Significance
Numerous cell death-controlling genes have been identified in fungi, especially in the context of conspecific nonself discrimination (allorecognition). However, our understanding of the molecular mechanisms by which these genes trigger programmed cell death (PCD) is limited, as is our knowledge about their relation to PCD pathways in other major eukaryotic kingdoms. Here, we show that the cell death-inducing RCD-1 protein from Neurospora crassa is related to the cytotoxic N-terminal domain of gasdermin, which is the executioner of inflammatory cell death reaction in mammals termed pyroptosis. Our work documents an evolutionary transkingdom relationship of cell death execution proteins between fungi and animals.
Keywords: innate immunity, programmed cell death, pyroptosis, Neurospora, gasdermin
Abstract
Programmed cell death (PCD) in filamentous fungi prevents cytoplasmic mixing following fusion between conspecific genetically distinct individuals (allorecognition) and serves as a defense mechanism against mycoparasitism, genome exploitation, and deleterious cytoplasmic elements (i.e., senescence plasmids). Recently, we identified regulator of cell death-1 (rcd-1), a gene controlling PCD in germinated asexual spores in the filamentous fungus Neurospora crassa. rcd-1 alleles are highly polymorphic and fall into two haplogroups in N. crassa populations. Coexpression of alleles from the two haplogroups, rcd-1–1 and rcd-1–2, is necessary and sufficient to trigger a cell death reaction. Here, we investigated the molecular bases of rcd-1-dependent cell death. Based on in silico analyses, we found that RCD-1 is a remote homolog of the N-terminal pore-forming domain of gasdermin, the executioner protein of a highly inflammatory cell death reaction termed pyroptosis, which plays a key role in mammalian innate immunity. We show that RCD-1 localizes to the cell periphery and that cellular localization of RCD-1 was correlated with conserved positively charged residues on predicted amphipathic α-helices, as shown for murine gasdermin-D. Similar to gasdermin, RCD-1 binds acidic phospholipids in vitro, notably, cardiolipin and phosphatidylserine, and interacts with liposomes containing such lipids. The RCD-1 incompatibility system was reconstituted in human 293T cells, where coexpression of incompatible rcd-1–1/rcd-1–2 alleles triggered pyroptotic-like cell death. Oligomers of RCD-1 were associated with the cell death reaction, further supporting the evolutionary relationship between gasdermin and rcd-1. This report documents an ancient transkingdom relationship of cell death execution modules involved in organismal defense.
Programmed cell death (PCD) is a key process in both metazoan and plant immune systems (1, 2). In mammals, intracellular infections are contained and eliminated through dedicated molecular pathways triggering inflammatory cell death responses (3). Pyroptosis is a lytic and proinflammatory form of cell death that is triggered in response to certain infections and other cellular stresses (4). These immunogenic cues induce the formation of multiprotein signaling complexes called inflammasomes, which frequently contain scaffolding proteins of the nucleotide-binding domain leucine-rich repeat (NLR) superfamily. Assembled inflammasomes recruit and activate downstream proinflammatory caspases, most notably, caspase-1. A proteolytic target of these caspases is a protein called Gasdermin-D, a member of the gasdermin family of proteins that serve as executioners of pyroptotic cell death (5–8). The proteolysis of gasdermin liberates a cytotoxic N-terminal domain (gasdermin-NT) from an inhibitory C-terminal domain (gasdermin-CT), which normally keeps the gasdermin molecules inactive (8). Gasdermin-NT binds to negatively charged phospholipids on the inner leaflet of mammalian cell membranes and assembles into ring-shaped pores (9–12). The nonselective pores disrupt ion gradients across the plasma membrane, which rapidly leads to lytic cell death with the pyroptotic cells acquiring a balloon-like shape shortly before cell lysis (6, 13).
In fungi, PCD can result from anastomoses (cellular fusions) between genetically incompatible strains of the same species (14). Cell death occurs exclusively in the heterokaryotic fusion cells and results in compartmentation to separate the hyphal networks of incompatible individuals and prevent cytoplasmic mixing (15, 16). The PCD reaction, referred to as heterokaryon incompatibility (HI), prevents the spread of deleterious cytoplasmic elements (i.e., mycoviruses), genome exploitation, and resources plundering (16–19). Thus, HI represents a defense mechanism against conspecific nonself (allorecognition). Some of the molecular determinants controlling HI in fungi have been characterized and are related to metazoan and plant NLRs (20, 21). In Neurospora crassa, an NLR-encoding gene (plp-1: NCU09244) controls allorecognition and PCD in germinating asexual spores (germlings) (20). We recently reported the identification of a second gene controlling germling-regulated death (GRD) in N. crassa (22). The rcd-1 (regulator of cell death-1) locus encodes two antagonistic alleles (rcd-1–1 and rcd-1–2), which induce PCD when coexpressed in the same cell (22). rcd-1 homologs are widespread in fungi, especially in the Ascomycota phylum (22). The latter finding suggests that rcd-1 may play a central role in the control of PCD in fungi.
Here, we identified in silico remote protein homology between RCD-1 and the cytotoxic N-terminal domain of mammalian gasdermin, suggesting that RCD-1 may function similarly to a gasdermin-like protein to induce cell death in N. crassa. To test this hypothesis, we assessed cellular localization of RCD-1 and tested whether localization to the cell periphery occurs through a similar mechanism as gasdermin proteins. To probe RCD-1 function, we produced recombinant RCD-1 and showed that RCD-1, similar to gasdermin, interacted with acidic phospholipids and liposomes in vitro and had a propensity to oligomerize. Furthermore, we reconstituted the rcd-1 incompatibility system by coexpressing rcd-1–1 and rcd-1–2 in human 293T kidney cells, which triggered pyroptotic-like cell death. During the cell death reaction, the RCD-1 allelic variants formed homo- and heterotypic protein–protein complexes. Overall, this study documents an evolutionary relationship of defense-related cell death-inducing proteins between fungi and animals.
Results
In Silico Analyses Uncovered Remote Homology between RCD-1 and Gasdermin.
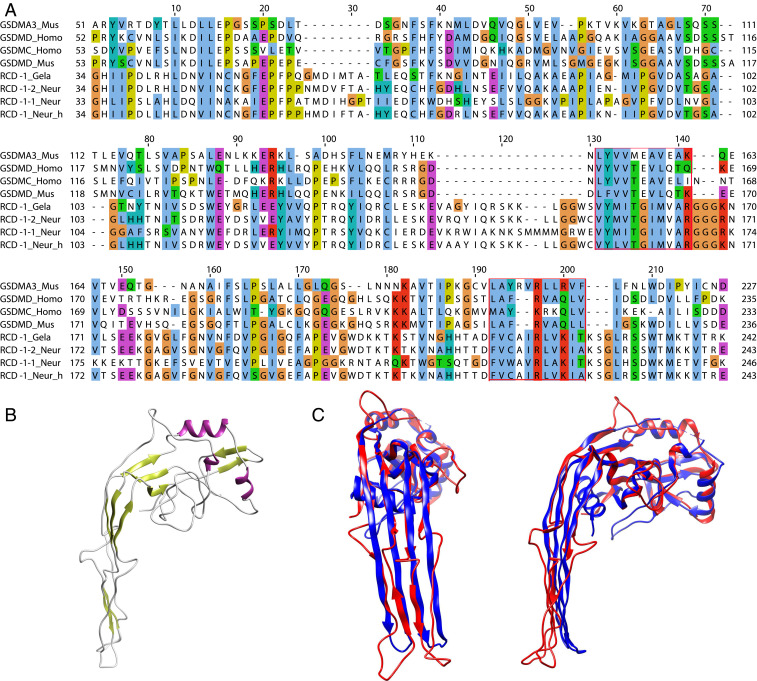
To identify distant RCD-1 homologs, we performed Hidden Markov Model (HMM) searches integrating primary sequence homology and secondary structure information using the online HHpred suite (23, 24). Searches with both allelic variants of RCD-1 (RCD-1–1 and RCD-1–2) from N. crassa resulted in hits (with probability scores close to 95%) matching the N-terminal domain of human (PDB: 6N9O) and murine (PDB: 6N9N) gasdermin-D (GSDMD) and murine gasdermin-A3 (GSDMA3, PDB: 5B5R, 6CB8). The HHpred-based homology extended over more than 140 amino acid residues, accounting for similarities in primary sequence and secondary structure between gasdermin and RCD-1 (Fig. 1A). The primary sequence homology between RCD-1 and gasdermin represented an overall conservation pattern of similar or identical amino acid residues that extended over almost the entire length of RCD-1 and contained two highly conserved blocks of ∼10 amino acids each (Fig. 1A). We did not identify significant hits outside of the gasdermin family, and similar results were obtained with 20 randomly tested RCD-1 homologs out of >900 homologs identified in various fungal species (22) (SI Appendix, Table S1). To further explore the evolutionary relationship between RCD-1 and gasdermin, we generated a molecular model of N. crassa RCD-1–2 with the RaptorX web server (25). The RaptorX template-based modeling approach of RCD-1–2 revealed that GSDMA3 (6CB8) is the best available template for the homology-based model of RCD-1, further supporting the relationship between the two protein families (Fig. 1 B and C). Homology modeling of RCD-1–1 showed that the best template for molecular modeling was murine GSDMA3 (PDB: 5B5R) (SI Appendix, Fig. S1). Based on our observations, we hypothesized that RCD-1 is a distant gasdermin homolog.
Fig. 1.
Protein alignment and molecular models of the pore-forming N-terminal domains of mammalian gasdermins and fungal RCD-1 proteins. (A) Alignment of protein sequences of murine (Mus) gasdermin-A3 (NP_001007462), gasdermin-D (NP_081236), human (Homo) gasdermin-D (NP_079012), gasdermin-C (NP_113603) with RCD-1–1 (XP_962898) and RCD-1–2 from N. crassa (Neur) and RCD-1 variants from N. hispaniola (Neur_h) and Gelasinospora tetrasperma. Conserved residues are shown in ClustalX colors, based on the nature of the amino acid residues: hydrophobic – blue, polar – green, negative charge – magenta, positive charge – red, aromatic – cyan, glycine – orange, proline – yellow, cysteine – pink, any/gap – white. Two highly conserved blocks between all sequences are boxed in red. (B) Template-based model of the RCD-1–2 allelic variant from Neurospora crassa. Murine GSDMA3 (6CB8) in the active pore-forming state was determined (P value = 7.22e-05) as the best template based on the sequence input (RCD-1–2). (C) Structural alignment of GSDMA3 (6CB8A) (blue) and the generated model of RCD-1–2 (red). Shown are a frontal view (Left) and side view (Right) of the two overlaid structures.
We previously identified several bacterial RCD-1 homologs (22). HMM-based sequence searches (26) with bacterial RCD-1 homologs as queries retrieved both fungal RCD-1 sequences and metazoan gasdermin sequences (SI Appendix, Fig. S1C). The region of sequence homology between all putative homologs (bacterial, fungal, and metazoan sequences) contained the two previously identified motifs of ∼10 amino acids (SI Appendix, Figs. S1D and S2). These results suggested that the genomes of some bacterial species also encode distant homologs of the cytotoxic N-terminal domain of gasdermin. When analyzed with HHpred, the bacterial proteins were found to be homologous with the N-terminal pore-forming domain of GSDMD/GSDMA3 with probability scores above 99% (SI Appendix, Table S2). Overall, these results indicated that the N-terminal domain of gasdermin is not exclusively found in Metazoa.
RCD-1 Shows Plasma Membrane Affinity In Vivo.
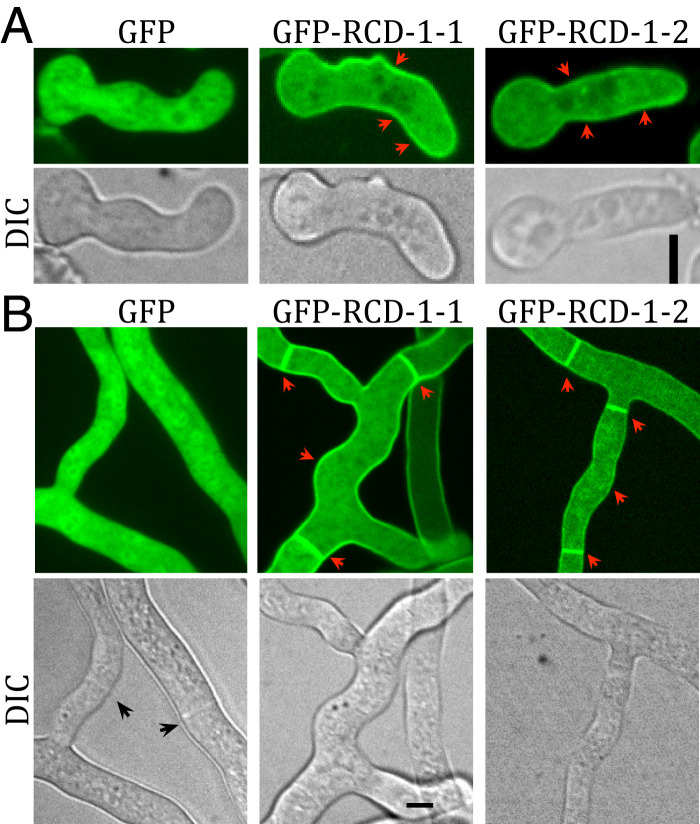
The N-terminal domain of gasdermin binds to phospholipids on the inner leaflet of mammalian cell membranes. We therefore examined the cellular localization of RCD-1 in germlings and hyphae of N. crassa. N-terminal GFP-rcd-1–1 and GFP-rcd-1–2 alleles induced PCD when coexpressed with rcd-1–2 or rcd-1–1, respectively (SI Appendix, Fig. S3D). GFP-RCD-1–1 and GFP-RCD-1–2 localized in the cytoplasm, but also at the cell periphery (Fig. 2), with both proteins showing strong septal localization in mature hyphae of N. crassa (Fig. 2B). The localization of RCD-1 remained unchanged when the protein was fused to a different fluorescent label (mCherry) (SI Appendix, Fig. S3 A and B).
Fig. 2.
Cellular localization of RCD-1. Fluorescent microscopy of GFP-RCD-1–1 and GFP-RCD-1–2 in germlings (A) and hyphae (B). Localization of RCD-1 to cell periphery and septa are shown by red arrowheads. Free GFP showed diffuse cytoplasmic localization in germlings and hyphae. (Scale bars, 5 µm.)
We confirmed the membrane affinity of RCD-1 by performing a density-gradient subcellular fractionation with a V5-tagged RCD-1. Using sucrose gradient ultracentrifugation, V5-RCD-1 was found in the denser sucrose fractions containing the cellular membranes, unlike the GFP alone, which remained in the lighter fractions with the cytoplasmic content (SI Appendix, Fig. S3C). Collectively, these data indicate that RCD-1 is associated with the plasma membrane in vivo in N. crassa.
Functional Analogy of Secondary Structure Motifs between RCD-1 and Gasdermin.
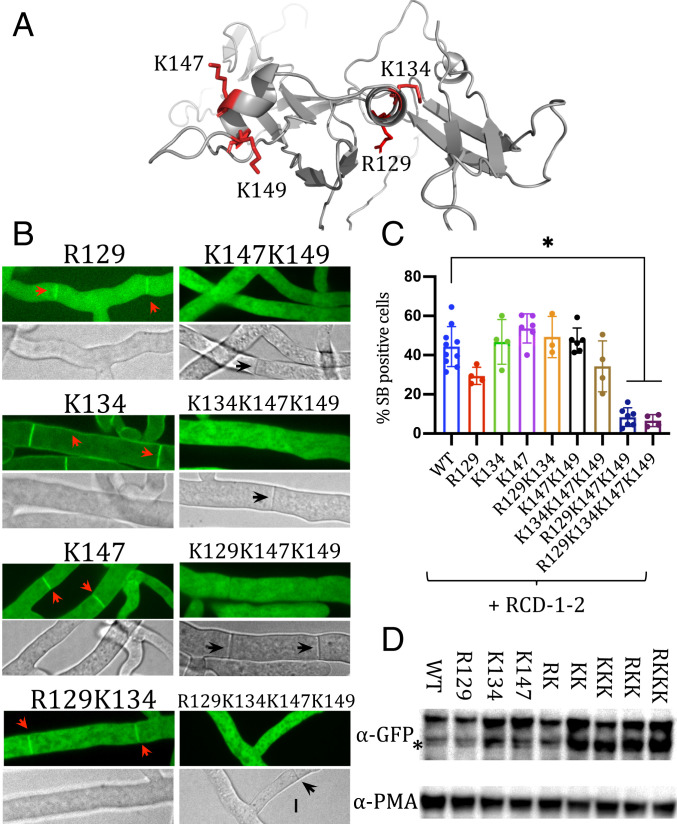
The N-terminal domain of murine gasdermin-D (GSDMD-NT) binds to acidic (negatively charged) phospholipids through a cluster of four positively charged residues, situated on a predicted pair of amphipathic α-helices (9). We reasoned that as a distant gasdermin homolog, RCD-1 could employ a similar mechanism for plasma membrane targeting. Our in silico analyses predicted a similar α-helical region, composed of two (or three) α-helices, on both RCD-1–1 and RCD-1–2 (Fig. 1 and SI Appendix, Fig. S1 A and B). Furthermore, the α-helical region of RCD-1 contained five conserved positively charged amino acid residues (Fig. 3A and SI Appendix, Fig. S4 A and B). We used site-directed mutagenesis to replace four of the five positively charged residues in the α-helical region of RCD-1–1 with alanine codons, obtaining a set of mutants with decreased positive charge in the predicted secondary structure motifs. Three single amino acid substitution mutants (R129A, K134A, K147A), two double mutants (R129A;K134A, K147A;K149A), two triple mutants (R129A;K147A;K149A, K134A;K147A;K149A), and one quadruple mutant (R129A;K134A;K147A;K149A) (SI Appendix, Table S3) were used to investigate both the cellular localization and GRD activity of these mutant RCD-1 proteins in vivo. The cellular localization of the single mutants appeared only slightly affected, with GFP-RCD-1–1R129A showing the most notable decrease in plasma membrane and septal localization (Fig. 3B). One double mutant (GFP-RCD-1–1R129A;K134A) retained some ability to localize to septa, but the remaining double, triple, and quadruple RCD-1–1 mutants lost septal and plasma membrane affinity and instead localized exclusively in the cytoplasm (Fig. 3B). The importance for membrane targeting of these conserved charged residues in the predicted α-helical region was also confirmed with the RCD-1–2 allelic variant (RCD-1–2K146A;K148A) (SI Appendix, Fig. S4C).
Fig. 3.
Cellular localization of RCD-1 is dependent on a set of conserved positively charged amino acid residues. (A) Molecular model of RCD-1–1, showing the predicted α-helical region. Conserved positively charged residues, subjected to site-directed mutagenesis altered to alanine residues, shown by red sticks and coordinates (of the RCD-1–1 allelic variant) near each residue. (B) Fluorescent microscopy of GFP-RCD-1–1 mutants. Red arrowheads show plasma membrane or septal localization, black arrowheads point to septa, where cellular localization of GFP-RCD-1–1 is not observed. (C) Uptake of the vital dye SYTOX Blue measured by flow cytometry in mixtures of germlings expressing wild-type or mutant GFP-RCD-1–1 variants and germlings expressing the antagonistic RCD-1–2 allelic variant. Experiments were performed at least in triplicate, with 20,000 events counted per experiment. *P < 0.0001, one-way ANOVA with Tukey’s multiple comparisons test. (D) Western blot using anti-GFP antibodies showing protein levels of wild-type and mutant GFP-RCD-1–1 variants. The asterisk symbol (*) indicates bands of likely partial degradation of the expressed molecular fusions. Abbreviated names are derived from the type of mutations presented in C. Excessive protein accumulation of mutant RCD-1 variants could lead to higher levels of degradation. Loading control is shown below with a blot of the plasma membrane ATPase PMA-1 (44).
Consistent with cellular localization of the RCD-1 mutant proteins, the alanine mutations affected GRD competency in a synergistic manner, with triple and quadruple RCD-1 mutants (RCD-1–1R129A;K147A;K149A and RCD-1–1R129A;K134A;K147A;K149A) completely abolishing cell death-inducing ability (Fig. 3C). We verified that cellular localization of RCD-1 and decreased GRD competency of mutant RCD-1 proteins were not caused by decreased RCD-1 protein levels (Fig. 3D). The results with RCD-1 were similar to those obtained for GSDMD-NT (9) and suggested that the two proteins share a functional secondary structure motif and mode of interaction with the plasma membrane.
Recombinant RCD-1 Forms Oligomers In Vitro.
The proteolysis of gasdermin results in oligomerization of N-terminal gasdermin monomers (9, 12, 27). While we have no evidence that proteolysis is required for RCD-1 function, we nevertheless reasoned that that RCD-1–1 and RCD-1–2 might also form oligomers. To test this hypothesis, we purified V5-RCD-1 that had been expressed in insect cells (See Materials and Methods and SI Appendix). Recombinant RCD-1 remained soluble, yet showed a tendency to form oligomers of higher molecular weight in samples of high protein concentration (5 mg.mL−1) and that were observed under denaturing conditions (presence of sodium dodecyl sulfate [SDS]) on a Coomassie-stained electrophoresis gel (SI Appendix, Fig. S5). To confirm the formation of RCD-1 oligomers in vitro, we separated the proteins under native conditions by NC-PAGE and Western blot analyses using anti-V5 antibody, which revealed the formation by recombinant V5-RCD-1-1 of various oligomeric states (SI Appendix, Fig. S5C). We observed a ladder-like banding pattern, likely corresponding to n + 1 composition of V5-RCD-1–1 oligomers, where n is a number of RCD-1–1 molecules to which an additional monomer of RCD-1 is added. Some oligomers were of high molecular weight (>300 kDa), likely containing more than 10 RCD-1 monomers. We used transmission electron microcopy to investigate the appearance of spontaneously formed protein aggregates by FPLC-purified recombinant RCD-1–1. We found that some RCD-1 aggregates produced arc-like and ring-like shapes, which exhibit a tendency to cluster together reminiscent of irregularly sized cells of a honeycomb (SI Appendix, Fig. S5D). These studies showed that RCD-1 has a tendency to oligomerize in vitro.
RCD-1 Interacts with Negatively Charged Phospholipids and Liposomes In Vitro.
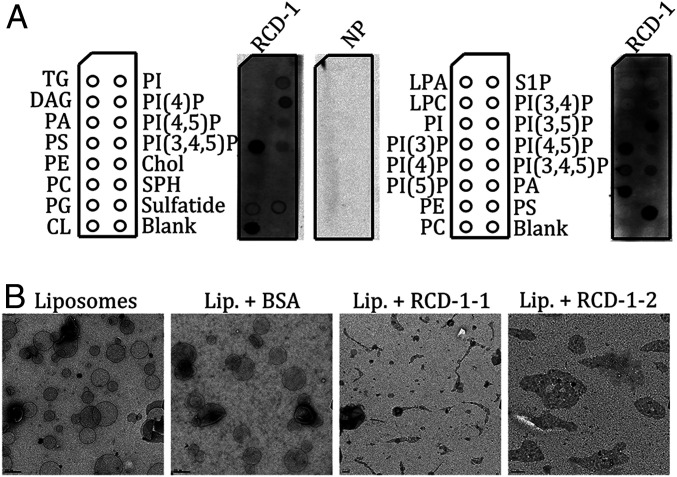
To test whether purified RCD-1 has an affinity for negatively charged lipids, we performed lipid-protein overlay assays using lipid strips (28). We found that RCD-1 showed in vitro affinity for negatively charged phospholipids, notably, cardiolipin (CL) and phosphatidylserine (PS) (Fig. 4A). RCD-1 also binds to phosphoinositides, preferentially to PI(3)P, PI(5)P and PI(3,5)P2 (Fig. 4A). Noteworthily, gasdermin also binds and permeabilizes cardiolipin and phosphoinositide-containing membranes (10, 27). We therefore investigated whether RCD-1 interacts with liposomes in vitro. When RCD-1 was incubated with liposomes containing acidic phospholipids (PS and/or CL), selected on the basis of the lipid-strip tests, the liposomes rapidly disintegrated or lost shape (Fig. 4B). Liposome shredding occurred with both RCD-1–1 and RCD-1–2 variants; we did not observe this effect when the liposomes were incubated by themselves or with bovine serum albumin (BSA) (Fig. 4B).
Fig. 4.
RCD-1 interacts in vitro with negatively charged phospholipids. (A) Lipid-protein overlay assays with FPLC-purified V5-tagged RCD-1–1. Bound protein is revealed by primary anti-V5 antibody. Lipids blotted on the strips are shown as abbreviations: TG, triglyceride; DAG, diacylglycerol; PA, phosphatidic acid; PS, phosphatidylserine; PE, phosphatidylethanolamine; PC, phosphatidylcholine; PG, phosphatidylglycerol; CL, cardiolipin; PI, phosphatidylinositol; Chol, cholesterol; SPH, sphingomyelin; LPA, lysophosphatidic acid; LPC, lysophosphocholine; S1P, sphingosine-1-phosphate; PI()P, phosphatidylinositol phosphate; NP, no protein control. (B) Electron micrographs of liposomes alone or in presence of BSA and fragmented liposomes in presence of RCD-1–1 or RCD-1–2.
We then explored the state of RCD-1–1 with or without liposome incubation. In the absence of liposomes, RCD-1–1 was enriched in the soluble fraction (S) relative to the pellet fraction (P) after centrifugation (SI Appendix, Fig. S6). In the presence of liposomes containing lipids not bound by RCD-1–1 (PC and PE) or containing 10% of CL or PS, RCD-1–1 was also enriched in the soluble fraction. In these samples or with RCD-1–1 alone, we observed in the soluble fractions the presence of a second band of higher molecular weight, which could correspond to a stable RCD-1–1 dimer. However, when the percentage of CL and PS in liposomes was increased (up to 50%), RCD-1–1 was significantly enriched in the pellet fractions (SI Appendix, Fig. S6). In these samples, two novel protein bands of even higher molecular size appeared exclusively in the pellet fractions (SI Appendix, Fig. S6). As FPLC-purified recombinant RCD-1–1 was used, these data indicated that the protein bands corresponded to a modified state of RCD-1, most likely SDS-resistant oligomers, induced by the presence (above 10%) of CL or PS in the liposomes. The percentage of acidic phospholipids (∼25%) in some liposome samples with which the protein interacted was in a similar range to what has been reported for the plasma membrane of N. crassa (29). Because incubation of RCD-1 with liposomes containing >10% CL and PS induced oligomerization, the presence of the protein in the pellet fractions (after incubation with the liposomes) is likely due to direct liposome binding and/or sedimentation of large RCD-1 oligomers.
Overall, these results indicate that RCD-1 binds to acidic phospholipids in vitro on lipid strips and liposomes with a similar profile to gasdermin. The observed destabilizing effect on liposomes containing such lipids and the observed RCD-1 oligomers induced in these conditions suggest that RCD-1 carries a membrane-disturbing or permeabilization function, similar to a pore-forming toxin like gasdermin.
RCD-1 Allelic Variants Induced Pyroptotic-Like Cell Death in Human 293T Cells.
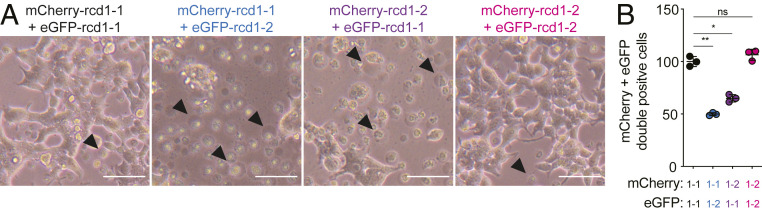
Due to the similarities between gasdermin and RCD-1 and considering that the N-terminal domain of gasdermin is sufficient to trigger cell death, we reasoned that RCD-1–1 and RCD-1–2 could potentially induce cell death when coexpressed in mammalian cells. To test this hypothesis, we reconstituted the incompatibility reaction by cotransfecting plasmids producing RCD-1–1 and/or RCD-1–2 in human 293T kidney cells. The expression of RCD-1–1 alone or RCD-1–2 alone in 293T cells did not result in cell death (Fig. 5). In contrast, the coexpression of RCD-1–1 and RCD-1–2 resulted in a lytic cell death reminiscent of pyroptosis (Fig. 5A). To quantify the cell death reaction, we used fluorescent proteins (GFP and mCherry) expressed from the same vector carrying the RCD-1–1 or RCD-1–2 allelic variants (using internal ribosomal entry site sequences) and measured the number of double-positive cells (GFP+, mCherry+) using flow cytometry (SI Appendix). The number of double-positive cells was significantly decreased in cells transfected with both RCD-1–1 and RCD-1–2 as compared to cell cultures cotransfected with GFP/mCherry-carrying vectors expressing the same RCD-1 variant (Fig. 5B). Thus, coexpression of incompatible RCD-1 proteins triggered a pyroptotic-like cell death in human cells, suggesting that RCD-1, as gasdermin, is the executioner of cell death.
Fig. 5.
Coexpression of antagonistic RCD-1–1 and RCD-1–2 variants triggers pyroptotic-like cell death in human 293T kidney cells. (A) Images of human 293T cells transfected with plasmids expressing mCherry- or eGFP-tagged RCD-1–1 or RCD1-2 in compatible combinations (RCD-1–1 + RCD-1–1 or RCD-1–2 + RCD-1–2) or antagonistic combinations (RCD-1–1 + RCD-1–2). Representative pyroptotic-like cells are indicated with an arrowhead. (Scale bar, 100 µm.) (B) Quantification of mCherry and eGFP double-positive cells for compatible and antagonistic RCD-1 combinations. Values are relative to cells transfected with mCherry-RCD-1–1 + eGFP-RCD-1–1. Welch’s unpaired t test *P < 0.01, **P < 0.001. ns, nonsignificant.
Oligomerization of RCD-1 Is Associated with GRD and Cell Death.
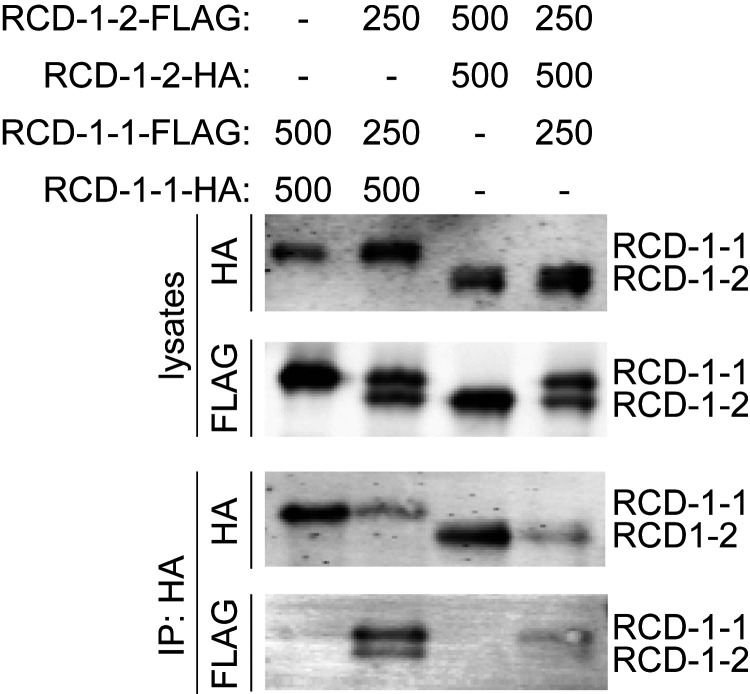
The homology between RCD-1 and gasdermin and the finding that RCD-1 induced cell death in mammalian cells prompted us to investigate whether RCD-1 self-assembles (like gasdermin) during the cell death reaction. Using human 293T kidney cells, HA and FLAG-tagged versions of RCD-1–1/RCD-1–2 were introduced, and coimmunoprecipitation (co-IP) experiments were performed to probe for homotypic and heterotypic protein–protein interactions. We did not observe “self” interactions in cells cotransfected with differentially tagged RCD-1–1 or RCD-1–2 (i.e., HA-RCD-1–1 did not bind to FLAG-RCD-1–1, and HA-RCD-1–2 did not bind to FLAG-RCD-1–2) (Fig. 6). In contrast, pull-down of HA-RCD-1–1 coimmunoprecipitated FLAG-RCD-1–1 in the presence of FLAG-RCD-1–2 (second column). Similarly, pull-down of HA-RCD-1–2 coimmunoprecipitated both FLAG-RCD-1–2 and FLAG-RCD-1–1 (fourth column) (Fig. 6). These results indicated that RCD-1–1 and RCD-1–2 variants interact and that heterotypic interactions were essential for the cell death reaction. As these interactions were detected in a heterologous system, where the conservation of protein partners of RCD-1 is unlikely, these results indicated that RCD-1–1 and RCD-1–2 variants interact directly.
Fig. 6.
Protein–protein interactions between antagonistic RCD-1 allelic variants in human 293T cells. co-IP experiments with HA- and FLAG-labeled RCD-1–1 and RCD-1–2 variants. Numbers indicate the amount of DNA vector (nanograms) used for each transfection reaction. Dashes indicate that the vector has not been used in a transfection reaction. Lysates are defined as input, where the presence of the HA-labeled RCD-1 variants (top blot) and the FLAG-labeled (second blot) RCD-1 variants are shown. The immunoprecipitation of HA-RCD-1–1 and HA-RCD-1–2 pulls down both FLAG-tagged RCD-1 variants (IP:HA blots).
In N. crassa, the formation of large, highly ordered fluorescent aggregates were observed in a small number of fusing germlings expressing incompatible GFP-RCD-1–1 and GFP-RCD-1–2 proteins (SI Appendix, Fig. S7). Such large, fibrillar aggregates appeared exclusively when both allelic variants carried the GFP tag and were not observed in fused RCD-1–1/RCD-1–2 germlings expressing cytoplasmic GFP. However, unlike the high death frequency observed in pairings between germlings carrying a single GFP-tagged rcd-1–1 or rcd-1–2 allele and an untagged incompatible rcd-1 variant (SI Appendix, Fig. S3), pairings between germlings carrying GFP-rcd-1–1 and GFP-rcd-1–2 did not show a significant increase in cell death as compared to compatible controls, yet strong vacuolization was observed in fused germlings coexpressing GFP-RCD-1–1/GFP-RCD-1–2 (SI Appendix, Fig. S7). These results and the GRD competency of both GFP-RCD-1–1 and GFP-RCD-1–2 suggest that both alleles are equivalent in the GRD reaction and likely play an active, cell death-inducing role.
Discussion
The rcd-1 locus controls programmed cell death in germinating asexual spores and hyphae of N. crassa (22). Here, we show that rcd-1 encodes a gasdermin-like fungal protein. We showed that RCD-1 localizes to the cell periphery and likely interacts with the plasma membrane of the cell through a secondary structure motif consisting of amphipathic α-helices carrying positively charged residues, similarly to GSDMD-NT. Recombinant RCD-1 bound in vitro to acidic phospholipids with similar specificity as gasdermin and formed SDS-resistant oligomers when incubated with liposomes containing cardiolipin, phosphatidylserine, and phosphoinositides. Remarkably, when antagonistic RCD-1 allelic variants (RCD-1–1 and RCD-1–2) were introduced into human 293T kidney cells, a PCD reaction reminiscent of pyroptosis was induced. The induction of pyroptotic-like cell death by RCD-1 in a heterologous system suggests that the protein exerts direct cytotoxic activity, as conservation of additional molecular partners and broader cell death pathways between fungi and animals appears unlikely.
The pyroptotic-like cell death induced by RCD-1 in mammalian cells also suggests that the mechanism of cytotoxicity between RCD-1 and gasdermin may share certain similarities and further supports the functional relatedness between the two proteins. While signaling domains from fungi have been shown to function in mammalian cells (30), we now reconstituted an entire fungal allorecognition system to function in mammalian cells. Furthermore, we determined that antagonistic RCD-1–1 and RCD-1–2 proteins interact exclusively during the cell death reaction in mammalian cells and that RCD-1 forms large supramolecular assemblies in N. crassa. However, it remains unclear if the RCD-1 interactions are strictly heterotypic or also homotypic. The observation that individual RCD-1 variants oligomerize in vitro suggests that homotypic interactions could occur in vivo. Further analyses of the RCD-1 oligomers will provide data on homo and/or heterotypic structural conformations of RCD-1, once PCD is induced by coexpression of RCD-1–1 and RCD-1–2 proteins.
Two models describing the RCD-1–1/RCD-1–2 allorecognition process and cell death induction are possible (SI Appendix, Fig. S8). In the first model, RCD-1–1 and RCD-1–2 form a cytotoxic heterooligomeric complex, when coexpressed in the same cell, in a similar fashion to the bicomponent leukocidins from Staphylococcus aureus (31, 32). In the second model, the RCD-1–1 and RCD-1–2 interactions initiate the formation of homooligomers of each allelic variant. Here, the heterotypic interaction represents an activation event similar to the one described for the het-s/het-S allorecognition system from Podospora anserina, where one allelic variant (HET-s), under the form of self-propagating protein aggregates, triggers the cytotoxicity of a second allelic variant, which is a pore-forming toxin (HET-S) (33, 34). Although additional experiments are needed to establish the molecular details of RCD-1–1 and RCD-1–2 interactions, the observation that both alleles are equivalent in the GRD reaction and likely play an active cell death-inducing role is an argument in favor of a model where homooligomers formation plays a key role in the GRD process. The heterologous experimental system in human 293T cell cultures will allow rapid exploration of molecular details regarding the rcd-1–1/rcd-1–2 cell death reaction and help establish additional functional parallels with gasdermin.
The RCD-1 protein does not have the C-terminal inhibitory domain of gasdermin. Thus, it is not clear how the RCD-1 cytotoxic activity is controlled. Our studies do not show evidence for the involvement of proteolytic activity in activation of RCD-1 as is the case for gasdermin. An alternative regulatory mechanism would consist of the recognition in pairings of antagonistic PCD-inducing RCD-1–1 and RCD-1–2 proteins. The latter mechanism of control appears to be directly suggested by the rcd-1–1/rcd-1–2 incompatibility system. Further experiments will illuminate specific regions of the RCD-1 proteins required for induction of PCD, or whether entire RCD-1–1 and RCD-1–2 proteins are required. If the latter is so, these data would suggest the formation of heterooligomers are essential for PCD.
We have previously shown that rcd-1 belongs to a large gene family in fungi (22). It is currently unknown if other members of this gene family are involved in allorecognition and PCD. In this context, allorecognition systems in fungi have been proposed to originate from broader pathways, evolutionarily related to innate immunity systems of plants and animals (14, 35, 36). It has been speculated that emergence of such allorecognition systems is due to sporadic mutations and subsequent cooptation of genes evolved primarily to mediate heterospecific nonself recognition and detection of modified self (37, 38). The rcd-1 locus has undergone extensive genomic rearrangements in the genus Neurospora, which have been proposed to be at the origin of the allorecognition system (22). In such a system, it is possible that mutations have “primed” the two rcd-1 alleles to cross-activate when coexpressed, explaining the plasma membrane affinity of RCD-1, which occurs before activation of the allorecognition reaction.
The homology between RCD-1 and the gasdermin protein family, the executioner proteins of pyroptosis, appears to establish an additional parallel between a fungal PCD reaction and metazoan innate immunity pathway. In particular, an evolutionary relationship between PCD pathways in fungi and metazoan necroptosis has also been reported (39). Because the allorecognition reaction in fungi can represent a cellular defense mechanism (14, 15), one could conclude that RCD-1 and gasdermin are involved in analogous cell death programs that correspond broadly to organismal defense. In light of the similar functional role these proteins play and the uncovered evolutionary relationship, our study suggests that pyroptosis and rcd-1–controlled cell death are of ancient evolutionary origin.
Materials and Methods
Sequence Analysis and Alignments.
Protein homology detection was carried out on HHpred (23) online server (https://toolkit.tuebingen.mpg.de/tools/hhpred). Protein sequences were aligned using MAFFT (40) or Muscle (41) with defaults settings, and sequence alignments were visualized with Jalview (42). Molecular modeling and structure alignment were performed on the online RaptorX server (raptorx.uchicago.edu/) (25) with the sequence of RCD-1–2 as a query, and visualization carried out with PyMOL (https://pymol.org/2/).
Confocal Microscopy.
Cellular localization of fluorescently tagged RCD-1–1/RCD-1–2 was performed on a Leica SD6000 microscope with 40× or 100×, 1.4-numerical aperture, oil-immersed objective, which was equipped with Yokogawa CSU-X1 spinning disk head. A 488-nm and 563-nm laser was used for GFP and mCherry fluorescence excitation, respectively. Lasers were controlled with Metamorph software (Molecular Devices), and images were processed with ImageJ (https://imagej.nih.gov/ij/).
Transmission Electron Microscopy.
Electron microscopy was performed on an FEI Tecnai 12 transmission electron microscope (120 kV) equipped with 2k × 2k CCD camera. FPLC-purified protein, liposomes, or mixtures of protein and liposomes were blotted on a formvar/carbon film-coated, 400-mesh copper grid and stained for 1 min with 1.5% (wt/vol) of uranyl acetate.
Protein-Lipid Overlay Assay.
FPLC-purified recombinant RCD-1 (0.5–2 µg/mL) was incubated with membrane lipid strips (Echelon Biosciences), following protocols from the manufacturer. Briefly, lipid strips were incubated with 3% (wt/vol) fatty acid free BSA in Tris-buffered saline (TBS) buffer (with 0.1% Tween-20). After the recombinant protein was incubated for 1 h at room temperature in 10 mL TBS 3% (wt/vol) BSA with gentle agitation, the lipid strips were washed three times (10 min/wash) with TBS. Western blotting was performed with primary and secondary antibodies in 3% BSA TBS, and lipid binding by RCD-1 was revealed using SuperSignal ELISA Femto Substrate chemiluminescence ECL kit (Thermo Fisher).
Cell Culture and Reconstituted Cell Death Assay.
Human 293T kidney cells were grown in DMEM supplemented with 10% FBS, 100 U/mL penicillin, 100 mg/mL streptomycin, and 2 mM l-glutamine. To reconstitute RCD-mediated cell death in 293T cells, constructs producing mCherry- or enhanced green fluorescent protein (eGFP)-tagged RCD-1–1 or RCD-1–2 were cotransfected using Lipofectamine 2000 (Invitrogen) following the manufacturer’s protocol. To quantify mCherry- and eGFP-positive cells, cells were harvested manually, washed in PBS, and analyzed using standard flow cytometry protocols.
Immunoprecipitations and Western Blotting.
Conidia of N. crassa were inoculated in 100 mL of liquid minimal media at a concentration of 1 × 106 cells per milliliter and incubated at 30 °C for 6 h (shaking at 220 rpm for 3 h and standing for 3 h). Proteins were extracted using 1 mL of lysis buffer per sample as described by Pandey et al. (43), without the addition of phosphatase inhibitors. Protein samples were run on a 4–12% Bis-Tris NuPAGE gel (Invitrogen) or Native PAGE 3–12% Bis-Tris (Invitrogen). Proteins were transferred to a polyvinylidene fluoride (PVDF) membrane via Western blotting, and blots were probed with α-PMA-1 (ab4645), α-V5 (Invitrogen R96025), α-GFP (Life Technologies), or α-6xHis (MA1-21315).
Transfected human 293T kidney cells were lysed in 50 mM Tris⋅HCl, 150 mM NaCl, 1% Triton X-100 for 2 h on ice, then clarified by spinning at ∼16,000 × g for 10 min at 4 °C. Clarified lysates were incubated with anti-HA–bound Protein G magnetic beads (Biorad) for 1 h, rotating at 4 °C. Beads were washed six times in lysis buffer and then denatured in 2× SDS loading buffer. Samples were separated on NuPAGE Bis-Tris 4–12% gradient gels (ThermoFisher) following the manufacturer’s protocol. Gels were transferred onto Immobilon-FL PVDF membranes at 35V for 90 min and blocked with Odyssey blocking buffer (Li-Cor). Proteins were detected on a Li-Cor Odyssey Blot Imager using the following primary and secondary antibodies: anti-HA clone 3F10 (Sigma), anti-Flag M2 (Sigma), Alexfluor-680–conjugated secondary antibodies (Invitrogen) (see SI Appendix, Materials and Methods).
Supplementary Material
Acknowledgments
We thank the Berkeley Flow Cytometry Facility at the Cancer Research Laboratory, the Robert D. Ogg Electron Microscope Laboratory, and the Biological Imaging Facility (University of California, Berkeley) for their technical support. This work was supported by an NIH R01 grant (GM060468), Laboratory Directed Research and Development Program of the Lawrence Berkeley National Laboratory under US Department of Energy Contract DE-AC02-05CH11231 and a Fred E. Dickenson Chair of Wood Science and Technology Award (to N.L.G.) P.S.M. is supported by a Jane Coffin Childs Memorial Fund postdoctoral fellowship.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Strains are publicly available from the Fungal Genetics Stock Center (http://www.fgsc.net/fgsc/search_form.php).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2004876117/-/DCSupplemental.
Data Availability.
All data from this work are included in the main text and the SI Appendix of the paper. Strains are publicly available from the Fungal Genetics Stock Center (http://www.fgsc.net/fgsc/search_form.php).
References
- 1.Nagata S., Tanaka M., Programmed cell death and the immune system. Nat. Rev. Immunol. 17, 333–340 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Coll N. S., Epple P., Dangl J. L., Programmed cell death in the plant immune system. Cell Death Differ. 18, 1247–1256 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jorgensen I., Rayamajhi M., Miao E. A., Programmed cell death as a defence against infection. Nat. Rev. Immunol. 17, 151–164 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shi J., Gao W., Shao F., Pyroptosis: Gasdermin-mediated programmed necrotic cell death. Trends Biochem. Sci. 42, 245–254 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Broz P., Pelegrín P., Shao F., The gasdermins, a protein family executing cell death and inflammation. Nat. Rev. Immunol. 20, 143–157 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Kovacs S. B., Miao E. A., Gasdermins: Effectors of pyroptosis. Trends Cell Biol. 27, 673–684 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kayagaki N. et al., Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 526, 666–671 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Shi J. et al., Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 526, 660–665 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Liu X. et al., Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature 535, 153–158 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding J. et al., Pore-forming activity and structural autoinhibition of the gasdermin family. Nature 535, 111–116 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Aglietti R. A. et al., GsdmD p30 elicited by caspase-11 during pyroptosis forms pores in membranes. Proc. Natl. Acad. Sci. U.S.A. 113, 7858–7863 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruan J., Xia S., Liu X., Lieberman J., Wu H., Cryo-EM structure of the gasdermin A3 membrane pore. Nature 557, 62–67 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen X. et al., Pyroptosis is driven by non-selective gasdermin-D pore and its morphology is different from MLKL channel-mediated necroptosis. Cell Res. 26, 1007–1020 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonçalves A. P., Heller J., Daskalov A., Videira A., Glass N. L., Regulated forms of cell death in fungi. Front. Microbiol. 8, 1837 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daskalov A., Heller J., Herzog S., Fleißner A., Glass N. L., Molecular mechanisms regulating cell fusion and heterokaryon formation in filamentous fungi. Microbiol. Spectr. 5 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Zhang D.-X., Spiering M. J., Dawe A. L., Nuss D. L., Vegetative incompatibility loci with dedicated roles in allorecognition restrict mycovirus transmission in chestnut blight fungus. Genetics 197, 701–714 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang D.-X., Nuss D. L., Engineering super mycovirus donor strains of chestnut blight fungus by systematic disruption of multilocus vic genes. Proc. Natl. Acad. Sci. U.S.A. 113, 2062–2067 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Debets A. J. M., Griffiths A. J. F., Polymorphism of het-genes prevents resource plundering in Neurospora crassa. Mycol. Res. 102, 1343–1349 (1998). [Google Scholar]
- 19.van Diepeningen A. D., Debets A. J., Hoekstra R. F., Heterokaryon incompatibility blocks virus transfer among natural isolates of black Aspergilli. Curr. Genet. 32, 209–217 (1997). [DOI] [PubMed] [Google Scholar]
- 20.Heller J., Clavé C., Gladieux P., Saupe S. J., Glass N. L., NLR surveillance of essential SEC-9 SNARE proteins induces programmed cell death upon allorecognition in filamentous fungi. Proc. Natl. Acad. Sci. U.S.A. 115, E2292–E2301 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chevanne D. et al., Identification of the het-r vegetative incompatibility gene of Podospora anserina as a member of the fast evolving HNWD gene family. Curr. Genet. 55, 93–102 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Daskalov A., Gladieux P., Heller J., Glass N. L., Programmed cell death in Neurospora crassa is controlled by the allorecognition determinant rcd-1. Genetics 213, 1387–1400 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Söding J., Biegert A., Lupas A. N., The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33, W244–W248 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zimmermann L. et al., A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J. Mol. Biol. 430, 2237–2243 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Källberg M., Margaryan G., Wang S., Ma J., Xu J., RaptorX server: A resource for template-based protein structure modeling. Methods Mol. Biol. 1137, 17–27 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Finn R. D., Clements J., Eddy S. R., HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mulvihill E. et al., Mechanism of membrane pore formation by human gasdermin-D. EMBO J. 37, e98321 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shirey C. M., Scott J. L., Stahelin R. V., Notes and tips for improving quality of lipid-protein overlay assays. Anal. Biochem. 516, 9–12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Friedman K. J., Glick D., Role of lipids in the Neurospora crassa membrane: III. Lipid composition and phase transition properties of the plasma membrane, and its components. J. Membr. Biol. 54, 183–190 (1980). [DOI] [PubMed] [Google Scholar]
- 30.Cai X. et al., Prion-like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell 156, 1207–1222 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spaan A. N., van Strijp J. A. G., Torres V. J., Leukocidins: Staphylococcal bi-component pore-forming toxins find their receptors. Nat. Rev. Microbiol. 15, 435–447 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seilie E. S., Bubeck Wardenburg J., Staphylococcus aureus pore-forming toxins: The interface of pathogen and host complexity. Semin. Cell Dev. Biol. 72, 101–116 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seuring C. et al., The mechanism of toxicity in HET-S/HET-s prion incompatibility. PLoS Biol. 10, e1001451 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saupe S. J., The [Het-s] prion of Podospora anserina and its role in heterokaryon incompatibility. Semin. Cell Dev. Biol. 22, 460–468 (2011). [DOI] [PubMed] [Google Scholar]
- 35.Dyrka W. et al., Diversity and variability of NOD-like receptors in fungi. Genome Biol. Evol. 6, 3137–3158 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uehling J., Deveau A., Paoletti M., Do fungi have an innate immune response? An NLR-based comparison to plant and animal immune systems. PLoS Pathog. 13, e1006578 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daskalov A., Saupe S. J., As a toxin dies a prion comes to life: A tentative natural history of the [Het-s] prion. Prion 9, 184–189 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paoletti M., Saupe S. J., Fungal incompatibility: Evolutionary origin in pathogen defense? BioEssays 31, 1201–1210 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Daskalov A. et al., Identification of a novel cell death-inducing domain reveals that fungal amyloid-controlled programmed cell death is related to necroptosis. Proc. Natl. Acad. Sci. U.S.A. 113, 2720–2725 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Katoh K., Kuma K., Toh H., Miyata T., MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33, 511–518 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edgar R. C., MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Waterhouse A. M., Procter J. B., Martin D. M. A., Clamp M., Barton G. J., Jalview Version 2–A multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pandey A., Roca M. G., Read N. D., Glass N. L., Role of a mitogen-activated protein kinase pathway during conidial germination and hyphal fusion in Neurospora crassa. Eukaryot. Cell 3, 348–358 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bowman B. J., Blasco F., Slayman C. W., Purification and characterization of the plasma membrane ATPase of Neurospora crassa. J. Biol. Chem. 256, 12343–12349 (1981). [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data from this work are included in the main text and the SI Appendix of the paper. Strains are publicly available from the Fungal Genetics Stock Center (http://www.fgsc.net/fgsc/search_form.php).