fig. 3H). However, the individual RE4 or RE5 fragments failed to drive reporter expression in the hearts of five transgenic embryos per fragment (SI Appendix, Fig. S10), indicating the function of RE4 and RE5 is interdependent. To further characterize this behavior, we tested the responsiveness of RE4-5 subfragments in luciferase assays (Fig. 3 I–K). RE5 was responsive to Tbx3-VP16 (Fig. 3J). In contrast, a fragment spanning most of RE4 was synergistically induced by Bmp signaling effectors and Gata4, but not by either of these factors alone. A 1.5-kbp fragment including the junction of RE4 and RE5 and most of RE5 could not be stimulated by any of the transfected effectors (Fig. 3K). These results confirm that RE4-5 together drive AV canal expression in vivo, and suggest the individual parts integrate different AV canal-associated transcriptional mechanisms.
Functional Analysis of a Bipartite AV Conduction System Regulatory Element In Vivo.
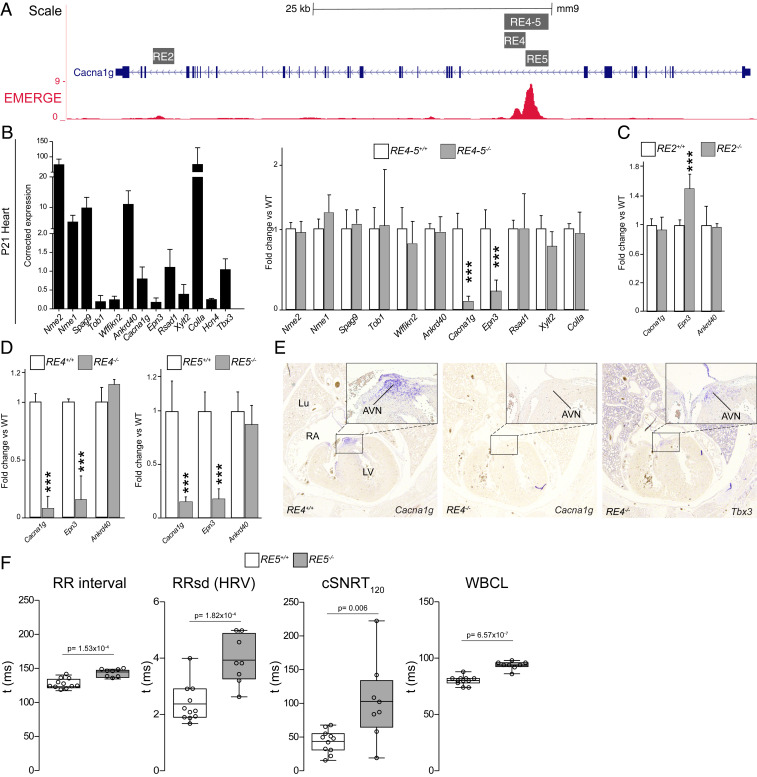
To determine if the Tbx3-dependent REs identified in the Cacan1g locus are required for expression in the AV conduction system, we deleted RE2 and RE4-5 from the mouse genome using CRISPR/Cas9 (Fig. 4A and see SI Appendix, Fig. S11A). Homozygous mice for all deletions developed and bred normally and were morphologically indistinguishable from their WT littermates. Because REs are known to regulate expression of distally located genes within a TAD (7–9), we analyzed cardiac transcript levels of genes within the same TAD as Cacna1g by qPCR (SI Appendix, Fig. S11A). Heart samples from P21 mice homozygous for a 3-kbp deletion spanning RE4-5 (RE4-5−/−) showed a 90 and 75% down-regulation of Cacna1g and Epn3 transcripts, respectively, when compared to WT littermates, while all other genes within the TAD were unaffected (Fig. 4B). In contrast, isolated brain samples showed no significant changes in transcript levels (SI Appendix, Fig. S11B), indicating that RE4-5 is tissue-specific. Deletion of RE2 resulted in modest up-regulation of Epn3 expression only, thus suggesting that this RE is dispensable for cardiac-specific Cacna1g expression (Fig. 4C).
Fig. 4.
Deletion of RE4-5, RE4, or RE5 abolishes expression of Cacna1g in the heart. (A) CRISPR/Cas9 deletions in the context of EMERGE signal. (B) Relative expression of TAD genes from P21 WT hearts (n = 7) determined by qPCR with Tbx3 and Hcn4 shown as cardiac conduction system markers (Left). Fold change expression levels of TAD genes in P21 RE4-5−/− (n = 8) vs. WT littermates (Right). (C and D) Relative expression of Cacna1g, Epn3, and Ankrd40 as a control in RE2−/− (n = 6) (C) and RE4−/− (n = 7) and RE5−/− (n = 8) (D). (E) Histological sections of WT and RE4−/− E17.5 mice stained for Cacna1g and Tbx3. (F) Boxplots visualizing the individually measured and average RR interval, RRsd, cSNRT, and WBCL. *P < 0.05 was considered significant one-way ANOVA (B–D) or Student’s t test (F) (**P < 0.01; ***P < 0.001); AVN, AV node; cSNRT, corrected sinus node recovery time; HRV, heart rate variation; Lu, lung; LV, left ventricle; RA, right atrium; RRsd, RR interval SD; WBCL, Wenckebach cycle length.
To test whether the function of RE4 and RE5 is interdependent, as suggested by the in vivo RE assays, we generated mouse lines in which RE4 and RE5 were deleted individually (Fig. 4A and see SI Appendix, Fig. S11A). Homozygous removal of either genomic region resulted in down-regulation of Cacna1g specifically in the heart to levels comparable to those observed in hearts of RE4-5−/− mice (Fig. 4D). To confirm this finding, we analyzed the expression pattern of Cacna1g by in situ hybridization. Sections of E17.5 RE4−/− or RE5−/− mice showed no staining for Cacna1g transcripts in the heart, whereas expression of Tbx3 was unchanged (Fig. 4E; not shown for RE5−/−). RE5−/− mice showed bradycardia (Fig. 4F), similar to Cacna1g homozygous mutants (17), further demonstrating functional loss of Cacna1g in the conduction system of RE5−/− mutants. Additionally, RE5−/− mice showed higher heart rate variation (HRV) when compared to WT littermates (Fig. 4F). We observed slower corrected sinus node recovery times (cSNRTs) and higher Wenckebach cycle length in RE5−/− mice, consistent with sinus and AV node dysfunction (Fig. 4F). Together, our data show that RE4-5 functions as a bipartite, AV conduction system-specific RE that is essential to drive expression of Cacna1g.
Discussion
We demonstrate that heterozygous loss of Tbx3 shortens AV nodal conduction (PR interval) and prolongs QRS duration. The haploinsufficiency causes modest changes in expression of genes associated with AV nodal identity and function. Additionally, Tbx3 heterozygous mice fail to maintain AV bundle-BBs structure after birth. We show that REs in the Ryr2 locus are active in the chambers and are repressed by Tbx3. Further, we found a bipartite RE in the Cacna1g locus that is active in the Tbx3+ AV conduction system where it selectively controls expression of Cacna1g and Epn3 in vivo and none of the >15 other genes in the TAD.
Genome-wide association studies have identified common noncoding variants associated with PR interval and QRS duration in a 1-Mbp TAD that only harbors TBX3 (2–6, 10, 12) (SI Appendix, Fig. S1); we infer that these variants affect the function of REs controlling TBX3 expression. Currently, this assumption is difficult to validate; expression quantitative trait loci analysis is not yet feasible due to the low number of human AV conduction system samples available, extremely low expression in other cardiac cells, and the unavailability of reliable human AV conduction system cell models.
Tbx3 mutants showed prolonged QRS, which may result from the hypoplastic AV bundle and BBs (49). Apoptosis does not seem to underlie the AV bundle-BBs hypoplasia, and less proliferation after birth is unlikely to be the culprit, because the AV bundle shows very limited proliferation from early fetal stages onwards (49–51), and cardiomyocyte proliferation rates, in general, decline to negligible levels shortly after birth (52–54).
Using a series of Tbx3 hypomorphic and deficient alleles, Frank et al. found that Tbx3 is required for the function of the conduction system in a dosage-sensitive manner (55). Mice expressing 30% or less of WT transcript levels developed severe arrhythmias, and surviving adults displayed PR shortening. Tbx3+/− mutants in our study show PR interval shortening in addition to AV bundle-BBs hypoplasia and prolonged QRS duration. We speculate that the PR interval shortening in Tbx3 haploinsufficient mice results from AV bundle hypoplasia in addition to the modest deregulation of many genes implicated in the electrophysiological properties of AV nodal cells (e.g., calcium channels/handling proteins, Hcn channels, gap junction subunits, sodium channels, potassium channels). Additional insight into the quantitative changes and functional interactions between the dysregulated targets is required to understand the observed net effect.
Tbx3 and its paralog and functional homolog Tbx2 have been implicated in accessory AV pathway formation (28, 55–57), which could result in conduction abnormalities consistent with the phenotypes found in Tbx3+/− mutants. However, we found no delta waves in ECG traces, and optical mapping showed no early breakthrough at the base of the left ventricles in controls and mutants (Fig. 1). Therefore, it is unlikely that the presence of an accessory pathway underlies the observed electrophysiological phenotypes in Tbx3+/− mutants. Instead, AV bundle-BBs hypoplasia is more likely to explain the conduction changes and is in line with the observed in Tbx3+/− mutants. The shorter PR interval in Tbx3+/− mutants was only present ex vivo, and neither in vivo nor with chemical ANS blockade. The autonomic nervous system has a large impact on AV conduction (58). We consider it possible that in Tbx3+/− mutants, the autonomic nervous system activity partially compensated for the modest defects caused by heterozygous loss of Tbx3, explaining the absence of PR interval changes in vivo. Therefore, we suggest that intrinsic AV nodal conduction is affected in Tbx3 mutants.
Many transcription factors in addition to Tbx3 have been implicated in the development and function of the AV conduction system, including Nkx2-5, Tbx5, Prox1, Hopx, Irx3, and Gata4/6 (reviewed in refs. 1, 13, and 14). Several of these factors target the same genes and interact with each other, both physically and genetically, thus substantiating that they form a transcriptional network in the AV conduction system. Tbx3 and Tbx5 are coexpressed in the entire AV conduction system, and both function in highly dose-sensitive manners (33, 49, 55, 58–62). Tbx3 likely competes with Tbx5 for DNA binding sites (41). Tbx5 stimulates a fast-conduction gene program (e.g., Scn5a and Gja5) in the AV bundle (61), whereas Tbx3 suppresses it, while stimulating the nodal (pacemaker-specific) program (e.g., Hcn4 and Cacna1g) (47, 63, 64), suggesting a finely tuned balance between these factors (51).
Unexpectedly, Tbx5 was down-regulated in Tbx3 haploinsufficient AV nodes. Whether Tbx5 directly regulates the expression of Tbx3 in the AV node, as it does in the sinus node, is unknown (60). Our data show that the expression of other transcription factors in the AV node is largely unaffected by Tbx3 haploinsufficiency. This suggests that while Tbx3 reduction causes deregulation of genes directly involved in cellular properties, such as ion channels, it has a minimal impact on the transcription factor networks in this tissue.
We mapped the accessible chromatin in fetal AV canals, which contain the embryonic AV nodal cells, and integrated these data with Tbx5 and Tbx3 occupancy profiles and EMERGE cardiac enhancer prediction signal to identify candidate Tbx3/5-regulated REs on a genome-wide scale. We then selected two example loci, Ryr2 and Cacna1g, for further examination. Ryr2 expression is chamber-enriched and induced in the AV node of Tbx3+/− mice, whereas Cacna1g is conduction system-specific and reduced in Tbx3+/− mutants, and both loci contain PR interval-associated variants (44). Two REs in close proximity to the Ryr2 promoter were active in the fish cardiac chambers and were active in an atrial-like cell line where their activity was repressed by Tbx3. The repressor function of Tbx3 is well described (65). This suggests that Tbx3 may directly control expression of Ryr2 and other chamber-enriched genes in the AV conduction system through these classes of REs.
Cacna1g is expressed specifically in the AV conduction system in a pattern resembling that of Tbx3, and it is important for AV node cell automaticity (17). Previously, Cacna1g expression was found to be induced by Tbx3 (47, 64) and was found to be down-regulated in Tbx3+/− AV nodes in this study. Congruently, we identified an intronic bipartite Cacna1g RE accessible in the Tbx3+ AV junction, occupied by Tbx5, Tbx3, Nkx2-5, and Gata4, and acetylated H3K27, a mark associated with active REs (SI Appendix, Fig. S9). The bipartite RE and both its individual components were required for cardiac expression of Cacna1g in vivo, whereas the entire RE but not its components was sufficient to drive AV conduction system expression in the mouse embryo. Furthermore, while REs have been observed to act redundantly in vivo (8, 66), this RE seems to integrate all of the regulatory information required to drive Cacna1g in the entire conduction system. The slower heart rates observed in RE5 mutants can be attributed to loss of Cacna1g in the sinus node—also demonstrated by slower cSNRTs (Fig. 4F) and studies of a Cacna1g knockout model (17). Nevertheless, our data are unable to correlate the observed heart rate variation in mutants solely to sinus or AV node dysfunction, and we speculate the phenotype could be a consequence of deficiencies in both compartments of the cardiac conduction system.
Finally, the activity of REs is usually limited to the TAD they share with potential target genes, in which they can act on multiple target promoters (7–9). Our study reveals that in the context of the TAD, which contains >15 genes, the RE selectively and dominantly regulates Cacna1g in vivo. The expression of only one distal gene, Epn3, was affected by the deletion of the REs. Epn3 (Epsin 3) is expressed at very low levels in cardiac tissue and its function has not been associated with the heart. In contrast, RE2, also in the Cacna1g gene, was dispensable for Cacna1g expression, but was involved in regulation of Epn3, further illustrating the complexity of the regulatory landscape.
How Tbx3/5-occupied AV conduction REs such as the one we identified in Cacna1g are stimulated by Tbx3 remains unclear. Previously, AV canal REs were found to function as switches activated by Gata4 and Bmp signaling effectors and histone H3K27 acetylation activity in the AV canal and repressed by Gata4, Hey1/2, and HDACs in the chambers (48). In vitro, one part of the bipartite RE was stimulated by Gata4 and Bmp signaling effectors, suggesting parts of this RE function similarly. The other part of the composite RE was responsive to Tbx3 and occupied by Tbx3 and Tbx5 based on cardiac ChIP-seq data, suggesting it mediates the response to the Tbx3/5 ratio in the AV conduction system. Together, these data suggest that while each part of the RE responds to a different AV conduction system-relevant regulatory signaling system, these signals need to be combined and integrated for the bipartite enhancer to function in vivo.
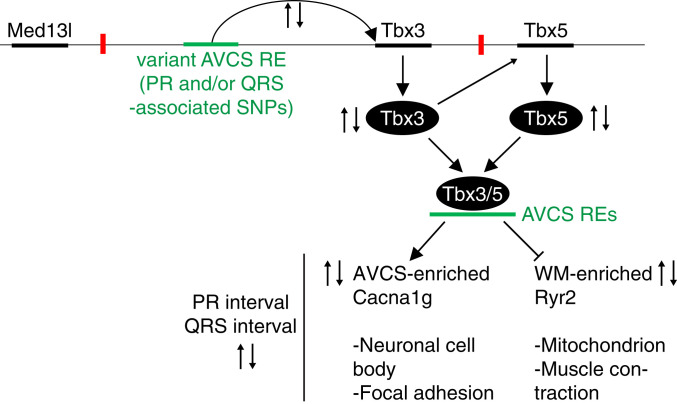
We conclude that Tbx3 dose-dependently governs a transcriptional program that provides an AV conduction system-typical identity. Additionally, our study contributes a possible explanation how individuals carrying common noncoding variants in the TBX3 locus have an increased risk for differences in the duration of PR and QRS intervals (Fig. 5).
Fig. 5.
A model illustrating how variants in REs that affect Tbx3 expression in the AV conduction system may regulate PR and QRS duration.
Materials and Methods
For detailed methods, see SI Appendix.
Cryosectioning and Laser Capture Microdissection.
Hearts from Tbx3+/+;BAC-Tbx3-Egfp and Tbx3+/CreERT2;BAC-Tbx3-Egfp ND8-10 pups were removed, the AV node region microdissected (based on fluorescence) and immediately embedded in O.C.T. (Tissue-Tek, 621232) in the −20 °C for 10 min. Cryosectioning was performed with a cryostat (Leica Biosystems, CM1950), and AV node tissue was sectioned at a thickness of 8 µm. Five consecutive sections were collected and mounted onto PEN-membrane coated slides (MembraneSlide 1.0 PEN, Carl Zeiss Microscopy, 415190-9041-001). Microdissection was performed immediately afterward with a Laser Microdissection Microscope (Leica Microsystems, LMD6), and the AV node was identified visually based on fluorescence. Laser power, aperture, and speed of the laser varied from experiment to experiment. Microdissected AV node tissue sections were collected in a tube containing the lysis buffer of the PicoPure RNA isolation kit (Thermo Fisher Scientific, KIT0204) and the first step of the manufacturer’s protocol conducted before the samples were stored at −80 °C.
RNA Isolation and Sequencing of Laser Capture Microdissected AV Node Sections.
Total RNA was isolated from neonatal day 8–10 AV node sections according to the manufacturer’s protocol of the PicoPure RNA isolation kit (Thermo Fisher Scientific, KIT0204) with an on-column DNase treatment (RNase-free DNase, Qiagen, 79254). RNA integrity and concentration were determined using the Bioanalyzer RNA 6000 Pico materials (Agilent Technologies, 5067-1513 and 5067-1535). The Ovation RNA-seq system V2 (Nugen, 7102-32) was used for cDNA synthesis with a RNA input between ∼300 and 2,000 pg. The quality of the cDNA library was determined with a Bioanalyzer DNA 7500 kit (Agilent Technologies, 5067-1506), the purity with the Nanodrop (Isogen Life Science, ND-1000) and the quantity with the Qubit dsDNA HS kit (Thermo Fisher Scientific, Q32854). RNA-seq libraries were made using Ovation Ultralow Library System V2 (Nugen, 0344-32) and sequenced on the HiSeq 4000 (Illumina). The raw data have been deposited with the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) under accession code GSE121465.
Assay for Transposase-Accessible Chromatin Sequencing.
Tbx3+/Venus hearts were dissected from embryonic day (E) 12.5–E14.5 embryos and enriched for AV conduction system tissues by microdissection. Tissue samples were dissociated into a single-cell suspension using 0.05% Trypsin/EDTA (Thermo Fisher Scientific, 25300-054) for 15 min at 37 °C. Dissociated cells were resuspended in DMEM (Thermo Fisher Scientific, 31966-021) supplemented with 10% fetal bovine serum (ThermoFisher Scientific, 10270-106) and FACS was performed. Samples were sorted on a FacsAria flow cytometer (BD Biosciences) and gated to exclude debris and cell clumps and sorted for Venus+ cells. Approximately 75,000 Venus+ cells were collected and used as input for ATAC-seq. Nucleus preparation, transposition reaction, and library amplification were performed as previously described (35). In short, cells were washed in phosphate buffered saline and lysed in cold lysis buffer (10 mM Tris·Cl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1% [vol/vol] Igepal CA-630). Transposition reaction was performed as described using the Nextera DNA Library Prep kit (Illumina, FC-121-1030) for 30 min, followed by cleanup using the MinElute PCR Purification Kit (Qiagen, 28004). Transposed DNA fragments were amplified using NEBNext High-Fidelity 2× PCR Master Mix (New England Biolabs, M0541) with custom barcoded PCR primers (sequences as described in ref. 35). The amplified library was purified using the MinElute PCR Purification Kit (Qiagen, 28004). The library was sequenced (paired-end 125 bp) and data collected on a HiSeq 2500.
Supplementary Material
Acknowledgments
We thank Jaco Hagoort for his assistance with the AMIRA 3D reconstructions, Dr. Vasanth Vedantham for advice on the laser capture microdissection protocol, and Joyce Man for help with figures. This work was supported by Fondation Leducq (Grant 14CVD01, to V.M.C.), and by the Dutch Heart Foundation (Grant 2016T047 to B.J.B. and Cardiovasculair Onderzoek Nederland Project 2014-18 CONCOR-genes, to J.B. and V.M.C.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The data have been deposited with the Gene Expression Omnibus, https://www.ncbi.nlm.nih.gov/geo/ (accession code GSE121465).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1919379117/-/DCSupplemental.
References
- 1.van Eif V. W. W., Devalla H. D., Boink G. J. J., Christoffels V. M., Transcriptional regulation of the cardiac conduction system. Nat. Rev. Cardiol. 15, 617–630 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Pfeufer A. et al., Genome-wide association study of PR interval. Nat. Genet. 42, 153–159 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sotoodehnia N. et al., Common variants in 22 loci are associated with QRS duration and cardiac ventricular conduction. Nat. Genet. 42, 1068–1076 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verweij N. et al., Genetic determinants of P wave duration and PR segment. Circ. Cardiovasc. Genet. 7, 475–481 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van der Harst P. et al., 52 genetic loci influencing Myocardial Mass. J. Am. Coll. Cardiol. 68, 1435–1448 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Setten J. et al., PR interval genome-wide association meta-analysis identifies 50 loci associated with atrial and atrioventricular electrical activity. Nat. Commun. 9, 2904 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dixon J. R. et al., Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Long H. K., Prescott S. L., Wysocka J., Ever-changing landscapes: Transcriptional enhancers in development and evolution. Cell 167, 1170–1187 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nora E. P. et al., Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381–385 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rao S. S. et al., A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krijger P. H., de Laat W., Regulation of disease-associated gene expression in the 3D genome. Nat. Rev. Mol. Cell Biol. 17, 771–782 (2016). [DOI] [PubMed] [Google Scholar]
- 12.van Weerd J. H. et al., A large permissive regulatory domain exclusively controls Tbx3 expression in the cardiac conduction system. Circ. Res. 115, 432–441 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Munshi N. V., Gene regulatory networks in cardiac conduction system development. Circ. Res. 110, 1525–1537 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park D. S., Fishman G. I., Development and function of the cardiac conduction system in health and disease. J. Cardiovasc. Dev. Dis. 4, 7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greener I. D. et al., Molecular architecture of the human specialised atrioventricular conduction axis. J. Mol. Cell. Cardiol. 50, 642–651 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Aanhaanen W. T. et al., Developmental origin, growth, and three-dimensional architecture of the atrioventricular conduction axis of the mouse heart. Circ. Res. 107, 728–736 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Mangoni M. E. et al., Bradycardia and slowing of the atrioventricular conduction in mice lacking CaV3.1/alpha1G T-type calcium channels. Circ. Res. 98, 1422–1430 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Liang X. et al., HCN4 dynamically marks the first heart field and conduction system precursors. Circ. Res. 113, 399–407 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu M., Peng S., Zhao Y., Inducible gene deletion in the entire cardiac conduction system using Hcn4-CreERT2 BAC transgenic mice. Genesis 52, 134–140 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miquerol L. et al., Architectural and functional asymmetry of the His-Purkinje system of the murine heart. Cardiovasc. Res. 63, 77–86 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Mohan R. A. et al., Embryonic Tbx3+ cardiomyocytes form the mature cardiac conduction system by progressive fate restriction. Development 145, 1–12 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Horsthuis T. et al., Gene expression profiling of the forming atrioventricular node using a novel tbx3-based node-specific transgenic reporter. Circ. Res. 105, 61–69 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Goodyer W. R. et al., Transcriptomic profiling of the developing cardiac conduction system at single-cell resolution. Circ. Res. 125, 379–397 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marionneau C. et al., Specific pattern of ionic channel gene expression associated with pacemaker activity in the mouse heart. J. Physiol. 562, 223–234 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miquerol L. et al., Resolving cell lineage contributions to the ventricular conduction system with a Cx40-GFP allele: A dual contribution of the first and second heart fields. Dev. Dyn. 242, 665–677 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Rentschler S. et al., Notch signaling regulates murine atrioventricular conduction and the formation of accessory pathways. J. Clin. Invest. 121, 525–533 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verhoeven M. C., Haase C., Christoffels V. M., Weidinger G., Bakkers J., Wnt signaling regulates atrioventricular canal formation upstream of BMP and Tbx2. Birth Defects Res. A Clin. Mol. Teratol. 91, 435–440 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Gillers B. S. et al., Canonical wnt signaling regulates atrioventricular junction programming and electrophysiological properties. Circ. Res. 116, 398–406 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Virágh S., Challice C. E., The development of the conduction system in the mouse embryo heart. Dev. Biol. 89, 25–40 (1982). [DOI] [PubMed] [Google Scholar]
- 30.Canale E. D., Campbell G. R., Smolich J. J., Campbell J. H., Cardiac muscle, (Springer Verlag, Berlin, 1986). [Google Scholar]
- 31.Dehaan R. L., Differentiation of the atrioventricular conducting system of the heart. Circulation 24, 458–470 (1961). [DOI] [PubMed] [Google Scholar]
- 32.Lambert S. A. et al., The human transcription factors. Cell 172, 650–665 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Moskowitz I. P. G. et al., The T-Box transcription factor Tbx5 is required for the patterning and maturation of the murine cardiac conduction system. Development 131, 4107–4116 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Risebro C. A. et al., Prox1 maintains muscle structure and growth in the developing heart. Development 136, 495–505 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buenrostro J. D., Giresi P. G., Zaba L. C., Chang H. Y., Greenleaf W. J., Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Preissl S. et al., Deciphering the epigenetic code of cardiac myocyte transcription. Circ. Res. 117, 413–423 (2015). [DOI] [PubMed] [Google Scholar]
- 37.van Duijvenboden K. et al., Conserved NPPB+ border zone switches from MEF2- to AP-1-driven gene program. Circulation 140, 864–879 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Singh R. et al., Tbx20 interacts with smads to confine tbx2 expression to the atrioventricular canal. Circ. Res. 105, 442–452 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Munshi N. V. et al., Cx30.2 enhancer analysis identifies Gata4 as a novel regulator of atrioventricular delay. Development 136, 2665–2674 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Ouwerkerk A. F. et al., Identification of atrial fibrillation associated genes and functional non-coding variants. Nat. Commun. 10, 4755 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van den Boogaard M. et al., Genetic variation in T-box binding element functionally affects SCN5A/SCN10A enhancer. J. Clin. Invest. 122, 2519–2530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luna-Zurita L. et al., Complex interdependence regulates heterotypic transcription factor distribution and coordinates Cardiogenesis. Cell 164, 999–1014 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Duijvenboden K., de Boer B. A., Capon N., Ruijter J. M., Christoffels V. M., EMERGE: A flexible modelling framework to predict genomic regulatory elements from genomic signatures. Nucleic Acids Res. 44, e42 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cartwright J. H., et al. , Multi-ancestry GWAS of the electrocardiographic PR interval identifies 210 loci underlying cardiac conduction. bioRxiv:10.1101/712398 (24 July 2019), p 53. [DOI] [PMC free article] [PubMed]
- 45.Landstrom A. P., Dobrev D., Wehrens X. H. T., Calcium signaling and cardiac arrhythmias. Circ. Res. 120, 1969–1993 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chandler N. J. et al., Molecular architecture of the human sinus node: Insights into the function of the cardiac pacemaker. Circulation 119, 1562–1575 (2009). [DOI] [PubMed] [Google Scholar]
- 47.Hoogaars W. M. et al., Tbx3 controls the sinoatrial node gene program and imposes pacemaker function on the atria. Genes Dev. 21, 1098–1112 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stefanovic S. et al., GATA-dependent regulatory switches establish atrioventricular canal specificity during heart development. Nat. Commun. 5, 3680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moskowitz I. P. et al., A molecular pathway including Id2, Tbx5, and Nkx2-5 required for cardiac conduction system development. Cell 129, 1365–1376 (2007). [DOI] [PubMed] [Google Scholar]
- 50.Cheng G. et al., Development of the cardiac conduction system involves recruitment within a multipotent cardiomyogenic lineage. Development 126, 5041–5049 (1999). [DOI] [PubMed] [Google Scholar]
- 51.Bakker M. L. et al., Transcription factor Tbx3 is required for the specification of the atrioventricular conduction system. Circ. Res. 102, 1340–1349 (2008). [DOI] [PubMed] [Google Scholar]
- 52.Soonpaa M. H., Kim K. K., Pajak L., Franklin M., Field L. J., Cardiomyocyte DNA synthesis and binucleation during murine development. Am. J. Physiol. 271, H2183–H2189 (1996). [DOI] [PubMed] [Google Scholar]
- 53.Alkass K. et al., No evidence for cardiomyocyte number expansion in preadolescent mice. Cell 163, 1026–1036 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Soonpaa M. H. et al., Cardiomyocyte cell-cycle activity during preadolescence. Cell 163, 781–782 (2015). [DOI] [PubMed] [Google Scholar]
- 55.Frank D. U. et al., Lethal arrhythmias in Tbx3-deficient mice reveal extreme dosage sensitivity of cardiac conduction system function and homeostasis. Proc. Natl. Acad. Sci. U.S.A. 109, E154–E163 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aanhaanen W. T. et al., Defective Tbx2-dependent patterning of the atrioventricular canal myocardium causes accessory pathway formation in mice. J. Clin. Invest. 121, 534–544 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nguyen H. H., Grange D. K., Johnson M. C., Van Hare G. F., Jay P. Y., Dichotomous roles of TBX3 in the establishment of atrioventricular conduction pathways in the human heart. HeartRhythm Case Rep. 5, 109–111 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fabritz L. et al., Autonomic modulation and antiarrhythmic therapy in a model of long QT syndrome type 3. Cardiovasc. Res. 87, 60–72 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hoogaars W. M. et al., The transcriptional repressor Tbx3 delineates the developing central conduction system of the heart. Cardiovasc. Res. 62, 489–499 (2004). [DOI] [PubMed] [Google Scholar]
- 60.Mori A. D. et al., Tbx5-dependent rheostatic control of cardiac gene expression and morphogenesis. Dev. Biol. 297, 566–586 (2006). [DOI] [PubMed] [Google Scholar]
- 61.Arnolds D. E. et al., TBX5 drives Scn5a expression to regulate cardiac conduction system function. J. Clin. Invest. 122, 2509–2518 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bamshad M. et al., Mutations in human TBX3 alter limb, apocrine and genital development in ulnar-mammary syndrome. Nat. Genet. 16, 311–315 (1997). [DOI] [PubMed] [Google Scholar]
- 63.Burnicka-Turek O. et al., Transcriptional patterning of the ventricular cardiac conduction system. Circ. Res., 10.1161/CIRCRESAHA.118.314460 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Singh R. et al., Tbx2 and Tbx3 induce atrioventricular myocardial development and endocardial cushion formation. Cell. Mol. Life Sci. 69, 1377–1389 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.He Ml., Wen L., Campbell C. E., Wu J. Y., Rao Y., Transcription repression by Xenopus ET and its human ortholog TBX3, a gene involved in ulnar-mammary syndrome. Proc. Natl. Acad. Sci. U.S.A. 96, 10212–10217 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Osterwalder M. et al., Enhancer redundancy provides phenotypic robustness in mammalian development. Nature 554, 239–243 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.