Significance
Feedback control of cholesterol metabolism is essential for cell viability and prevention of heart attacks. Cells acquire cholesterol from receptor-mediated uptake of low-density lipoprotein, which delivers cholesterol to lysosomes. To exert feedback control, cholesterol must reach the endoplasmic reticulum (ER). Here we use a CRISPR screen to show that lysosome-derived cholesterol moves first to the plasma membrane and then to the ER. The last movement requires an enzyme that produces phosphatidylserine. This demonstrates that transmembrane movement of one lipid (cholesterol) requires another lipid (phosphatidylserine). Our results explain how one organelle (ER) monitors the cholesterol content of another organelle (plasma membrane), thereby maintaining membrane integrity and ensuring cell survival.
Keywords: cholesterol, plasma membrane, CRISPR screen, PTDSS1, phosphatidylserine
Abstract
Animal cells acquire cholesterol from receptor-mediated uptake of low-density lipoprotein (LDL), which releases cholesterol in lysosomes. The cholesterol moves to the endoplasmic reticulum (ER), where it inhibits production of LDL receptors, completing a feedback loop. Here we performed a CRISPR-Cas9 screen in human SV589 cells for genes required for LDL-derived cholesterol to reach the ER. We identified the gene encoding PTDSS1, an enzyme that synthesizes phosphatidylserine (PS), a phospholipid constituent of the inner layer of the plasma membrane (PM). In PTDSS1-deficient cells where PS is low, LDL cholesterol leaves lysosomes but fails to reach the ER, instead accumulating in the PM. The addition of PS restores cholesterol transport to the ER. We conclude that LDL cholesterol normally moves from lysosomes to the PM. When the PM cholesterol exceeds a threshold, excess cholesterol moves to the ER in a process requiring PS. In the ER, excess cholesterol acts to reduce cholesterol uptake, preventing toxic cholesterol accumulation. These studies reveal that one lipid—PS—controls the movement of another lipid—cholesterol—between cell membranes. We relate these findings to recent evidence indicating that PM-to-ER cholesterol transport is mediated by GRAMD1/Aster proteins that bind PS and cholesterol.
Animal cells maintain cholesterol homeostasis by transporting cholesterol from one membrane to another. Cholesterol derived from low-density lipoprotein (LDL) is taken into cells through endocytosis mediated by LDL receptors (LDLRs) (1). The LDL-derived cholesterol is released in lysosomes and then transported to the plasma membrane (PM), where it plays a structural role, and to the endoplasmic reticulum (ER) membrane, where it serves two functions: it turns off the transcriptional program for cholesterol synthesis and uptake by blocking activation of sterol regulatory element-binding proteins (SREBPs) (2) and is esterified with a fatty acid for storage as cholesteryl esters in lipid droplets (3, 4).
The path that cholesterol takes from lysosomes to the ER remains elusive, but two recent developments have begun to provide some insight. First, Niemann–Pick C1 (NPC1) and Niemann–Pick C2 (NPC2), two lysosomal proteins, have been shown to act in tandem to export cholesterol from lysosomes (5, 6). Second, studies with cholesterol-binding toxins have provided evidence that LDL-derived cholesterol travels from lysosomes to the PM before moving to the ER (7, 8). Other studies have provided evidence indicating that cholesterol moves directly from lysosomes to the ER and other membranes (9–12).
To clarify this issue and to identify proteins required for lysosome-to-ER cholesterol transport, in the present study we used a CRISPR (clustered regularly-interspaced short palindromic repeats) library and CRISPR-associated Cas9 to screen human cells for genes required for cholesterol trafficking from lysosome to ER. This screen revealed a requirement for PTDSS1, which encodes an enzyme that produces phosphatidylserine (PS) (13–15). In cells lacking PTDSS1, cellular PS levels fall, and LDL-derived cholesterol is sequestered in PMs and fails to accumulate in the ER. As a result, SREBPs are not inhibited, and cholesteryl ester synthesis is decreased. These results reveal a specific phospholipid requirement for cholesterol transport from the PM to the ER, and they advance our understanding of cholesterol traffic among lysosomes, the ER, and the PM.
Results
Strategy for CRISPR-Cas9 Screen for Genes Required for Transport of LDL-Derived Cholesterol.
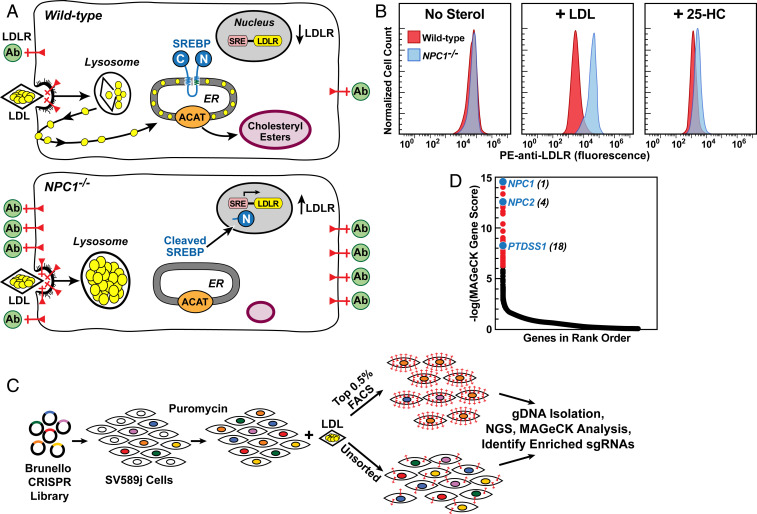
Our CRISPR-Cas9 screen was designed to identify genes whose products are required for cholesterol transport from lysosomes to the ER. When cells are incubated with LDL, cholesterol is released from lysosomes and transported to the ER, where it blocks the proteolytic processing of SREBPs, which are required for transcription of the LDLR gene. As a result, the number of LDLRs is low (Fig. 1 A, Upper). In cells with mutations in NPC1, LDL-derived cholesterol is retained in lysosomes, SREBPs remain active, and the number of LDLRs is high (Fig. 1 A, Lower). To screen for other genes required for the transport of LDL-derived cholesterol to the ER, we incubated human SV589 cells in cholesterol-depleting medium devoid of LDL and containing the HMG-CoA reductase inhibitor compactin to activate SREBPs and induce the production of LDLRs. We then incubated the cells with LDL for 24 h. After this incubation, we washed the cells and incubated them with a monoclonal antibody to LDLRs tagged with fluorescent phycoerythrin (PE-anti-LDLR). The cells were then sorted by fluorescence-activated cell sorting (FACS). Cells with blocks in cholesterol transport are predicted to have high fluorescence compared with wild-type (WT) cells.
Fig. 1.
Strategy for CRISPR-Cas9 screen for genes required for transport of LDL-derived cholesterol. (A) Overall strategy. In WT cells (Upper), LDL delivers cholesterol to lysosomes, and NPC2 and NPC1 transport cholesterol out of lysosomes. Cholesterol reaches the ER, where it blocks SREBP cleavage, leading to a reduction in LDLRs (red) and decreased binding of fluorescent anti-LDLR antibodies (green). Excess ER cholesterol is esterified by ACAT and stored in cholesteryl ester droplets. In NPC−/− cells (Lower), LDL-derived cholesterol is trapped in lysosomes, SREBP is cleaved, LDLRs increase and anti-LDLR binding is high. Cells fail to synthesize cholesteryl esters. (B) Measurement of LDLRs by flow cytometry in WT and NPC1−/− SV589 cells incubated with LDL or 25-hydroxycholesterol (25-HC). On day 0, cells were set up in medium A with 5% FCS. On day 1, cells were switched to cholesterol-depletion medium A. After 16 h, cells were refed with the same cholesterol-depletion medium in the absence (Left) or presence of 50 µg protein/mL human LDL (Middle) or 0.3 µg/mL 25-HC (Right). After 24 h, cells were harvested, incubated with PE-anti-LDLR, and assessed by flow cytometry (SI Appendix, Materials and Methods). (C) CRISPR-Cas9 screen for genes required for transport of LDL-derived cholesterol to ER. SV589j cells were infected with the Brunello CRISPR Knockout Library in lentiCRISPRv2 at a low multiplicity of infection and subjected to puromycin selection for approximately 10 d. Surviving cells were cultured in the presence of LDL, incubated with PE-anti-LDLR, and subjected to FACS (SI Appendix, Materials and Methods). Genomic DNA was isolated from sorted cells expressing the most LDLRs (top 0.5% of cells) and from ∼40 × 106 unsorted cells. DNA was subjected to next-generation sequencing), and the data were analyzed by MAGeCK to identify sgRNAs overrepresented in the top 0.5% of cells (SI Appendix, Materials and Methods and Fig. S1). (D) Ranking of genes (19,114 total) in sorted cells expressing the most LDLRs compared with unsorted cells. The y-axis denotes negative log of the robust rank aggregation score as calculated by MAGeCK (Dataset S1). NPC1 was the gene with the highest rank (i.e., greatest enrichment).
To validate our screen, we used flow cytometry to measure the binding of PE-anti-LDLR to WT SV589 cells and to cells lacking NPC1 (Fig. 1B). When the cells were preincubated in the absence of sterols, WT and NPC1−/− cells bound the same amount of PE-anti-LDLR as indicated by the peak fluorescence (Fig. 1 B, Left). When the cells were preincubated with LDL, the reduction in LDLRs was much more significant in WT cells than in NPC1−/− cells. As a result, the NPC1-/− cells bound approximately 10-fold more PE-anti-LDLR than the WT cells (Fig. 1 B, Middle). When the cells were preincubated with 25-hydroxycholesterol, which inhibits SREBP processing without traversing the lysosome (16), the LDLR reduction was similar in WT and NPC1−/− cells (Fig. 1 B, Right).
To identify additional genes required for transport of LDL-derived cholesterol to the ER, we used the Brunello genome-wide lentiviral CRISPR library, which contains four single guide RNAs (sgRNAs) directed against each of 19,114 human genes plus cassettes for Cas9 and puromycin resistance (17). The lentiviral library was used to infect human SV589j cells, a clonal line of SV589 cells selected by LDLR flow cytometry. After growth in puromycin for 10 d (Fig. 1C), the surviving SV589j cells were depleted of cholesterol and incubated with LDL to suppress LDLRs, followed by incubation with PE-anti-LDLR and FACS (SI Appendix, Materials and Methods). Cells that bound the most PE-anti-LDLR (brightest 0.5% of cells) were collected by FACS (SI Appendix, Fig. S1). The sgRNAs from the top 0.5% of cells and an unsorted population of cells were sequenced. sgRNAs enriched in the top 0.5% were identified by MAGeCK (model-based analysis of genome-wide CRISPR/Cas9 knockout) (18). Fig. 1D shows the spectrum of MAGeCK scores for genes with identified sgRNAs. The genes with the highest MAGeCK scores are the ones whose sgRNAs were most highly enriched in the cells in the top 0.5% of PE-anti-LDLR binding. NPC1 was the gene whose sgRNAs were most enriched in the top 0.5% of cells, and NPC2 was in fourth place. The complete list of scores for all 19,114 genes is shown in Dataset S1.
PTDSS1 and NPC1 Are Required for Transport of LDL-Derived Cholesterol to ER.
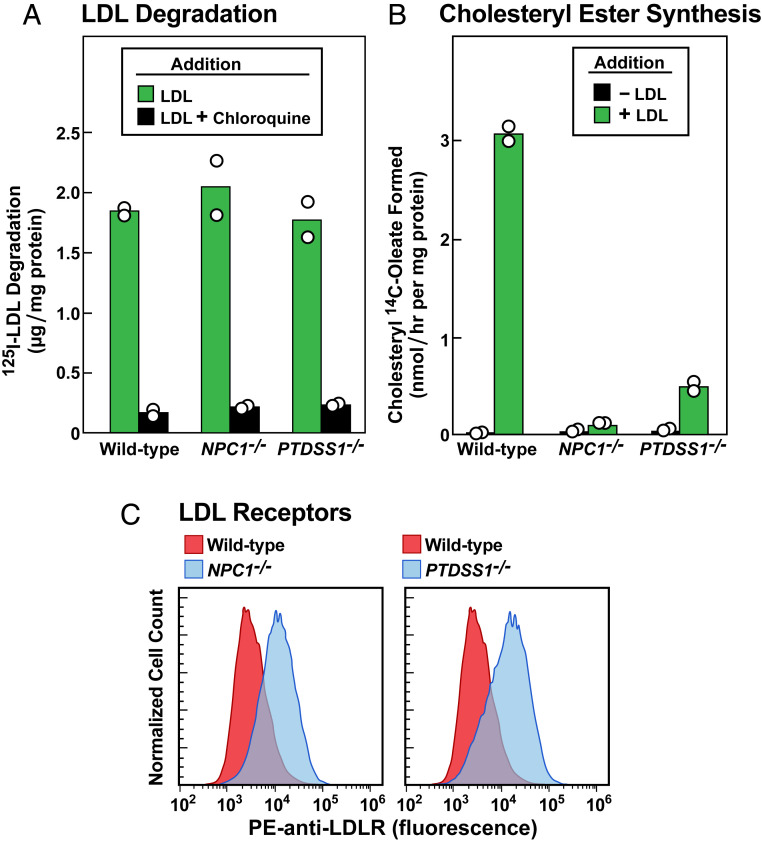
As shown in Fig. 1D, PTDSS1 was among the genes that scored highest in our CRISPR screen. PTDSS1 encodes an enzyme that exchanges serine for choline in phosphatidylcholine (PC), thereby synthesizing PS (13). Inasmuch as PS is a component of cholesterol-containing cell membranes (14), we chose to do further studies of PTDSS1-deficient cells. To create PTDSS1-deficient SV589j cells, we coinfected two CRISPR-Cas9 lentiviruses, each encoding one of the four sgRNAs that targeted PTDSS1 in the original screen. For comparison, we used the same method to generate SV589j cells lacking NPC1 (SI Appendix, Materials and Methods). To confirm that the PTDSS1−/− cells had normal LDLR function, we depleted the cells of cholesterol to induce LDLRs, incubated them with 125I-LDL, and measured the amount of 125I-monoiodotyrosine released into the culture medium (Fig. 2A). We also studied WT and NPC1−/− cells in the same experiment. All three cell lines took up and degraded similar amounts of 125I-LDL. Degradation was blocked by chloroquine, confirming that it occurred in lysosomes (19).
Fig. 2.
PTDSS1 and NPC1 are required for transport of LDL-derived cholesterol to the ER. (A and B) LDL degradation and LDL-mediated stimulation of cholesteryl ester synthesis in WT, NPC1−/−, and PTDSS1−/− SV589j cells. On day 0, lentiviral-generated KO cells were set up in medium A with 10% FCS. On day 2, cells were switched to cholesterol-depletion medium A (SI Appendix, Materials and Methods). (A) 125I-LDL degradation. On day 3, one group of cells were refed with cholesterol-depletion medium A containing 10 µg protein/mL 125I-LDL (33 cpm/ng protein) in the absence or presence of 100 μM chloroquine. After 5 h, the amount of 125I-monoiodotyrosine in the medium was measured (SI Appendix, Materials and Methods). (B) Cholesteryl ester synthesis. On day 3, another group of cells were refed with cholesterol-depletion medium A in the absence or presence of 50 µg protein/mL of LDL. After incubation for 4 h, these cells were pulse-labeled for 2 h with 0.1 mM sodium [14C]oleate (7939 dpm/nmol), after which the cellular content of cholesteryl [14C]oleate was measured (SI Appendix, Materials and Methods). Each bar in A and B represents the average of duplicate incubations, with individual values shown as circles. (C) Measurement of LDLRs by flow cytometry in WT, NPC1−/−, and PTDSS1−/− SV589j cells incubated with LDL. On day 0, the indicated cells were set up in medium A with 10% FCS. On day 1, cells were switched to cholesterol-depletion medium A. After 16 h, cells then received the above cholesterol-depletion medium containing 50 µg protein/mL of LDL. After 24 h, the cells were harvested by incubation with EDTA, washed, incubated with PE-anti-LDLR, and subjected to flow cytometry (SI Appendix, Materials and Methods). The same WT control histogram is shown in both panels for reference.
When LDL-derived cholesterol reaches the ER, some of it is esterified by acyl-CoA:cholesterol acyltransferase (ACAT) (Fig. 1A). Esterification can be followed by measuring the incorporation of [14C]oleate into cholesteryl [14C]oleate. After incubation with LDL, WT cells synthesized abundant cholesteryl [14C]oleate, but much less synthesis occurred in the PTDSS1−/− and NPC1−/− cells (Fig. 2B). After a 24-h incubation with LDL, the PTDSS1−/− and NPC1−/− cells bound more PE-anti-LDLR than WT cells as determined by flow cytometry, indicating that the LDL-derived cholesterol had not blocked SREBP processing (Fig. 2C). Considered together, the data in Fig. 2 suggest that LDL-derived cholesterol fails to reach the ER in PTDSS1−/− cells just as it does in NPC1−/− cells.
Of note, IDOL (also called MYLIP), the second-ranked gene in our screen, showed no defect in LDL-mediated cholesteryl ester synthesis (SI Appendix, Fig. S2A), owing to its function as the E3-ubiquitin ligase that promotes degradation of the LDLR (20). Cells lacking IDOL are expected to have high LDLRs (SI Appendix, Fig. S2B) owing to reduced degradation, and thus they scored positive in our screen.
PTDSS1 cDNA Restores Transport of LDL-Derived Cholesterol in PTDSS1−/− Cells.
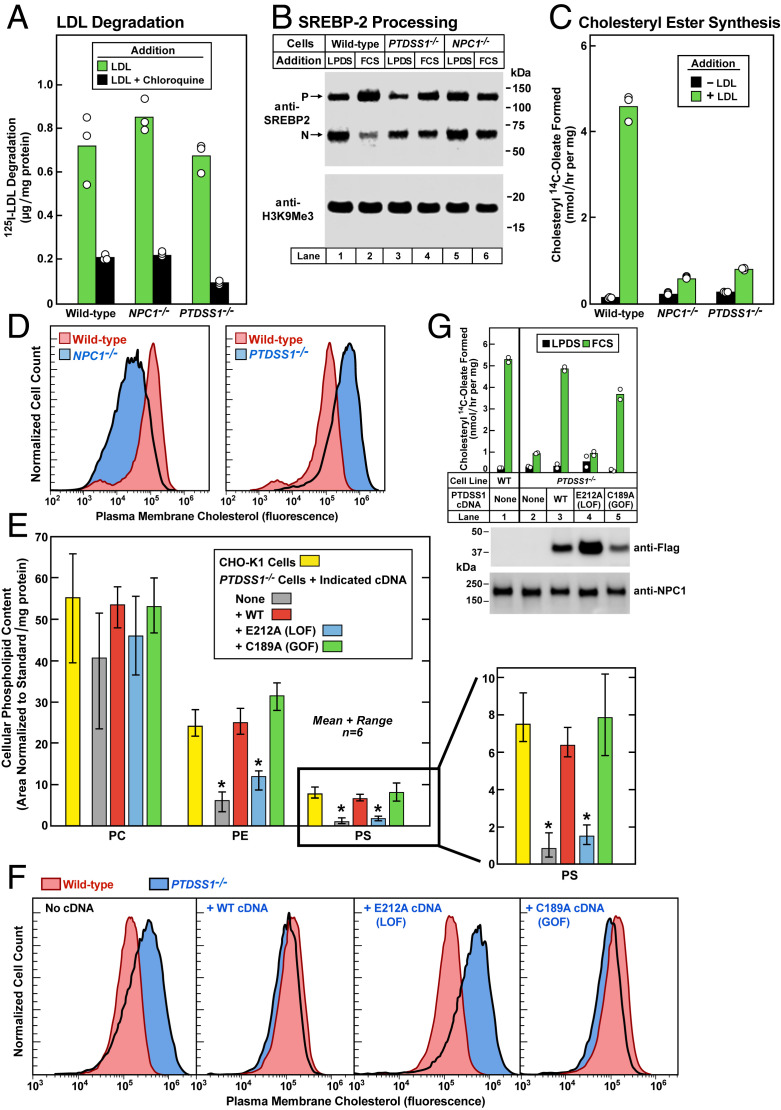
To further study the role of PTDSS1 in cholesterol transport, we created a clonal line of PTDSS1-deficient CHO-K1 cells using CRISPR-Cas9 technology. The sgRNAs flank exon 4, whose deletion results in a frameshift with a premature stop codon corresponding to amino acid 124, as determined by DNA sequencing of the surrounding genomic DNA. The truncated protein lacks the region required for catalytic activity (SI Appendix, Materials and Methods and Fig. S3). These PTDSS1−/− cells were then compared with NPC1−/− cells created with the same CRISPR technology (SI Appendix, Materials and Methods). After preincubation in cholesterol-depleting medium, the PTDSS1−/− and NPC1−/− cells degraded comparable amounts of 125I-LDL to that degraded by WT cells (Fig. 3A). However, when the PTDSS1−/− cells and NPC−/− cells were depleted of cholesterol and then incubated for 6 h with fetal calf serum (FCS) containing LDL, there was no significant inhibition of SREBP cleavage (Fig. 3B) and no increase in cholesterol esterification (Fig. 3C), indicating that both cell lines had blocks in the delivery of LDL-derived cholesterol to the ER.
Fig. 3.
PTDSS1 cDNA restores transport of LDL-derived cholesterol in PTDSS1−/− cells. (A–C) 125I-LDL degradation, LDL-mediated regulation of SREBP processing, and LDL-mediated stimulation of cholesteryl ester synthesis in WT, NPC1−/−, and PTDSS1−/− CHO-K1 cells. On day 0, cells were set up in medium C with 5% FCS. Each bar in A–C represents the average of duplicate incubations, with individual values shown. (A) 125I-LDL degradation. On day 2, cells were switched to cholesterol-depletion medium C. After a 16-h incubation, cells were refed with cholesterol-depletion medium D containing 30 μg protein/mL of 125I-LDL (45 cpm/ng protein) in the absence or presence of 100 μM chloroquine. After 5 h, the amount of 125I-monoiodotyrosine in the medium was measured. (B) SREBP processing. On day 2, cells were switched to cholesterol-depletion medium C. After a 16-h incubation, the cells received medium D containing (CPN/MEV) and either 5% calf LPDS or 10% FCS as indicated. After 6 h, cells were harvested for immunoblotting of SREBP-2 and H3K9Me3 (SI Appendix, Materials and Methods). P, precursor. N, nuclear. (C) Cholesteryl ester synthesis. On day 2, cells were switched to cholesterol-depletion medium C. After incubation for 12 h, the cells received cholesterol-depletion medium D in the absence or presence of 50 μg protein/mL LDL. After incubation for 4 h, the cells were pulse-labeled for 2 h with 0.1 mM sodium [14C]oleate (10,379 dpm/nmol), after which the cellular content of cholesteryl [14C]oleate was measured. (D) Measurement of PM cholesterol by flow cytometry in WT, NPC1−/−, and PTDSS1−/− CHO-K1 cells incubated with serum containing LDL. On day 0, cells were set up in medium C with 5% FCS. On day 2, cells were switched to cholesterol-depletion medium C. After a 16-h incubation, cells received medium D containing compactin plus mevalonate (CPN/MEV) and 10% FCS. After 6 h, cells were harvested for flow cytometry analysis using AF488-labeled PFO*. The same WT control histogram is shown in both panels for reference. (E) Phospholipid content in WT and PTDSS1−/− CHO-K1 cells expressing lentiviral-transduced cDNAs encoding WT, E212A, and C189A variants of human PTDSS1. Cells were plated as in D. After 3 d, cells were harvested for analysis of phospholipid content by liquid chromatography-tandem mass spectrometry (LC-MS/MS) (SI Appendix, Materials and Methods) and normalized to protein content. Each bar represents mean and range of six incubations. Statistical analysis was performed using Student’s t test; *P < 0.0001, when the indicated bar is compared with CHO-K1 cells. An expanded view of the PS content is shown at the right (SI Appendix, Table S2). (F) Measurement of LDL-derived cholesterol by flow cytometry in PMs of WT and PTDSS1−/− CHO-K1 cells expressing lentiviral-transduced cDNAs encoding WT, E212A, and C189A variants of human PTDSS1. On day 0, cells were set up in medium C with 5% FCS. On day 2, cells were switched to cholesterol-depletion medium C. After a 16-h incubation, the cells received medium D containing CPN/MEV supplemented with 10% FCS. After 6 h, cells were harvested, incubated with AF488-labeled PFO*, and examined by flow cytometry. The same WT control histogram is shown in all panels for reference. LOF, loss-of-function; GOF, gain-of-function. (G) Cholesterol esterification activity (Top) and immunoblots (Bottom) of WT and PTDSS1−/− CHO-K1 cells expressing cDNAs encoding Flag-tagged human WT, E212A, and C189A PTDSS1. On day 0, cells were set up in medium C with 5% FCS. On day 2, cells were switched to cholesterol-depletion medium C. After a 12-h incubation, cells received medium D with CPN/MEV supplemented with either 5% calf LPDS (black bar) or 10% FCS (green bar). After 4 h, cells were pulse-labeled for 2 h with 0.1 mM sodium [14C]oleate (8699 dpm/nmol), after which the cellular content of cholesteryl [14C]oleate was measured. Each bar represents average of duplicate incubations with individual values shown. (Bottom) Representative immunoblots of Flag-tagged PTDSS1 in the above cell lines.
To estimate the amount of toxin-accessible cholesterol in PM, we incubated cells with perfringolysin O* (PFO*), a mutant bacterial toxin that binds cholesterol (21, 22). The mutation preserves binding but prevents cell lysis at 4 °C. We tagged PFO* with AF488, a fluorescent dye, and analyzed the cells by flow cytometry. When incubated with LDL, the NPC1−/− cells exhibited less AF488-PFO* binding than WT cells, consistent with the failure of LDL-derived cholesterol to reach the PM (Fig. 3 D, Left). In direct contrast, the PTDSS1−/− cells bound more AF488-PFO* than WT cells (Fig. 3 D, Right).
Mass spectrometry analysis indicated that the PTDSS1−/− cells have a deficiency in PS and PE (Fig. 3E), both of which are produced by PTDSS1. PS is synthesized by PTDSS1 in the ER, after which a portion of PS is decarboxylated by PS decarboxylase 1 (PSD1) in mitochondria to form phosphatidylethanolamine (PE) (14). PC, the most abundant phospholipid, was present in normal amounts in the PTDSS1−/− cells (Fig. 3E). The deficiencies of PS and PE in the PTDSS1−/− cells were corrected when the cells were infected with a lentivirus expressing a cDNA encoding WT PTDSS1 or a version with a gain-of-function mutation that renders the enzyme resistant to feedback inhibition by PS (23) (Fig. 3E). Conversely, no correction occurred when the cells were infected with a cDNA encoding PTSDD1 containing a loss-of-function mutation that renders the enzyme catalytically inactive (23).
When cells were cultured in FCS, which contains LDL, PM cholesterol was higher in PTDSS1−/− cells than in WT cells, as indicated by flow cytometry after incubation with AF488-PFO* (Fig. 3 F, Left). PM cholesterol was normalized by lentiviral expression of cDNAs encoding WT PTDSS1 or the gain-of-function mutant, but not the loss-of-function mutant (Fig. 3 F, Right). When the PTDSS1−/− cells were incubated with LDL-containing FCS, cholesterol esterification was low, indicating that cholesterol was not reaching the ER (Fig. 3 G, Upper). cDNAs encoding Flag-tagged WT PTDSS1 or the gain-of-function mutant restored cholesterol esterification, but the loss-of-function mutant did not. Expression of all three transfected versions of PTDSS1 was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting with anti-FLAG antibody (Fig. 3 G, Lower).
PS Restores Plasma Membrane-to-ER Transport of LDL-Derived Cholesterol in PTDSS1−/− Cells.
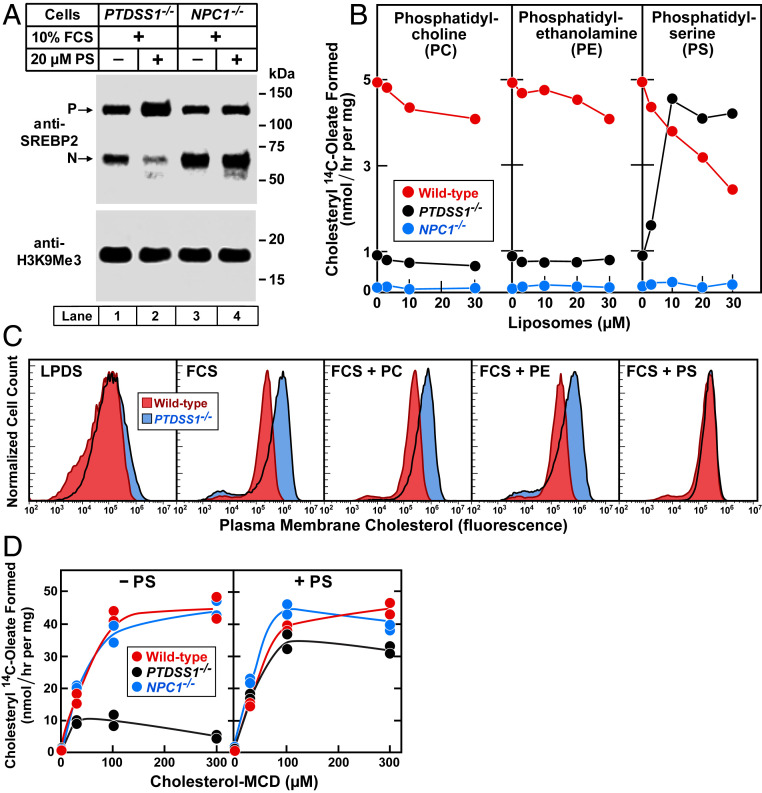
We next sought to determine whether the cholesterol transport defect in PTDSS1−/− cells can be reversed by incubating the cells with PS in the form of liposomes. For this purpose, we first depleted the cells of cholesterol to induce SREBP-2 processing and then incubated the cells for 6 h with FCS in the absence or presence of PS liposomes. As shown in Fig. 4A, in the absence of PS, the PTDSS1−/− cells continued to exhibit nuclear SREBP-2 despite the presence of LDL-containing FCS (lane 1). When the FCS was added in the presence of PS (lane 2), the amount of nuclear SREBP-2 was reduced, consistent with the conclusion that LDL cholesterol was reaching the ER. As a control, we included NPC1−/− cells. Like the PTDSS1−/− cells, the NPC1−/− cells exhibited nuclear SREBP-2 despite the presence of FCS (lane 3), but in sharp contrast, there was no significant reduction when PS liposomes were present (lane 4).
Fig. 4.
PS restores plasma membrane-to-ER transport of LDL-derived cholesterol in PTDSS1−/− cells. (A) SREBP-2 processing in PTDSS1−/− and NPC1−/− CHO-K1 cells incubated with PS. On day 0, cells were set up in medium C with 5% FCS. On day 2, cells were switched to cholesterol-depletion medium C in the absence or presence of 20 μM PS. After 16 h, cells were switched to medium D containing CPN/MEV and 10% FCS in the absence or presence of 20 μM PS. After 6 h, cells were harvested for immunoblotting of SREBP-2 and H3K9Me3. (B) Cholesteryl ester synthesis in cells incubated with phospholipids. On day 0, cells were set up in medium C with 5% FCS. On day 2, cells were switched to cholesterol-depletion medium C, and the indicated liposomes. After a 12-h incubation, cells were switched to medium D containing CPN/MEV, 10% FCS, and the indicated liposomes. After 4 h, cells were pulse-labeled for 2 h with 0.1 mM sodium [14C]oleate (9495 dpm/nmol), after which the cellular content of cholesteryl [14C]oleate was measured. Each value represents an average of duplicate incubations. (C) Measurement of PM cholesterol by flow cytometry. On day 0, cells were set up in medium C with 5% FCS. On day 2, cells were switched to cholesterol-depletion medium C in the absence or presence of 20 µM of the indicated liposomes. After a 16-h incubation, cells were switched to medium D with CPN/MEV and supplemented with either 5% calf LPDS or 10% FCS as indicated and 20 µM of the indicated liposomes. After 6 h, cells were harvested for flow cytometry using AF-488-conjugated-PFO*. (D) Stimulation of cholesteryl ester synthesis by cholesterol-MCD in cells incubated with or without PS. On day 0, cells were set up in medium C with 5% FCS. On day 2, cells were refed with cholesterol-depletion medium C in the absence or presence of 10 µM PS liposomes. After a 12-h incubation, cells received medium D containing CPN/MEV and the indicated amount of cholesterol-MCD in the absence (Left) or presence (Right) of 10 µM PS liposomes. After 4 h, cells were pulse-labeled for 2 h with 0.1 mM sodium [14C]oleate (4588 dpm/nmol), after which the cellular content of cholesteryl [14C]oleate was measured. Each value represents the average of duplicate incubations, with individual values shown.
Fig. 4B shows the effect of PS liposomes on cholesterol transport to the ER using another assay involving the esterification of LDL-derived cholesterol by the ER enzyme ACAT. PTDSS1−/− cells were depleted of cholesterol and then incubated with FCS to deliver LDL cholesterol. The incubations were conducted in the presence of varying concentrations of liposomes composed of PC, PE, or PS. After 4 h, the cells were pulse-labeled with [14C]oleate, and the amount of cholesteryl [14C]oleate was measured. In the absence of liposomes, WT CHO-K1 cells synthesized abundant cholesteryl [14C]oleate (red lines in Fig. 4B) whereas the PTDSS1−/− cells produced much less (black lines). The addition of PC or PE liposomes had no effect on WT or PTDSS1−/− cells (Fig. 4B, Left and Middle). In contrast, PS liposomes led to marked esterification of cholesterol in the PTDSS1−/− cells (Fig. 4B, Right). At higher concentrations, PS decreased cholesteryl ester synthesis in WT cells. We believe that this effect is related to the ability of PS to directly inhibit the ACAT enzyme, which has been well demonstrated in cell-free assays (24).
When PTDSS1−/− cells were incubated with lipoprotein-deficient serum (LPDS), PM cholesterol levels were the same as in WT cells, as indicated by AF488-PFO* binding and flow cytometry (Fig. 4C). When incubated with LDL-containing FCS, PM cholesterol was elevated in the PTDSS1−/− cells. It was normalized when the cells were incubated with PS, but not when the cells were incubated with PC or PE.
An important difference between PTDSS1−/− cells and NPC1−/− cells was apparent when we delivered cholesterol not in the form of LDL, but rather in a complex with methyl β-cyclodextrin (MCD), which delivers cholesterol not to lysosomes, but directly to the PM (7). Cholesterol-MCD increased cholesteryl ester synthesis in NPC1−/− cells as much as in WT cells, indicating that the cholesterol had traveled from the PM to the ER (Fig. 4D, Left). Cholesteryl ester synthesis was much less in the PTDSS1−/− cells. When the PTDSS1−/− cells were preincubated with PS liposomes, cholesterol-MCD delivered cholesterol to the ER normally, as indicated by the normal stimulation of cholesteryl ester synthesis (Fig. 4D, Right). PS had no effect in NPC1−/− cells.
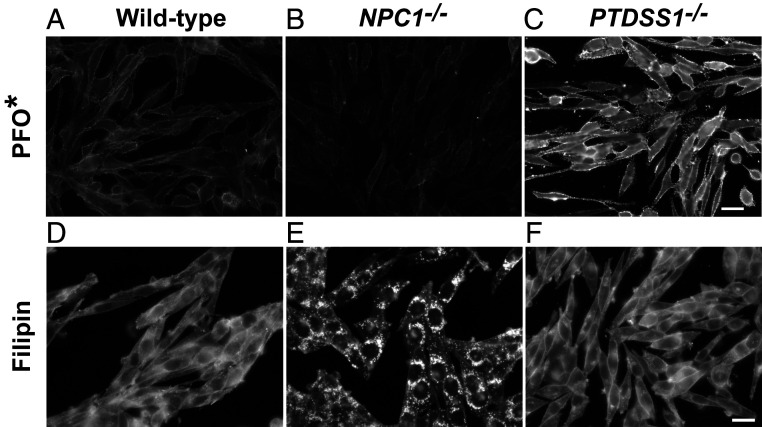
Difference in Cholesterol Distribution in PTDSS1−/− and NPC1−/− Cells as Assessed by Fluorescence Microscopy.
To demonstrate the cholesterol transport defect in PTDSS1−/− cells morphologically, we incubated cell monolayers with FCS, stained the cells with AF488-PFO*, and examined them with a fluorescence microscope (Fig. 5). The images revealed intense staining of the PM in the PTDSS1−/− cells but not in the NPC1−/− cells (Fig. 5 A–C). A different result was obtained when we stained the cells with filipin, a cell-permeable fluorophore that binds cholesterol (Fig. 5 D–F). Filipin showed intense staining of lysosomes in NPC1−/− cells but not in PTDSS1−/− cells, confirming that the PTDSS1−/− cells have no block in the release of cholesterol from lysosomes.
Fig. 5.
Difference in cholesterol distribution in NPC1−/− and PTDSS1−/− cells as assessed by fluorescence microscopy. (A–C) PM cholesterol. On day 0, WT, NPC1−/−, and PTDSS1−/− CHO-K1 cells were plated on 12-mm glass coverslips in medium C with 5% FCS. On day 2, cells were switched to cholesterol-depletion medium C. After a 12-h incubation, cells were switched to medium D containing CPN/MEV and supplemented with 10% FCS. After 6 h, cells were imaged using AF-488–conjugated PFO* (Scale bar: 20 µm.) (D–F) Total cellular cholesterol. Cells were set up and incubated as above and then imaged with filipin (SI Appendix, Materials and Methods) (Scale bar: 20 µm.)
PTDSS2: A Second Enzyme That Synthesizes PS.
In addition to PTDSS1, animal cells can synthesize PS through the action of PTDSS2 (13, 15). In contrast to PTDSS1, which exchanges serine for choline, thereby converting PC to PS, PTDSS2 exchanges serine for ethanolamine, thereby converting PE to PS. In contrast to the PTDSS1 gene, which was among the highest-scoring genes in our CRISPR screen (ranking 18th out of 19,114 genes), the PTDSS2 gene was among the lowest (ranking 18,611), suggesting that PTDSS2 is not essential for transport of LDL-derived cholesterol in human SV589 cells (Dataset S1). To confirm these results, we used CRISPR-Cas 9 to inactivate the PTDSS2 gene in CHO-K1 cells and compared these cells with CHO-K1 cells lacking PTDSS1. In contrast to the PTDSS1-deficient cells, in cells lacking PTDSS2, LDL did not produce an increase in PM cholesterol, and the LDL-derived cholesterol was esterified normally (SI Appendix, Fig. S4). These results indicate that PTDSS1 is the only enzyme in human SV589 cells or CHO-K1 cells that can synthesize the PS required for transport of PM cholesterol to the ER.
Discussion
Our present results have broad implications for the control of cholesterol metabolism in animal cells. First, they reveal a specific requirement for PS in the transport of cholesterol from the PM to the ER. Second, they support the previously reported conclusion that LDL-derived cholesterol moves first from lysosomes to the PM and that it reaches the ER only after traversing the PM. When the PM is depleted of PS, LDL-derived cholesterol is trapped in the PM and cannot exert its regulatory functions in the ER.
To reach these conclusions, we conducted a CRISPR-Cas9 screen to search for genes whose inactivation leads to a failure of LDL-derived cholesterol to inhibit the processing of SREBP-2 in the ER. We measured SREBP-2 processing indirectly by incubating cells with a fluorescent anti-LDLR antibody and sorting for cells that expressed excess LDLRs after incubation with LDL. We were gratified that NPC1 and NPC2 were among the genes scoring the highest in this screen (Fig. 1D). Mutations in these genes are already known to lead to sequestration of LDL-derived cholesterol in lysosomes, thereby preventing inhibition of SREBP-2 processing (25).
Among the highest-scoring genes was PTDSS1, whose product, PS synthase-1, is a major source of PS in cell membranes (13, 14). In cells lacking PTDSS1, total cellular PS levels were low (Fig. 3E). LDL uptake and degradation were normal (Figs. 2 and 3), but the cholesterol failed to reach the ER, as indicated by its failure to inhibit SREBP-2 processing and to be esterified with [14C]oleate (Figs. 2 and 3). Instead, LDL-derived cholesterol accumulated on the PM, as revealed by increased binding of PFO*, a cholesterol-binding toxin (Fig. 3). These abnormalities were reversed by infecting cells with a lentivirus encoding PTDSS1 or by incubating cells with PS liposomes. Inasmuch as PS is the major source of PE, the PTDSS1-deficient cells had low levels of PE as well as PS; however, PE liposomes failed to restore cholesterol transport, indicating that the block is caused by PS deficiency (Fig. 4).
The PS requirement for cholesterol transport is particularly relevant in light of recent studies with a family of animal proteins known as GRAMD1s (26, 27) or Asters (28). These proteins are embedded in the ER membrane and cluster at sites where the ER membrane contacts the PM (26). They contain two functional domains: the StART-like domain (also known as the Aster domain) that binds cholesterol (27, 28) and the GRAM domain that binds anionic lipids, including PS (27, 28). The GRAM domain of these ER proteins binds to anionic lipids in the PM, linking the ER to the PM. The StART/Aster domain then transfers cholesterol from the PM to the ER.
PM cholesterol exists in three pools (7). One pool is termed “accessible” because its cholesterol is accessible to the cholesterol-binding toxins PFO* and anthrolysin O (7, 8). A second pool is bound to sphingomyelin and thus is inaccessible to these toxins. The third pool is also inaccessible and is essential for cell viability (7). Cholesterol released from LDL in lysosomes adds to the accessible pool. Treatment with sphingomyelinase releases cholesterol from sphingomyelin, thereby increasing the accessible pool. The accessible pool of cholesterol is the only pool that is free to move to the ER to exert regulatory actions (7, 8, 29).
It is striking that sphingomyelin and PS have opposite effects on cholesterol movement from the PM. Sphingomyelin is concentrated in the outer leaflet of the PM and it traps cholesterol, preventing its movement to the ER (7, 30). PS is concentrated in the inner leaflet (15), and it is essential for cholesterol movement to the ER. Taken together, these observations suggest the fundamental principle that cells control the cholesterol content of their PM by controlling the concentrations of sphingomyelin and PS.
Two recent studies have demonstrated that GRAMD1/Aster proteins are essential for movement of the accessible pool of cholesterol from PM to ER. In one study, Sandhu et al. (28) used high-density lipoproteins (HDLs) to deliver cholesterol to the PM, increasing the accessible pool. In the other study, Naito et al. (27) treated cells with sphingomyelinase to increase the accessible pool. In both studies, movement of this cholesterol to the ER was blocked when GRAMD1/Aster proteins were eliminated. Inasmuch as GRAMD1/Aster proteins are known to bind to PS, it is likely that the PS deficiency caused by deletion of PTDSS1 prevents the recruitment of GRAMD1/Aster proteins to the PM, thereby accounting for the trapping of LDL-derived cholesterol in the PM of PTDSS1-deficient cells (Figs. 3 and 4).
Sandhu et al. (28) demonstrated that GRAMD1 proteins are essential for delivery of HDL cholesterol to the adrenal cortex, where it is used in the synthesis of steroid hormones. Mice lacking one of the Aster proteins (Aster B) failed to store sufficient cholesterol in the adrenal cortex and had defects in steroidogenesis. Mice and other rodents have very low levels of LDL and use HDL to deliver cholesterol to the adrenal (31, 32); however, in other species, including humans, LDL is the predominant cholesterol-carrying lipoprotein. In these species, the adrenal cortex expresses high levels of LDLRs (33). Indeed, LDLRs were originally purified from cow adrenal glands because they were the richest source (34, 35). Our present results suggest that GRAMD1/Aster proteins may be essential for the delivery of LDL cholesterol as well as HDL cholesterol for storage in the adrenal gland.
LDLR-mediated cholesterol homeostasis was discovered nearly 50 y ago (early review in ref. 36). The addition of LDL to cells reduced the activity of 3-hydroxy-3-methylglutaryl CoA reductase and blocked cholesterol synthesis. Over the ensuing decades, individual pieces were added stepwise until the regulatory mechanism became clear. LDL-derived cholesterol is taken up by LDLRs and released in lysosomes (1). It reaches the ER, where it binds and inhibits Scap, an escort protein for SREBP-2, the transcription factor that activates genes for cholesterol synthesis and LDL uptake (2). The discovery that SREBP-2 resides in the ER was puzzling (37). Most cellular cholesterol is in the PM, yet the Scap/SREBP-2 sensor that adjusts PM cholesterol levels is located in the ER (38). How does the ER detect an excess or deficiency of cholesterol in the PM? The answer lies in the observation that LDL-derived cholesterol moves first to the PM, where it expands the accessible cholesterol pool. Only when this pool exceeds an optimal level does the excess cholesterol move from the PM to the ER in a process that requires PS and GRAMD1/Aster proteins. Once in the ER, the excess cholesterol inhibits Scap/SREBP-2, thereby completing the feedback loop. Although additional elements are likely to be added in the future, the framework for regulation has been established, and progress is sure to follow.
Materials and Methods
Reagents, cell lines and knockouts, plasmids, CRISPR-Cas9 screening, lentiviral-mediated transduction of PTDSS1 cDNA, flow cytometry, unilamellar liposomes, metabolic assays, fluorescence microscopy, measurement of phospholipids, immunoblot analysis, and reproducibility are described in detail in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank John Doench (Broad Institute) for providing the CRISPR library; our colleagues Ryan Golden, Xiaoqiang Zhu, Jong-Sun Lee, Jian Xu, Jose Rizo-Rey, and Arun Radhakrishnan for helpful discussions; Hannah Powell, William Salter, and Erin Ruhlman for excellent technical assistance; and Lisa Beatty, Briana Carter, Camille Harry, and Ije Dukes for invaluable help with tissue culture. This research was supported by grants from the NIH (HL20948, R35CA197311, and GM096070) and the Welch Foundation (I-1961 and I-1910). M.N.T. was supported by the NIH Medical Scientist Training Program (GM008014). J.H. is a recipient of a postdoctoral fellowship from the American Heart Association (19POST34380222). J.T.M. is an Investigator of the HHMI.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2010682117/-/DCSupplemental.
Data Availability.
The results of the CRISPR-Cas9 screen involving 19,114 human genes are provided in Dataset S1. There are no restrictions on these data.
References
- 1.Brown M. S., Goldstein J. L., A receptor-mediated pathway for cholesterol homeostasis. Science 232, 34–47 (1986). [DOI] [PubMed] [Google Scholar]
- 2.Brown M. S., Goldstein J. L., The SREBP pathway: Regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89, 331–340 (1997). [DOI] [PubMed] [Google Scholar]
- 3.Goldstein J. L., Brown M. S., The low-density lipoprotein pathway and its relation to atherosclerosis. Annu. Rev. Biochem. 46, 897–930 (1977). [DOI] [PubMed] [Google Scholar]
- 4.Chang T. Y., Chang C. C. Y., Cheng D., Acyl-coenzyme A:cholesterol acyltransferase. Annu. Rev. Biochem. 66, 613–638 (1997). [DOI] [PubMed] [Google Scholar]
- 5.Infante R. E. et al., NPC2 facilitates bidirectional transfer of cholesterol between NPC1 and lipid bilayers, a step in cholesterol egress from lysosomes. Proc. Natl. Acad. Sci. U.S.A. 105, 15287–15292 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwon H. J. et al., Structure of N-terminal domain of NPC1 reveals distinct subdomains for binding and transfer of cholesterol. Cell 137, 1213–1224 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Das A., Brown M. S., Anderson D. D., Goldstein J. L., Radhakrishnan A., Three pools of plasma membrane cholesterol and their relation to cholesterol homeostasis. eLife 3, e02882 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Infante R. E., Radhakrishnan A., Continuous transport of a small fraction of plasma membrane cholesterol to endoplasmic reticulum regulates total cellular cholesterol. eLife 6, e25466 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heybrock S. et al., Lysosomal integral membrane protein-2 (LIMP-2/SCARB2) is involved in lysosomal cholesterol export. Nat. Commun. 10, 3521 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao K., Ridgway N. D., Oxysterol-binding protein-related protein 1L regulates cholesterol egress from the endo-lysosomal system. Cell Rep. 19, 1807–1818 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Chu B.-B. et al., Cholesterol transport through lysosome-peroxisome membrane contacts. Cell 161, 291–306 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Höglinger D. et al., NPC1 regulates ER contacts with endocytic organelles to mediate cholesterol egress. Nat. Commun. 10, 4276 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuge O., Nishijima M., Phosphatidylserine synthase I and II of mammalian cells. Biochim. Biophys. Acta 1348, 151–156 (1997). [DOI] [PubMed] [Google Scholar]
- 14.Leventis P. A., Grinstein S., The distribution and function of phosphatidylserine in cellular membranes. Annu. Rev. Biophys. 39, 407–427 (2010). [DOI] [PubMed] [Google Scholar]
- 15.Vance J. E., Tasseva G., Formation and function of phosphatidylserine and phosphatidylethanolamine in mammalian cells. Biochim. Biophys. Acta 1831, 543–554 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Radhakrishnan A., Ikeda Y., Kwon H. J., Brown M. S., Goldstein J. L., Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: Oxysterols block transport by binding to Insig. Proc. Natl. Acad. Sci. U.S.A. 104, 6511–6518 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doench J. G. et al., Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34, 184–191 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li W. et al., MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 15, 554 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldstein J. L., Brown M. S., Binding and degradation of low-density lipoproteins by cultured human fibroblasts. Comparison of cells from a normal subject and from a patient with homozygous familial hypercholesterolemia. J. Biol. Chem. 249, 5153–5162 (1974). [PubMed] [Google Scholar]
- 20.Zelcer N., Hong C., Boyadjian R., Tontonoz P., LXR regulates cholesterol uptake through Idol-dependent ubiquitination of the LDL receptor. Science 325, 100–104 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flanagan J. J., Tweten R. K., Johnson A. E., Heuck A. P., Cholesterol exposure at the membrane surface is necessary and sufficient to trigger perfringolysin O binding. Biochemistry 48, 3977–3987 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Das A., Goldstein J. L., Anderson D. D., Brown M. S., Radhakrishnan A., Use of mutant 125I-perfringolysin O to probe transport and organization of cholesterol in membranes of animal cells. Proc. Natl. Acad. Sci. U.S.A. 110, 10580–10585 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohsawa T., Nishijima M., Kuge O., Functional analysis of Chinese hamster phosphatidylserine synthase 1 through systematic alanine mutagenesis. Biochem. J. 381, 853–859 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doolittle G. M., Chang T.-Y., Solubilization, partial purification, and reconstitution in phosphatidylcholine-cholesterol liposomes of acyl-CoA:cholesterol acyltransferase. Biochemistry 21, 674–679 (1982). [DOI] [PubMed] [Google Scholar]
- 25.Abi-Mosleh L., Infante R. E., Radhakrishnan A., Goldstein J. L., Brown M. S., Cyclodextrin overcomes deficient lysosome-to-endoplasmic reticulum transport of cholesterol in Niemann-Pick type C cells. Proc. Natl. Acad. Sci. U.S.A. 106, 19316–19321 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Besprozvannaya M. et al., GRAM domain proteins specialize functionally distinct ER-PM contact sites in human cells. eLife 7, e31019 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naito T. et al., Movement of accessible plasma membrane cholesterol by the GRAMD1 lipid transfer protein complex. eLife 8, e51401 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sandhu J. et al., Aster proteins facilitate nonvesicular plasma membrane to ER cholesterol transport in mamalian cells. Cell 175, 514–529.e20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson K. A., Endapally S., Vazquez D. C., Infante R. E., Radhakrishnan A., Ostreolysin A and anthrolysin O use different mechanisms to control movement of cholesterol from the plasma membrane to the endoplasmic reticulum. J. Biol. Chem. 294, 17289–17300 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Endapally S. et al., Molecular discrimination between two conformations of sphingomyelin in plasma membranes. Cell 176, 1040–1053.e17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kovanen P. T., Schneider W. J., Hillman G. M., Goldstein J. L., Brown M. S., Separate mechanisms for the uptake of high- and low-density lipoproteins by mouse adrenal gland in vivo. J. Biol. Chem. 254, 5498–5505 (1979). [PubMed] [Google Scholar]
- 32.Andersen J. M., Dietschy J. M., Kinetic parameters of the lipoprotein transport systems in the adrenal gland of the rat determined in vivo. Comparison of low- and high-density lipoproteins of human and rat origin. J. Biol. Chem. 256, 7362–7370 (1981). [PubMed] [Google Scholar]
- 33.Kovanen P. T., Basu S. K., Goldstein J. L., Brown M. S., Low-density lipoprotein receptors in bovine adrenal cortex, II: Low-density lipoprotein binding to membranes prepared from fresh tissue. Endocrinology 104, 610–616 (1979). [DOI] [PubMed] [Google Scholar]
- 34.Brown M. S., Kovanen P. T., Goldstein J. L., Receptor-mediated uptake of lipoprotein-cholesterol and its utilization for steroid synthesis in the adrenal cortex. Recent Prog. Horm. Res. 35, 215–257 (1979). [DOI] [PubMed] [Google Scholar]
- 35.Schneider W. J., Beisiegel U., Goldstein J. L., Brown M. S., Purification of the low-density lipoprotein receptor, an acidic glycoprotein of 164,000 molecular weight. J. Biol. Chem. 257, 2664–2673 (1982). [PubMed] [Google Scholar]
- 36.Brown M. S., Goldstein J. L., Receptor-mediated control of cholesterol metabolism. Science 191, 150–154 (1976). [DOI] [PubMed] [Google Scholar]
- 37.Wang X., Sato R., Brown M. S., Hua X., Goldstein J. L., SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell 77, 53–62 (1994). [DOI] [PubMed] [Google Scholar]
- 38.Radhakrishnan A., Goldstein J. L., McDonald J. G., Brown M. S., Switch-like control of SREBP-2 transport triggered by small changes in ER cholesterol: A delicate balance. Cell Metab. 8, 512–521 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The results of the CRISPR-Cas9 screen involving 19,114 human genes are provided in Dataset S1. There are no restrictions on these data.