Significance
The assembly of β-barrel outer membrane proteins (OMPs) is broadly conserved in diderm bacteria as well as in mitochondria and chloroplasts. The β-barrel assembly machine (BAM), which assembles OMPs into the outer membrane of gram-negative microbes, contains two essential proteins, BamA and BamD. Here we identify a genetic background in which BamD is nonessential, indicating that BamD does not function in OMP catalysis, but rather plays a regulatory role in OMP assembly. BamD is not conserved in the complexes that assemble OMPs in chloroplasts and mitochondria, likely because these organelles, unlike bacteria, inhabit a carefully controlled cytoplasmic environment.
Keywords: OMP assembly, BAM complex, outer membrane biogenesis
Abstract
The outer membrane (OM) of gram-negative bacteria confers innate resistance to toxins and antibiotics. Integral β-barrel outer membrane proteins (OMPs) function to establish and maintain the selective permeability of the OM. OMPs are assembled into the OM by the β-barrel assembly machine (BAM), which is composed of one OMP—BamA—and four lipoproteins—BamB, C, D, and E. BamB, C, and E can be removed individually with only minor effects on barrier function; however, depletion of either BamA or BamD causes a global defect in OMP assembly and results in cell death. We have identified a gain-of-function mutation, bamAE470K, that bypasses the requirement for BamD. Although bamD::kan bamAE470K cells exhibit growth and OM barrier defects, they assemble OMPs with surprising robustness. Our results demonstrate that BamD does not play a catalytic role in OMP assembly, but rather functions to regulate the activity of BamA.
The outer membrane (OM) of gram-negative bacteria, including Escherichia coli, is a uniquely asymmetric bilayer that functions as a selective permeability barrier. The OM allows small and hydrophilic compounds to cross while excluding large and hydrophobic molecules (1). β-barrel outer membrane proteins (OMPs) not only function to construct and maintain the OM, but also contribute to the properties of the selective barrier as porins and as the channel constituents of efflux pumps. Accordingly, the essential cellular components required for OMP biogenesis have generated considerable interest as drug targets (2).
In gram-negative microbes, OMPs are assembled into the OM by the heteropentomeric β-barrel assembly machine (BAM complex). The BAM complex is composed of two essential proteins—BamA and D—and three nonessential proteins—BamB, C, and E. BamA is conserved in all diderm bacteria, and homologs can be found in the OM of mitochondria and chloroplasts (3–6). BamD is not conserved in organelles; however, it is ubiquitous in diderm bacteria, including endosymbionts with a greatly reduced genome (7). Due to this conservation, BamA and BamD have been proposed to be the ancestral BAM complex of proteobacteria, which represents the most phenotypically diverse phylum of prokaryotes (8).
Although OMPs share a common structure, they differ significantly in size and architecture. OMPs can have a β-barrel domain containing between 8 and 26 β-strands (9), exhibit different oligomeric states, and can be composite barrels made up of different subunits (10, 11). Despite this diversity, all are BAM complex substrates.
Recent studies have investigated the mechanism by which OMP substrates are assembled into the OM. Assembly begins with the presentation of an unfolded OMP to the BAM complex by periplasmic chaperones, such as SurA (11). Substrate binding to BamD is communicated to BamA through the BamAD binding interface (12–14). Proper coordination between BamA and BamD is required for OMP assembly, as mutations that disrupt BamAD binding are lethal (12–14). BamA then undergoes conformational changes that prime it for the physical folding and insertion of the OMP substrate into the OM (12, 15–18). Assembly does not proceed if substrates bind aberrantly to BamD, indicating that BamAD coordination serves as an important assembly checkpoint that regulates the insertion of OMPs into the OM (12, 19).
Here we probe the mechanistic role of the two essential BAM complex proteins, BamA and BamD. We show that the previously identified gain-of-function mutation bamAE470K (20) bypasses the requirement for BamD. While the suppressed bamD null cells exhibit OM barrier and growth defects, BamAE470K is able to robustly assemble OMPs in the absence of BamD. Thus, BamD does not play a catalytic role in OMP assembly, but rather functions to regulate the activity of BamA.
Results
BamD Is Nonessential in a bamAE470K Background.
We recently characterized bamAE470K, a mutation that confers resistance to the BamA inhibitor MRL-494 (20) (SI Appendix, Fig. S1). Intriguingly, we also found that bamAE470K is an intragenic suppressor of bamAE373K, a mutation that disrupts the interaction between BamA and BamD, resulting in a loss of cell viability (13, 20). bamAE373K likely causes a defect not in BamA, but rather in BamD function, as the lethality can be suppressed by the activating mutation bamDR197L (13). The viability of bamAE373K/E470K demonstrates that OMP assembly can proceed in the double mutant without the need for the normally critical interaction between BamA and BamD. We hypothesized that BamAE470K might bypass the conformational coordination checkpoint that typically regulates the insertion of OMPs into the OM and perhaps bypasses the requirement for BamD altogether.
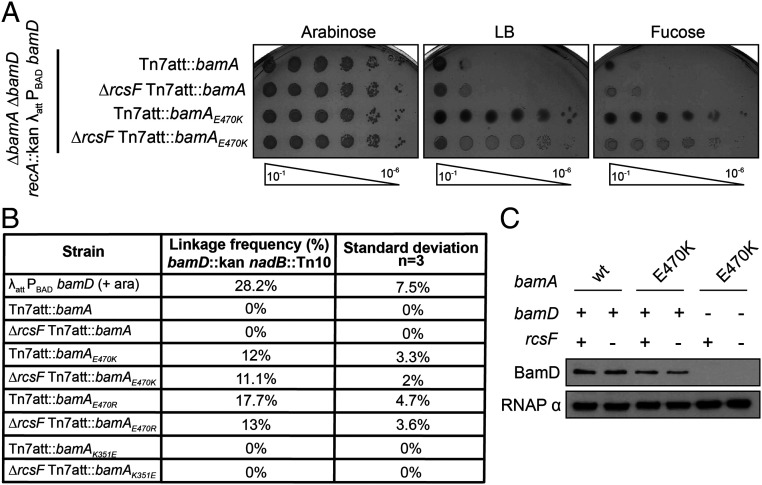
To test this model, we attempted to deplete or delete bamD in a bamAE470K background. BamA was expressed from the Tn7 attachment site, as described previously (20–22), in backgrounds in which the native locus was deleted. We assayed survival in both the presence and absence of the surface-exposed lipoprotein RcsF, the amino terminus of which is threaded through the lumen of an abundant OMP (23, 24). This novel, interlocked complex is known to be a very difficult to assemble BAM substrate. Indeed, many synthetic BAM phenotypes, including the conditional lethal phenotype of a ΔbamBΔbamE double mutant, are caused by the stalled assembly of RcsF/OMP complexes (22, 25). To deplete BamD, we used an arabinose-inducible system in which bamD is placed under the control of the PBAD promoter in a strain lacking the native bamD locus (26). Cells expressing wild-type BamA were not viable when grown on the anti-inducer fucose due to the depletion of BamD. In contrast, cells expressing BamAE470K in either the presence or absence of RcsF survived depletion of BamD (Fig. 1A). Cells carrying the bamAE470K mutation, then, are viable under conditions of BamD depletion.
Fig. 1.
BamD is nonessential in a bamAE470K background. (A) Depletion of BamD from an arabinose-inducible PBAD promoter (26). Cultures were serially diluted and spotted onto LB, arabinose (inducer), or fucose (anti-inducer) media. (B) Linkage disruption using P1 phage carrying bamD::kan nadB::Tn10 was quantified by calculating the number of kanamycin-resistant transductants out of the total number of transductants. (C) α BamD immunoblot analysis of bamD::kan bamAE470K (+/−ΔrcsF) strains and bamD+ control strains. αRpoA (RNAP α) served as a loading control.
To investigate whether BamAE470K can assemble OMPs in the absence of BamD altogether, we quantitated the ability of cells to inherit a bamD null allele using linkage disruption. P1 phage were grown on a strain expressing an ectopic bamD+ gene with a bamD::kan (confers resistance to kanamycin) allele at the native chromosomal locus linked to the nearby nadB::Tn10 (confers resistance to tetracycline) allele. This lysate was then used to infect bamA+ and bamAE470K cells. Following selection for tetracycline-resistant transductants, the percentage of cells that inherited the linked bamD::kan allele was determined. As a control, we used a bamD diploid strain, which carries a complementing copy of bamD at an ectopic location.
The bamD diploid control strain was able to inherit both the bamD::kan and nadB::Tn10 alleles at a frequency of approximately 28%. As expected, the bamA+ strains could not receive the bamD::kan allele, reflecting the essential nature of bamD in these strains (Fig. 1B); however, the bamD::kan allele was cotransduced with the linked marker into a bamAE470K background with a frequency of 12% in the presence of rcsF and 11.1% in the absence of rcsF (Fig. 1B). Similarly, the bamD::kan allele could be cotransduced with the linked marker into a bamAE470R background at a frequency of 17.7% in the presence of rcsF and 13% in the absence of rcsF (Fig. 1B), demonstrating that this result is not specific to the glutamate-to-lysine substitution.
To definitively demonstrate that BamD is nonessential in the bamAE470K background, we constructed strains in which bamD was directly disrupted using a bamD::kan insertion deletion in cells expressing BamAE470K. Appropriate PCR analysis, immunoblot analysis, and mass spectrometry confirmed that these strains lacked both the structural gene and the BamD protein (Fig. 1C and Fig. 3 C and D) (27). The survival of bamD::kan bamAE470K cells confirms that this gain-of-function mutation bypasses the requirement for BamD.
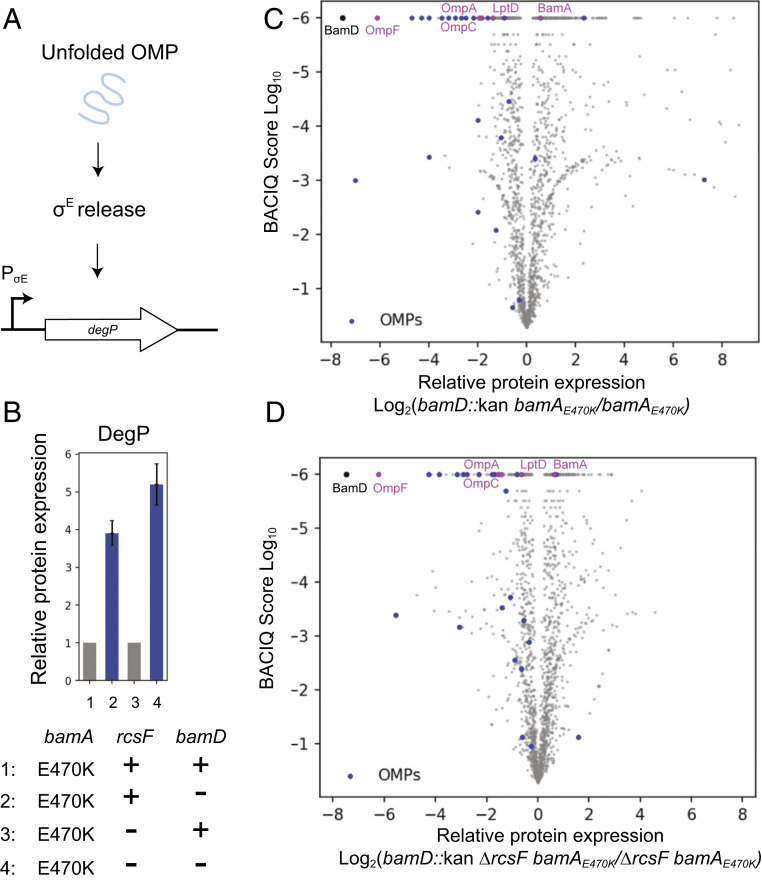
Fig. 3.
OMP assembly profiles of ΔbamD strains. (A) Schematic of σE activation. Unfolded OMPs in the periplasm trigger a proteolytic cascade that releases σE from the IM to activate regulon gene expression. (B) Relative fold change of DegP levels in the bamD::kan bamAE470K strain compared with the bamAE470K control and in the bamD::kan ΔrcsF bamAE470K strain compared with the ΔrcsF bamAE470K control. Whiskers represent 95% confidence intervals (27, 31). (C and D) Volcano plot of protein expression in the bamD::kan bamAE470K (27) (C) and bamD::kan ΔrcsF bamAE470K (D) strains compared with their respective isogenic controls, as indicated. The vertical axis represents the probability of observing differential protein expression between the two strains (31), and the horizontal axis is log2- fold change of protein abundance. Each dot represents an individual protein, with β-barrel OMPs in blue, commonly studied β-barrel OMPs in magenta, and BamD in black.
Bypass of BamD Is Unique to bamAE470K.
Previous work has identified bamAK351E as an intragenic suppressor of bamAE373K (14). bamAK351E harbors a mutation near bamAE373K at the BamAD binding interface. This interface is formed by a network of electrostatic interactions in which charged residues in this region dynamically interchange salt bridges (13, 14, 28). The E373K charge change mutation lethally disrupts one of these residues at the essential binding interface (13). The compensatory K351E mutation promotes the viability of bamAE373K without restoring BamAD binding (14). We hypothesized that the ability of bamAK351E to suppress bamAE373K indicated that this mutant would also bypass the essentiality BamD.
We used linkage disruption to investigate bamAK351E viability in the absence of BamD. Linkage disruption showed that bamD could not be deleted in the bamAK351E background (Fig. 1B). While both bamAE470K and bamAK351E promote cell survival despite disruption of BamAD coordination, only bamAE470K bypasses the requirement for bamD completely. Intragenic suppression of bamAE373K, then, is not predictive of BamD bypass. It is noteworthy that bamAK351E, unlike bamAE470K, does not confer resistance to MRL-494 either (20).
BamAE470K Robustly Assembles OMPs in the Absence of BamD.
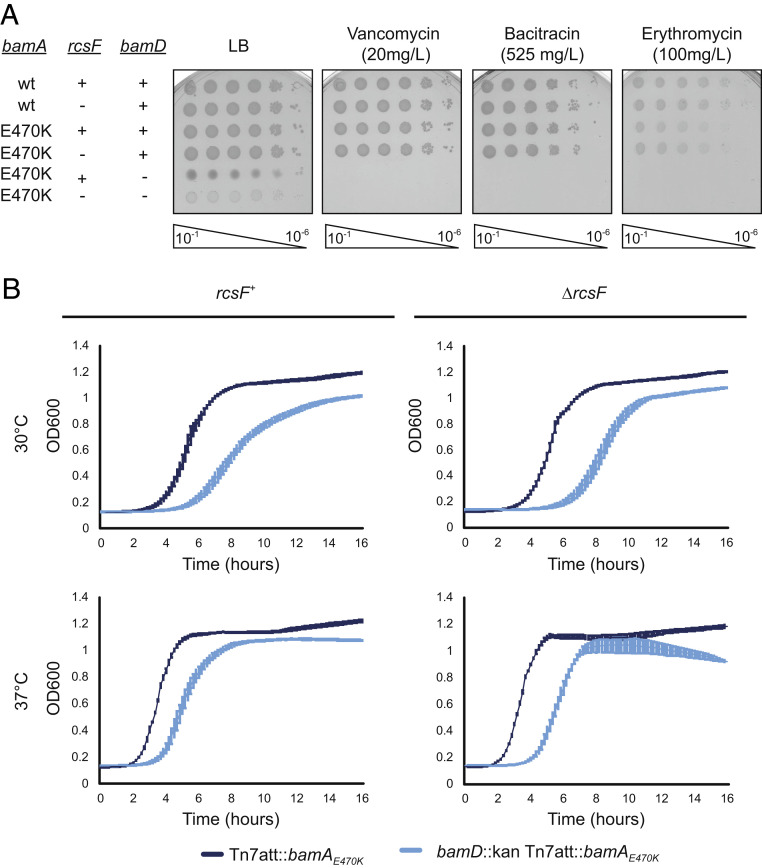
We characterized the bamD null mutants to evaluate viability and membrane integrity in the absence of BamD. Phenotypic analysis of the bamD null mutants expressing bamAE470K in both the presence and the absence of RcsF revealed that cells lacking BamD have significant OM permeability defects (Fig. 2A). Furthermore, bamD::kan bamAE470K +/− ΔrcsF strains exhibit growth defects at both 30 °C and 37 °C (Fig. 2B); bamD null strains showed reduced growth rates, especially in the presence of RcsF. The mutants’ observed difficulty in exiting lag phase is likely due, at least in part, to fewer viable cells in the inoculum as a consequence of reduced survival in stationary phase (Table 1 and SI Appendix, Table S1).
Fig. 2.
Growth of bamD null mutants. (A) The indicated strains were serially diluted and spotted onto LB, vancomycin, bacitracin, and erythromycin. (B) Growth curves of bamD mutants compared with the isogenic control. Strains were normalized by OD600, inoculated into LB media, and grown at the indicated temperature.
Table 1.
Doubling rate calculation for the indicated strains
| Strain | Doubling rate, min | |
| 30 °C (SD) | 37 °C (SD) | |
| Tn7att::bamA | 67 (4) | 55 (2) |
| Tn7att::bamAE470K | 74 (10) | 61(9) |
| ΔrcsF Tn7att::bamA | 60 (10) | 50 (4) |
| ΔrcsF Tn7att::bamAE470K | 66 (4) | 55 (2) |
| bamD::kan Tn7att::bamAE470K | 119 (7) | 73 (8) |
| bamD::kan ΔrcsF Tn7att::bamAE470K | 97 (3) | 69 (4) |
OMP assembly is monitored by the σE stress response, which detects unfolded OMPs in the periplasm and up-regulates production of proteases, such as the periplasmic protease DegP, to combat envelope damage (Fig. 3A) (29). We compared the proteomes of bamAE470K and ΔrcsF bamAE470K cells in the presence or absence of bamD using the TMTc+ multiplexed relative proteomics approach (27, 30). The expression of DegP increases four-fold when BamD is removed from the bamAE470K mutant, and the presence of RcsF did not markedly change this induction (an approximate fivefold increase) (Fig. 3B). Although bamAE470K clearly bypasses the requirement for BamD, the decreased efficiency of the heterotetrameric BAM complex likely explains the phenotypes observed.
Our quantitative proteomic analysis also shows that in the bamD::kan ΔrcsF bamAE470K strain, most of the OMPs, including the major porins OmpC and OmpA, are reduced by fivefold or less (Fig. 3D and SI Appendix, Table S6) (27). LptD, a large OMP that functions as an essential component of the lipopolysaccharide transport machine (32, 33) and is assembled with the lipoprotein LptE in its lumen (34), is reduced by only one-third. Even in the presence of RcsF, many OMP levels were reduced only slightly more (Fig. 3C and SI Appendix, Table S6). Across all β-barrel OMPs detected in the proteomics dataset, the median reduction in level is 3.9-fold in the presence of rcsF and 2.7-fold in the absence of rcsF. Thus, with the exception of the RcsF/OMP interlocked complex, the defects in OMP assembly are not limited to a subset of OMP substrates, indicating that BamAE470K is competent to fold nearly the entire scope of OMP substrates independently of BamD.
Other protein groups found to be up-regulated or down-regulated in the absence of BamD were largely related to changes to metabolism and gene regulation, as determined by Gene Ontology (GO) term enrichment analysis (SI Appendix, Tables S2 to S5) (27). We attribute these global changes to a reduction in cell fitness and the activation of envelope stress responses caused by reduced OMP assembly.
Discussion
Here we investigate the roles of the BAM complex essential proteins, BamA and BamD. We demonstrate that the bamAE470K mutation bypasses the requirement for BamD altogether. This indicates that BamD does not catalyze an essential folding or OM insertion step in the β-barrel assembly reaction, as OMP assembly can occur in its absence. Rather, we suggest that the essential role of BamD in wild-type cells is solely to regulate the ability of BamA to actively engage unfolded OMP substrates (12, 13, 15, 19, 35, 36). BamAE470K is a gain-of-function mutation that bypasses this requirement and actively engages OMP substrates without regulation by BamD.
In wild-type cells, BamD functions as an assembly checkpoint to prevent the attempted assembly of defective substrates, which would likely compromise the BAM complex, the OM permeability barrier, or both. LptD has proven to be a useful model substrate because it folds more slowly than other OMPs (37, 38), and several lptD mutations that affect the assembly process have been characterized. Indeed, assembly of the defective substrate LptD4213, which partially escapes this checkpoint, profoundly disrupts the permeability barrier (39). In contrast, the defective substrate lptDY721D, which binds BamD abnormally, is not engaged efficiently by BamA and is largely rejected from the BAM complex (12).
Our results suggest that all of the reactions necessary to fold and insert OMPs into the OM are catalyzed by BamA. Early structural studies suggested the presence of an opening of the BamA barrel at the seam between the first and last β-strands (16, 40), and recent studies with stalled BAM-substrate complexes suggest that the first β-strand of BamA provides a template for β-strand formation of the C-terminal substrate β-strand (41). Studies with the stalled substrate LptD4213 indicate that substantial substrate folding occurs inside the BamA barrel (35, 41, 42). Strikingly, the mutation bamAE470G suppresses lptDΔD330, a mutation that alters the disposition of LptD loop 4 in a less severe manner than the lptD4213 mutation, which is a 23-codon deletion (35). The bamAE470G mutation also restores function to BamA chimeras composed of the E. coli β-barrel domain and four of the five periplasmic POTRA domains from Pseudomonas aeruginosa (43). The ability of a substitution within the BamA β-barrel to overcome incompatibility with the periplasmic domain suggests that substitutions at codon 470 may bypass at least some of the functions of the POTRA domains as well. Our results are consistent with the proposal that residues located in the interior of the barrel, far from the seam, are critically important for barrel folding (35).
The OM of mitochondria and chloroplasts contains β-barrel proteins that are assembled by the BamA homologs Sam50 and OEP80, respectively (3–6). These β-barrel assembly complexes do not have a protein with sequence similarity to BamD (44). The mitochondrial system does have an additional essential protein, Sam35; however, this component is located on the cytosolic side of the organelle and thus likely has a function distinct from that of BamD (45). As such, sequence alignment of the Omp85 family homologs BamA, Sam50, and OEP80 revealed a low level of conservation at the residue corresponding to the E. coli E470 (SI Appendix, Fig. S2). Intriguingly, the chloroplast homolog OEP80 contains an arginine at the corresponding residue, and E470R renders the E. coli BamD nonessential. Since mitochondria and chloroplasts have a simpler, less complex set of substrates and exist in a controlled cytosolic environment, a regulatory checkpoint similar to that provided by BamD likely is not needed. Gram-negative bacteria, which inhabit diverse environments, require additional regulation to maintain the efficient assembly of diverse substrates and the remarkably effective innate immunity conferred by the OM permeability barrier.
Materials and Methods
Bacterial Strains and Growth Conditions.
All strains, plasmids, and oligonucleotides used in this study are presented in SI Appendix, Tables S1 to S3, respectively. Strains were constructed using standard microbiological techniques, as described previously (46). Strains were grown in LB medium supplemented with 50 mg/L kanamycin, 25 mg/L ampicillin, 25 mg/L tetracycline, and 20 mg/L chloramphenicol, 0.2% arabinose, and 0.05% fucose, when appropriate. Unless stated otherwise, all strains were grown at 30 °C. The bamD and bamA deletion alleles originated from previous studies (26, 47). Other deletion alleles used originated from the Keio collection and the Carol Gross Tn10 collection (48, 49). FRT-flanked resistance cassettes were removed using the Flp recombinase system, as described previously (50). bamA alleles were inserted at the Tn7 attachment site, as described previously (21), under control of the native bamA promoter (20, 22) in backgrounds in which the native bamA locus was deleted. Depletion of BamD was performed in backgrounds in which the native bamD locus was deleted.
Efficiency of Plating Assay.
Stationary phase cultures were normalized by OD600 and serially diluted into LB medium. Cells were spotted onto the indicated medium and incubated at 30 °C overnight.
Linkage Disruption Assay.
P1 phage carrying a bamD::kan disruption allele linked to a nearby nadB::Tn10 marker (48, 49) was prepared as described previously (46). The lysate was used to transduce the indicated strains and plated on LB medium containing 25 mg/L tetracycline. The tetracycline-resistant transductants were patched to LB medium containing 50 mg/L kanamycin to test for integration of the bamD::kan allele. The number of kanamycin-resistant transductants over the total number of tetracycline transductants was used to quantify linkage frequency. Linkage frequencies were calculated from transductions performed in biological triplicate.
Immunoblot Analysis.
Stationary phase cultures were normalized by OD600 and lysed in a mixture of BugBuster (Millipore), protease inhibitor mixture (1:100, Sigma-Aldrich), benzonase (1:100, Sigma-Aldrich), and 1 M MgCl2 (1:100) for 10 min at room temperature. Samples were boiled for 10 min and electrophoresed on a 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel. Proteins were transferred to a nitrocellulose membrane, and immunoblotting was performed using rabbit polyclonal primary antibody probing for αBamD (1:10,000) or αRpoA (RNAP α) (1:50,000). Donkey anti-rabbit IgG horseradish peroxidase secondary antibody (GE Healthcare) was used at 1:10,000.
Growth Curves.
Overnight cultures of the indicated strains were normalized by OD600 and inoculated into LB medium. Cells were grown shaking in a 24-well plate with a 2 mL volume (Sarstedt) for 16 h at 30 °C or 37 °C in a BioTek Synergy H1 plate reader. Growth was determined based on OD600 measurement. Error bars represent ± SEM.
Doubling Rate and Viability Assays.
Viability was assessed at the indicated temperatures by plating stationary-phase bacteria on LB medium and counting the number of colonies for calculating CFUs. The doubling rate of strains was calculated during the exponential phase of growth using OD600 values. The values are averages of at least three biological replicates.
Quantitative Proteomics Sample Preparation.
Samples were prepared as described previously (51). Cells were grown in suspension to exponential phase (OD600 ∼0.5 to 0.8) in LB at 30 °C. Samples were normalized by OD600. Cells were harvested by pelleting at room temperature and flash-frozen. Each pellet containing ∼200 μg (∼9 × 108 cells) of total protein was resuspended in 200 μL of lysis buffer containing 50 mM Hepes pH 7.2, 2% CTAB (hexadecyltrimethylammonium bromide), 6 M GuaCl (guanidine hydrochloride), and 5 mM DTT. Cells were lysed by sonication, with 10 pulses of 30 s at 60% amplitude, and the lysate was further heated at 60 °C for 20 min.
Next, 200 μL of lysate from every condition was precipitated with methanol-chloroform (52). Protein concentration was determined using the bicinchoninic acid protein assay (Thermo Fisher Scientific). The samples were resuspended in 6 M guanidine chloride in 10 mM EPPS [4-(2-hydroxyethyl)-1-piperazinepropanesulfonic acid] pH 8.5, with a subsequent dilution to 2 M guanidine chloride in 10 mM EPPS pH 8.5 for digestion with Lys-C (Wako) at room temperature with 20 ng/μL Lys-C overnight. The samples were further diluted to 0.5 mM guanidine chloride in 10 mM EPPS pH 8.5 in 10 mM EPPS pH 8.5 and then digested with 20 ng/μL Lys-C and 10 ng/μL trypsin at 37 °C overnight.
The digested samples were dried using a vacuum evaporator at room temperature and taken up in 200 mM EPPS pH 8.0 for a pH shift, which is necessary for optimal labeling conditions. Then 10 μL of total material from each condition was labeled with 3 μL of 20 ng/μL tandem mass tags (TMT). TMT reagents were dissolved in anhydrous acetonitrile. TMT samples were labeled for 2 h at room temperature. Labeled samples were quenched with 0.5% hydroxylamine solution (Sigma-Aldrich). Samples from all conditions were combined into one tube, acidified with 5% phosphoric acid (pH <2), and subjected to a subsequent spin at 16,000 relative centrifugal force for 10 min at 4 °C. The samples were dried using a vacuum evaporator at room temperature. Dry samples were taken up in high- pressure liquid chromatography (HPLC)-grade water and stage-tipped for desalting (53). The samples were resuspended in 1% formic acid to 1 μg/μL, and 1 μg of the total combined sample was analyzed with the TMTc+ approach (30).
Liquid Chromatography-Mass Spectrometry Analysis.
Approximately 1 µL of each sample was analyzed by liquid chromatography-mass spectrometry (LC-MS). LC-MS experiments were performed on Orbitrap Fusion Lumos (Thermo Fisher Scientific). The instrument was equipped with Easy-nLC 1200 HPLC pump (Thermo Fisher Scientific). For each run, peptides were separated on a 100-μm i.d. microcapillary column, packed first with ∼0.5 cm of 5-µm BEH C18 packing material (Waters), followed by 30 cm of 1.7-µm BEH C18 (Waters) and then 30 cm of 1.7-μm BEH C18 (Waters). Separation was achieved by applying a 4.8% to 24% acetonitrile gradient in 0.125% formic acid and 2% DMSO over 120 min at 350 nL/min at 60 °C. Electrospray ionization was enabled by applying a voltage of 2.6 kV through a MicroTee at the inlet of the microcapillary column. We used the Orbitrap Fusion Lumos with a TMTc+ method (30). The instrument was operated in data-dependent mode with a scan range of 500 to 1,400 m/z with an RF lens (%) of 60, automatic gain control (AGC) target of 1.0e6, and a maximum injection time of 100 ms. Only charge states of 2_ were included. A dynamic exclusion window of 60 s with a mass tolerance of 10 ppm was used. Peptides with a minimum intensity of 3e6 or higher were subjected to an MS2 scan using an isolation window of 0.4 Th (or of different size if indicated) using the quadrupole. Peptides were fragmented using a higher energy collisional dissociation (HCD) collision energy of 32%, and a mass spectrum was acquired using the Orbitrap with a resolution of 60,000 with an AGC target of 5.0e5 and a maximum injection time of 120 ms. The scan range of the Orbitrap wasset to 500–2,000 m/z.
MS Data Analysis.
A suite of software tools developed in-house was used to convert mass spectrometry data from the Thermo RAW file to the mzXML format, as well as to correct erroneous assignments of peptide ion charge state and monoisotopic m/z (54). Assignment of MS2 spectra was performed using the SEQUEST algorithm by searching the data against the appropriate proteome reference dataset acquired from UniProt, including common contaminants like human keratins and trypsin (55, 56). This forward database component was followed by a decoy component that included all listed protein sequences in reverse order (57). Searches were performed using a 20-ppm precursor ion tolerance, in which both peptide termini were required to be consistent with trypsin or LysC specificity while allowing one missed cleavage. Fragment ion tolerance in the MS2 spectrum was set at 0.02 Th (TMTc+), TMT was set as a static modification of lysine residues and peptides’ N termini (+229.162932 Da), oxidation of methionine residues (+15.99492 Da) was set as as a variable modification. An MS2 spectral assignment false discovery rate (FDR) of 0.5% was achieved by applying the target decoy database search strategy (57). Filtering was performed using a linear discrimination analysis with the following features: SEQUEST parameters XCorr and unique ΔXCorr, absolute peptide ion mass accuracy, peptide length, and charge state. Forward peptides within 3 SD of the theoretical m/z of the precursor served as a positive training set; all reverse peptides served as a negative training set. Linear discrimination scores were used to sort peptides with at least seven residues and to filter with the desired cutoff. Furthermore, we performed a filtering step toward on the protein level by the “picked” protein FDR approach (58). Protein redundancy was removed by assigning peptides to the minimal number of proteins which can explain all observed peptides, with above described filtering criteria (59). We did not use isolation specificity filtering for the TMTc+ method, as co-isolation of other peptides does not perturb the measurement results for this method. The probabilities of differential expression were calculated based on agreement between the underlying peptides and signal level of the peptides for every protein quantified in the experiment using BACIQ software (60).
We carried out GO term enrichment analysis of significantly up-regulated proteins from the quantitative proteomic dataset (significance cutoff score = 0.95) using DAVID bioinformatics software (31, 61). Biological process GO terms were ranked according to fold enrichment, with P values listed as determined by DAVID.
Sequence Alignment.
Sequence alignment of BamA from E. coli (62, 63), Sam50 from Saccharomyces cerevisiae (64), and OEP80 from Arabidopsis thaliana (65) was performed using the ClustalW program (66), and images were created using Jalview software (67).
Data and Material Availability.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (http://proteomecentral.proteomexchange.org/cgi/GetDataset) with the dataset identifier PXD017962.
Supplementary Material
Acknowledgments
We thank former and current members of the T.J.S and M.W. laboratories for helpful discussions and editing of the manuscript and thank Lillia Ryazanova for technical assistance. The research performed for this study was supported by the National Institute of General Medical Sciences of the NIH under Grants R35-GM128813 (to M.W.), R35-GM118024 (to T.J.S.), and R01-GM034821 (to T.J.S.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. This work was also supported by the Lewis–Sigler collaboration fund.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The proteomic dataset has been deposited at the ProteomeXchange Consortium via the PRIDE partner repository. Data can be accessed at: http://proteomecentral.proteomexchange.org/cgi/GetDataset with the dataset identifier PXD017962.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2007696117/-/DCSupplemental.
References
- 1.Silhavy T. J., Kahne D., Walker S., The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2, a000414 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sousa M. C., New antibiotics target the outer membrane of bacteria. Nature 576, 389–390 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Gentle I., Gabriel K., Beech P., Waller R., Lithgow T., The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. J. Cell Biol. 164, 19–24 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kozjak V. et al., An essential role of Sam50 in the protein sorting and assembly machinery of the mitochondrial outer membrane. J. Biol. Chem. 278, 48520–48523 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Paschen S. A. et al., Evolutionary conservation of biogenesis of β-barrel membrane proteins. Nature 426, 862–866 (2003). [DOI] [PubMed] [Google Scholar]
- 6.Sommer M. S. et al., Chloroplast Omp85 proteins change orientation during evolution. Proc. Natl. Acad. Sci. U.S.A. 108, 13841–13846 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruiz N., Gronenberg L. S., Kahne D., Silhavy T. J., Identification of two inner-membrane proteins required for the transport of lipopolysaccharide to the outer membrane of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 105, 5537–5542 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anwari K. et al., The evolution of new lipoprotein subunits of the bacterial outer membrane BAM complex. Mol. Microbiol. 84, 832–844 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koebnik R., Locher K. P., Van Gelder P., Structure and function of bacterial outer membrane proteins: Barrels in a nutshell. Mol. Microbiol. 37, 239–253 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Hagan C. L., Silhavy T. J., Kahne D., β-barrel membrane protein assembly by the Bam complex. Annu. Rev. Biochem. 80, 189–210 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Konovalova A., Kahne D. E., Silhavy T. J., Outer membrane biogenesis. Annu. Rev. Microbiol. 71, 539–556 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee J. et al., Substrate binding to BamD triggers a conformational change in BamA to control membrane insertion. Proc. Natl. Acad. Sci. U.S.A. 115, 2359–2364 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ricci D. P., Hagan C. L., Kahne D., Silhavy T. J., Activation of the Escherichia coli β-barrel assembly machine (Bam) is required for essential components to interact properly with substrate. Proc. Natl. Acad. Sci. U.S.A. 109, 3487–3491 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCabe A. L., Ricci D., Adetunji M., Silhavy T. J., Conformational changes that coordinate the activity of BamA and BamD allowing β-barrel assembly. J. Bacteriol. 199, e00373-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rigel N. W., Ricci D. P., Silhavy T. J., Conformation-specific labeling of BamA and suppressor analysis suggest a cyclic mechanism for β-barrel assembly in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 110, 5151–5156 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noinaj N., Kuszak A. J., Balusek C., Gumbart J. C., Buchanan S. K., Lateral opening and exit pore formation are required for BamA function. Structure 22, 1055–1062 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Misra R., Stikeleather R., Gabriele R., In vivo roles of BamA, BamB and BamD in the biogenesis of BamA, a core protein of the β-barrel assembly machine of Escherichia coli. J. Mol. Biol. 427, 1061–1074 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pavlova O., Peterson J. H., Ieva R., Bernstein H. D., Mechanistic link between β barrel assembly and the initiation of autotransporter secretion. Proc. Natl. Acad. Sci. U.S.A. 110, E938–E947 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hagan C. L., Wzorek J. S., Kahne D., Inhibition of the β-barrel assembly machine by a peptide that binds BamD. Proc. Natl. Acad. Sci. U.S.A. 112, 2011–2016 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hart E. M. et al., A small-molecule inhibitor of BamA impervious to efflux and the outer membrane permeability barrier. Proc. Natl. Acad. Sci. U.S.A. 116, 21748–21757 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKenzie G. J., Craig N. L., Fast, easy and efficient: Site-specific insertion of transgenes into enterobacterial chromosomes using Tn7 without need for selection of the insertion event. BMC Microbiol. 6, 39 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hart E. M., Gupta M., Wühr M., Silhavy T. J., The synthetic phenotype of ΔbamB ΔbamE double mutants results from a lethal jamming of the bam complex by the lipoprotein RcsF. MBio 10, e00662-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Konovalova A., Mitchell A. M., Silhavy T. J., A lipoprotein/β-barrel complex monitors lipopolysaccharide integrity transducing information across the outer membrane. eLife 5, e15276 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Konovalova A., Perlman D. H., Cowles C. E., Silhavy T. J., Transmembrane domain of surface-exposed outer membrane lipoprotein RcsF is threaded through the lumen of β-barrel proteins. Proc. Natl. Acad. Sci. U.S.A. 111, E4350–E4358 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tata M., Konovalova A., Improper coordination of BamA and BamD results in bam complex jamming by a lipoprotein substrate. MBio 10, e00660-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malinverni J. C. et al., YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol. Microbiol. 61, 151–164 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Hart E. M., Gupta M., Wühr M., Silhavy T. J., The gain-of-function allele bamAE470K bypasses the essential function for BamD in β-barrel outer membrane protein assembly. ProteomeXchange. http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD017962. Deposited 10 March 2020. [DOI] [PMC free article] [PubMed]
- 28.Sinnige T. et al., Solid-state NMR studies of full-length BamA in lipid bilayers suggest limited overall POTRA mobility. J. Mol. Biol. 426, 2009–2021 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Mitchell A. M., Silhavy T. J., Envelope stress responses: Balancing damage repair and toxicity. Nat. Rev. Microbiol. 17, 417–428 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sonnett M., Yeung E., Wühr M., Accurate, sensitive, and precise multiplexed proteomics using the complement reporter ion cluster. Anal. Chem. 90, 5032–5039 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang W., Sherman B. T., Lempicki R. A., Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bos M. P., Tefsen B., Geurtsen J., Tommassen J., Identification of an outer membrane protein required for the transport of lipopolysaccharide to the bacterial cell surface. Proc. Natl. Acad. Sci. U.S.A. 101, 9417–9422 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Braun M., Silhavy T. J., Imp/OstA is required for cell envelope biogenesis in Escherichia coli. Mol. Microbiol. 45, 1289–1302 (2002). [DOI] [PubMed] [Google Scholar]
- 34.Wu T. et al., Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 103, 11754–11759 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee J. et al., Formation of a β-barrel membrane protein is catalyzed by the interior surface of the assembly machine protein BamA. eLife 8, e49787 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wzorek J. S., Lee J., Tomasek D., Hagan C. L., Kahne D. E., Membrane integration of an essential β-barrel protein prerequires burial of an extracellular loop. Proc. Natl. Acad. Sci. U.S.A. 114, 2598–2603 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chng S. S. et al., Disulfide rearrangement triggered by translocon assembly controls lipopolysaccharide export. Science 337, 1665–1668 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ureta A. R., Endres R. G., Wingreen N. S., Silhavy T. J., Kinetic analysis of the assembly of the outer membrane protein LamB in Escherichia coli mutants each lacking a secretion or targeting factor in a different cellular compartment. J. Bacteriol. 189, 446–454 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee J. et al., Characterization of a stalled complex on the β-barrel assembly machine. Proc. Natl. Acad. Sci. U.S.A. 113, 8717–8722 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doerner P. A., Sousa M. C., Extreme dynamics in the BamA β-barrel seam. Biochemistry 56, 3142–3149 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doyle M. T., Bernstein H. D., Bacterial outer membrane proteins assemble via asymmetric interactions with the BamA β-barrel. Nat. Comms. 10, 3358 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tomasek D. et al., Structure of a nascent membrane protein as it folds on the BAM complex. Nature, 10.1038/s41586-020-2370-1 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Browning D. F. et al., Cross-species chimeras reveal BamA POTRA and β-barrel domains must be fine-tuned for efficient OMP insertion. Mol. Microbiol. 97, 646–659 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Misra R., Assembly of the β-barrel outer membrane proteins in gram-negative bacteria, mitochondria, and chloroplasts. ISRN Mol. Biol. 2012, 708203 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Milenkovic D. et al., Sam35 of the mitochondrial protein sorting and assembly machinery is a peripheral outer membrane protein essential for cell viability. J. Biol. Chem. 279, 22781–22785 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Silhavy T. J., Berman M. L., Enquist L. W., Experiments with Gene Fusions, (Cold Spring Harbor Laboratory, 1984). [Google Scholar]
- 47.Wu T. et al., Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell 121, 235–245 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Baba T. et al., Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2, 2006.0008 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singer M. et al., A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol. Rev. 53, 1–24 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Datsenko K. A., Wanner B. L., One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gupta M., Sonnett M., Ryazanova L., Presler M., Wühr M., “Quantitative proteomics of xenopus embryos I, sample preparation methods and protocols” in Protein Nanotechnology, Vleminckx K., Ed. (Springer, 2018), pp. 175–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wessel D., Flügge U. I., A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138, 141–143 (1984). [DOI] [PubMed] [Google Scholar]
- 53.Rappsilber J., Mann M., Ishihama Y., Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 (2007). [DOI] [PubMed] [Google Scholar]
- 54.Huttlin E. L. et al., A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143, 1174–1189 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eng J. K., McCormack A. L., Yates J. R., An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989 (1994). [DOI] [PubMed] [Google Scholar]
- 56.The UniProt Consortium , UniProt: The universal protein knowledgebase. Nucleic Acids Res. 45, D158–D169 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Elias J. E., Gygi S. P., Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 (2007). [DOI] [PubMed] [Google Scholar]
- 58.Savitski M. M., Wilhelm M., Hahne H., Kuster B., Bantscheff M., A scalable approach for protein false discovery rate estimation in large proteomic data sets. Mol. Cell. Proteomics 14, 2394–2404 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chvatal V., A greedy heuristic for the set-covering problem. Math. Oper. Res. 4, 209–302 (1979). [Google Scholar]
- 60.Peshkin L., Gupta M., Ryazanova L., Wühr M., Bayesian confidence intervals for multiplexed proteomics integrate ion-statistics with peptide quantification concordance. Mol. Cell. Proteomics 18, 2108–2120 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang W., Sherman B. T., Lempicki R. A., Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- 62.Keseler I. M. et al., The EcoCyc database: Reflecting new knowledge about Escherichia coli K-12. Nucleic Acids Res. 45, D543–D550 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Blattner F. R. et al., The complete genome sequence of Escherichia coli K-12. Science 277, 1453–1462 (1997). [DOI] [PubMed] [Google Scholar]
- 64.Philippsen P. et al., The nucleotide sequence of Saccharomyces cerevisiae chromosome XIV and its evolutionary implications. Nature 387, 93–98 (1997). [PubMed] [Google Scholar]
- 65.Tabata S. et al.; Kazusa DNA Research Institute; Cold Spring Harbor and Washington University in St Louis Sequencing Consortium; European Union Arabidopsis Genome Sequencing Consortium , Sequence and analysis of chromosome 5 of the plant Arabidopsis thaliana. Nature 408, 823–826 (2000). [DOI] [PubMed] [Google Scholar]
- 66.Madeira F. et al., The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47, W636–W641 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Waterhouse A. M., Procter J. B., Martin D. M. A., Clamp M., Barton G. J., Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.