Significance
Despite clinically effective antiretroviral therapy (ART), replication-competent HIV-1 persists as latent proviral DNA capable of rekindling viral replication when ART is stopped. To cure the infection, therapies are being developed to eliminate latent HIV-1. Accurate assays for intact or rebound-competent HIV-1 are important to this effort. We previously developed the intact proviral DNA assay (IPDA) as a more accurate and scalable assay for intact HIV-1 proviruses. Here we present IPDA analysis in a diverse cohort of 400 ART-treated individuals. We confirm that intact proviruses, although vastly outnumbered by defective proviruses, are present at significantly higher frequencies than previously detected by viral outgrowth assays. We also show that IPDA amplicon signal issues, observed in 6.3% of samples, result from sequence polymorphisms.
Keywords: IPDA, HIV, reservoir, latency, cure
Abstract
A scalable approach for quantifying intact HIV-1 proviruses is critical for basic research and clinical trials directed at HIV-1 cure. The intact proviral DNA assay (IPDA) is a novel approach to characterizing the HIV-1 reservoir, focusing on the genetic integrity of individual proviruses independent of transcriptional status. It uses multiplex digital droplet PCR to distinguish and separately quantify intact proviruses, defined by a lack of overt fatal defects such as large deletions and APOBEC3G-mediated hypermutation, from the majority of proviruses that have such defects. This distinction is important because only intact proviruses cause viral rebound on ART interruption. To evaluate IPDA performance and provide benchmark data to support its implementation, we analyzed peripheral blood samples from 400 HIV-1+ adults on ART from several diverse cohorts, representing a robust sample of treated HIV-1 infection in the United States. We provide direct quantitative evidence that defective proviruses greatly outnumber intact proviruses (by >12.5 fold). However, intact proviruses are present at substantially higher frequencies (median, 54/106 CD4+ T cells) than proviruses detected by the quantitative viral outgrowth assay, which requires induction and in vitro growth (∼1/106 CD4+ T cells). IPDA amplicon signal issues resulting from sequence polymorphisms were observed in only 6.3% of individuals and were readily apparent and easily distinguished from low proviral frequency, an advantage of the IPDA over standard PCR assays which generate false-negative results in such situations. The large IPDA dataset provided here gives the clearest quantitative picture to date of HIV-1 proviral persistence on ART.
In individuals with HIV-1, a latent form of the virus persists in resting CD4+ T cells despite effective antiretroviral therapy (ART) that suppresses the level of free virus in the plasma to below the limit of detection of clinical assays (1–3). This latent reservoir is a major barrier to cure and the subject of an intense international research effort. Accurate measurement of the reservoir is essential for evaluating cure strategies (4). The reservoir was originally defined using a quantitative viral outgrowth assay (QVOA) for latently infected cells that release replication-competent virus after reversal of latency by a single round of in vitro T cell activation (1). This assay provides a definitive minimal estimate for the frequency of latently infected cells; however, it is costly, requires large blood volumes, and misses many replication-competent proviruses that are not induced following a single round of T cell activation (5, 6). In addition, an elegant recent study has shown that extinction events in culture may lead to a further underestimation of the total number of replication-competent proviruses (7).
Owing to the complexity of the QVOA, most studies use simpler PCR assays that measure proviral DNA (4). However, these assays substantially overestimate reservoir size because they do not distinguish intact HIV-1 proviruses from the vast excess of proviruses with overt fatal defects, including large deletions and/or APOBEC3G-mediated hypermutation (8, 9). Other assays measure viral RNA or protein expression before or after a single round of T cell activation (10–13); however, these assays not only miss proviruses not induced by a single round of T cell activation (as with the QVOA), but also may detect defective proviruses still capable of generating both viral RNA and proteins, leading to overestimation of reservoir size (14, 15).
A novel approach to reservoir analysis is the quantification of genetically intact proviruses irrespective of their transcriptional status at any given time. This can be done by near-full-length proviral sequencing (5, 8, 16–18), but such single genome sequencing approaches are not scalable. In addition, they do not provide absolute quantitation because they depend on an initial near-full-length (9 kb) PCR, which is inefficient. The recently described intact proviral DNA assay (IPDA) provides a scalable way to determine the frequency of cells carrying intact proviruses (9). The IPDA leverages droplet digital PCR to simultaneously interrogate individual proviruses with two strategically chosen and highly informative short amplicons placed in conserved regions of the proviral genome. Interrogation at these regions enables the assay to distinguish and separately quantify apparently intact proviruses from the overwhelming majority of defective proviruses, which harbor large deletions and/or hypermutation.
The IPDA has already led to novel insights. For example, the assay has been used to provide more accurate estimates of the decay rate of the latent reservoir (19), to demonstrate differential decay rates for cells carrying intact and defective proviruses (9, 19, 20), to document low reservoir size in elite suppressors (21), and to explore the frequency and inducibility of intact HIV-1 proviruses in different subsets of CD4+ T cells (22).
The analysis of large and diverse cohorts is an important step in defining the performance of a novel assay. However, such systematic analysis has not been widely undertaken for assays for persistent HIV-1, including the IPDA. Owing to its scalability and sensitivity, the IPDA can be applied to large donor cohorts. In this study, we present the IPDA analysis of samples from a large and diverse group of individuals with HIV-1 who are on ART. This dataset will serve as a quantitative benchmark for the frequency and composition of proviral DNA in treated individuals. In addition, this analysis of a large group of people with HIV-1 defines the frequency with which HIV-1 sequence polymorphisms interfere with amplification in the IPDA, which has rarely been defined for other molecular assays for HIV-1 persistence. Furthermore, in this study we describe how the design of the IPDA allows for the detection of such amplification problems, which is not possible with other typical PCR-based approaches for quantifying proviral DNA. We also describe how these problems can be mitigated to determine the frequency of cells with intact proviruses. We suggest that this information will further support the implementation of a novel and scalable approach for measuring intact HIV-1 proviruses in cure studies.
Results
Independent Cohorts Included in This Study.
We evaluated 400 individuals with HIV-1 who are on ART, making this one of the largest cross-sectional analyses of intact and defective proviruses to be undertaken. This effort required significant collaboration from multiple independent groups. A summary of the source of samples is provided in Table 1. All participants included in the study were HIV-1 positive adults on ART living in the United States who had suppression of detectable viremia for >6 mo. The aggregated study population was diverse with respect to age, sex, race, HIV risk factor, and other clinical parameters and thus represents a reasonable sample of treated HIV-1 infection in the United States. Demographic data for each contributing cohort are provided in Table 1.
Table 1.
Summary of the source of samples in this study
| Sample source | Institution | N* | Age, y, median | Male sex, % | Race, %† | Risk‡ | ART duration, y | CD4 count, cells/µL, median | Reference | |
| Nadir | Proximal | |||||||||
| A5321/A5341S | AIDS Clinical Trials Group | 74 | 46 | 73.0 | W 50 | Mix | 7.1 | 319 | 748 | R. Gandhi et al.§ |
| B 20 | ||||||||||
| H 30 | ||||||||||
| SCOPE | University of California, San Francisco | 81 | 49 | 95.1 | W 74 | MSM (>84%) | 10.8 | 183 | 584 | (19) |
| B 12 | ||||||||||
| H 8 | ||||||||||
| O 6 | ||||||||||
| BEAT-HIV | Wistar Institute | 63 | 44 | 89.0 | W 14 | Mix | >1 | NA¶ | 786 | (23) |
| B 79 | ||||||||||
| O 7 | ||||||||||
| ALIVE | Johns Hopkins | 96 | 53 | 70.8 | W, H 4 | IVDU | 7.8# | 180 | 464 | G. D. Kirk et al.‖ |
| B 96 | ||||||||||
| Bartlett Clinic | Johns Hopkins | 20 | 59 | 63.6 | W 9 | Mix | 5.0 | NA¶ | 670 | (21, 24) |
| B 91 | ||||||||||
| ACDX sample collection | AccelvirDx | 66 | 56 | 73.5 | W 18 | Mix | 9.3 | NA¶ | 746 | This study |
| B 72 | ||||||||||
| Total | 400 | |||||||||
N, number of unique donors.
W, White non-Hispanic; B, Black non-Hispanic; H, Hispanic (regardless of race); O, other.
Predominant risk factor for HIV-1 acquisition for the cohort. Mix, cohort includes participants with different risk factors for acquisition; MSM, men who have sex with men; IVDU, intravenous drug use.
R. Gandhi et al., Conference on Retroviruses and Opportunistic Infections, March 8–11, 2020, Boston, MA.
NA, not available; data on CD4 nadir not available from patient records.
Time since first introduction of ART. Due to nonstructured treatment interruption, the time from last detectable HIV-1 RNA was 3.4 y.
G. D. Kirk et al., Conference on Retroviruses and Opportunistic Infections, March 8-11, 2020, Boston, MA.
Overview of the IPDA.
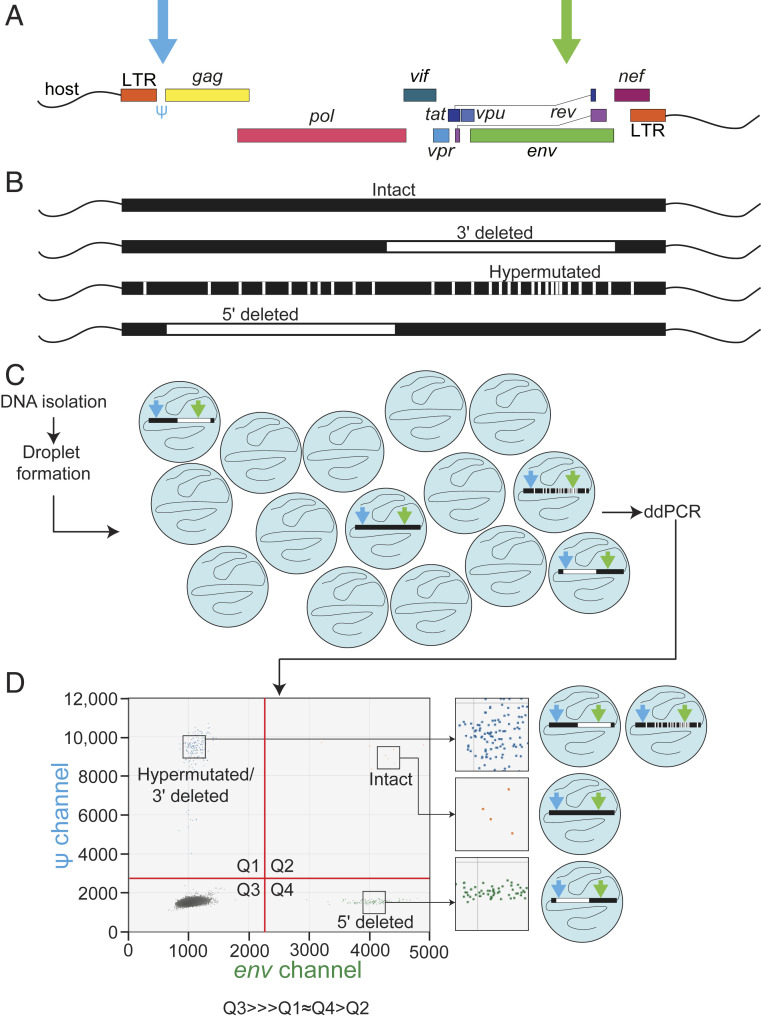
An overview of the IPDA is provided in Fig. 1. DNA isolated from donor samples is distributed into nanoliter-size droplets such that individual proviruses are contained within single droplets, enabling multiplex analysis of each provirus. In each droplet containing a provirus, duplex PCR reactions occurring in each droplet interrogate two carefully chosen regions of the HIV-1 genome to distinguish intact from defective proviruses (Fig. 1 A and B). These regions are the packaging signal (Ψ) and the Rev-response element of the env gene (9). Previous studies have shown that the majority of defective proviruses harbor large internal deletions (average size, 49.6% of genome) (9), and thus intact proviruses can be distinguished from most deleted proviruses based on successful amplification at just two positions in the genome. Amplification occurs in nanoliter-sized droplets containing all of the components necessary for the duplex PCR reactions (Fig. 1C). The intact proviruses are positive for both the Ψ and env amplicons. In addition to large internal deletions, defective proviruses are often hypermutated, predominately by APOBEC3G (5, 8, 9, 25, 26). To distinguish hypermutated proviruses, the env amplicon targets a region containing adjacent, frequently mutated APOBEC3G consensus sites and uses labeled wild-type and unlabeled hypermutated probes. As a result, the majority of hypermutated proviruses fail to give a signal for the env amplicon (9).
Fig. 1.
Overview of the IPDA (A) Map of the HIV-1 genome indicating positions of the Ψ (blue arrow) and env (green arrow) amplicons. (B) Illustration of the common types of fatal defects detected by the IPDA. Precision maps of sequenced proviruses are available elsewhere (5, 8, 9). (C) Overview of the IPDA. In addition to the HIV-1 proviral discrimination reaction illustrated here, the IPDA includes a parallel ddPCR reaction targeting a host gene to allow determination of input cell number and correction for shearing between Ψ and env amplicons (9, 19). (D) Representative 2D ddPCR plot from the HIV-1 proviral discrimination component reaction of the IPDA. The typical droplet counts follow the pattern Q3 >>> Q1 ∼ Q4 > Q2.
In the IPDA, digital droplet PCR (ddPCR) results are displayed on a 2D dot plot with fluorescence from the Ψ and env probes on the y- and x-axes, respectively. Measurement of intact proviruses with the IPDA depends on the presence of droplets in all four quadrants of the plot (Fig. 1D). Proviruses with 3′ deletions encompassing the env amplicon and hypermutated proviruses give a fluorescent signal only for the Ψ amplicon and are found in the upper left quadrant (Q1). Proviruses with 5′ deletions encompassing the Ψ region give a signal only for the env amplicon and are found in the lower right quadrant (Q4). Droplets with no proviruses occupy the lower left quadrant (Q3), along with a small number of proviruses defective at both amplicons. Intact proviruses are counted directly as double-positive droplets in the upper right quadrant (Q2). In infected individuals who started ART during chronic infection, defective proviruses greatly outnumber intact proviruses. This is reflected in the typical droplet distribution observed in an IPDA 2D dot plot: Q3 (empty)>>> Q1 ∼ Q4 > Q2. Data demonstrating the sensitivity, specificity, and reproducibility of the assay have been reported previously (9). As described previously, a separate ddPCR for a cellular gene quantitates input cell number and allows correction for DNA shearing (9).
IPDA Analysis of Peripheral Blood Samples from Individuals with HIV-1 on ART.
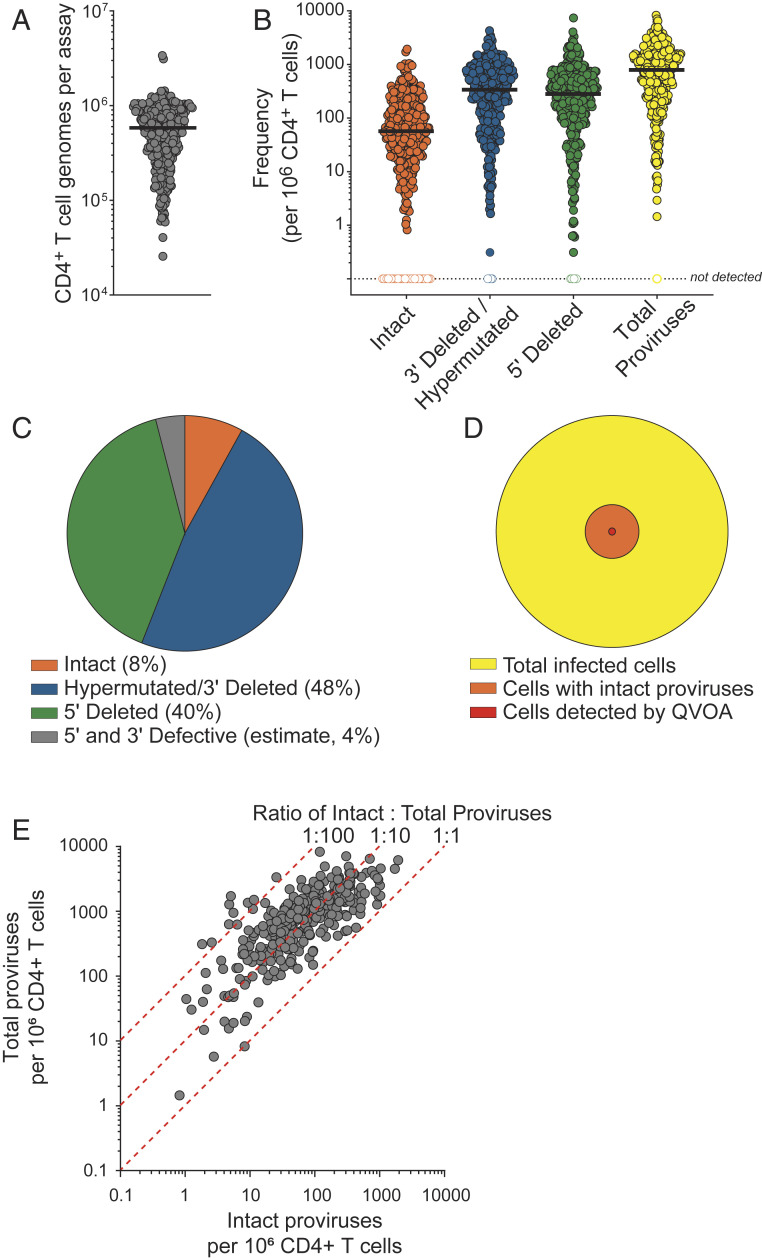
In this study, IPDA was performed on DNA isolated from peripheral blood CD4+ T cells from 400 individuals with HIV-1 who were on ART. The number of CD4+ T cells analyzed per sample varied depending on initial cryopreserved cell availability and the number of total CD4+ T cells recovered (Fig. 2A). For most samples, ∼6 × 105 cells were analyzed. The output value of intact proviruses/106 cells showed little or no dependence on input cell number (Spearman’s ρ = −0.1551) or the degree of DNA shearing (Spearman’s ρ = 0.18).
Fig. 2.
IPDA analysis of peripheral blood CD4+ T cells from 400 HIV-1–infected individuals on ART provides absolute frequencies and relative composition of the population of persistent proviruses. (A) Number of CD4+ T cell genomes analyzed per assay, as measured by the copy reference/DNA shearing component reaction of the IPDA. The horizontal line indicates median value. n = 400 (B) Absolute frequencies of intact and defective proviruses per 106 total CD4+ T cells as measured by the IPDA. Total proviruses were determined as the sum of intact, 5′ defective, and 3′ defective proviruses from each donor. Horizontal line indicated median values. n = 400 (C) Average relative composition of persistent proviruses as measured by the IPDA. (D) Landscape of persistent proviruses in ART-treated individuals with HIV-1. The QVOA detects an average of 1/106 resting CD4+ T cells, based on previous studies (1, 4, 20–22). The value for the frequency of cells with intact proviruses (38/106 resting CD4+ T cells) is based on the mean frequency measured in B, corrected for the fraction of cells with small defects not detected by the IPDA (∼30% of Q2 droplets) (9). The value for total infected cells is based on data from B, corrected for the fraction of proviruses (4%) not detected by either of the IPDA amplicons. (E) Frequency of intact and total proviruses per million CD4+ T cells, as measured by the IPDA. The dashed lines reference relative ratios of intact:total proviruses. Correlation was performed on log-transformed data (Spearman’s ρ = 0.76; P < 0.0001).
A summary of the proviral frequencies measured by the IPDA is shown in Fig. 2B. The median proviral frequencies per million cells as measured by the IPDA in this study were as follows: intact, 54; 3′ defective, 322; 5′ defective, 269; and total (calculated on a per donor basis), 755. The results are consistent with IPDA values reported initially from a smaller cohort (9). There were no significant differences in the frequency of intact proviruses between the groups in Table 1 using one-way ANOVA with Tukey’s multiple-comparisons test. Analysis of this large cohort revealed that the frequencies of intact and defective proviruses can vary across a multi-log10 range. Based on our previous analysis with near-full-length genome sequences (9), we predicted that only 4% of proviruses are likely to have defects at both IPDA amplicons and thus would not be directly quantified by the IPDA. In other words, we predict that 96% of proviruses would be captured and scored as intact or defective by the IPDA. Further analysis of the proviral landscape is needed to verify this estimate.
The relative composition of the population of persistent proviruses as measured by the IPDA is shown in Fig. 2C. Intact proviruses composed 8% of the total proviruses on average. Thus, our results provide direct evidence from a large diverse population of infected individuals that the frequency of cells with intact proviruses is substantially smaller than the frequency of cells with defective proviruses.
Previous analyses of the proviral landscape in ART-treated individuals with HIV-1 have relied on near-full-length genome sequencing, an approach that is not quantitative, due in part to the inefficiency of the long-distance PCR reactions involved (5, 8, 16–18). In contrast, the IPDA provides direct quantitation and the scalability to interrogate large cohorts. Our results also establish in a large and diverse population of HIV-1–positive individuals that intact proviruses are present at a substantially higher frequency than that observed in the QVOA (mean frequency, ∼1/106 CD4+ T cells) (27–29). This difference reflects the fact that most intact proviruses are not induced following a single round of in vitro T cell activation (5, 6, 22). In addition, some cells carrying intact proviruses do not give rise to a sufficient amount of virus to establish a spreading in vitro infection (7). Finally, we have reported that some proviruses characterized as intact by the IPDA contain minor defects affecting viral fitness (9).
Considering these factors and taking advantage of the direct quantitation provided by the IPDA, we provide an updated picture of the proviral landscape in 400 individuals with HIV-1 receiving ART (Fig. 2D). Proviruses scored as intact are present at a frequency of 50 to 60/106 CD4+ T cells, 12.5-fold less than the total frequency of infected cells and more than 50-fold greater than the frequency of cells detected by QVOA in previous studies (1, 4, 20–22). This is consistent with multiple previously published near-full-length proviral sequencing studies demonstrating that the QVOA underestimates the size of the latent reservoir (5, 8, 16–18). We observed a cross-sectional association between the frequency per million CD4+ T cells of intact proviruses and total proviruses (Spearman’s ρ = 0.76; P < 0.0001) . The ratio of intact to total proviruses for the aggregate dataset showed considerable interparticipant variability (Fig. 2E), consistent with published analysis for the SCOPE subgroup (19).
IPDA Amplicon Signal Failure due to Polymorphism Is Infrequent and Readily Distinguished.
For all PCR-based HIV assays, sequence variation in the primer or probe binding sites can prevent the generation of a signal in samples from some individuals. The frequency at which this occurs has not been systematically investigated for commonly used molecular assays for persistent HIV-1. The IPDA uses primers and probes designed to bind to conserved regions of the HIV-1 genome, based on clade B alignments (9). The ddPCR format and the primer/probe design are tolerant of minor sequence mismatches. Nevertheless, within large populations with HIV-1, some amplification problems due to sequence polymorphism are expected.
Importantly, the design of the IPDA allows such problems to be immediately identified. Polymorphisms preventing signal generation from the Ψ amplicon will result in plots with no droplets in Q1 and no droplets in Q2 (Fig. 1D). Similarly, polymorphisms preventing signal generation from the env amplicon will result in plots with no droplets in Q2 and no droplets in Q4. In such situations, intact proviruses cannot be quantitated unless alternative primers or probes are used (see below). Except in rare cases of failure of both amplicons, the two-amplicon format of the assay automatically distinguishes signal problems from the absence of proviral templates, thus preventing the reporting of false-negative results. This is another important advantage of the IPDA over single-amplicon quantitative PCR assays for proviral DNA, which cannot identify poor amplification or signal failure due to polymorphisms.
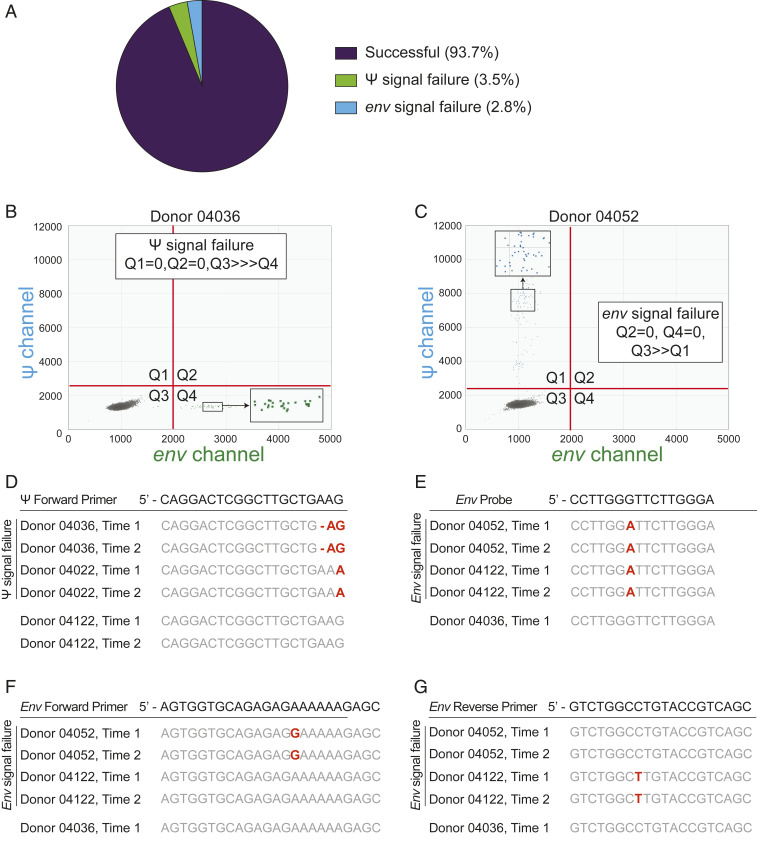
In this study, we evaluated signal failure by identifying samples for which no fluorescence-positive droplets were discernible for one of the amplicons, while the other amplicon exhibited normal amplification and droplet fluorescence. Of note, signal failure has not been evaluated for any other PCR-based reservoir assay. In this cohort of 400 individuals with HIV-1, samples from 3.5% of individuals exhibited signal failure in the Ψ amplicon, and samples from 2.8% of individuals exhibited signal failure in the env amplicon (Fig. 3A), while 93.7% of the samples from 400 donors exhibited no signal issues. Examples of signal failures are shown in Fig. 3 B and C. As each IPDA run included both positive controls (J-Lat 6.3 cells) and negative controls (CD4+ T cells from HIV-1–negative donors), we ensured that all assay runs were successful in specifically amplifying both the Ψ and env regions of control HIV-1 proviral DNA.
Fig. 3.
IPDA amplicon signal failure due to polymorphisms in primer or probe binding sites is infrequent. (A) Frequency of amplicon signal failure for the Ψ and env amplicons. Of the 400 individuals analyzed in this study, we observed Ψ amplicon signal failure in samples from 3.5% and env amplicon signal failure in samples from 2.8%. (B) Representative 2D ddPCR plot from an HIV-1 proviral discrimination reaction illustrating Ψ amplicon signal failure. Ψ amplicon signal failure is easily identified by the absence of droplets in Q1 and Q2. (C) Representative 2D ddPCR plot from an HIV-1 proviral discrimination reaction illustrating env amplicon signal failure, which is readily identified by the absence of droplets in Q2 and in Q4. (D–G) Targeted sequencing (Methods) in individuals with signal failure revealing polymorphisms that account for a lack of signal. Polymorphisms at the 3′ end of the Ψ forward primer account for the Ψ signal failure observed in donor 04036 (B) and another donor, 04022. Donor 04052 (C) and another donor, 04122, showed normal Ψ amplification but failed env amplification. In these cases, polymorphisms in the env primers and probe were responsible. Donor 04036 showed no polymorphisms in this region and a normal droplet distribution.
Using independent longitudinal samples collected several years apart, we confirmed that signal failures were consistently observed within the same donor. For example, in two donors (04036 and 04022) whose samples exhibited signal failure in the Ψ amplicon, signal failure was consistent across two timepoints separated by multiple years of suppressive ART. The IPDA plot for donor 04036 is shown in Fig. 3B. In addition, we performed targeted amplicon sequencing on samples from selected donors with signal failures. In these samples, we observed polymorphisms in the IPDA oligo-binding sites, which would account for signal failure. In each of these samples, we identified the source of amplification failure as a polymorphism at the critical 3′ end of the forward Ψ primer that is predicted to prevent PCR extension (Fig. 3D). Similarly, for donors 04052 and 04122, whose samples showed signal failure for the env amplicon, we identified polymorphisms which could account for that signal failure (Fig. 3 E–G).
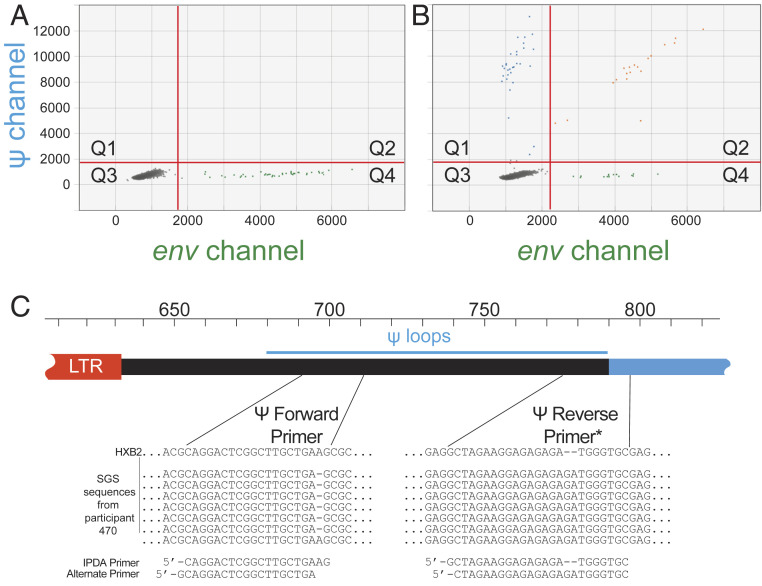
In cases where sequence polymorphisms cause signal failure for the Ψ or env amplicons, minor adjustments in the primer or probe sequences can restore the signal, producing the expected droplet pattern (Q3 >>> Q1 ∼ Q4 > Q2) and allowing for quantitation of intact proviruses. An example is shown in Fig. 4.
Fig. 4.
Custom IPDA oligos can resolve problematic mismatches, salvage amplification in the affected amplicon, and allow detection of intact proviruses. (A) IPDA dot plot showing ψ amplicon failure. (B) Restoration of the normal droplet pattern using custom primers based on single genome sequencing (SGS) of the ψ region. (C) SGS sequences of the ψ forward and reverse primer regions with custom primer design. A single nucleotide length polymorphism at the 3′ end of the ψ forward primer may have precluded amplification. There was also a two-nucleotide insertion in the ψ reverse primer. The use of alternate primers allowed successful ψ amplification shown in B. *Sequences are shown as sense strand nucleotides. The ψ reverse primers used in the IPDA are the reverse complements of the sequences shown.
Discussion
In this large multicohort study, we used a novel reservoir assay, the IPDA, to provide a quantitative picture of the landscape of persistent proviruses that preclude the cure of HIV-1 infection with ART. Previous studies using near-full-length sequencing of individual proviruses showed that the landscape of persistent proviruses is dominated by those with very large deletions and/or APOBEC3G-mediated hypermutation (5, 8, 9, 16–18). Most of these defective proviruses are defective in most viral genes (9) and thus cannot contribute to viral rebound, and these should not be included in reservoir measurements. The IPDA allows direct quantitation of intact proviruses, defined here as those lacking the common fatal defects (large deletions and hypermutation). As discussed below, some proviruses scored as intact by the IPDA contain smaller defects that affect viral fitness (9). Nevertheless, the ability of the IPDA to distinguish intact proviruses from those with the most common fatal defects allows for more accurate measurement of the proviruses that preclude cure.
Because the IPDA separately quantifies intact and defective proviruses, the assay provides information on the composition of the total pool of persistent proviruses. Furthermore, tracking defective proviruses may be of additional value because some defective proviruses may be transcribed in vivo, which could lead to the production of viral proteins that may contribute to persistent immune activation (13–16). Initial studies using the IPDA in a small convenience cohort indicated that defective proviruses outnumbered intact proviruses (9). The present study in a large and diverse group of people with HIV-1 allows us to clearly define the mean and range of frequency values for intact and defective proviruses that persist during ART.
In this study, intact proviruses were detected at a median frequency of 54/106 CD4+ T cells. As with virtually all reservoir assays (4), there was considerable variation between different individuals, with values spanning a 3-log range. Importantly, our results confirm in a large and diverse cohort that overtly defective proviruses outnumber intact proviruses. Indeed, after correction for the fraction of proviruses estimated to contain smaller defects not captured by the IPDA, the ratio of defective to intact proviruses is ∼12.5:1 (Fig. 2D). These results are in general agreement with results from near-full-length sequencing (5, 8, 9, 16–18). However, it must be noted that these sequencing methods all rely on an initial 9-kb PCR; proviruses with large deletions may be detected more readily. Thus, such methods provide qualitative information but cannot be considered quantitative unless the efficiency of single molecule detection in the 9-kb PCR is rigorously verified. In contrast, the IPDA directly counts individual proviruses using digital PCR with highly efficient short amplicons, and the initial performance of the assay has been reported (9). Thus, we can now definitively confirm that the proviral landscape is dominated by highly defective proviruses.
The ratio of intact to total proviruses showed considerable interparticipant variability (Fig. 2E) that is not strictly related to time on ART. We have previously shown using near-full-length genome sequencing that defective proviruses accumulate during the first few weeks of infection, rapidly reaching the levels seen in patients who start ART during chronic infection (8). We have also shown that the fraction of intact to total proviruses is significantly higher in treated SIV infection relative to HIV-1 infection, a difference that may reflect both host and viral factors as well as time on ART (30). The critical factors remain to be identified for patients who start ART during chronic infection. One possibility is that expansion of particular clones of infected cells carrying either intact or defective proviruses could affect the ratio. Dramatic clonal expansions have been documented in previous studies (6, 31–35).
The most important feature of the proviral landscape (shown in Fig. 2D) is the large difference (>50 fold) between intact provirus levels measured by IPDA and levels of replication-competent proviruses determined by the QVOA. The QVOA, previously considered the gold standard assay for the latent reservoir, provides a definitive minimal estimate of the frequency of cells carrying inducible, replication-competent proviruses; however, it is now clear that the QVOA underestimates reservoir size. A recent elegant study showed that this difference is due in part to extinction events in which replication-competent virions released from an infected cell following latency reversal are unable to establish a spreading infection in the assay (7). Previous studies have shown that multiple rounds of T cell activation are required to induce some replication-competent proviruses to produce sufficient virus to establish a spreading infection in vitro (5, 6, 36), which may reflect proviral silencing by epigenetic mechanisms (37, 38) or transcriptional interference from surrounding host genes (39, 40). The degree to which these mechanisms influence reactivation of latent proviruses in vivo, and whether such silencing may be permanent for some proviruses, remain unclear. The IPDA represents a novel approach to reservoir evaluation in that it quantitates intact proviruses regardless of their transcriptional status in any in vitro assay.
For the reasons stated above, the IPDA represents a significant advance in reservoir measurement. It has already proven useful in documenting factors associated with more rapid reservoir decay (19), differential decay rates of intact vs. defective proviruses (9, 19), and an inflection in the decay curve in individuals with HIV-1 on long term ART (19). As with all PCR assays for HIV-1, sequence polymorphisms that affect performance of the IPDA will be observed. In the present study, 6.3% of individuals exhibited amplicon signal failure resulting in nonreportable IPDA results. Through targeted sequencing, we were able to demonstrate that these amplicon signal failures are a result of polymorphisms. Importantly, we show that these polymorphisms are stable over time in individuals on suppressive ART.
An advantage of the IPDA is that problems created by these sequence polymorphisms are readily apparent as an absence or dramatically reduced number of positive droplets for either the ψ or env amplicons. Importantly, the IPDA does not generate a false-negative result in these situations, because the presence of infected cells is indicated by the other amplicon. In contrast, for single amplicon assays, it is impossible to distinguish amplicon failure due to polymorphisms from low template concentrations.
While continued interrogation of large and diverse cohorts of infected individuals with the IPDA is likely to reveal additional unique sequence polymorphisms that interfere with amplification, this study provides an important performance benchmark for the assay. Alternative sets of primers and probes allow measurement of intact proviruses in cases of amplicon failure, as illustrated in Fig. 4. Alternative primers and probes that will allow successful amplification in individuals with the common polymorphisms are currently under development and should alleviate this problem in most cases.
In conclusion, a scalable assay for quantifying intact HIV-1 proviruses is essential for the evaluation of cure strategies. The IPDA provides a novel approach for reservoir quantitation that allows for a much more accurate assessment of reservoir size than standard PCR assays and does not depend on inefficient long-distance PCR. Analysis of samples from a large and diverse set of infected individuals shows than the frequency of proviruses lacking major common defects is on average 50 to 60 per 106 CD4+ T cells, 1 to 2 logs higher than the frequency detected with the QVOA and consistent with previous studies demonstrating that the QVOA underestimates the true size of the latent reservoir.
Methods
Study Participants and Samples.
Samples were obtained from six independent cohorts of adults with HIV-1 on ART living in the United States (Table 1). The aggregated study population was diverse with respect to age, sex, race, HIV risk factors, and other clinical parameters. Details of each contributing cohort will be reported separately. Deidentified, previously collected samples were provided to AccelevirDx in the form of cryopreserved peripheral blood mononuclear cells (PBMCs). Each sample source confirmed that Institutional Review Board approval had been obtained for each study cohort, and that each individual had provided informed consent at the time of enrollment.
IPDA Measurements.
In-depth descriptions of the IPDA rationale and procedure have been published previously (9, 19). In this study, the IPDA was performed by AccelevirDx under company standard operating procedures on DNA from CD4+ T cells isolated from cryopreserved PBMCs. As described previously (9), each IPDA consists of two multiplex ddPCR reactions performed in parallel: the HIV-1 Proviral Discrimination reaction, which distinguishes intact from defective proviruses (Fig. 1A); and the Copy Reference/Shearing reaction, which quantifies DNA shearing and input diploid cell equivalents. In this study, we observed a mean and median DNA shearing index (DSI) of 0.38, consistent with our previous report (9). The shearing correction is applied to both IPDA component reactions to account for the measured intra-amplicon shearing using the DSI, and final IPDA results are reported as proviral frequencies per million CD4+ T cells. ddPCR reactions were assembled via automated liquid handlers to maximize reproducibility. Samples were batch-processed and analyzed. For each batch, CD4+ T cells from uninfected donors were parallel-processed and analyzed as negative controls, while cells of the JLat full-length clone 6.3 served as positive controls. Clone 6.3 from E. Verdin was obtained through the NIH AIDS Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases. Operators were blinded to donor identity and time point.
Targeted Proviral Sequencing.
Targeted proviral sequencing was performed to obtain IPDA amplicon sequences. To obtain material for sequencing, bulk subgenomic PCR was performed on gDNA extracted from CD4+ T cells of HIV-1–infected individuals of interest to amplify regions of the proviral genome containing each IPDA amplicon. Sequencing was performed by independent approaches to verify results.
In some instances, a nested PCR approach was used to amplify these regions. For nested PCR reactions, amplification was performed with the Q5 High-Fidelity PCR Kit (New England BioLabs) with primer concentrations of 0.5 µM. The outer PCR product was diluted 1:1,000 in 10 mM Tris before inner PCR amplification. PCR cleanup of inner PCR reactions was performed using the PureLink PCR Purification Kit (Invitrogen), and the presence of a single PCR amplicon of correct size was verified by agarose gel electrophoresis. Proviral sequencing of inner PCR products was performed by either traditional Sanger sequencing or short-read next-generation sequencing (Amplicon-EZ; GENEWIZ). Sequences were aligned to the HXB2 reference genome (Los Alamos National Laboratory HIV Sequence Database; accession no. K03455.1 GI:1906382) using Geneious Prime to identify polymorphisms.
In other instances, nested PCR amplification was performed with the Platinium Taq DNA Polymerase High-Fidelity PCR Kit (Invitrogen) with primer concentrations of 0.5 µM. The outer PCR product was diluted 1:45 in 5 mM Tris before inner PCR amplification. PCR cleanup of inner PCR reactions was performed using the KAPA Pure Beads Kit (Roche), and the presence of a single PCR amplicon of correct size was verified by agarose gel electrophoresis. Proviral sequencing of inner PCR products was performed by Sanger sequencing (GENEWIZ). Sequences were aligned and analyzed for quality using Sequencher 5.4.
Acknowledgments
We acknowledge the immense contributions of Drs. Deborah McMahon and Joseph Eron, co-chairs of the AIDS Clinical Trials Group (ACTG) A5321 study, to the creation and study of this cohort. This work was supported by the NIH Martin Delaney I4C (UM1AI126603), BEAT-HIV (UM1AI126620), and DARE (UM1AI12661) Collaboratories; the Johns Hopkins Center for AIDS Research (P30AI094189), the HHMI and the Bill and Melinda Gates Foundation (OPP1115715), NIH Grant 5R01 AI140789 (to J.N.B.); the Philadelphia Foundation Robert I. Jacobs Fund; the National Institute on Drug Abuse (R61DA047022); the NIH Small Business Innovation Research (SBIR) Grant Program (R43AI142866 and R44AI124996); and the NSF SBIR Grant Program (1738428). Grant support for the ACTG A5321 study is provided by NIH NIAID Grants UM1AI068636, UM1AI106701, UM1AI068634, and UM1AI069481.
Footnotes
Competing interest statement: Aspects of the IPDA are the subject of patent application PCT/US16/28822 filed by Johns Hopkins University with R.F.S. as an inventor and licensed to AccelevirDx. R.F.S. holds no equity interest in AccelevirDx. R.F.S. consults for Merck and AbbVie on HIV cure-related issues. K.D.R. and M.C. are employees of AccelevirDx. A.M. and G.M.L. are employees of and equity holders in AccelevirDx. B.J.H. is an employee of Merck.
Data Availability.
All experimental results described in the paper are presented in the table and figures. The IPDA method has been previously described in detail (9, 19). A description of study cohorts is given in Table 1. Detailed descriptions of each cohort have been published (19, 21, 24) or will be published separately and can be made available on request.
References
- 1.Finzi D. et al., Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278, 1295–1300 (1997). [DOI] [PubMed] [Google Scholar]
- 2.Chun T. W. et al., Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. U.S.A. 94, 13193–13197 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wong J. K. et al., Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 278, 1291–1295 (1997). [DOI] [PubMed] [Google Scholar]
- 4.Eriksson S. et al., Comparative analysis of measures of viral reservoirs in HIV-1 eradication studies. PLoS Pathog. 9, e1003174 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ho Y. C. et al., Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 155, 540–551 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hosmane N. N. et al., Proliferation of latently infected CD4+ T cells carrying replication-competent HIV-1: Potential role in latent reservoir dynamics. J. Exp. Med. 214, 959–972 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hataye J. M. et al., Principles governing establishment versus collapse of HIV-1 cellular spread. Cell Host Microbe 26, 748–763.e20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bruner K. M. et al., Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat. Med. 22, 1043–1049 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bruner K. M. et al., A quantitative approach for measuring the reservoir of latent HIV-1 proviruses. Nature 566, 120–125 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Procopio F. A. et al., A novel assay to measure the magnitude of the inducible viral reservoir in HIV-infected individuals. EBioMedicine 2, 874–883 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baxter A. E. et al., Multiparametric characterization of rare HIV-infected cells using an RNA-flow FISH technique. Nat. Protoc. 12, 2029–2049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beliakova-Bethell N. et al., Relative efficacy of T cell stimuli as inducers of productive HIV-1 replication in latently infected CD4 lymphocytes from patients on suppressive cART. Virology 508, 127–133 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu G. et al., HDAC inhibition induces HIV-1 protein and enables immune-based clearance following latency reversal. JCI Insight 2, 92901 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pollack R. A. et al., Defective HIV-1 proviruses are expressed and can be recognized by cytotoxic T lymphocytes, which shape the proviral landscape. Cell Host Microbe 21, 494–506.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imamichi H. et al., Defective HIV-1 proviruses produce viral proteins. Proc. Natl. Acad. Sci. U.S.A. 117, 3704–3710 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imamichi H. et al., Defective HIV-1 proviruses produce novel protein-coding RNA species in HIV-infected patients on combination antiretroviral therapy. Proc. Natl. Acad. Sci. U.S.A. 113, 8783–8788 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hiener B. et al., Identification of genetically intact HIV-1 proviruses in specific CD4(+) T cells from effectively treated participants. Cell Rep. 21, 813–822 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee G. Q. et al., Clonal expansion of genome-intact HIV-1 in functionally polarized Th1 CD4+ T cells. J. Clin. Invest. 127, 2689–2696 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peluso M. J. et al., Differential decay of intact and defective proviral DNA in HIV-1 infected individuals on suppressive antiretroviral therapy. JCI Insight 5, 132997 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Antar A. A. R. et al., Longitudinal study reveals HIV-1–infected CD4+ T cell dynamics during long-term antiretroviral therapy. J. Clin. Invest., 10.1172/JCI135953 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwaa A. K., Garliss C. C., Ritter K. D., Laird G. M., Blankson J. N., Elite suppressors have low frequencies of intact HIV-1 proviral DNA. AIDS 34, 641–643 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwon K. J. et al., Different human resting memory CD4+ T cell subsets show similar low inducibility of latent HIV-1 proviruses. Sci. Transl. Med. 12, eaax6795 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Papasavvas E., et al. , Intact HIV reservoir estimated by the intact proviral DNA assay correlates with levels of total and integrated DNA in the blood during suppressive antiretroviral therapy. Clin Infect Dis. 2020 Jun 18:ciaa809. doi: 10.1093/cid/ciaa809. [DOI] [PMC free article] [PubMed]
- 24.Garliss C. C., Kwaa A. K., Blankson J. N., A comparison of different immune activation strategies to reverse HIV-1 latency. Open Forum Infect. Dis. 7, ofaa082 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheehy A. M., Gaddis N. C., Choi J. D., Malim M. H., Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418, 646–650 (2002). [DOI] [PubMed] [Google Scholar]
- 26.Harris R. S., Liddament M. T., Retroviral restriction by APOBEC proteins. Nat. Rev. Immunol. 4, 868–877 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Finzi D. et al., Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat. Med. 5, 512–517 (1999). [DOI] [PubMed] [Google Scholar]
- 28.Siliciano J. D. et al., Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat. Med. 9, 727–728 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Crooks A. M. et al., Precise quantitation of the latent HIV-1 reservoir: Implications for eradication strategies. J. Infect. Dis. 212, 1361–1365 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bender A. M. et al., The landscape of persistent viral genomes in ART-treated SIV, SHIV, and HIV-2 infections. Cell Host Microbe 26, 73–85.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maldarelli F. et al., HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science 345, 179–183 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wagner T. A. et al., HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science 345, 570–573 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simonetti F. R. et al., Clonally expanded CD4+ T cells can produce infectious HIV-1 in vivo. Proc. Natl. Acad. Sci. U.S.A. 113, 1883–1888 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lorenzi J. C. et al., Paired quantitative and qualitative assessment of the replication-competent HIV-1 reservoir and comparison with integrated proviral DNA. Proc. Natl. Acad. Sci. U.S.A. 113, E7908–E7916 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bui J. K. et al., Proviruses with identical sequences comprise a large fraction of the replication-competent HIV reservoir. PLoS Pathog. 13, e1006283 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Z. et al., Expanded cellular clones carrying replication-competent HIV-1 persist, wax, and wane. Proc. Natl. Acad. Sci. U.S.A. 115, E2575–E2584 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jordan A., Bisgrove D., Verdin E., HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J. 22, 1868–1877 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tyagi M., Pearson R. J., Karn J., Establishment of HIV latency in primary CD4+ cells is due to epigenetic transcriptional silencing and P-TEFb restriction. J. Virol. 84, 6425–6437 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lenasi T., Contreras X., Peterlin B. M., Transcriptional interference antagonizes proviral gene expression to promote HIV latency. Cell Host Microbe 4, 123–133 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han Y. et al., Orientation-dependent regulation of integrated HIV-1 expression by host gene transcriptional readthrough. Cell Host Microbe 4, 134–146 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All experimental results described in the paper are presented in the table and figures. The IPDA method has been previously described in detail (9, 19). A description of study cohorts is given in Table 1. Detailed descriptions of each cohort have been published (19, 21, 24) or will be published separately and can be made available on request.