Significance
DNA methylation and histone modifications are two key epigenetic mechanisms in regulating gene expression, heterochromatin assembly, and genome stability. In mammals, maintenance of DNA methylation is mainly mediated by DNA methyltransferase 1 (DNMT1) in a replication-dependent manner. The spatiotemporal regulation of DNMT1 is essential for faithful propagation of DNA methylation patterns between cell generations. Here, we report the direct readout of the heterochromatic mark H3K9me3 by the RFTS domain of DNMT1, which serves to enhance the enzymatic stimulation of DNMT1 by previously characterized H3 ubiquitylation and mediates the cellular colocalization of DNMT1 and H3K9me3. This study uncovers a direct link between the repressive histone modification and DNMT1-mediated DNA methylation.
Keywords: DNA methylation, H3K9me3, DNMT1, allosteric regulation
Abstract
In mammals, repressive histone modifications such as trimethylation of histone H3 Lys9 (H3K9me3), frequently coexist with DNA methylation, producing a more stable and silenced chromatin state. However, it remains elusive how these epigenetic modifications crosstalk. Here, through structural and biochemical characterizations, we identified the replication foci targeting sequence (RFTS) domain of maintenance DNA methyltransferase DNMT1, a module known to bind the ubiquitylated H3 (H3Ub), as a specific reader for H3K9me3/H3Ub, with the recognition mode distinct from the typical trimethyl-lysine reader. Disruption of the interaction between RFTS and the H3K9me3Ub affects the localization of DNMT1 in stem cells and profoundly impairs the global DNA methylation and genomic stability. Together, this study reveals a previously unappreciated pathway through which H3K9me3 directly reinforces DNMT1-mediated maintenance DNA methylation.
DNA methylation is an evolutionarily conserved epigenetic mechanism that critically influences chromatin structure and function (1). In mammals, DNA methylation predominantly occurs at the C-5 position of cytosine within the CpG dinucleotide context, which regulates the silencing of retrotransposons (2), allele-specific genomic imprinting (3), X-chromosome inactivation (4), and tissue-specific gene expression that underlies cell fate commitment (5). During DNA replication, DNA methylation is stably propagated by DNA methyltransferase 1 (DNMT1), which replicates the DNA methylation patterns from parental DNA strands to the newly synthesized strands in a replication-dependent manner (6, 7). Faithful propagation of DNA methylation patterns is essential for clonal transmission of epigenetic regulation between cell generations.
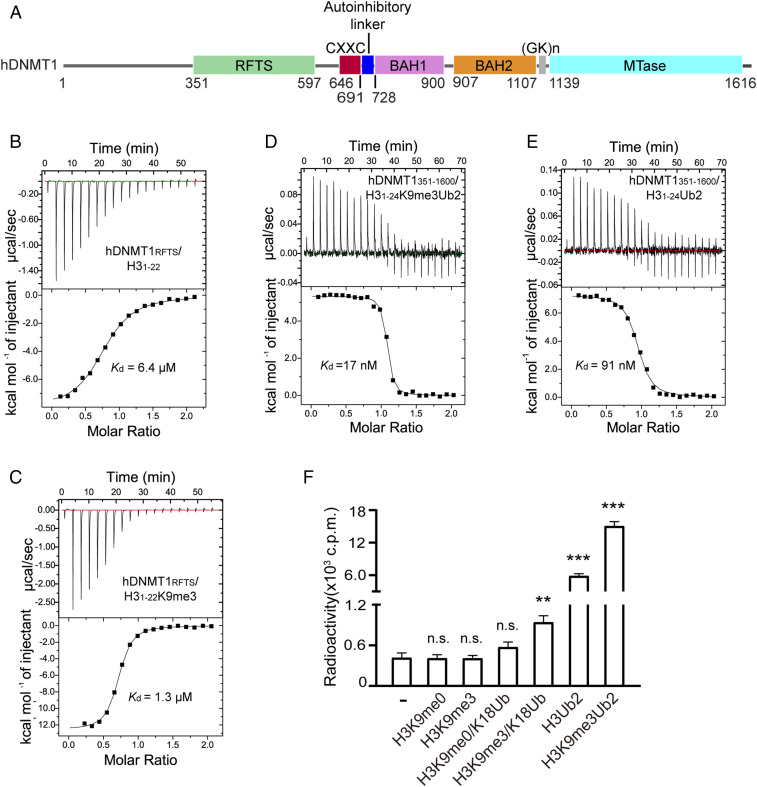
DNMT1 is a multidomain protein, comprised of a large N-terminal regulatory region and a C-terminal methyltransferase (MTase) domain, linked via a conserved (GK)n dipeptide repeat (Fig. 1A). The regulatory region contains a replication foci targeting sequence (RFTS), a CXXC zinc finger domain and a pair of bromo-adjacent homology (BAH) domains (8–11). Recent structural and biochemical evidence has revealed that both the RFTS and CXXC domains regulate the activity of DNMT1 through autoinhibitory mechanisms: the RFTS domain directly interacts with the MTase domain to inhibit DNA binding (10–14), whereas the CXXC domain specifically recognizes unmethylated CpG nucleotides, which in turn blocks the de novo methylation activity of DNMT1 (8, 15). These N-terminal domain-mediated allosteric regulations, together with an inherent enzymatic preference of the MTase domain for hemimethylated CpG sites (9, 16), shape the enzymatic specificity of DNMT1 in maintaining DNA methylation.
Fig. 1.
Specific interaction between the DNMT1 RFTS domain and H3K9me3Ub. (A) Domain architecture of human DNMT1 (hDNMT1), with individual domains delimited by residue numbers. (B and C) ITC-binding assays of hDNMT1RFTS over H31–22 (B) and H31–22K9m3 (C) peptides. (D and E) ITC-binding assays of hDNMT1351–1600 over H31–24K9me3Ub2 (D) and H31–24Ub2 (E) peptides. (F) In vitro DNA methylation assays for hDNMT1351–1600 in the absence or presence of H3 peptides with the indicated modification. Mean and SD were derived from three independent measurements. (**P < 0.01, ***P < 0.001, n.s., not significant, Student’s t test).
The crosstalk between DNA methylation and other gene-silencing mechanisms, such as histone H3K9 methylation, is widely observed across evolution, ensuring proper chromatin assembly and loci-specific gene suppression (17, 18). The H3K9 methylation-dependent DNA methylation in Neurospora and Arabidopsis has been elucidated, which is attributed to a direct or indirect H3K9me2/H3K9me3 readout mechanism of DNA methyltransferases (19, 20). In mammals, H3K9me3 is strongly correlated with DNA methylation at pericentric heterochromatin (21). However, the mechanism by which H3K9 methylation is translated into mammalian DNA methylation remains far from being fully understood. Nevertheless, it has been established that DNMT1-mediated maintenance DNA methylation is critically regulated by Ubiquitin-like, containing PHD and RING Finger domains, 1 (UHRF1) (22, 23). During the S phase, UHRF1 is recruited to replicating heterochromatin through its association with both hemimethylated CpG DNA and H3K9me3 (24), where it stochastically catalyzes the monoubiquitylation of histone H3 at lysine 14 (H3K14Ub), lysine 18 (H3K18Ub) (25), and/or lysine 23 (H3K23Ub) (25–28), and PCNA-associated factor 15 (PAF15) at lysine 15 and 24 (29, 30). The DNMT1 RFTS domain recognizes all these modifications, with a preference for the two-monoubiquitin marks (i.e., H3K18Ub/H3K23Ub), leading to allosteric stimulation of DNMT1 (26). The structure of DNMT1 RFTS domain in complex with H3K18Ub/K23Ub revealed that the H3K18Ub/K23Ub binding leads to structural rearrangement of RFTS and its dissociation with the C-terminal linker, thereby facilitating the conformational transition of DNMT1 from an autoinhibitory state to an active state (26, 31). These observations suggest a UHRF1-bridged link between H3K9me3 and DNA methylation. However, whether H3K9me3 directly interacts with DNMT1 to regulate maintenance DNA methylation remains unknown.
To determine how H3K9me3 affects DNMT1-mediated maintenance DNA methylation, we examined the histone-binding activity of DNMT1 RFTS domain and its relationship to the chromatin association and enzymatic activity of DNMT1. Through isothermal titration calorimetry (ITC) and in vitro enzymatic assays, we identified that the RFTS domain binds preferably to H3K9me3 over H3K9me0, which serves to strengthen the enzymatic stimulation of DNMT1 by H3 ubiquitylation (H3Ub). Furthermore, we determined the crystal structure of bovine RFTS domain complexed with H3K9me3 peptide and two ubiquitins, providing the molecular basis for the H3K9me3 recognition. In addition, our cellular and genomic methylation analysis demonstrated that impairment of the RFTS-H3K9me3Ub recognition led to reduced colocalization of DNMT1 with H3K9me3, a global loss of DNA methylation patterns and genome instability in mouse embryonic stem (ES) cells. Together, this study provides a mechanism by which H3K9me3 directly regulates DNMT1-mediated maintenance DNA methylation in mammalian cells.
Results
The RFTS Domain of DNMT1 Is an H3K9me3Ub Reader Module.
Recent studies have indicated that UHRF1-mediated recognition of H3K9me3 and hemimethylated CpG DNA stimulates its E3 ubiquitin ligase activity on histone H3 (25, 32), thus providing a linkage between H3K9me3 and H3 ubiquitylation during UHRF1/DNMT1-mediated DNA methylation. This observation prompted us to ask whether or not H3K9me3 directly influences the interaction between the RFTS domain of DNMT1 and ubiquitylated H3. To this end, we performed ITC assays using the purified human DNMT1 RFTS domain (hDNMT1RFTS) and histone H3 peptides (residues 1–22, H31–22), either unmodified or with H3K9me3 modification (H31–22K9me3) (SI Appendix, Table S1). Titration of hDNMT1RFTS with H31–22 gives a dissociation constant (Kd) of 6.4 μM (Fig. 1B). In comparison, titration of hDNMT1RFTS with H31–22K9me3 gives a Kd of 1.3 μM, suggesting that hDNMT1RFTS has an ∼5-fold binding preference for H3K9me3 over unmodified H3 tails (Fig. 1C). Furthermore, we probed the interaction of hDNMT1RFTS with the H31–22 peptide acetylated at K9 (H31–22K9Ac) and other histone trimethylations (H3K4me3, H3K27me3, H3K36me3, and H4K20me3) and observed a much weaker binding (SI Appendix, Fig. S1 A–E). These observations not only confirm the previously observed interaction between hDNMT1RFTS and H3 (26), but also support a preferential binding of H3K9me3 over H3K9me0 by hDNMT1RFTS.
Next, we asked whether H3K9me3 can enhance the interaction between H3K18Ub/K23Ub and the RFTS domain in the context of full-length DNMT1. Following a previously established approach (26, 33), we installed dual ubiquitin marks onto a K18C/K23C mutant form of H31–24 (H31–24Ub2) or H31–24K9me3 (H31–24K9me3Ub2) peptides through dichloroacetone (DCA) linkage (33), followed by ITC-binding assay against a hDNMT1 fragment (residues 351–1600, hDNMT351–1600) spanning from the RFTS domain to the C-terminal MTase domain (Fig. 1A). We found that hDNMT351–1600 binds to the H31–24K9me3Ub2 peptide with a Kd of 17 nM (Fig. 1D), exhibiting an ∼5-fold binding preference over the H31–24Ub2 peptide (Kd = 91 nM) (Fig. 1E). These data support that, together with H3Ub, H3K9me3 provides a signal for tethering DNMT1 onto chromatin via its RFTS domain.
The H3K9me3 Readout Boosts the Stimulation Effect of H3Ub on DNMT1 Activity.
In light of a previous study showing that recognition of H3Ub by the DNMT1 RFTS domain results in enhanced enzymatic activity of DNMT1 (26), we have further evaluated the effect of H3K9me3 on the enzymatic stimulation of hDNMT1. Here, we found that, consistent with the previous observation (26), incubation of hDNMT1351–1600 with equimolar H3K18Ub and H3Ub2 led to enhanced hDNMT1351–1600 activity by 1.3- and 14-fold, respectively (Fig. 1F). In contrast, incubation with equimolar H3K9me3/K18Ub and H3K9me3Ub2 promoted the enzymatic activity of hDNMT1351–1600 by 2.2- and 36-fold, respectively. Note that incubation of hDNMT1351–1600 with equimolar H3K9me0 or H3K9me3 did not change its activity appreciably (Fig. 1F), although at increased peptide concentrations, the activity of hDNMT1351–1600 is significantly higher when incubated with H3K9me3, relative to H3K9me0 peptide controls (SI Appendix, Fig. S1F). Together, these results show the specific recognition of H3K9me3 over H3K9me0 by hDNMT1RFTS elevates the stimulation effect of H3Ub, a transient ligand of hDNMT1RFTS as previously shown (26, 27).
The Structure of RFTS Domain in Complex with H3K9me3 Peptide and Ubiquitin.
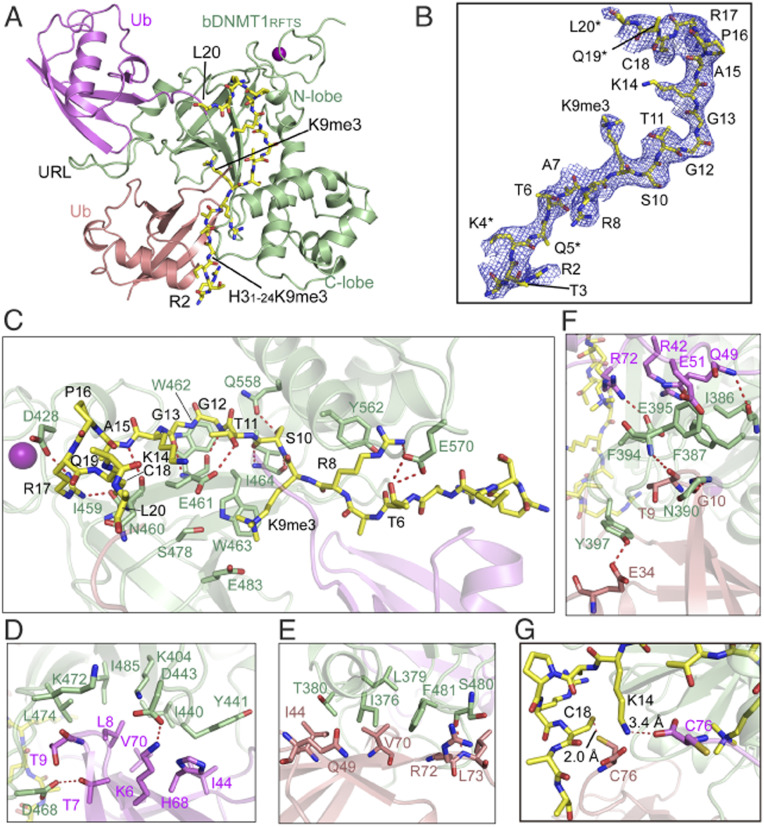
To gain a molecular understanding of the RFTS-H3K9me3 recognition, we next crystallized the complex of the RFTS domain from bovine DNMT1 (bDNMT1RFTS), which shares ∼87% sequence identity with hDNMT1RFTS (SI Appendix, Fig. S2A), with both the H31–24K9me3/K18C/K23C peptide and G76C-mutated ubiquitin, and solved the structure at 3.0-Å resolution (Fig. 2A and SI Appendix, Table S2). The structure reveals a bDNMT1RFTS-H3K9me3-Ub2 complex, with clearly traced H3 peptide from Arg2 to Leu20 (Fig. 2B). The bDNMT1RFTS domain folds into a two-lobe architecture, with an N-lobe harboring a zinc finger and a C-lobe dominated by a helical bundle (Fig. 2A), resembling that of hDNMT1RFTS (11, 14, 26, 31). Like the previously reported hDNMT1RFTS-H3K18Ub/K23Ub complex (26), the H3K9me3 peptide traverses across the surface of the C-lobe and N-lobe of the bDNMT1RFTS domain and engages extensive intermolecular interactions, with the side chain of H3K9me3 residue embraced by a surface groove at the interface between bDNMT1RFTS and one of the ubiquitin molecules (Fig. 2 B and C). The two ubiquitin molecules are mostly positioned at the N-lobe of the RFTS domain, separated by the ubiquitin recognition loop (URL; residues 386–398), and interact with the RFTS domain and H3 in a fashion similar to what was previously observed for the hDNMT1RFTS-H3K18Ub/K23Ub complex (Fig. 2 C–F) (26). As with the hDNTM1RFTS-H3K18Ub/K23Ub complex, binding of the H3K9me3 peptide to bDNMT1RFTS induces partial closure of the cleft between the N- and C-lobe of the RFTS domain, while binding of ubiquitins induces a disorder-to-order transition of the RFTS URL (SI Appendix, Fig. S2 B and C).
Fig. 2.
Structural details for the bDNMT1RFTS domain in complex with H3K9me3K18C/K23C and ubiquitin. (A) Crystal structure of bovine DNMT1RFTS (light green) in complex with H31–24K9me3/K18C/K23C peptide (yellow) and G76C-mutated ubiquitin (magenta and salmon). The zinc ions are shown as purple spheres. (B) The Fo-Fc omit map (blue) of the H31–24K9me3/K18C/K23C peptide (yellow sticks) in the complex, contoured at the 1.5-σ level, with individual amino acids labeled. Partial side-chain modeling due to lack of electron density is indicated by asterisk. (C) Close-up views of the intermolecular interactions between bDNMT1 RFTS (light green) and the H31–24K9me3K18CK23C peptide (yellow stick). The two ubiquitin molecules are colored in salmon and magenta, respectively. Hydrogen bonds are shown as dashed lines. The zinc ion is shown as purple sphere. (D and E) Close-up views of the intermolecular interactions between the conserved I44 patch of two ubiquitin molecules (magenta and salmon) and bDNMT1 RFTS (light green). (F) Close-up views of the intermolecular interactions between URL of bDNMT1 RFTS (light green) and two ubiquitin molecules (magenta and salmon). (G) Close-up views of the close proximity between the H31–24K9me3K18CK23C peptide (yellow stick) and the ubiquitin molecules (salmon and magenta).
Of particular note, residue G76C of one ubiquitin and H3 K18C are positioned in a distance that allows disulfide bond formation, while the C terminus of the other ubiquitin is near to the side chain of H3K14 and also likely accessible to the disordered H3K23 (Fig. 2G), reminiscent of the close proximity of these moieties in the hDNMT1RFTS-H3K18Ub/K23Ub complex. Note that in the hDNMT1RFTS-H3K18Ub/K23Ub complex, the H3 peptide was conjugated with two G76C-mutated ubiquitins through disulfide linkages before mixing with hDNMT1RFTS, whereas in the current complex, no covalent linkage was involved between the H3K9me3 peptide and the ubiquitins prior to the complex assembly. In this regard, the similar positioning of H3 and ubiquitins between the two complexes, crystallized under different conditions, reinforces the notion that H3 and ubiquitin molecules are able to engage DNMT1RFTS via independent intermolecular interactions (26). These results also indicate that H3K9me3 and H3Ub2 may act synergistically or independently for associating DNMT1 onto chromatin.
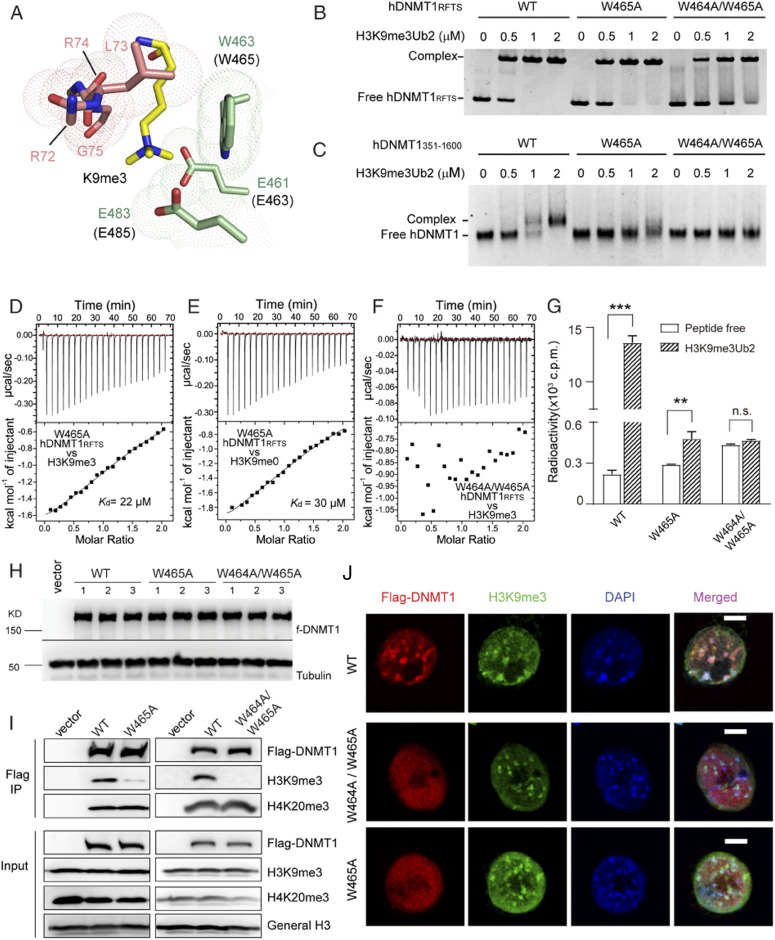
Strikingly, in our structure, we found that both DNMT1 and H3Ub contribute to H3K9me3 engagement. First, H3K9me3 stacks its side chain against the indole ring of bDNMT1RFTS W463, with the quaternary ammonium group interacting with the side-chain carboxylates of bDNMT1RFTS E461 and E483 through electrostatic attractions (Fig. 3A). In addition, residues R72-G75 of one of the bound ubiquitins make both main-chain and side-chain van der Waals contacts with H3K9me3 from an opposite direction, leading to formation of an H3K9me3-binding pocket (Fig. 3A). Such an H3K9me3-binding site of DNMT1RFTS is distinct from the multiwalled aromatic cage typical for a trimethyl-lysine (Kme3) reader (34), but is reminiscent of the interaction between the PHD finger domain of E3 ubiquitin ligase TRIM33 and H3K9me3 (SI Appendix, Fig. S2D) (35). Through superimposition with the previously determined hDNTM1RFTS-H3K18Ub/K23Ub structure (26), we found that those two structures overlap very well, with an RMSD of 0.6 Å over 384 aligned Cα atoms (SI Appendix, Fig. S2C). However, in comparison to the H3K9me0 binding, association of H3K9me3 with bDNMT1RFTS W463 leads to an increase in buried surface area of the H3K9-engaging pocket by 8–9 Å2 (SI Appendix, Fig. S2C).
Fig. 3.
Biochemical and cellular analysis of the RFTS-H3K9me3Ub2 interaction. (A) Close-up view of the residues forming the H3K9me3-binding pocket, in the same color scheme as in Fig. 2. For clarity, the side chains of ubiquitin R72 and R74 are not shown. (B) EMSA analysis of the interaction between hDNMT1351–597, either WT, W465A, or W464A/W465A, and the H31–24K9me3Ub2 peptide. (C) EMSA analysis of the interaction between hDNMT1351–1600, either WT, W465A, or W464A/W465A, and the H31–24K9me3Ub2 peptide. (D and E) ITC-binding assays of hDNMT1RFTS W465A mutant over H31–22K9me3 (D) and H31–22 (E) peptides. (F) ITC-binding assays of hDNMT1RFTS W464A/W465A mutant over H31–22K9me3 peptide. (G) DNA methylation activities of hDNMT1351–1600, either WT, W465A, or W464A/W465A, in the absence of presence of H31–24K9me3Ub2 peptide. Mean and SD were derived from three independent measurements (**P < 0.01, ***P < 0.001, n.s., not significant, Student’s t test). (H) Immunoblots of the indicated Flag-tagged DNMT1 after stable reconstitution into the independently derived 1KO-ESC lines. (I) CoIP (Top) detecting association of the indicated Flag-tagged DNMT1 with H3K9me3 or H4K20me3; and (Bottom) immunoblots of input. (J) Representative confocal immunofluorescence images revealing localization of the indicated DNMT1 (Flag-tagged, red), H3K9me3 (green), and chromatin (stained by DAPI, blue) in the 1KO-ESC stable expression lines synchronized at S phase. (Scale bar, 5 micrometers.)
Sequence analysis of the DNMT1 RFTS domains reveals that both the H3K9me3- and ubiquitin-binding sites are highly conserved across evolution (SI Appendix, Fig. S2A). Among these, a di-tryptophan motif (W462 and W463 in bDNMT1) is unvaried from zebrafish to human (SI Appendix, Fig. S2A), in line with their important roles in binding to H3K9me3 and surrounding H3 tail residues (Figs. 2C and 3A).
Mutational Analysis of the DNMT1-H3K9me3Ub2 Binding.
To test the above structural observations, we selected hDNMT1 W464 and W465, corresponding to bDNMT1 W462 and W463, respectively (SI Appendix, Fig. S2A), for mutagenesis. Using electrophoretic mobility shift assays (EMSA), we found that the mutation of hDNMT1 W465 into alanine (W465A) leads to a significant reduction in the H3K9me3Ub2-binding affinity of hDNMT1RFTS and hDNMT1351–1600 (Fig. 3 B and C and SI Appendix, Fig. S3A), while the W464A/W465A mutation reduced the hDNMT1RFTS-H3K9me3Ub2 binding even further (Fig. 3B) and led to nearly undetectable binding between hDNMT1351–1600 and H3K9me3Ub2 (Fig. 3C). Likewise, the ITC assays indicated that the W465A mutation reduced the H3K9me0- and H3K9me3-binding affinities of hDNMT1RFTS by ∼5- and ∼17-fold, respectively (Fig. 3 D and E); introduction of a W464A/W465A mutation largely abolished the hDNMT1RFTS-H3K9me3 binding (Fig. 3F). Furthermore, we performed in vitro DNA methylation assays to evaluate the effect of these mutations on the enzymatic activity of DNMT1. Unlike wild-type hDNMT1351–1600, which shows substantial increase in methylation efficiency in the presence of H3K9me3K18Ub and H3K9me3Ub2 (Fig. 1F) as well as an enzymatic preference for H3K9me3 over H3K9me0 (SI Appendix, Fig. S1F), introduction of the W465A or W464A/W465A mutation into hDNMT1351–1600 greatly dampened the methylation-stimulating effects of H3K9me3, H3K9me3K18Ub, or H3K9me3Ub2 (Fig. 3G and SI Appendix, Fig. S3 B and C). These data support the important roles of the W464 and W465 residues in mediating the DNMT1-H3K9me3/H3Ub interaction and the consequent enzymatic activation of DNMT1.
Given that H3Ub is reportedly a transient mark in cells (36), we have focused on examining the requirement of the DNMT1RFTS domain for H3K9me3 binding in cells. To this end, we used the Dnmt1-knockout mouse embryonic stem cells (1KO-ESC) (37) and generated multiple 1KO-ESC lines with comparable, stable expression of exogenous DNMT1, either wild-type (DNMT1WT) or an H3K9me3-binding–defective mutant (DNMT1W465A or DNMT1W464A/W465A) (Fig. 3H). By coimmunoprecipitation (CoIP), we found that both the W465A and W464A/W465A mutations interfere with efficient binding of DNMT1 to H3K9me3, but not H4K20me3, in cells (Fig. 3I). Furthermore, in the ES cells synchronized at S phase, confocal immunofluorescence microscopy showed punctate nuclear foci of DNMT1WT that overlapped with the H3K9me3-marked, DAPI-dense regions of chromatin (Fig. 3J), whereas the RFTS-mutant DNMT1 proteins show a more diffuse distribution in the nucleus and lose their colocalization with H3K9me3 (Fig. 3J). Together, these data support that the histone-engaging activity of the RFTS domain is critical for both the enzymatic stimulation and chromatin occupancy of DNMT1.
The RFTS Domain of DNMT1 Is Crucial for Global DNA Methylation in Cells.
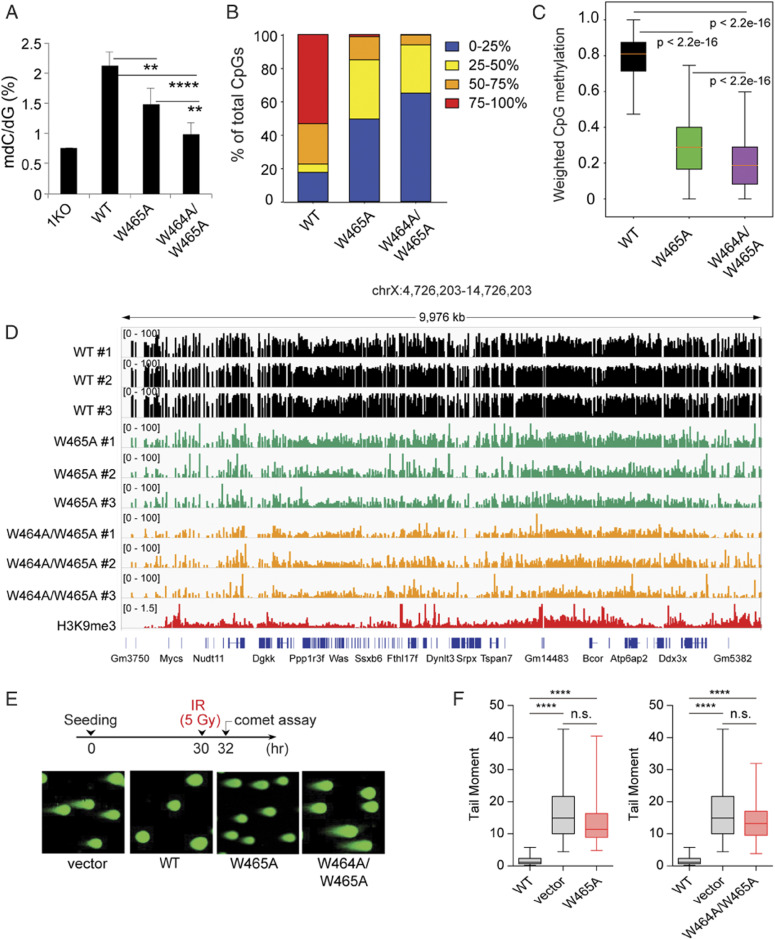
Next, we sought to examine the role of the DNMT1 RFTS domain in maintenance DNA methylation in cells. First, we used liquid chromatography–mass spectrometry (LC-MS) to quantify global levels of methylated cytosine (5mC) in 1KO-ESC cells reconstituted with DNMT1WT or the RFTS-defective mutant. As expected, stable transduction of DNMT1WT led to an increase in overall 5mC level (Fig. 4A, WT vs. 1KO). However, such an increase was found compromised by the W465A or W464A/W465A mutation, with the latter exhibiting a more severe DNA methylation defect, lacking significant methylation-stimulating effect relative to 1KO-ESC cells (Fig. 4A). Further, we carried out genome-wide methylation profiling with enhanced reduced representation bisulfite sequencing (eRRBS). Our eRRBS data showed the desired high bisulfite conversion rates (SI Appendix, Fig. S4A, 99.79–99.83%; and Dataset S1), with at least fivefold coverage for ∼5–8 million of CpG sites in all samples. Relative to 1KO-ESCs reconstituted with DNMT1WT, those with DNMT1W465A or DNMT1W464A/W465A showed a marked decrease in overall CpG methylation, with a complete loss of most heavily methylated CpG sites that are typically seen at heterochromatic repetitive elements (38) (Fig. 4B, red; and SI Appendix, Fig. S4 B and C). In particular, there is a significantly decreased level of CpG methylation at the H3K9me3-decorated genomic regions in cells reconstituted with either DNMT1W465A or DNMT1W464A/W465A, relative to DNMT1WT controls (Fig. 4C), as demonstrated by subtelomeric regions located in chromosomes 1 and X (Fig. 4D and SI Appendix, Fig. S4D). Again, these cellular assays show that the double RFTS mutant (DNMT1W464A/W465A) produced more severe CpG methylation defects, relative to DNMT1W465A (Fig. 4 A–D and SI Appendix, Fig. S4 A–D), which is consistent with our in vitro biochemical and enzymatic observations. Collectively, we have demonstrated an essential role of the histone-engaging RFTS domain in maintenance of CpG methylation at the H3K9me3-associated heterochromatin in cells.
Fig. 4.
The effects of RFTS mutations on cellular CpG methylation pattern and IR response. (A) LC-MS analysis of global 5-methyl-2'-deoxycytidine (5-mdC) levels (calculated as 5-mdC/2'-deoxyguanosine on the y axis) in 1KO-ESC lines after stable transduction of empty vector (1KO) or the indicated DNMT1 (n = 3–6 biological replicates). Data are mean ± SD. (B) Bar plots showing the CpG methylation levels in 1KO-ESC lines with stable expression of the indicated DNMT1. (C) Box plot shows weighted methylation levels of CpGs located within H3K9me3 peaks in 1KO-ESC lines with stable expression of the indicated DNMT1. Scores are calculated after aggregating data from three replicated samples per group. Only H3K9me3 peaks with the mapped reads are included (n = 103,603 for WT, 101,791 for W465, and 102,892 for W464A/W465A). In the plot, the box depicts the 25th to 75th percentiles, with the band in the box representing the median. (D) Representative IGV view shows CpG methylation at an H3K9me3-marked genomic region located in chromosome X among three replicated 1KO-ESC lines with stable expression of the indicated DNMT1. Cytosines covered by at least five reads according to eRRBS data are shown, with each site designated by a vertical line. (E and F) Neutral comet assays revealing DNA breaks (DNA breaks quantified in F) after IR treatment of 1KO-ESC cells reconstituted with vector control or the indicated DNMT1. Box-and-whisker plots in F depict 25–75% in the box; whiskers are 10–90%, and median is indicated. Data represent the mean ± SEM from >100 cells (n = 3 biologically independent replicates) (****P < 0.0001, n.s., not significant, Student’s t test).
Maintenance of proper DNA methylation levels in cells is crucial for genome stability (39). To investigate the role of DNMT1-mediated DNA methylation in genome stabilization, we further challenged 1KO-ESC cells, reconstituted with WT or mutant DNMT1, with ionizing radiation (IR) treatment. By using the neutral comet assay (Fig. 4E), a surrogate method for scoring DNA double-strand break (DSB) lesions, we found that loss of DNMT1 rendered ESC cells hypersensitivity to IR treatment, reflecting a possible change in chromatin structure or impairment in DSB repair (Fig. 4 E and F), a phenotype that can be rescued by complementation with WT DNMT1. Importantly, cells with the RFTS single mutant (DNMT1W465A) and double mutant (DNMT1W464A/W465A) exhibited modest and severe impairment of IR resistance (Fig. 4 E and F), respectively, confirming the role of DNMT1-mediated DNA methylation in genomic stability maintenance.
Together, the above results strongly indicated that the recognition of H3K9 trimethylation by DNMT1 is important for maintenance DNA methylation, and also genome stability and radiation resistance of ES cells.
Discussion
The crosstalk between H3K9 methylation and DNA methylation, two of the major epigenetic silencing mechanisms, critically influences gene silencing and heterochromatin formation (17, 18). For instance, previous studies have demonstrated that Suv39h-mediated H3K9-trimethylation promotes the enrichment of DNA methylation at major satellite repeats of pericentromeric heterochromatin (40), which is essential for maintaining heterochromatic assembly and genome stability (41). Whereas the mechanism by which H3K9 methylation and DNA methylation crosstalk has been established in fungi and plants (19, 20), how H3K9me3 is translated into DNA methylation in mammals remains elusive. Through a set of structural, biochemical, and cellular analyses, this study shows that the RFTS domain of DNMT1 specifically recognizes H3K9me3 over H3K9me0, in conjunction with the previously identified H3Ub mark. The readout of H3K9me3 not only enhances the stimulation effect of H3Ub on DNMT1 activity but also regulates the genome targeting of DNMT1. This study therefore establishes a direct link between histone H3K9me3 modification and DNMT1-mediated maintenance DNA methylation, which influences the global DNA methylation patterns and genome stability.
Previous studies from others and us demonstrated that DNMT1 assumes autoinhibitory conformations either in the DNA-free state (10, 11) or in the presence of unmethylated CpG DNA (8), in which the autoinhibitory linker located between the CXXC and BAH1 domains serves as a key inhibition-enforcing element in both regulations (8, 10). This study reveals that the recognition between the RFTS domain and H3K9me3 strengthens the RFTS interaction with histone H3 tails that carry either one or two monoubiquitin marks, directly contributing to the relief of the autoinhibition of DNMT1. The specific recognition of H3K9me3 by the RFTS domains of DNMT1 presumably helps transduce the H3K9me3 signal into DNA methylation, thereby ensuring the epigenetic fidelity of DNA methylation in heterochromatin domains.
DNMT1-mediated maintenance DNA methylation is subjected to a cell cycle-dependent regulation by multiple chromatin regulators, such as UHRF1 (22, 23, 42, 43), Ubiquitin specific protease 7 (USP7) (36, 42–45), PCNA (46, 47), and PAF15 (29, 30). The RFTS-H3K9me3 interaction reinforces the previously identified UHRF1-H3K9me3 axis on chromatin targeting of DNMT1. UHRF1 harbors a tandem TUDOR domain that recognizes H3K9me3 (48–53) and a RING finger domain that mediates H3 ubiquitylation for DNMT1 targeting (26–28), thereby serving as a platform for the functional crosstalk between H3K9me3 and DNA methylation (54). However, disruption of the interaction between H3K9me3 and UHRF1 via the TUDOR domain mutation only leads to a modest (∼10%) reduction of DNA methylation. In this regard, the direct readout of H3K9me3Ub by the RFTS domain of DNMT1 provides a potentially redundant mechanism in transducing H3K9me3 into maintenance DNA methylation. Note that the RFTS-H3K9me3Ub readout does not involve the discrimination of the methylation state of DNA substrates, therefore providing a mechanism in supporting the region-specific methylation maintenance by DNMT1, as opposed to site-specific methylation maintenance (55, 56). Consistently, impairment of this interaction in cells compromises the DNMT1-mediated CpG methylation, leading to an aberrant landscape of DNA methylation and defects in maintenance of genome stability. It remains to be determined whether the DNMT1 mutations introduced in this study also affect the interaction of DNMT1 with other regulatory factors, such as PAF15. This targeting-coupled allosteric stimulation mechanism is reminiscent of the role of histone H3K4me0 in DNMT3A-mediated de novo DNA methylation, in which the specific recognition of the DNMT3A ADD domain with H3K4me0 allosterically stimulates its enzymatic activity, thereby providing a mechanism of locus-specific DNA methylation establishment (57).
The DNMT1 RFTS domain adds to the reader modules that offer interpretation of specific histone modifications (34), but deviates from the typical Kme3 readout that depends on aromatic or other hydrophobic residues (34): the RFTS domain presents a single tryptophan to stack against H3K9me3, unlike the typical Kme3 readout involving a hydrophobic cage composed of multiple aromatic residues (34). These observations highlight the evolutionary divergence of histone modification-binding mechanisms.
Materials and Methods
Details of the materials and methods including plasmids, protein expression and purification, chemical modifications of histones, crystallization and structure determination, ITC-binding assay, DNA methylation kinetics assay, electrophoretic mobility shift assay, cell lines and tissue culture, antibodies and Western blotting, quantification of 5-methyl-2′-deoxycytidine in genomic DNA, confocal immunofluorescence, coimmunoprecipitation, preparation of eRRBS libraries, eRRBS data processing, eRRBS data analysis, and neutral comet assay are provided in SI Appendix. The two-tailed Student t tests were performed to compare distributions between different groups. The P value lower than 0.01 was considered to be statistically significant. Coordinates and structure factors for the bDNMT1RFTS-H3K9me3-Ub2 complex have been deposited in the Protein Data Bank under accession code 6PZV (58), while the data collection and structure refinement statistics are provided in SI Appendix. The eRRBS data have been deposited in Gene Expression Omnibus under accession code GSE145698 (59).
Supplementary Material
Acknowledgments
We thank staff members at the Advanced Light Source, Lawrence Berkeley National Laboratory for access to X-ray beamlines. We are also grateful for professional support of Drs. D. Chen and H. Uryu in the Wang Laboratory and the University of North Carolina facilities including Genomics Core, which are partly supported by the UNC Cancer Center Core Support Grant P30-CA016086. This work was supported by NIH grants (1R35GM119721 to J.S., 5R01 CA210072 to Y.W., R01GM079641 to O.G., 1R35GM126900 to B.D.S., 1R01CA215284 and 1R01CA211336 to G.G.W., and 1R01CA198279 and 1R01CA201268 to K.M.M.), NIH T32 Training Fellowship for Integrated Training in Cancer Model Systems (T32CA009156) and an American Cancer Society Postdoctoral Fellowship (PF-19-027-01-DMC) to C.J.P. and grants from When Everyone Survives Leukemia Research Foundation (to G.G.W.) and Gabrielle’s Angel Foundation for Cancer Research (to G.G.W.). D.J.P. was supported by a Specialized Center of Research grant by the Leukemia and Lymphoma Society and by the Memorial Sloan-Kettering Cancer Center Core Grant (P30CA008748). This work was also supported in part by the Intramural Research Program of the National Institute of Environmental Health Sciences, NIH (ES101965 to P.A.W.). J.P.C. is supported in part by T32 CA09302. G.G.W. and K.M.M. are American Cancer Society (ACS) Research Scholars, C.J.P. is an ACS postdoctoral fellow, and G.G.W. is a Leukemia and Lymphoma Society Scholar.
Footnotes
Competing interest statement: O.G. and B.D.S are cofounders of EpiCypher, Inc.
Data deposition: Coordinates and structure factors for the bDNMT1RFTS-H3K9me3-Ub2 complex have been deposited in the Protein Data Bank under accession code 6PZV. The enhanced reduced representation bisulfite sequencing data have been deposited in Gene Expression Omnibus under accession code GSE145698.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2009316117/-/DCSupplemental.
References
- 1.Smith Z. D., Meissner A., DNA methylation: Roles in mammalian development. Nat. Rev. Genet. 14, 204–220 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Walsh C. P., Chaillet J. R., Bestor T. H., Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 20, 116–117 (1998). [DOI] [PubMed] [Google Scholar]
- 3.Li E., Beard C., Jaenisch R., Role for DNA methylation in genomic imprinting. Nature 366, 362–365 (1993). [DOI] [PubMed] [Google Scholar]
- 4.Panning B., Jaenisch R., RNA and the epigenetic regulation of X chromosome inactivation. Cell 93, 305–308 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Yagi S. et al., DNA methylation profile of tissue-dependent and differentially methylated regions (T-DMRs) in mouse promoter regions demonstrating tissue-specific gene expression. Genome Res. 18, 1969–1978 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goll M. G., Bestor T. H., Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 74, 481–514 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Jeltsch A., On the enzymatic properties of Dnmt1: Specificity, processivity, mechanism of linear diffusion and allosteric regulation of the enzyme. Epigenetics 1, 63–66 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Song J., Rechkoblit O., Bestor T. H., Patel D. J., Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation. Science 331, 1036–1040 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song J., Teplova M., Ishibe-Murakami S., Patel D. J., Structure-based mechanistic insights into DNMT1-mediated maintenance DNA methylation. Science 335, 709–712 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takeshita K. et al., Structural insight into maintenance methylation by mouse DNA methyltransferase 1 (Dnmt1). Proc. Natl. Acad. Sci. U.S.A. 108, 9055–9059 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Z. M. et al., Crystal structure of human DNA methyltransferase 1. J. Mol. Biol. 427, 2520–2531 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bashtrykov P. et al., Targeted mutagenesis results in an activation of DNA methyltransferase 1 and confirms an autoinhibitory role of its RFTS domain. ChemBioChem 15, 743–748 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Berkyurek A. C. et al., The DNA methyltransferase Dnmt1 directly interacts with the SET and RING finger-associated (SRA) domain of the multifunctional protein Uhrf1 to facilitate accession of the catalytic center to hemi-methylated DNA. J. Biol. Chem. 289, 379–386 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Syeda F. et al., The replication focus targeting sequence (RFTS) domain is a DNA-competitive inhibitor of Dnmt1. J. Biol. Chem. 286, 15344–15351 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Svedruzić Z. M., Reich N. O., Mechanism of allosteric regulation of Dnmt1’s processivity. Biochemistry 44, 14977–14988 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Yoder J. A., Soman N. S., Verdine G. L., Bestor T. H., DNA (cytosine-5)-methyltransferases in mouse cells and tissues. Studies with a mechanism-based probe. J. Mol. Biol. 270, 385–395 (1997). [DOI] [PubMed] [Google Scholar]
- 17.Cheng X., Blumenthal R. M., Coordinated chromatin control: Structural and functional linkage of DNA and histone methylation. Biochemistry 49, 2999–3008 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du J., Johnson L. M., Jacobsen S. E., Patel D. J., DNA methylation pathways and their crosstalk with histone methylation. Nat. Rev. Mol. Cell Biol. 16, 519–532 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du J. et al., Dual binding of chromomethylase domains to H3K9me2-containing nucleosomes directs DNA methylation in plants. Cell 151, 167–180 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freitag M., Hickey P. C., Khlafallah T. K., Read N. D., Selker E. U., HP1 is essential for DNA methylation in neurospora. Mol. Cell 13, 427–434 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Cedar H., Bergman Y., Linking DNA methylation and histone modification: Patterns and paradigms. Nat. Rev. Genet. 10, 295–304 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Bostick M. et al., UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 317, 1760–1764 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Sharif J. et al., The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 450, 908–912 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Liu X. et al., UHRF1 targets DNMT1 for DNA methylation through cooperative binding of hemi-methylated DNA and methylated H3K9. Nat. Commun. 4, 1563 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Harrison J. S. et al., Hemi-methylated DNA regulates DNA methylation inheritance through allosteric activation of H3 ubiquitylation by UHRF1. eLife 5, e17101 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ishiyama S. et al., Structure of the Dnmt1 reader module complexed with a unique two-mono-ubiquitin mark on histone H3 reveals the basis for DNA methylation maintenance. Mol. Cell 68, 350–360.e7 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Nishiyama A. et al., Uhrf1-dependent H3K23 ubiquitylation couples maintenance DNA methylation and replication. Nature 502, 249–253 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Qin W. et al., DNA methylation requires a DNMT1 ubiquitin interacting motif (UIM) and histone ubiquitination. Cell Res. 25, 911–929 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishiyama A. et al., Two distinct modes of DNMT1 recruitment ensure stable maintenance DNA methylation. Nat. Commun. 11, 1222 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.González-Magaña A. et al., Double monoubiquitination modifies the molecular recognition properties of p15PAF promoting binding to the reader module of Dnmt1. ACS Chem. Biol. 14, 2315–2326 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Li T. et al., Structural and mechanistic insights into UHRF1-mediated DNMT1 activation in the maintenance DNA methylation. Nucleic Acids Res. 46, 3218–3231 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fang J. et al., Hemi-methylated DNA opens a closed conformation of UHRF1 to facilitate its histone recognition. Nat. Commun. 7, 11197 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morgan M. T. et al., Structural basis for histone H2B deubiquitination by the SAGA DUB module. Science 351, 725–728 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taverna S. D., Li H., Ruthenburg A. J., Allis C. D., Patel D. J., How chromatin-binding modules interpret histone modifications: Lessons from professional pocket pickers. Nat. Struct. Mol. Biol. 14, 1025–1040 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xi Q. et al., A poised chromatin platform for TGF-β access to master regulators. Cell 147, 1511–1524 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamaguchi L. et al., Usp7-dependent histone H3 deubiquitylation regulates maintenance of DNA methylation. Sci. Rep. 7, 55 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsumura A. et al., Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells 11, 805–814 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Saksouk N., Simboeck E., Déjardin J., Constitutive heterochromatin formation and transcription in mammals. Epigenet. Chromatin 8, 3 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robertson K. D., DNA methylation and human disease. Nat. Rev. Genet. 6, 597–610 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Lehnertz B. et al., Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 13, 1192–1200 (2003). [DOI] [PubMed] [Google Scholar]
- 41.Peters A. H. et al., Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107, 323–337 (2001). [DOI] [PubMed] [Google Scholar]
- 42.Du Z. et al., DNMT1 stability is regulated by proteins coordinating deubiquitination and acetylation-driven ubiquitination. Sci. Signal. 3, ra80 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qin W., Leonhardt H., Spada F., Usp7 and Uhrf1 control ubiquitination and stability of the maintenance DNA methyltransferase Dnmt1. J. Cell. Biochem. 112, 439–444 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Cheng J. et al., Molecular mechanism for USP7-mediated DNMT1 stabilization by acetylation. Nat. Commun. 6, 7023 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Felle M. et al., The USP7/Dnmt1 complex stimulates the DNA methylation activity of Dnmt1 and regulates the stability of UHRF1. Nucleic Acids Res. 39, 8355–8365 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chuang L. S. et al., Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science 277, 1996–2000 (1997). [DOI] [PubMed] [Google Scholar]
- 47.Jimenji T., Matsumura R., Kori S., Arita K., Structure of PCNA in complex with DNMT1 PIP box reveals the basis for the molecular mechanism of the interaction. Biochem. Biophys. Res. Commun. 516, 578–583 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Arita K. et al., Recognition of modification status on a histone H3 tail by linked histone reader modules of the epigenetic regulator UHRF1. Proc. Natl. Acad. Sci. U.S.A. 109, 12950–12955 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheng J. et al., Structural insight into coordinated recognition of trimethylated histone H3 lysine 9 (H3K9me3) by the plant homeodomain (PHD) and tandem tudor domain (TTD) of UHRF1 (ubiquitin-like, containing PHD and RING finger domains, 1) protein. J. Biol. Chem. 288, 1329–1339 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Karagianni P., Amazit L., Qin J., Wong J., ICBP90, a novel methyl K9 H3 binding protein linking protein ubiquitination with heterochromatin formation. Mol. Cell. Biol. 28, 705–717 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rothbart S. B. et al., Multivalent histone engagement by the linked tandem Tudor and PHD domains of UHRF1 is required for the epigenetic inheritance of DNA methylation. Genes Dev. 27, 1288–1298 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rothbart S. B. et al., Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nat. Struct. Mol. Biol. 19, 1155–1160 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xie S., Jakoncic J., Qian C., UHRF1 double tudor domain and the adjacent PHD finger act together to recognize K9me3-containing histone H3 tail. J. Mol. Biol. 415, 318–328 (2012). [DOI] [PubMed] [Google Scholar]
- 54.Hashimoto H., Horton J. R., Zhang X., Cheng X., UHRF1, a modular multi-domain protein, regulates replication-coupled crosstalk between DNA methylation and histone modifications. Epigenetics 4, 8–14 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jones P. A., Liang G., Rethinking how DNA methylation patterns are maintained. Nat. Rev. Genet. 10, 805–811 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jurkowska R. Z., Jurkowski T. P., Jeltsch A., Structure and function of mammalian DNA methyltransferases. ChemBioChem 12, 206–222 (2011). [DOI] [PubMed] [Google Scholar]
- 57.Guo X. et al., Structural insight into autoinhibition and histone H3-induced activation of DNMT3A. Nature 517, 640–644 (2015). [DOI] [PubMed] [Google Scholar]
- 58.Ren W., Song J., Crystal Structure of Bovine DNMT1 RFTS domain in complex with H3K9me3 and Ubiquitin. Protein Data Bank. https://www.rcsb.org/structure/unreleased/6PZV. Deposited 31 July 2019.
- 59.Wang G. G., Grimm S. A., Readout of heterochromatic H3K9me3 and H4K20me3 regulates DNMT1-mediated maintenance of DNA. National Center for Biotechnology Information. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE145698. Deposited 20 February 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.