Significance
Studies of eukaryotic cell division have focused on the actomyosin ring, whose filaments of F-actin and myosin-II are hypothesized to generate the contractile force for ingression of the cleavage furrow. However, myosin-II has a very limited taxonomic distribution, whereas division by furrowing is much more widespread. We used the green alga Chlamydomonas reinhardtii to investigate how a furrow can form without myosin-II and the potential roles of F-actin in this process. Although F-actin was associated with ingressing furrows, its complete removal only modestly delayed furrowing, suggesting that an actin-independent mechanism (possibly involving microtubules) drives furrow ingression. Such a mechanism presumably emerged early in eukaryotic evolution and may still underlie cell division in a diverse range of modern species.
Keywords: cell division, chloroplast division, cytokinesis, microtubules, myosin
Abstract
It is widely believed that cleavage-furrow formation during cytokinesis is driven by the contraction of a ring containing F-actin and type-II myosin. However, even in cells that have such rings, they are not always essential for furrow formation. Moreover, many taxonomically diverse eukaryotic cells divide by furrowing but have no type-II myosin, making it unlikely that an actomyosin ring drives furrowing. To explore this issue further, we have used one such organism, the green alga Chlamydomonas reinhardtii. We found that although F-actin is associated with the furrow region, none of the three myosins (of types VIII and XI) is localized there. Moreover, when F-actin was eliminated through a combination of a mutation and a drug, furrows still formed and the cells divided, although somewhat less efficiently than normal. Unexpectedly, division of the large Chlamydomonas chloroplast was delayed in the cells lacking F-actin; as this organelle lies directly in the path of the cleavage furrow, this delay may explain, at least in part, the delay in cytokinesis itself. Earlier studies had shown an association of microtubules with the cleavage furrow, and we used a fluorescently tagged EB1 protein to show that microtubules are still associated with the furrows in the absence of F-actin, consistent with the possibility that the microtubules are important for furrow formation. We suggest that the actomyosin ring evolved as one way to improve the efficiency of a core process for furrow formation that was already present in ancestral eukaryotes.
Cytokinesis is the final stage in the cell division cycle in which the cytoplasms and plasma membranes of the daughter cells become separated. In unikonts (animals, fungi, slime molds, and their close relatives), cytokinesis occurs by the symmetric or asymmetric ingression of a “cleavage furrow” from the periphery of the cell. For 50 y, thinking about cleavage-furrow ingression in these cells has been dominated by the contractile actomyosin ring (CAR) model, in which bipolar filaments of myosin-II walk along actin filaments (F-actin), much as in muscle, to produce the force that pulls the plasma membrane in to form the furrow (1–4). Actin, myosin-II, and functionally related proteins are clearly present in a ring that constricts during furrow ingression in unikont cells (2–11), and there is good evidence both that this ring produces contractile force (12, 13) and that this force is required for normal cytokinesis in at least some cell types (14–18).
However, there are also multiple observations that are difficult to reconcile with the CAR model, at least in its simplest forms. For example, in mammalian NRK (rat kidney) cells, local application of the actin-depolymerizing agent cytochalasin D to the furrow region accelerated, rather than delayed, furrowing (19, 20). Moreover, equatorial furrows could form in NRK cells while myosin-II was inhibited by blebbistatin, so long as the cells were attached to a substratum (21), and a motor-impaired myosin-II supported a normal rate of furrow ingression in mammalian COS-7 (monkey kidney) cells (22). In addition, myosin-II-null mutants are viable and can divide in some microorganisms. In Dictyostelium discoideum amoebae, such mutants form equatorial cleavage furrows when growing on a solid substratum (23–26), and, in the budding yeast Saccharomyces cerevisiae, the mutant cells complete division even though they fail to assemble an actin ring at the division site (10). Although cytokinesis of the yeast mutants is inefficient, it can be almost completely rescued by expression of the myosin-II tail domain, which is incapable of generating force by myosin−actin interaction (18, 27). Moreover, in the fission yeast Schizosaccharomyces pombe, modeling indicates that the actomyosin ring cannot provide more than a small fraction of the force needed to drive furrow ingression in the face of intracellular turgor pressure, and pharmacological disassembly of F-actin after initiation of furrowing did not inhibit further furrow ingression (28).
The limitations of the CAR model become even more apparent when cytokinesis is viewed in a phylogenetic and evolutionary perspective. For example, most cells in plants divide by a mechanism [centrifugal cell plate growth mediated by the microtubule (MT)-based phragmoplast (29–32)] that seems completely different from the cleavage-furrow ingression of unikonts, although the two groups have a common ancestor. Moreover, except for the plants, some types of algal cells, and some intracellular parasites, all non-unikont eukaryotic cells that have been examined divide by cleavage-furrow ingression (33–41) although they lack a myosin-II, which appears to be present only in the unikont lineage (42–45) [with the interesting exception of a myosin-II in the Excavate Naegleria gruberi (46)]. There is very little information about the mechanisms by which such furrows form, although some non-unikont cells have been reported to have actin localized in the developing furrows (34, 36, 37, 39, 47–49), raising the possibility that actin might have a role that predates and is independent of myosin-II.
Taken together, these and other observations (30) suggest that the earliest eukaryotes had a mechanism for cleavage-furrow formation, presumably inherited from their prokaryotic forebears, that did not involve a CAR, although it might have involved actin. Importantly, such an ancestral mechanism might still exist as the underpinning for many or all of the seemingly diverse modes of cytokinesis seen today. To explore the nature of this postulated mechanism, we are studying the green alga Chlamydomonas reinhardtii, which divides by furrow formation (refs. 33 and 50; this study) but has no myosin-II (51).
During growth under a daily light−dark cycle, the Chlamydomonas cell cycle consists of a long G1 phase during the light period, followed, during the dark period, by one or more rapid binary division cycles of S phase, M phase, and cytokinesis (52). In each cycle, a cleavage furrow appears to ingress asymmetrically to separate the daughter nuclei, daughter chloroplasts, and associated cytoplasm (refs. 33 and 53; also see figure 5 of ref. 52). It remains unclear whether (and, if so, in what form) extracellular matrix/cell wall deposition occurs external to the plasma membrane in the ingressing furrow (see images in refs. 33 and 53). At least two sets of MTs are associated with the furrow and may be involved in determining its position and/or in promoting its ingression (33, 50, 54–56). In addition, immunofluorescence and phalloidin-staining studies have suggested that actin localizes to the furrow region (48, 50, 57, 58) in an MT-dependent manner (50), suggesting that actin might have a myosin-II-independent role(s) in Chlamydomonas cytokinesis. Testing this possibility has been challenging because actin is both essential for viability in Chlamydomonas and encoded by two genes, either of whose products can provide the essential function(s). However, in this study, we have been able to use a combination of a mutation and a drug to eliminate F-actin. We found that cleavage-furrow formation in Chlamydomonas does not require F-actin but is facilitated by it, apparently, at least in part, because of a previously unappreciated role for F-actin in chloroplast division. Our observations on MT behavior in the absence of F-actin are also consistent with the hypothesis that the MTs play a role in furrow formation.
Results
Live-Cell Observations of Cleavage-Furrow Ingression.
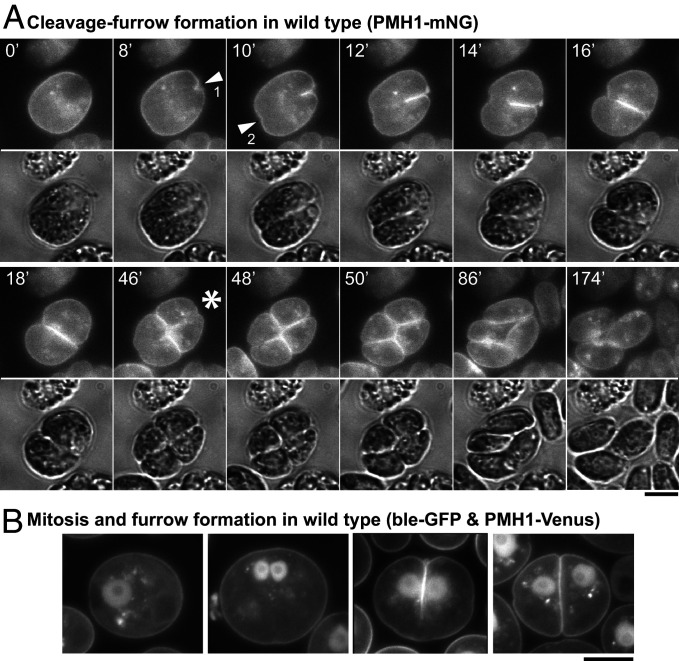
Previous descriptions of cytokinesis in Chlamydomonas have been based on light and electron micrographs of fixed cells (33, 50, 54). Because such images do not provide dynamic information, such as the rate of furrow ingression, we expressed the plasma membrane ATPase PMH1 (59) tagged with mNeonGreen (mNG) and observed living cells by time-lapse microscopy. As expected, a cleavage furrow ingressed primarily from one pole of the cell (Fig. 1A, arrowhead 1, and Movies S1 and S2); it reached the opposite side of the cell in 16 ± 3 min (n = 13). However, the earlier appearance of a small notch at the opposite side (Fig. 1A, arrowhead 2) suggested that a “lateral ingression” formed a groove around the entire perimeter of the plane of cleavage well before ingression of the medial furrow was complete, consistent with observations by differential-interference-contrast (DIC) (Fig. 1A, 10 min and 12 min) and electron (33) microscopy. When the imaged cells also expressed ble-GFP [a marker for the nucleus (60)], it was apparent that the medial furrow began to form at the anterior pole of the cell and progressed between the daughter nuclei (Fig. 1B), consistent with previous reports (33, 54). In most cells, furrow ingression was accompanied by cytoplasmic rotation (Fig. 1A, 0 min to 16 min), as reported previously (33).
Fig. 1.
Live-cell observations of cleavage-furrow formation in Chlamydomonas. (A) Wild-type cells expressing the plasma membrane ATPase PMH1 tagged with mNG were synchronized using the 12L:12D/TAP agar method, mounted on TAP + 1.5% low-melting agarose, and imaged over several hours at ∼25 °C. Selected images are shown (times in minutes); the full series is presented in Movies S1 and S2. (Top) The mNG fluorescence (YFP channel); (Bottom) Differential-interference-contrast (DIC). Arrowheads, positions of the initial appearance of furrow ingression visible in this focal plane in the anterior (1) and posterior (2) poles of the cell. Asterisk, onset of second cleavage. (B) Wild-type cells coexpressing PMH1-Venus and the nuclear marker ble-GFP were imaged using a YFP filter set during growth on TAP medium at 26 °C. Cells at different stages in mitosis and cytokinesis are shown. (Scale bars, 5 µm.)
Under the growth conditions used, most cells underwent two or three rapid divisions, then hatched from the mother cell wall as four or eight daughter cells (Movie S2). The second cleavage followed the first by 38 ± 4 min (n = 11) and was often not clearly visible because of its angle relative to the imaging plane. However, when it could be seen clearly, it always initiated at the center of the previous cleavage (Fig. 1A, 46 min, asterisk, and Movies S1 and S2)
Association of F-Actin, but Not Myosins, with the Cleavage Furrow.
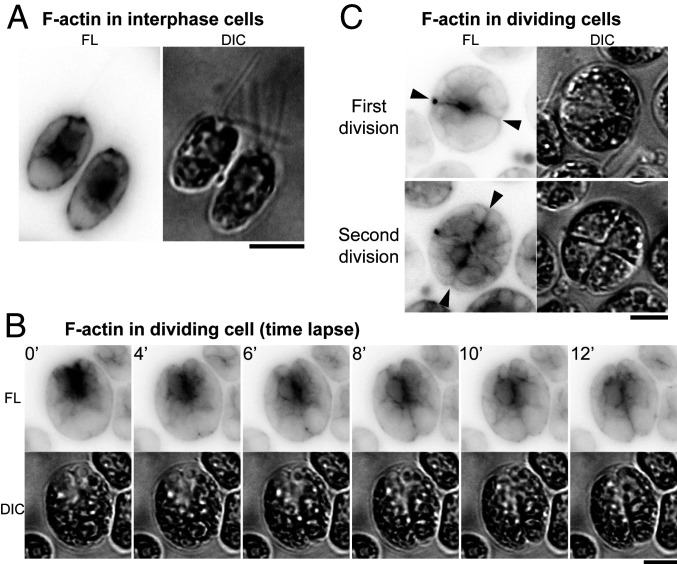
Despite the lack of a type-II myosin in Chlamydomonas, actin might still have a role in cleavage-furrow formation. To explore this possibility, we first expressed the F-actin−binding peptide Lifeact (61) as a fusion with mNG. In interphase cells, F-actin localized around the nucleus, at the basal body region, and in the cortex, as observed previously (Fig. 2A; refs. 62–64). In dividing cells, F-actin showed a transient but strong enrichment at the anterior pole (Fig. 2B, 0 min to 4 min) and then appeared to be associated with the furrow throughout its ingression (Fig. 2B, 4 min to 12 min, and C, “First division,” and SI Appendix, Fig. S1). F-actin was also associated with the new furrows (but not the old ones) in cells undergoing their second round of cytokinesis (Fig. 2 C, “Second division”).
Fig. 2.
F-actin localization to the region of the cleavage furrow. Wild-type cells expressing Lifeact-mNG were imaged; fluorescence (FL) and DIC images are shown. Fluorescence images are presented with contrast inverted for greater clarity. (A and C) Still images of (A) interphase and (C) dividing cells grown on TAP medium at 26 °C. (B) Time-lapse images (as in Fig. 1A) showing association of F-actin with the region of the cleavage furrow. Arrowheads, ingressing furrows. (Scale bars, 5 µm.)
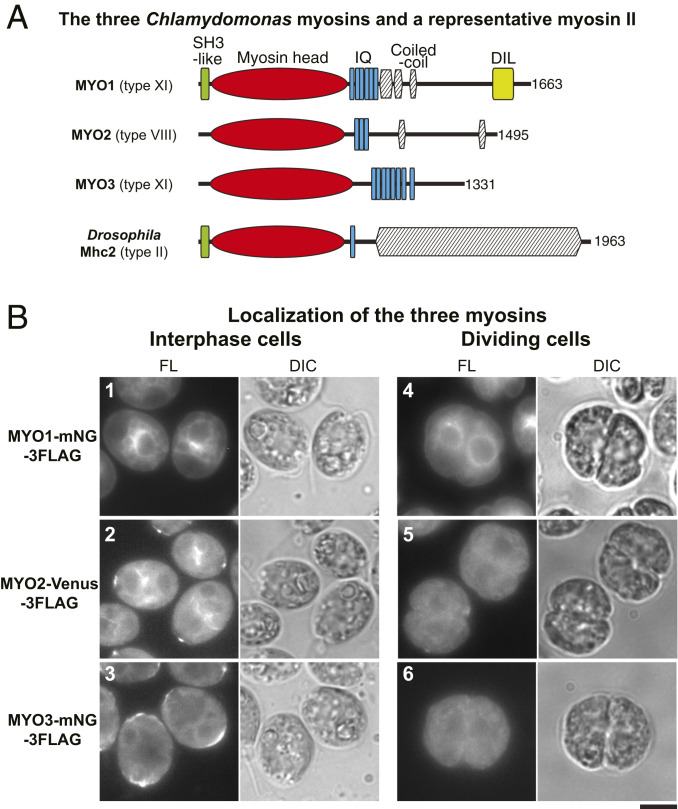
If the F-actin associated with the furrow region has a role in cleavage-furrow formation, one or more myosins might also be involved. A basic local alignment search tool (BLAST) search of the Chlamydomonas genome using the motor domain of Drosophila melanogaster type-II myosin detected only the three myosin genes reported previously (62). A phylogenetic analysis indicated that MYO1 and MYO3 encode type-XI myosins, whereas MYO2 encodes a type-VIII myosin (Fig. 3A and SI Appendix, Fig. S2A). Importantly, none of these myosins has an extended C-terminal coiled-coil domain such as those that allow type-II myosins to form bipolar filaments (65).
Fig. 3.
Lack of myosin localization to the region of the cleavage furrow. (A) Domain structures of the three Chlamydomonas myosins; a typical type-II myosin with a long coiled-coil tail (Drosophila Mhc2) is included for comparison. Domains were predicted using the HMMER (hmmer.org) and COILS (119) programs; total numbers of amino acids are indicated. (B) Localization of fluorescently tagged myosins in interphase and dividing cells; cells were grown on TAP medium at 26 °C. Fluorescence (FL) and DIC images are shown. (Scale bar, 5 µm.)
To ask whether any of these myosins might be involved in cytokinesis, we expressed each protein in wild-type cells with mNG-3FLAG or Venus-3FLAG fused at its C terminus; in each case, the fusion protein was detected at or near the expected molecular weight by Western blotting (SI Appendix, Fig. S2B). In interphase cells, MYO1 was enriched in the perinuclear region (Fig. 3 B, 1, and SI Appendix, Fig. S2 C, 4) in a pattern resembling one aspect of F-actin localization as seen with the tagged Lifeact probe (Fig. 2A, and SI Appendix, Fig. S2 C, 1). MYO2 also localized to the perinuclear region as well as to dots at the cell-anterior region near the basal bodies (Fig. 3 B, 2, and SI Appendix, Fig. S2 C, 7; ref. 62), again resembling aspects of F-actin localization. MYO3 also localized to the cell-anterior region, as well as to the cortex in the cell posterior (Fig. 3 B, 3, and SI Appendix, Fig. S2 C, 10). Importantly, in dividing cells, none of the myosins showed any detectable association with the cleavage furrow (Fig. 3 B, 4–6).
The similarity in localization of the myosins to that of F-actin in interphase cells (see above) suggests that the tagged myosins interact normally with actin. Further evidence for this conclusion was obtained in experiments that exploited the Chlamydomonas system for F-actin homeostasis (57, 63, 64, 66). In vegetative wild-type cells, only the conventional actin IDA5 is expressed. Exposure of cells to the F-actin-depolymerizing drug latrunculin B (LatB) leads to a rapid disassembly of F-IDA5 (SI Appendix, Fig. S2 C, 2), degradation of the monomeric IDA5, and upregulation of the divergent actin NAP1, which provides actin function by assembling into LatB-resistant filaments (SI Appendix, Fig. S2 C, 3). Similarly, the localization signals for MYO1 and MYO3 were largely lost during a short incubation with LatB but subsequently recovered (SI Appendix, Fig. S2 C, 4–6 and 10–12), suggesting that these tagged myosins can bind to both F-IDA5 and F-NAP1. The perinuclear signal for MYO2 was also sensitive to LatB but did not recover (SI Appendix, Fig. S2 C, 7–9; ref. 62), suggesting that this myosin binds only to F-IDA5.
Thus, although the current lack of null mutations for any of the myosin genes precludes a definitive test of the function of the tagged proteins, they at least localize as expected in an F-IDA5-dependent or F-NAP1-dependent manner, so that their apparent absence from the furrow region suggests that any function of actin in furrowing does not involve the myosins (e.g., in some type of noncanonical actomyosin ring).
Cleavage-Furrow Ingression in the Absence of F-Actin.
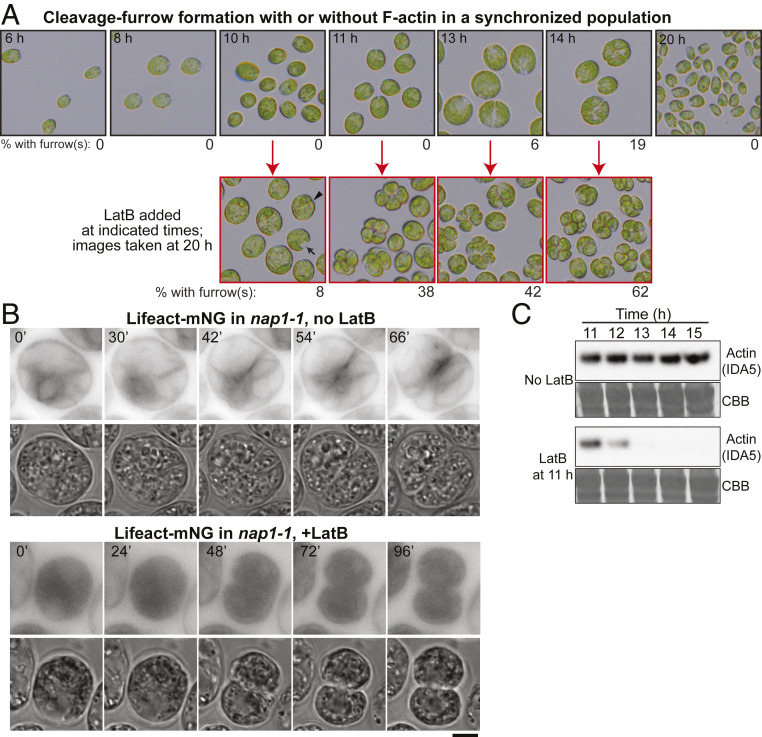
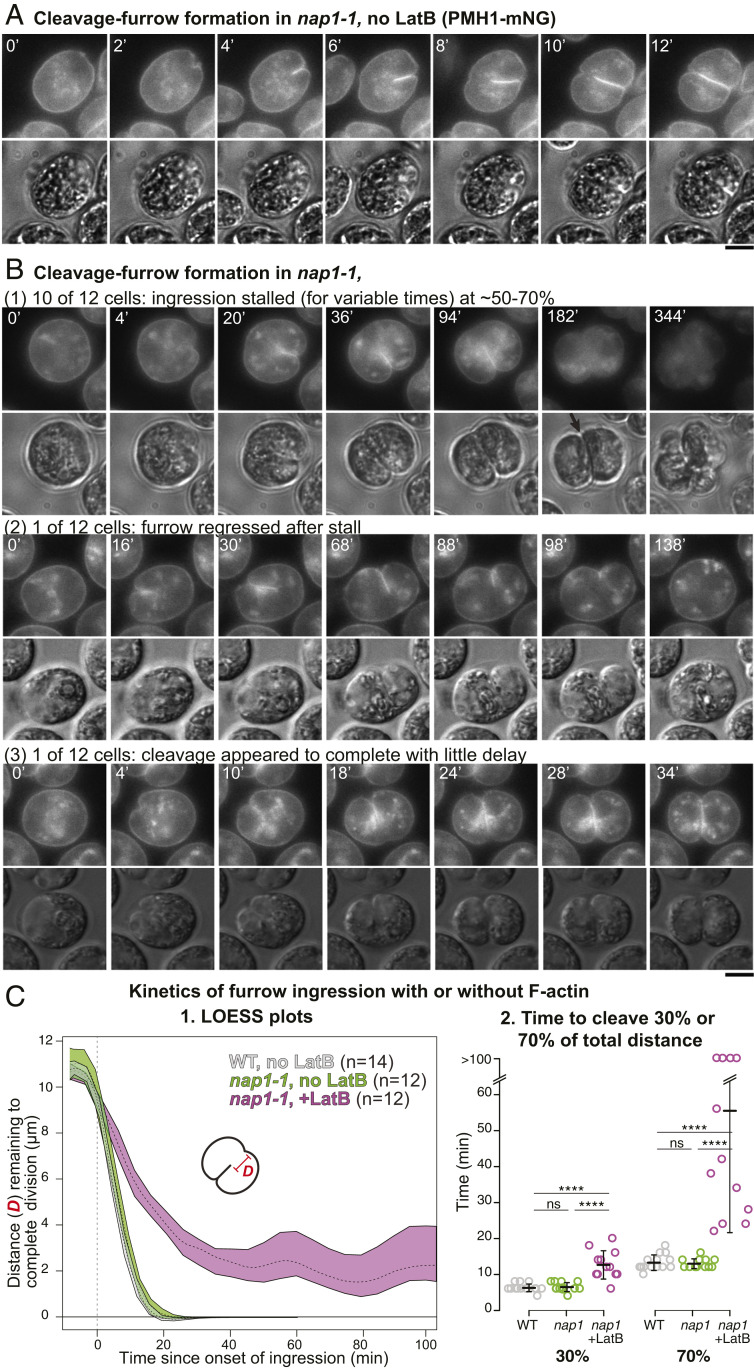
To ask whether actin (with or without myosin) plays a role in cleavage-furrow formation, we took advantage of our prior isolation of a null mutation (nap1-1) in the NAP1 gene (63). Because F-IDA5 is highly sensitive to LatB (see above; refs. 64 and 67), treatment of a nap1-1 strain with the drug results in a rapid and seemingly complete loss of F-actin, which is lethal (64, 67), indicating that F-actin is directly or indirectly important for various cellular processes in Chlamydomonas. The LatB-treated nap1-1 cells appear to cease growth and (probably as a consequence) DNA replication and cell-cycle progression. Thus, to evaluate a possible specific role of actin in cytokinesis, we synchronized the cell cycle of nap1-1 cells using a 12 h:12 h light:dark cycle that yielded populations enriched in cells that were close to initiation of division at the time of LatB application (52, 68). Under the conditions used, the cells grew in size throughout the light phase, began to divide at ∼13 h (i.e., 1 h into the dark phase), and hatched out as small daughter cells at ≤20 h (Fig. 4 A, Top). We then treated aliquots of the culture with LatB at different times before and during the onset of cytokinesis and examined the cells several hours later (Fig. 4 A, Bottom). When LatB was added at ≤9 h, the cells ceased to grow in size and never detectably initiated cytokinesis. Flow-cytometry analysis in separate but similar experiments indicated that most of these cells had arrested before DNA replication, as expected (63). When LatB was added at 10 h, most cells remained round and did not begin furrow formation, but a few formed what appeared to be normal cleavage furrows (Fig. 4A, arrowhead) or “notch”-like structures (Fig. 4A, arrow). A similar experiment using cells expressing PMH1-Venus and ble-GFP indicated that the cells with notches had not undergone mitosis (SI Appendix, Fig. S3 A, Left). In contrast, when LatB was added at ≥11 h, many cells appeared to have gone through two or more rounds of cleavage-furrow ingression, forming clusters of four to eight cells, each of which contained a nucleus (Fig. 4A, and SI Appendix, Fig. S3 A, Right). In a separate but similar experiment, we noted a rough positive correlation between the sizes of the undivided cells at the time of LatB addition and their likelihood of subsequently forming a furrow (SI Appendix, Fig. S3B), suggesting that there is either a direct size requirement for cytokinesis or a requirement for some size-correlated prerequisite cell-cycle event.
Fig. 4.
Cleavage-furrow formation in the absence of F-actin. (A) (Top) The nap1-1 cells were synchronized using the 12L:12D/liquid TP method at 26 °C, incubated for up to 20 h, and imaged at intervals. (Bottom) At the indicated times (red arrows), samples were plated on TAP agar containing 3 µM LatB, incubated at 26 °C, and then imaged at 20 h (i.e., 8 h into the dark period). The percentages of cells with visible cleavage furrows are shown below the images. (B) F-actin localization in nap1-1 cells with or without LatB treatment. The nap1-1 cells expressing Lifeact-mNG were synchronized using the 12L:12L/TAP agar method at 26 °C, mounted on TAP + 1.5% low-melting agarose with or without 3 µM LatB, and observed during growth at 26 °C. Selected images are shown; contrast is inverted for greater clarity. The LatB-treated cell had been incubated with the drug for ∼4.5 h before the first frame shown. Note that dispersion of Lifeact-mNG into the entire cytoplasm contributed to the strong apparent background in these cells. (Scale bar, 5 µm.) (C) Rapid degradation of IDA5 upon LatB addition to a synchronized culture. The nap1-1 cells were synchronized as in A, and the culture was split; 3 µM LatB was added to one culture at 11 h, and samples were drawn at the indicated times and subjected to Western blotting using an anti-actin antibody. Thirty µg of total protein were loaded in each lane. CBB, the membrane stained with Coomassie Brilliant Blue, shown as a loading control.
Although our prior work had suggested that LatB-treated nap1-1 cells contained no residual F-actin, it seemed possible that there might be a special population of drug-resistant filaments in the cleavage-furrow region. However, no such filaments were observed when time-lapse observations were made on LatB-treated nap1-1 cells expressing Lifeact-mNG (Fig. 4B). Moreover, consistent with our prior observations indicating rapid proteasomal degradation of LatB-depolymerized IDA5 (64), IDA5 was also largely or entirely degraded by the time such cells began furrow formation (Fig. 4C).
Taken together, these results appear to establish that F-actin is not required for cleavage-furrow formation in Chlamydomonas.
Reduced Efficiency of Furrow Ingression in Cells Lacking F-Actin.
To investigate the efficiency of cleavage-furrow formation in the absence of F-actin, we performed time-lapse microscopy on nap1-1 cells expressing PMH1-mNG. In the absence of LatB, furrow formation in these cells proceeded to completion in 17 ± 4 min (n = 12) (Fig. 5 A and C, 1, and SI Appendix, Fig. S3 C, 2, and Movie S3), not significantly different from the rate in wild-type cells (16 ± 3 min; n = 13) (Figs. 1 and 5 C, 1 and SI Appendix, Fig. S3 C, 1). In contrast, in the presence of LatB, although the rates of furrow ingression varied considerably in individual cells, they were slower in all cells examined than in control cells even during the early stages of furrow ingression, and more so during its later stages (Fig. 5 B and C, 1 and 2, and SI Appendix, Fig. S3 C, 3). The time gap between the formation of a small cell-anterior notch and the detectable ingression of the medial furrow was expanded from ∼2 min in control cells to 5–15 min. Moreover, although the medial furrow typically ingressed smoothly into about the middle of the cell, the second notch at the cell-posterior end did not appear normally at that time, and, in most cells, the medial furrow stalled at that point for an extended period before eventually appearing to complete its growth (10 of the 12 cells examined: Fig. 5 B, 1, arrow and Movie S4) or regressing (one of the 12 cells examined: Fig. 5 B, 2 and Movie S5). More rarely (one of the 12 cells examined), the furrow appeared to progress across the cell without interruption (Fig. 5 B, 3 and Movie S6). In all cells examined, the daughter cells remained clustered without hatching, and the fluorescence of PMH1-mNG became quite dim, making it difficult to determine when (or whether) the plasma membranes were fully resolved. Indeed, when the clusters of LatB-treated nap1-1 cells (Fig. 4A) were treated with the cell-wall-digesting enzyme autolysin, ∼5% of the cells remained connected by an intercellular bridge between the pairs (SI Appendix, Fig. S3D). Taken together, these results suggest that, although actin is not required for furrow ingression per se, it plays some ancillary role(s) that facilitates the early stages of furrowing and becomes more important during the later stages of furrow ingression and/or abscission.
Fig. 5.
Slower cleavage-furrow ingression and delay in furrow completion in the absence of F-actin. (A) The nap1-1 cells expressing PMH1-mNG were synchronized using the 12L:12D/TAP agar method and observed by time-lapse microscopy at 26 °C. The full series is presented in Movie S3. (Scale bar, 5 µm.) (B) As in A except that 3 µM LatB was added 120 min to 160 min before the first frames shown. Selected images are shown to illustrate the different time scales; full series are presented in Movies S4–S6. (Scale bar, 5 µm.) (C) Kinetics of furrow ingression in wild-type cells without LatB (Fig. 1A and Movie S2) and in nap1-1 cells with or without 3 µM LatB. Data are from the experiments shown in A and B. (1) Distance of the leading edge of the medial furrow from the opposite side of the cell as a function of time since the onset of furrow ingression (set at 0). Means and 95% CIs of 1,000×-bootstrapped locally estimated scatterplot smoothing (LOESS) curves are shown (see Materials and Methods). The curves for individual cells are shown in SI Appendix, Fig. S3 C. (2) Times for the furrow to reach 30% or 70% of the total distance across the cells. Because of mNG bleaching after prolonged time-lapse observations, cleavage times were capped at 100 min for these analyses. Bars indicate means and SDs. Statistical analyses were performed using one-way ANOVA and Tukey’s post hoc multiple comparisons (ns, not significant; ****P < 0.0001).
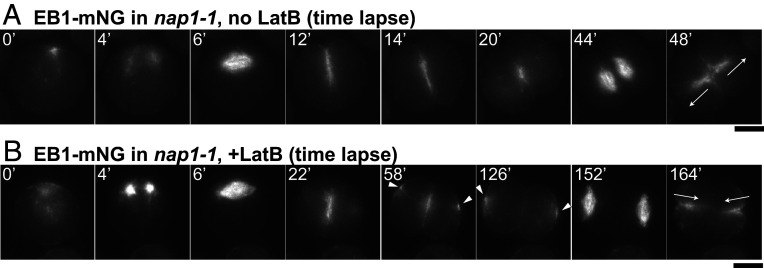
Association of MTs with the Cleavage Furrow in the Absence of F-Actin.
It has long been known that MTs are associated with the cleavage furrows in Chlamydomonas (33, 50, 54). To ask whether this association is maintained in the absence of F-actin, we first tried, but failed, to visualize the furrow-associated MTs by time-lapse imaging using a fluorescently tagged tubulin. However, we had better results upon expressing an mNG-tagged version of the plus-end-binding protein EB1 (69). In both wild-type cells and nap1-1 cells not treated with LatB (Fig. 6A, 0 min and Movie S7), EB1-mNG was concentrated in the basal body region of premitotic cells. When the cells entered mitosis, EB1-mNG disappeared from the cell pole and appeared in the mitotic spindle (Fig. 6A, 4 min to 6 min and Movie S7). After mitosis, the EB1-mNG signal reappeared at the apical pole of the cell, from which it moved into and across the cell body as the cleavage furrow formed (Fig. 6A, 12 min to 14 min and Movie S7) and remained concentrated in the middle of the division plane after cytokinesis (Fig. 6A, 20 min and Movie S7). Each daughter cell then formed an EB1-labeled spindle in the region proximal to this site, and the new furrows (as marked by EB1-mNG) grew outward from the center of the cell to the surface (Fig. 6A, 44 min and 48 min, arrows). In nap1-1 cells treated with LatB, EB1-mNG localization was nearly normal during the first division (Fig. 6B, 0 min to 22 min and Movie S8), except for a slower-than-normal progression through the cytoplasm, consistent with the PMH1-mNG data (see above). However, a fraction of the EB1-mNG appeared to leave the division plane prematurely and form foci at the far sides of the daughter cells (Fig. 6B, 58 min to 126 min, arrowheads), suggesting a defect in polarity maintenance caused by the loss of F-actin. Probably as a consequence, the spindles for the second mitosis were positioned distal from the previous division plane, and the cleavage furrows subsequently grew inward (Fig. 6B, 152 min to 164 min, arrows and Movie S8), although the localization of EB1-mNG, and thus presumably of both the spindle and furrow-associated MTs, otherwise appeared essentially normal. This preservation of nearly normal association of MTs with the furrows is consistent with the hypothesis that the MTs are involved in furrow ingression in both the presence and absence of F-actin.
Fig. 6.
Persistent association of MTs with cleavage furrows, but changes in polarity during the second division, in the absence of F-actin. The nap1-1 cells expressing EB1-mNG to visualize MTs were synchronized using the 12L:12D/TAP agar method and observed by time-lapse microscopy at 26 °C. Selected images are shown; full series are presented in Movies S7 and S8. (A) A cell not treated with LatB. (B) A cell treated with 3 µM LatB beginning ∼20 min before the first frame shown. Arrowheads, aberrant foci of EB1-mNG at positions distal to the cleavage furrow in cells lacking F-actin; arrows, the directions of furrowing during the second division of each cell. (Scale bars, 5 µm.)
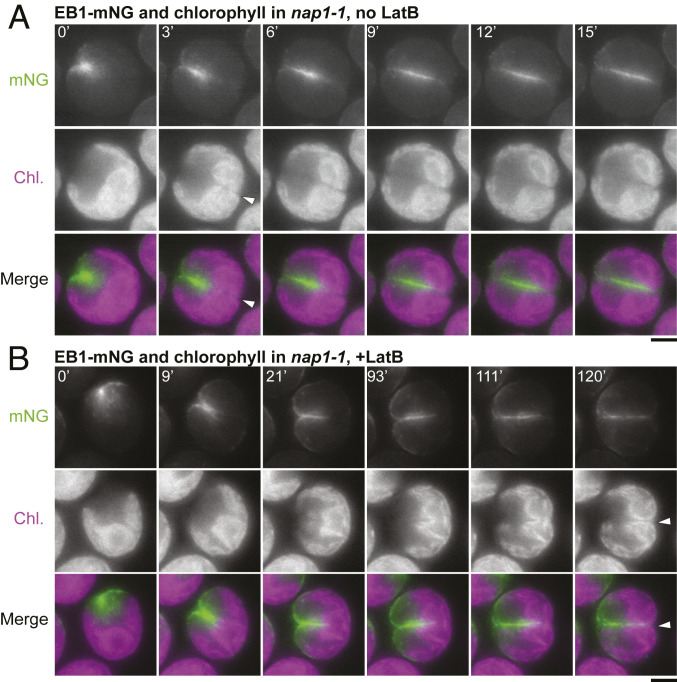
Defective Chloroplast Division in Cells without F-Actin.
The EB1-mNG studies also revealed a possible cause of the delay in furrow completion in cells lacking F-actin. In normal Chlamydomonas cells, the large, cup-shaped chloroplast is centered on the posterior pole of the cell (Fig. 7A, 0 min), so that the organelle lies squarely in the path of the ingressing cleavage furrow (53, 70). In control cells, division of the chloroplast (as visualized by chlorophyll autofluorescence) appeared to have occurred by the time the furrow (or at least its associated EB1) reached the organelle (Fig. 7A, 3 min, arrowhead), consistent with previous observations suggesting that the chloroplast has a division machinery that is independent of, although temporally and spatially coordinated with, cleavage-furrow ingression (53, 70). In contrast, in cells lacking F-actin, the furrow appeared to partially penetrate into the undivided chloroplast over an extended period (Fig. 7B, 21 min to 111 min), before finally moving through a gap formed in the chloroplast (Fig. 7B, arrowhead). These results suggest that efficient chloroplast division requires F-actin, that coordination of division between the cell and the plastid requires F-actin, or both, and that the physical barrier posed by the undivided chloroplast may explain the delay in furrow completion in cells lacking F-actin.
Fig. 7.
Delayed chloroplast division in the absence of F-actin. Chlorophyll autofluorescence and EB1-mNG fluorescence (to visualize the ingressing cleavage furrow) are shown in a nap1-1 cell not treated with LatB (A) and a nap1-1 cell treated with 3 µM LatB beginning ∼20 min before the first frame shown (B). Cells had been synchronized prior to imaging using the 12L:12D/TAP agar method. Arrowheads, times of apparent completion of chloroplast division. (Scale bars, 5 µm.)
Discussion
Rate of Cleavage-Furrow Ingression.
Both electron microscopy (33, 54, 71) and light microscopy on fixed cells (50, 72) had suggested that Chlamydomonas divides by means of an asymmetrically ingressing cleavage furrow (52), despite its lack of a type-II myosin, and we confirmed this model by live-cell imaging using a fluorescent plasma-membrane marker. The time-lapse imaging also allowed us to determine the rate of furrow ingression. At its maximum (∼0.85 µm/min; Fig. 5C), this rate is comparable to those in small- to medium-sized cells that have type-II myosins and form actomyosin rings at their furrow sites, such as S. pombe [∼0.15 µm/min (28, 73, 74)], Neurospora crassa [1.3 μm/min to 3.2 µm/min (75)], and various mammalian somatic cells [3 μm/min to 4 µm/min (22, 76–79)]. Thus, the presence of a CAR is not necessary to produce a rate of furrow ingression in this range.
Localization of F-Actin, but Not Myosin, to the Cleavage Furrow.
Our live-cell imaging also clarified the spatial relationships of the furrow, F-actin, and the three Chlamydomonas myosins. Although immunostaining had indicated that there was actin in the furrow region (48, 50, 66), it was not clear whether this actin was in filamentous form (48). However, imaging of cells expressing the F-actin-specific probe Lifeact showed clearly that F-actin is associated with the furrow during most or all of the period of furrow ingression. In contrast, although a previous report had suggested (based on immunostaining with an antibody to Dictyostelium type-II myosin) that myosin is also localized to the furrow region (50), fluorescence tagging of the three Chlamydomonas myosins (two type XI and one type VIII) showed no association with this region. Although this conclusion is subject to the caveat that we do not yet have definitive evidence that the tagged myosins are fully functional (see Results), the likelihood of a myosin role in Chlamydomonas furrow formation is also reduced by our finding that F-actin itself is not essential for this process.
Cleavage-Furrow Ingression without F-Actin.
Despite the apparent absence of myosins from the furrow region in Chlamydomonas, it seemed possible that the F-actin there might play an essential role in furrow formation. However, when we used a combination of a mutation (to eliminate the drug-resistant actin NAP1) and a drug (to depolymerize the drug-sensitive actin IDA5), the cells were still able to form cleavage furrows and divide. It seemed possible that a population of drug-resistant IDA5 filaments might remain specifically in the cleavage furrow. However, we could detect no such filaments by Lifeact staining. Moreover, Western blotting revealed that, as observed previously in asynchronous cells (64), depolymerization of F-IDA5 in dividing cells was followed quickly by degradation of IDA5 itself. Taken together, our results appear to establish that the actin cytoskeleton does not play a principal role in generating the force for cleavage-furrow ingression in Chlamydomonas.
Nonetheless, several observations also indicate that actin does contribute significantly to cytokinesis in Chlamydomonas. First, in the absence of F-actin, the rate of early furrow ingression was approximately twofold slower than normal. It seems possible that actin forms a contractile structure, not dependent on myosin, that contributes some force for furrow ingression, that it is necessary for the trafficking of Golgi-derived vesicles that ultimately provide the new cell surface material for the growing furrow, or that it has some other unknown role. Second, the last ∼30% of cleavage was slowed even more (fivefold or more), and some cells appeared to fail (or at least have long delays in) the final abscission of the daughters. In addition, EB1-mNG signal disappeared prematurely from the furrow region when F-actin was missing, presumably reflecting a failure to maintain MT organization during the final phase of cleavage. Finally, the directions of the second cleavages were inverted, probably because the polarities of the two daughter cells produced by the first cleavage were also inverted. The precise mechanisms by which F-actin facilitates cytokinesis are not yet clear, but the observed slowness of late furrow ingression in its absence appears to be due, at least in part, to a delay in chloroplast division (see below). Taken together, our findings also highlight the value of using Chlamydomonas to explore non-CAR-related roles of the actin cytoskeleton in cytokinesis.
An Apparent Role for F-Actin in Chloroplast Division.
In most cells lacking F-actin, there was a delay in chloroplast division of ≥120 min. This observation was surprising, because, despite early reports of an actin role in plastid division in both charophyte and red algae (80, 81), no such role has been recognized (to our knowledge) in plants or other organisms. As the Chlamydomonas chloroplast lies directly in the path of the ingressing cleavage furrow, the delay in chloroplast division may explain much or all of the delay also seen in furrow completion in these cells. Although we cannot currently test this model due to the lack of a method for clearing the chloroplast from the division path, a similar apparent obstruction of cytokinesis by an undivided chloroplast was observed after expression of a dominant-negative dynamin mutant in the red microalga Cyanidioschyzon merolae (82). Most other unicellular algae also contain only one or a few chloroplasts, whose division must presumably be coordinated both temporally and spatially (i.e., division in the same plane) with that of the cell. The temporal coordination is achieved, at least in part, by cell cycle control of the expression of the proteins (such as FtsZ and dynamin) directly involved in chloroplast division (68, 83). However, the spatial coordination seems to require a local, structure-based signal. Our results suggest that the furrow-associated F-actin may provide this spatial cue to the chloroplast division machinery and, in so doing, might also dictate the precise timing of its action.
Possible Role for MTs in Cleavage-Furrow Formation.
It has long been thought that MTs may be involved in cleavage-furrow positioning and/or ingression in Chlamydomonas. Electron microscopy and immunofluorescence studies have shown that two of the four “rootlet” MTs that run from the basal body along the cortex align with the division plane (like the preprophase band in plant cells), while an array of many MTs, the “phycoplast,” runs along the developing furrow (33, 50, 54–56). Moreover, pharmacological or genetic disruption of the MTs appears to inhibit cytokinesis (50). Our observations on cells expressing a fluorescently tagged EB1 protein have now added the information that dynamic MTs are associated with the furrow throughout its ingression and that this association is maintained even in the absence of F-actin. Thus, it seems likely that the MTs have a direct, actin-independent role in promoting furrow ingression, possibly by guiding the deposition of new plasma membrane and extracellular matrix/cell wall in the growing furrow as they do in plant cell phragmoplasts. Because studies of cell wall composition and biogenesis in Chlamydomonas have mostly been done on nondividing cells (84), little is known about how extracellular matrix/cell wall assembly might be regulated during cytokinesis. Testing this postulated MT role and elucidating the mechanisms involved will be major goals of future studies.
Cytokinesis in Phylogenetic and Evolutionary Perspective.
Our study also sheds some light on the evolutionary origins and underlying basal mechanisms of eukaryotic cytokinesis. The wide phylogenetic distribution of division by cleavage-furrow ingression, together with the near-universal absence of myosin-II outside the unikonts, had already made clear that a conventional CAR model cannot account generally for furrow formation. Moreover, we have shown here that even F-actin is not essential for the formation of cleavage furrows in Chlamydomonas. This observation has some precedents and parallels. Even within the unikonts, it is clear that F-actin is not always essential for furrow ingression (10, 19, 28, 85–87), and this may be the rule, rather than the exception, in non-unikonts. For example, in the ciliate Tetrahymena pyriformis, latrunculin A did not block cell division, despite a loss of F-actin and a consequent disruption of actin-dependent processes such as food-vacuole formation (41); in the Diplomonad parasite Giardia lamblia, which has one actin but no myosin (40), furrowing occurred efficiently when actin expression was knocked down with a morpholino (88), although the cells were delayed in abscission; and, in the red alga C. merolae, which also has no myosin, cells divide by furrowing, even though actin is not expressed under normal growth conditions (89). Taken together, these and related observations suggest that, in the earliest eukaryotes, the last eukaryotic common ancestor (LECA), and most branches of the modern eukaryotic phylogeny, actin and myosin are not primarily responsible for the force that produces cleavage-furrow ingression. Moreover, as prominent roles for actin and myosin evolved in the unikonts, the underlying ancestral mechanisms for driving furrow ingression may have remained in place, accounting for at least some of the cases in which unikonts form furrows without their normal CARs.
In thinking about these ancestral mechanisms, it should be instructive to consider the mechanisms of cytokinesis in modern prokaryotes. Nearly all bacteria divide using a furrowing mechanism in which the tubulin-like FtsZ plays a central role (90–95); an FtsZ-dependent mechanism appears to be used by many archaea as well (96–99). Thus, the immediate prokaryotic ancestor of the first eukaryotes probably also divided by such a mechanism. Current information about FtsZ action suggests that it functions both to bend the inner membrane and to organize the symmetric deposition of cell wall, which drives in the membrane to produce the division furrow (91, 92, 94, 95, 100). Thus, if an ancient FtsZ was the evolutionary progenitor of modern tubulin, the major role of MTs in cleavage-furrow formation in distantly related modern eukaryotes such as Chlamydomonas, G. lamblia (88), Penium margaritaceum (101), Trypanosoma brucei (102), Tetrahymena thermophila (103), and Toxoplasma gondii (104) may reflect the persistence of an ancient mechanism for adding cell-surface material to form a furrow. The same interpretation might apply even to the association of parallel arrays of MTs with cleavage furrows observed in some animal cells [e.g., embryos of Xenopus (105, 106), zebrafish (107), and Drosophila (108)] or to the MTs of the midbody during abscission in metazoan cells (30); in most or all of these cases, the MTs appear to play an important role in targeting vesicles containing new membrane to the furrow. In any case, the hypothesis of a primordial role for MTs in eukaryotic cytokinesis seems to make it easier to understand the central role of MTs in cytokinesis in modern plants, where both the preprophase band (which marks the future division plane) and the phragmoplast (which organizes the centrifugal deposition of new cell membrane and cell wall by fusion of post-Golgi vesicles) are MT based (29, 31, 32).
At the same time, it should also be noted that there is now good evidence that the prokaryotic ancestor of modern eukaryotes also had an actin-like protein (109, 110), and there is even some evidence that this protein might have been associated with division sites (111) despite the lack of evidence for any myosin in such organisms. Thus, the association of actin with the furrow regions both in Chlamydomonas and in many other eukaryotes without a myosin II may also be a conserved primordial trait. In this regard, it is interesting that the Escherichia coli actin-like protein FtsA serves as an anchor between the membrane and FtsZ during cytokinesis (95, 112). Moreover, the preprophase band in plants involves actin as well as MTs, conceivably reflecting an earlier stage in plant evolution in which MTs and actin functioned together to bring about ingression of a furrow (113). If both the division mechanisms of modern plants and those of modern unikonts evolved from such an ancestral state (by recruitment of intracellular MTs to form the phragmoplast in plants and by reduction of the MT role in furrow formation in favor of an actomyosin system in unikonts), then continuing studies of Chlamydomonas should help to elucidate both of these evolutionary paths as well as both of the modern mechanisms.
In summary, we suggest that a full understanding of eukaryotic cytokinesis, even in the intensively studied animal cells, will remain elusive unless a greater effort is made to incorporate the lessons about the evolution of this process that can be learned by studying it in the full diversity of modern eukaryotes.
Materials and Methods
Strains and Growth Conditions.
C. reinhardtii wild-type strains CC-124 (mt–) and iso10 (mt+, congenic to CC-124) were the parental strains. The nap1-1 mutant had previously been isolated and backcrossed three times in the CC-124 background (63). The ble-GFP and PMH1-Venus strains are progeny of previously established transgenic strains (59, 60, 114).
Routine cell culture was done in Tris-acetate-phosphate (TAP) medium (115) at ∼26 °C under constant illumination at 50–100 μmol photons⋅m−2⋅s−1. The same medium without acetate (TP) was used in one method for cell cycle synchronization (see below). Except for synchronized cultures, liquid cultures were in exponential phase when experiments were performed. LatB was purchased from Adipogen (AG-CN2-0031, Lots A00143/I and A00143/J), and dilutions into TAP or TP medium were made from a 10-mM stock in dimethyl sulfoxide.
Cell Cycle Synchronization.
Three different methods were used for cell cycle synchronization in this study: 1) The 12L:12D/liquid TP method was essentially as described by Fang et al. (116) except that TP medium at 26 °C was used in place of high-salt medium; 2) the 12L:12D/TAP agar method was as described previously (117), except that it was carried out at 26 °C; and 3) the −N method was exactly as described previously using a combination of 21 °C and 33 °C (118). Although overall synchrony and the timing of mitosis and cytokinesis as determined by microscopic examination varied slightly depending on the method, we observed no significant qualitative or quantitative difference in the cells’ response to F-actin perturbation introduced by LatB addition before the onset of cytokinesis.
Other Methods.
SI Appendix, Supplementary Materials and Methods includes information on genetic analysis, plasmids and transformation, light microscopy, Western blotting, and phylogenetic analysis.
Supplementary Material
Acknowledgments
We thank Arthur Grossman, Frej Tulin, Kresti Pecani, Geng Sa, Howard Berg, Heather Cartwright, David Ehrhardt, Devaki Bhaya, Takako Kato-Minoura, Ritsu Kamiya, Martin Jonikas, Luke Mackinder, Karl Lechtreck, Prachee Avasthi, Silvia Ramundo, Alex Paredez, Ryuichi Nishihama, and other members of our laboratories for valuable discussions, the provision of valuable reagents, or both. We also thank the Chlamydomonas Resource Center for providing essential strains and reagents. This work was supported by NSF Grants EAGER 1548533 (to J.R.P.), MCB 1818383 (to J.R.P.), and MCB 1515220 (to J.G.U.), by NIH Grants R01GM126557 (to J.G.U.) and 5R01GM078153 (to F.R.C.), and by Duke University.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1920337117/-/DCSupplemental.
Data Availability.
All data for this paper are provided in the main text, SI Appendix, or Movies S1–S8.
References
- 1.Schroeder T. E., Cytokinesis: Filaments in the cleavage furrow. Exp. Cell Res. 53, 272–276 (1968). [DOI] [PubMed] [Google Scholar]
- 2.Robinson D. N., Spudich J. A., Mechanics and regulation of cytokinesis. Curr. Opin. Cell Biol. 16, 182–188 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eggert U. S., Mitchison T. J., Field C. M., Animal cytokinesis: From parts list to mechanisms. Annu. Rev. Biochem. 75, 543–566 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Pollard T. D., O’Shaughnessy B., Molecular mechanism of cytokinesis. Annu. Rev. Biochem. 88, 661–689 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fujiwara K., Pollard T. D., Fluorescent antibody localization of myosin in the cytoplasm, cleavage furrow, and mitotic spindle of human cells. J. Cell Biol. 71, 848–875 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yumura S., Mori H., Fukui Y., Localization of actin and myosin for the study of ameboid movement in Dictyostelium using improved immunofluorescence. J. Cell Biol. 99, 894–899 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alfa C. E., Hyams J. S., Distribution of tubulin and actin through the cell division cycle of the fission yeast Schizosaccharomyces japonicus var. versatilis: A comparison with Schizosaccharomyces pombe. J. Cell Sci. 96, 71–77 (1990). [DOI] [PubMed] [Google Scholar]
- 8.Kitayama C., Sugimoto A., Yamamoto M., Type II myosin heavy chain encoded by the myo2 gene composes the contractile ring during cytokinesis in Schizosaccharomyces pombe. J. Cell Biol. 137, 1309–1319 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu J. Q., Pollard T. D., Counting cytokinesis proteins globally and locally in fission yeast. Science 310, 310–314 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Bi E. et al., Involvement of an actomyosin contractile ring in Saccharomyces cerevisiae cytokinesis. J. Cell Biol. 142, 1301–1312 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lippincott J., Li R., Sequential assembly of myosin II, an IQGAP-like protein, and filamentous actin to a ring structure involved in budding yeast cytokinesis. J. Cell Biol. 140, 355–366 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mishra M. et al., In vitro contraction of cytokinetic ring depends on myosin II but not on actin dynamics. Nat. Cell Biol. 15, 853–859 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Young B. A., Buser C., Drubin D. G., Isolation and partial purification of the Saccharomyces cerevisiae cytokinetic apparatus. Cytoskeleton (Hoboken) 67, 13–22 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mabuchi I., Okuno M., The effect of myosin antibody on the division of starfish blastomeres. J. Cell Biol. 74, 251–263 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mabuchi I., Cleavage furrow: Timing of emergence of contractile ring actin filaments and establishment of the contractile ring by filament bundling in sea urchin eggs. J. Cell Sci. 107, 1853–1862 (1994). [DOI] [PubMed] [Google Scholar]
- 16.Kiehart D. P., Mabuchi I., Inoué S., Evidence that myosin does not contribute to force production in chromosome movement. J. Cell Biol. 94, 165–178 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Straight A. F. et al., Dissecting temporal and spatial control of cytokinesis with a myosin II Inhibitor. Science 299, 1743–1747 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Lord M., Laves E., Pollard T. D., Cytokinesis depends on the motor domains of myosin-II in fission yeast but not in budding yeast. Mol. Biol. Cell 16, 5346–5355 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Connell C. B., Warner A. K., Wang Y., Distinct roles of the equatorial and polar cortices in the cleavage of adherent cells. Curr. Biol. 11, 702–707 (2001). [DOI] [PubMed] [Google Scholar]
- 20.Wang Y. L., The mechanism of cortical ingression during early cytokinesis: Thinking beyond the contractile ring hypothesis. Trends Cell Biol. 15, 581–588 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Kanada M., Nagasaki A., Uyeda T. Q., Adhesion-dependent and contractile ring-independent equatorial furrowing during cytokinesis in mammalian cells. Mol. Biol. Cell 16, 3865–3872 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma X. et al., Nonmuscle myosin II exerts tension but does not translocate actin in vertebrate cytokinesis. Proc. Natl. Acad. Sci. U.S.A. 109, 4509–4514 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Lozanne A., Spudich J. A., Disruption of the Dictyostelium myosin heavy chain gene by homologous recombination. Science 236, 1086–1091 (1987). [DOI] [PubMed] [Google Scholar]
- 24.Neujahr R., Heizer C., Gerisch G., Myosin II-independent processes in mitotic cells of Dictyostelium discoideum: Redistribution of the nuclei, re-arrangement of the actin system and formation of the cleavage furrow. J. Cell Sci. 110, 123–137 (1997). [DOI] [PubMed] [Google Scholar]
- 25.Gerisch G., Weber I., Cytokinesis without myosin II. Curr. Opin. Cell Biol. 12, 126–132 (2000). [DOI] [PubMed] [Google Scholar]
- 26.Nagasaki A., de Hostos E. L., Uyeda T. Q., Genetic and morphological evidence for two parallel pathways of cell-cycle-coupled cytokinesis in Dictyostelium. J. Cell Sci. 115, 2241–2251 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Fang X. et al., Biphasic targeting and cleavage furrow ingression directed by the tail of a myosin II. J. Cell Biol. 191, 1333–1350 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Proctor S. A., Minc N., Boudaoud A., Chang F., Contributions of turgor pressure, the contractile ring, and septum assembly to forces in cytokinesis in fission yeast. Curr. Biol. 22, 1601–1608 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jürgens G., Plant cytokinesis: Fission by fusion. Trends Cell Biol. 15, 277–283 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Otegui M. S., Verbrugghe K. J., Skop A. R., Midbodies and phragmoplasts: Analogous structures involved in cytokinesis. Trends Cell Biol. 15, 404–413 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McMichael C. M., Bednarek S. Y., Cytoskeletal and membrane dynamics during higher plant cytokinesis. New Phytol. 197, 1039–1057 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Smertenko A. et al., Plant cytokinesis: Terminology for structures and processes. Trends Cell Biol. 27, 885–894 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Johnson U. G., Porter K. R., Fine structure of cell division in Chlamydomonas reinhardi. Basal bodies and microtubules. J. Cell Biol. 38, 403–425 (1968). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamagishi T., Kawai H., Cytoskeleton organization during the cell cycle in two stramenopile microalgae, Ochromonas danica (Chrysophyceae) and Heterosigma akashiwo (Raphidophyceae), with special reference to F-actin organization and its role in cytokinesis. Protist 163, 686–700 (2012). [DOI] [PubMed] [Google Scholar]
- 35.Wordeman L., Cande W. Z., Cytokinesis by furrowing in diatoms. Ann. N. Y. Acad. Sci. 582, 252–259 (1990). [DOI] [PubMed] [Google Scholar]
- 36.Sawitzky H., Grolig F., Phragmoplast of the green alga Spirogyra is functionally distinct from the higher plant phragmoplast. J. Cell Biol. 130, 1359–1371 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takahashi H. et al., A possible role for actin dots in the formation of the contractile ring in the ultra-micro alga Cyanidium caldarium RK-1. Protoplasma 202, 91–104 (1998). [Google Scholar]
- 38.Pickett-Heaps J. D., Gunning B. E., Brown R. C., Lemmon B. E., Cleary A. L., The cytoplast concept in dividing plant cells: Cytoplasmic domains and the evolution of spatially organized cell. Am. J. Bot. 86, 153–172 (1999). [PubMed] [Google Scholar]
- 39.Sehring I. M., Reiner C., Mansfeld J., Plattner H., Kissmehl R., A broad spectrum of actin paralogs in Paramecium tetraurelia cells display differential localization and function. J. Cell Sci. 120, 177–190 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Paredez A. R. et al., An actin cytoskeleton with evolutionarily conserved functions in the absence of canonical actin-binding proteins. Proc. Natl. Acad. Sci. U.S.A. 108, 6151–6156 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shimizu Y., Kushida Y., Kiriyama S., Nakano K., Numata O., Formation and ingression of division furrow can progress under the inhibitory condition of actin polymerization in ciliate Tetrahymena pyriformis. Zool. Sci. 30, 1044–1049 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Odronitz F., Kollmar M., Drawing the tree of eukaryotic life based on the analysis of 2,269 manually annotated myosins from 328 species. Genome Biol. 8, R196 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Richards T. A., Cavalier-Smith T., Myosin domain evolution and the primary divergence of eukaryotes. Nature 436, 1113–1118 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Sebé-Pedrós A., Grau-Bové X., Richards T. A., Ruiz-Trillo I., Evolution and classification of myosins, a paneukaryotic whole-genome approach. Genome Biol. Evol. 6, 290–305 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cavalier-Smith T. et al., Multigene eukaryote phylogeny reveals the likely protozoan ancestors of opisthokonts (animals, fungi, choanozoans) and Amoebozoa. Mol. Phylogenet. Evol. 81, 71–85 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Fritz-Laylin L. K. et al., The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell 140, 631–642 (2010). [DOI] [PubMed] [Google Scholar]
- 47.Brawley S. H., Robinson K. R., Cytochalasin treatment disrupts the endogenous currents associated with cell polarization in fucoid zygotes: Studies of the role of F-actin in embryogenesis. J. Cell Biol. 100, 1173–1184 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harper J. D., McCurdy D. W., Sanders M. A., Salisbury J. L., John P. C., Actin dynamics during the cell cycle in Chlamydomonas reinhardtii. Cell Motil. Cytoskeleton 22, 117–126 (1992). [DOI] [PubMed] [Google Scholar]
- 49.Hirono M. et al., Tetrahymena actin: Localization and possible biological roles of actin in Tetrahymena cells. J. Biochem. 102, 537–545 (1987). [DOI] [PubMed] [Google Scholar]
- 50.Ehler L. L., Dutcher S. K., Pharmacological and genetic evidence for a role of rootlet and phycoplast microtubules in the positioning and assembly of cleavage furrows in Chlamydomonas reinhardtii. Cell Motil. Cytoskeleton 40, 193–207 (1998). [DOI] [PubMed] [Google Scholar]
- 51.Merchant S. S. et al., The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318, 245–250 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cross F. R., Umen J. G., The Chlamydomonas cell cycle. Plant J. 82, 370–392 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goodenough U. W., Chloroplast division and pyrenoid formation in Chlamydomonas reinhardi. J. Phycol. 6, 1–6 (1970). [Google Scholar]
- 54.O’Toole E. T., Dutcher S. K., Site-specific basal body duplication in Chlamydomonas. Cytoskeleton (Hoboken) 71, 108–118 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Doonan J. H., Grief C., Microtubule cycle in Chlamydomonas reinhardtii: An immunofluorescence study. Cell Motil. Cytoskeleton 7, 381–392 (1987). [Google Scholar]
- 56.Gaffal K. P., el-Gammal S., Elucidation of the enigma of the “metaphase band” of Chlamydomonas reinhardtii. Protoplasma 156, 139–148 (1990). [Google Scholar]
- 57.Kato-Minoura T., Uryu S., Hirono M., Kamiya R., Highly divergent actin expressed in a Chlamydomonas mutant lacking the conventional actin gene. Biochem. Biophys. Res. Commun. 251, 71–76 (1998). [DOI] [PubMed] [Google Scholar]
- 58.Craig E. W. et al., The elusive actin cytoskeleton of a green alga expressing both conventional and divergent actins. Mol. Biol. Cell 30, 2827–2837 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Onishi M., Pringle J. R., Robust transgene expression from bicistronic mRNA in the green alga Chlamydomonas reinhardtii. G3 (Bethesda) 6, 4115–4125 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fuhrmann M., Oertel W., Hegemann P., A synthetic gene coding for the green fluorescent protein (GFP) is a versatile reporter in Chlamydomonas reinhardtii. Plant J. 19, 353–361 (1999). [DOI] [PubMed] [Google Scholar]
- 61.Riedl J. et al., Lifeact: A versatile marker to visualize F-actin. Nat. Methods 5, 605–607 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Avasthi P. et al., Actin is required for IFT regulation in Chlamydomonas reinhardtii. Curr. Biol. 24, 2025–2032 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Onishi M., Pringle J. R., Cross F. R., Evidence that an unconventional actin can provide essential F-actin function and that a surveillance system monitors F-actin integrity in Chlamydomonas. Genetics 202, 977–996 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Onishi M., Pecani K., Jones T. 4th, Pringle J. R., Cross F. R., F-actin homeostasis through transcriptional regulation and proteasome-mediated proteolysis. Proc. Natl. Acad. Sci. U.S.A. 115, E6487–E6496 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Conti M. A., Kawamoto S., Adelstein R. S., “Non-muscle myosin II” in Myosins, A Superfamily of Molecular Motors, Coluccio L. M., Ed. (Springer Netherlands, 2008), pp. 223–264. [Google Scholar]
- 66.Hirono M., Uryu S., Ohara A., Kato-Minoura T., Kamiya R., Expression of conventional and unconventional actins in Chlamydomonas reinhardtii upon deflagellation and sexual adhesion. Eukaryot. Cell 2, 486–493 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jack B., Mueller D. M., Fee A. C., Tetlow A. L., Avasthi P., Partially redundant actin genes in Chlamydomonas control transition zone organization and flagellum-directed traffic. Cell Rep. 27, 2459–2467.e3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zones J. M., Blaby I. K., Merchant S. S., Umen J. G., High-resolution profiling of a synchronized diurnal transcriptome from Chlamydomonas reinhardtii reveals continuous cell and metabolic differentiation. Plant Cell 27, 2743–2769 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Harris J. A., Liu Y., Yang P., Kner P., Lechtreck K. F., Single-particle imaging reveals intraflagellar transport-independent transport and accumulation of EB1 in Chlamydomonas flagella. Mol. Biol. Cell 27, 295–307 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gaffal K. P., Arnold C. G., Friedrichs G. J., Gemple W., Morphodynamical changes of the chloroplast of Chlamydomonas reinhardtii during the 1st round of division. Arch. Protistenkd. 145, 10–23 (1995). [Google Scholar]
- 71.Harper J. D. I., John P. C. L., Coordination of division events in the Chlamydomonas cell cycle. Protoplasma 131, 118–130 (1986). [Google Scholar]
- 72.Tulin F., Cross F. R., A microbial avenue to cell cycle control in the plant superkingdom. Plant Cell 26, 4019–4038 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pollard L. W., Onishi M., Pringle J. R., Lord M., Fission yeast Cyk3p is a transglutaminase-like protein that participates in cytokinesis and cell morphogenesis. Mol. Biol. Cell 23, 2433–2444 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Okada H., Wloka C., Wu J. Q., Bi E., Distinct roles of myosin-II isoforms in cytokinesis under normal and stressed conditions. iScience 14, 69–87 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Calvert M. E. et al., Myosin concentration underlies cell size-dependent scalability of actomyosin ring constriction. J. Cell Biol. 195, 799–813 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Field C. M., Coughlin M., Doberstein S., Marty T., Sullivan W., Characterization of anillin mutants reveals essential roles in septin localization and plasma membrane integrity. Development 132, 2849–2860 (2005). [DOI] [PubMed] [Google Scholar]
- 77.Kanada M., Nagasaki A., Uyeda T. Q., Novel functions of Ect2 in polar lamellipodia formation and polarity maintenance during “contractile ring-independent” cytokinesis in adherent cells. Mol. Biol. Cell 19, 8–16 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Beach J. R., Licate L. S., Crish J. F., Egelhoff T. T., Analysis of the role of Ser1/Ser2/Thr9 phosphorylation on myosin II assembly and function in live cells. BMC Cell Biol. 12, 52 (2011). Correction in: BMC Cell Biol.13, 11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wagner E., Glotzer M., Local RhoA activation induces cytokinetic furrows independent of spindle position and cell cycle stage. J. Cell Biol. 213, 641–649 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mita T., Kuroiwa T., “Division of plastids by a plastid-dividing ring in Cyanidium caldarium” in Cell Dynamics, Tazawa M., Ed. (Springer Vienna, Vienna, Austria, 1988), Vol. chap. 16, pp. 133–152. [Google Scholar]
- 81.Hashimoto H., Involvement of actin filaments in chloroplast division of the alga Closterium ehrenbergii. Protoplasma 167, 88–96 (1992). [Google Scholar]
- 82.Sumiya N., Fujiwara T., Era A., Miyagishima S. Y., Chloroplast division checkpoint in eukaryotic algae. Proc. Natl. Acad. Sci. U.S.A. 113, E7629–E7638 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Miyagishima S. Y., Suzuki K., Okazaki K., Kabeya Y., Expression of the nucleus-encoded chloroplast division genes and proteins regulated by the algal cell cycle. Mol. Biol. Evol. 29, 2957–2970 (2012). [DOI] [PubMed] [Google Scholar]
- 84.Imam S. H., Buchanan M. J., Shin H. C., Snell W. J., The Chlamydomonas cell wall: Characterization of the wall framework. J. Cell Biol. 101, 1599–1607 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ayscough K. R. et al., High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J. Cell Biol. 137, 399–416 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Carvalho A., Desai A., Oegema K., Structural memory in the contractile ring makes the duration of cytokinesis independent of cell size. Cell 137, 926–937 (2009). [DOI] [PubMed] [Google Scholar]
- 87.Davies T. et al., Cell-intrinsic and -extrinsic mechanisms promote cell-type-specific cytokinetic diversity. eLife 7, e36204 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hardin W. R. et al., Myosin-independent cytokinesis in Giardia utilizes flagella to coordinate force generation and direct membrane trafficking. Proc. Natl. Acad. Sci. U.S.A. 114, E5854–E5863 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Matsuzaki M. et al., Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 428, 653–657 (2004). [DOI] [PubMed] [Google Scholar]
- 90.Bi E. F., Lutkenhaus J., FtsZ ring structure associated with division in Escherichia coli. Nature 354, 161–164 (1991). [DOI] [PubMed] [Google Scholar]
- 91.Bisson-Filho A. W. et al., Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science 355, 739–743 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Margolin W., FtsZ and the division of prokaryotic cells and organelles. Nat. Rev. Mol. Cell Biol. 6, 862–871 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Osawa M., Anderson D. E., Erickson H. P., Reconstitution of contractile FtsZ rings in liposomes. Science 320, 792–794 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yang X. et al., GTPase activity-coupled treadmilling of the bacterial tubulin FtsZ organizes septal cell wall synthesis. Science 355, 744–747 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Xiao J., Goley E. D., Redefining the roles of the FtsZ-ring in bacterial cytokinesis. Curr. Opin. Microbiol. 34, 90–96 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Balasubramanian M. K., Srinivasan R., Huang Y., Ng K. H., Comparing contractile apparatus-driven cytokinesis mechanisms across kingdoms. Cytoskeleton (Hoboken) 69, 942–956 (2012). [DOI] [PubMed] [Google Scholar]
- 97.Duggin I. G. et al., CetZ tubulin-like proteins control archaeal cell shape. Nature 519, 362–365 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Walsh J. C. et al., Division plane placement in pleomorphic archaea is dynamically coupled to cell shape. Mol. Microbiol. 112, 785–799 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zaremba-Niedzwiedzka K. et al., Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature 541, 353–358 (2017). [DOI] [PubMed] [Google Scholar]
- 100.Erickson H. P., Modeling the physics of FtsZ assembly and force generation. Proc. Natl. Acad. Sci. U.S.A. 106, 9238–9243 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ochs J., LaRue T., Tinaz B., Yongue C., Domozych D. S., The cortical cytoskeletal network and cell-wall dynamics in the unicellular charophycean green alga Penium margaritaceum. Ann. Bot. 114, 1237–1249 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Farr H., Gull K., Cytokinesis in trypanosomes. Cytoskeleton (Hoboken) 69, 931–941 (2012). [DOI] [PubMed] [Google Scholar]
- 103.Brown J. M., Marsala C., Kosoy R., Gaertig J., Kinesin-II is preferentially targeted to assembling cilia and is required for ciliogenesis and normal cytokinesis in Tetrahymena. Mol. Biol. Cell 10, 3081–3096 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shaw M. K., Compton H. L., Roos D. S., Tilney L. G., Microtubules, but not actin filaments, drive daughter cell budding and cell division in Toxoplasma gondii. J. Cell Sci. 113, 1241–1254 (2000). [DOI] [PubMed] [Google Scholar]
- 105.Danilchik M. V., Bedrick S. D., Brown E. E., Ray K., Furrow microtubules and localized exocytosis in cleaving Xenopus laevis embryos. J. Cell Sci. 116, 273–283 (2003). [DOI] [PubMed] [Google Scholar]
- 106.Danilchik M. V., Funk W. C., Brown E. E., Larkin K., Requirement for microtubules in new membrane formation during cytokinesis of Xenopus embryos. Dev. Biol. 194, 47–60 (1998). [DOI] [PubMed] [Google Scholar]
- 107.Jesuthasan S., Furrow-associated microtubule arrays are required for the cohesion of zebrafish blastomeres following cytokinesis. J. Cell Sci. 111, 3695–3703 (1998). [DOI] [PubMed] [Google Scholar]
- 108.Albertson R., Cao J., Hsieh T. S., Sullivan W., Vesicles and actin are targeted to the cleavage furrow via furrow microtubules and the central spindle. J. Cell Biol. 181, 777–790 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lindås A. C., Valegård K., Ettema T. J. G., Archaeal actin-family filament systems. Subcell. Biochem. 84, 379–392 (2017). [DOI] [PubMed] [Google Scholar]
- 110.van den Ent F., Amos L. A., Löwe J., Prokaryotic origin of the actin cytoskeleton. Nature 413, 39–44 (2001). [DOI] [PubMed] [Google Scholar]
- 111.Ettema T. J., Lindås A. C., Bernander R., An actin-based cytoskeleton in archaea. Mol. Microbiol. 80, 1052–1061 (2011). [DOI] [PubMed] [Google Scholar]
- 112.Szwedziak P., Wang Q., Freund S. M., Löwe J., FtsA forms actin-like protofilaments. EMBO J. 31, 2249–2260 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Buschmann H., Zachgo S., The evolution of cell division: From streptophyte algae to land plants. Trends Plant Sci. 21, 872–883 (2016). [DOI] [PubMed] [Google Scholar]
- 114.Li Y., Liu D., López-Paz C., Olson B. J., Umen J. G., A new class of cyclin dependent kinase in Chlamydomonas is required for coupling cell size to cell division. eLife 5, e10767 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gorman D. S., Levine R. P., Cytochrome f and plastocyanin: Their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardi. Proc. Natl. Acad. Sci. U.S.A. 54, 1665–1669 (1965). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fang S. C., de los Reyes C., Umen J. G., Cell size checkpoint control by the retinoblastoma tumor suppressor pathway. PLoS Genet. 2, e167 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tulin F., Cross F. R., Cyclin-dependent kinase regulation of diurnal transcription in Chlamydomonas. Plant Cell 27, 2727–2742 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Atkins K. C., Cross F. R., Interregulation of CDKA/CDK1 and the plant-specific cyclin-dependent kinase CDKB in control of the Chlamydomonas cell cycle. Plant Cell 30, 429–446 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lupas A., Van Dyke M., Stock J., Predicting coiled coils from protein sequences. Science 252, 1162–1164 (1991). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data for this paper are provided in the main text, SI Appendix, or Movies S1–S8.