Significance
Membrane proteins (MPs), encoded by ∼30% of the coding genes, play vital roles in numerous physiological processes. MPs are targets of more than half of the FDA-approved drugs. High-resolution structural studies of functional membrane proteins under near-physiological conditions are required to provide an in-depth mechanistic understanding and to facilitate drug discovery. To embed the proteins into liposomes represents a strategy to mimic native membrane conditions. Here we present a convenient workflow for cryo-EM analysis of liposome-embedded MPs using the prototypal protein AcrB. Our method sets the foundation for future investigation of MPs in the presence of electrochemical gradients and for the understanding of the interdependence of integral or peripheral MPs and various membrane properties.
Keywords: structural biology, proteoliposome, graphene grids, membrane protein, cryo-EM
Abstract
Membrane proteins (MPs) used to be the most difficult targets for structural biology when X-ray crystallography was the mainstream approach. With the resolution revolution of single-particle electron cryo-microscopy (cryo-EM), rapid progress has been made for structural elucidation of isolated MPs. The next challenge is to preserve the electrochemical gradients and membrane curvature for a comprehensive structural elucidation of MPs that rely on these chemical and physical properties for their biological functions. Toward this goal, here we present a convenient workflow for cryo-EM structural analysis of MPs embedded in liposomes, using the well-characterized AcrB as a prototype. Combining optimized proteoliposome isolation, cryo-sample preparation on graphene grids, and an efficient particle selection strategy, the three-dimensional (3D) reconstruction of AcrB embedded in liposomes was obtained at 3.9 Å resolution. The conformation of the homotrimeric AcrB remains the same when the surrounding membranes display different curvatures. Our approach, which can be widely applied to cryo-EM analysis of MPs with distinctive soluble domains, lays out the foundation for cryo-EM analysis of integral or peripheral MPs whose functions are affected by transmembrane electrochemical gradients or/and membrane curvatures.
Biological membranes enclose topologically insulated compartments, including cells and organelles, and afford the habitats for various integral and peripheral membrane proteins (MPs) (1). These physical barriers enable the generation and maintenance of life-essential electrochemical gradients, which result from the asymmetric distribution of ions and chemicals across the impermeable membranes (2). A variety of physiological processes depend on these gradients, such as adenosine triphosphate (ATP) synthesis that is driven by the proton gradient (the proton motive force) and the action potential that relies on the presence of the transmembrane electric field (3). Consequently, many MPs, exemplified by voltage-gated ion channels (VGICs) and primary and secondary active transporters, depend on the transmembrane electrochemical gradients to perform their biological functions (4).
In addition to residing within or on the surface of the membrane, the interplay between MPs and membranes also has a profound impact on cellular life (5). For instance, many peripheral MPs define the membrane contour for organelle formation (6, 7). Dimerization of the FoF1 ATP synthase plays an important role for shaping the mitochondria cristae (8). The mechanosensitive channels are gated by the mechanical force in part exerted by the deformation of the membrane (9). Therefore, the presence of the membrane is required for a comprehensive understanding of the function, structure, and mechanism of many MPs.
Structural analysis of integral MPs used to be extremely challenging when X-ray crystallography was the primary method for structural determination. Highly homogeneous MPs have to be purified from disrupted membranes and replaced with carefully selected detergents for crystallization. Since 2013, cryogenic electron microscopy (cryo-EM) single particle analysis (SPA) has become a mainstream means for high-resolution structural elucidation of MPs (10, 11). Multiple reagents have been applied to “solubilize” the MPs to single particles for analysis. In addition to detergent micelles, amphipol, nanodiscs, and styrene maleic acid lipid particle (SMALP)-enclosed nanodiscs with native membranes have been employed for successful cryo-EM structural analysis of integral MPs (12–20).
Notwithstanding these advances, all of the aforementioned MP isolation approaches disrupt the topology of the membrane, even in the case of SMALP-surrounded nanodiscs with native membrane patches, hence abolishing any existing electrochemical gradients and membrane curvature. To reserve these important properties, in situ structural analysis using electron cryo-tomography (cryo-ET) may be the ultimate solution (21, 22). However, the current technical hurdles prevent high-resolution in situ structural determination using cryo-ET. An alternative strategy is to study the structure of MPs embedded in liposomes, which has been widely used for functional analysis of MPs (23–26).
Despite extensive functional characterizations using proteoliposomes, there were only limited attempts to employ this system for structural elucidation of MPs. In the past decade, methods like random spherically constrained (RSC) have been developed to study the proteoliposome system with modified SPA strategy (27). More recently, a method using signal subtraction was reported to resolve the structure of the hBK channel in liposomes at near-atomic resolution (28). However, to perform accurate angular assignment or signal subtraction in both reports, the targeted proteoliposome must be near-perfect spheres, a prerequisite that is difficult to obtain. Extra preprocessing steps for raw images are also required in both methods. Additionally, previous results show that proteoliposomes tend to distribute unevenly on the cryo-grids, setting additional roadblocks for efficient data collection.
To apply cryo-EM for structural analysis of MPs embedded or attached to liposomes, it is necessary to develop a highly reproducible and convenient workflow for membrane protein incorporation, cryo-sample preparation, and cryo-EM data processing. For this purpose, we chose the well-studied multidrug-resistant transporter AcrB from Escherichia coli as a prototype for method development (29). The proton gradient-driven AcrB is a homo-trimer with a molecular weight of ∼350 kDa (30, 31). It is one of the MPs that are most prone to crystallization even with low purity and at low concentration. Therefore, the structure of AcrB was determined in early days (32, 33). Thus far, there are >100 AcrB structures in Protein Data Bank (PDB) determined using both X-ray crystallography and single-particle cryo-EM, providing excellent references for structural validation.
In this study, we report a workflow with optimized proteoliposome separation, graphene grid-supported cryo-sample preparation, and a convenient extra step, deep two-dimensional (2D) classification, for data processing. Using our simplified method, a 3.9 Å-resolution reconstruction of AcrB in proteoliposome was obtained. This workflow can be easily generalized for structural determination of MPs in proteoliposomes and establish a foundation for structural analysis of MPs in the presence of controlled electrochemical potentials or membrane curvatures using the proteoliposome system.
Results
Reconstitution of AcrB into Liposomes.
Multiple methods for proteoliposome reconstitution have been reported (34, 35). After trials, we have adapted and optimized the protocol for detergent-mediated proteoliposome reconstitution, followed by size-exclusion chromatography (SEC) separation (Fig. 1).
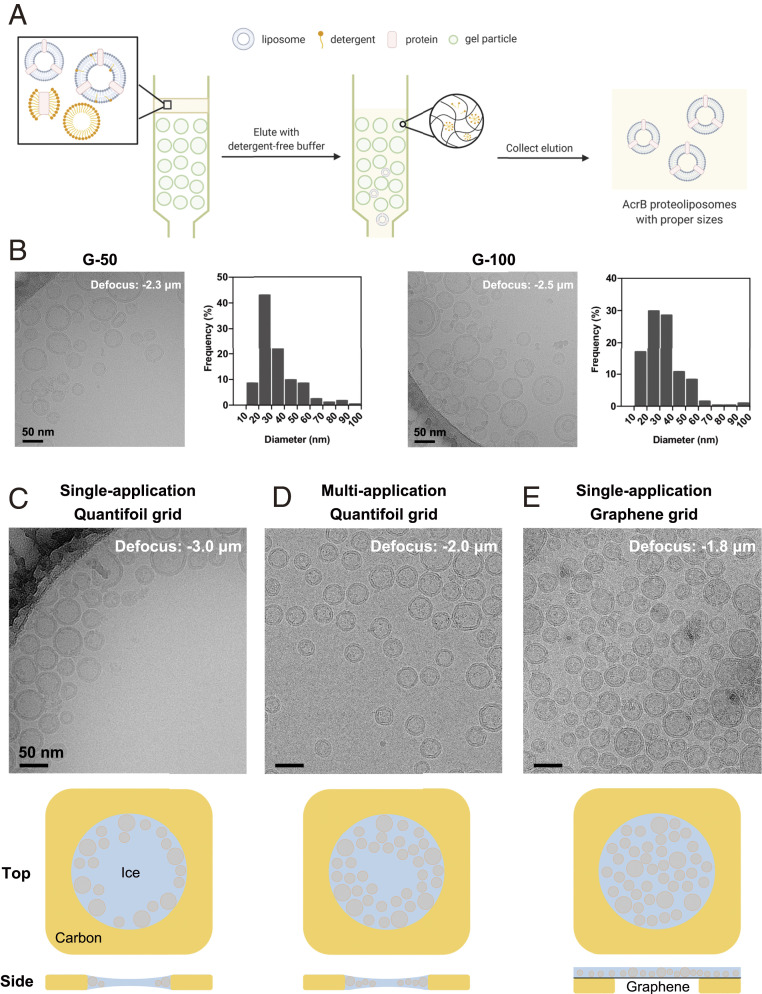
Fig. 1.
Optimization of proteoliposome cryo-sample preparation. (A) A schematic illustration of size selection of proteoliposomes using SEC. When the mixture of proteoliposomes and detergent micelles pass through different resins for SEC in the absence of supplemented detergents, liposomes are separated based on their diameters and detergents are removed. Please refer to SI Appendix, Fig. S1 (SI Appendix) for more trials of proteoliposome optimization. (B) Representative cryo-EM micrographs and size distribution of AcrB proteoliposomes prepared using manually packed Sephadex G-50 (Left) and G-100 columns (Right). Histograms are calculated from five holes of each sample. (C–E) Micrographs of AcrB proteoliposomes loaded on commercial Qauntifoil grids with (C) single- and (D) multiapplication approaches, and (E) on a graphene with single-application. The cartoon illustration for proteoliposome distribution in the holes by various sample preparation methods are presented below the corresponding panel.
Details are presented in Methods. Briefly, detergent-solubilized 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine (POPC) lipid was mixed with purified AcrB with a protein-to-lipid molar ratio of 1:1000. Removal of the residual detergents and size control of the proteoliposomes are the critical steps. Bio-beads were first tested to remove the undesired detergent (36). After repeated extrusion through 100 nm filters, the proteoliposomes were subject to cryo-EM examination. These proteoliposomes, whose diameters are generally larger than 100 nm, appeared to be flattened during Vitrobot blotting and stacked badly on the grids. Proteoliposomes prepared in this way cannot fulfill the requirement for single particle cryo-EM analysis (SI Appendix, Fig. S1A).
SEC was then applied to remove detergents (28, 37) (Fig. 1A). Reasoning that the rate and efficiency of detergent removal could influence the proteoliposome properties, such as size, shape, and protein insertion, we set out to test several SEC resins. The lipid-protein-detergent mixture was parallelly loaded to Sephadex G-10, G-50, and G-100 columns. Both G-50 and G-100 yielded unilamellar vesicles with diameters ranging from 10 to 100 nm, and the size of the proteoliposomes prepared with G-50 appeared to be more homogenous (Fig. 1B). In contrast, G-10 mostly yielded multilamellar proteoliposomes (SI Appendix, Fig. S1B). The protein recovery efficiency was estimated by quantifying the amount of protein after SEC compared to the input. Among the three tested resins, G-50 gave rise to the highest recovery rate of ∼52%, compared to ∼31% by G-10 and ∼42% by G-100 (SI Appendix, Fig. S1C). G-50 was thereby chosen for proteoliposome preparation.
Next, we compared the effect of lipid composition and choice of detergent for liposome solubilization. Only the combination of POPC and n-decyl-β-D-maltopyranoside (DM) yielded adequate proteoliposomes that were intact and homogenous. n-Octyl-β-D-glucopyranoside (OG)-treated POPC samples had rare protein insertion into the liposomes, with more AcrB present in micelles or aggregates (SI Appendix, Fig. S1D). Interestingly, DM appeared to be incompatible with the liposomes reconstituted from E. coli polar lipids. A majority of the liposomes was of large size and, even worse, burst, indicating poor separation of DM from E. coli polar extract liposomes through G-50 column. (SI Appendix, Fig. S1E).
Additional tests for detergent and lipid combinations were suspended because of COVID-19. Nevertheless, our trials suggested that multiple parameters, including the detergent used for protein purification, lipid composition, and means of liposome isolation, are all important parameters for optimization of cryo-sample preparation.
Graphene Grid for Cryo-Preparation of Proteoliposomes.
After successful reconstitution of AcrB into a relatively homogenous population of proteoliposomes with diameters of 50 nm or smaller, the next challenge was to obtain high-quality cryo-samples with well-distributed proteoliposomes.
We started with Vitrobot cryo-sample preparation for proteoliposomes loaded on Quantifoil grids using the routine single-application and blotting method. However, the proteoliposomes tended to cluster near the edge of the supporting carbon film, leaving the center of the hole vacant (Fig. 1C and SI Appendix, Fig. S2A). To increase the proteoliposome density, we implemented a multiapplication method to add protein solution repetitively before final blotting (38). This approach indeed improved the distribution of proteoliposomes, but did not mitigate the off-center accumulation (Fig. 1D and SI Appendix, Fig. S2B). In both single- and multiapplication approaches, smaller liposomes were likely to get closer to the center (Fig. 1 C and D). This was probably related to the concave thickness distribution of the vitreous ice in the hole caused by surface tension.
To produce cryo-samples with evenly distributed proteoliposomes, graphene grids, freshly prepared following our recently reported protocol (39), were used for cryo-sample preparation. With the same sample concentration, the density and distribution of proteoliposomes were evidently improved on graphene grids. As opposed to samples on regular Quantifoil grids (Fig. 1 C and D), the proteoliposomes on the graphene grids were evenly distributed throughout the entire hole (Fig. 1E). There was no discernible difference in distribution of proteoliposomes on graphene surface between the hole and the carbon foil area (Fig. 1E and SI Appendix, Fig. S2C). Proteoliposomes with various sizes were randomly distributed without position preference, consistent with the uniform ice thickness on graphene grids. Areas with densely distributed proteoliposomes were visible even at low magnification in low-dose search/view mode (SI Appendix, Fig. S2C), hence conveniently facilitating selection of positions for data collection.
High-Resolution Reconstruction of AcrB in Proteoliposomes.
After obtaining graphene cryo-grids with well-distributed proteoliposomes, we collected 5,757 raw movies in areas with proteoliposome features (Fig. 2 and SI Appendix, Fig. S2C) using the Titan Krios equipped with a Cs-corrector and a Gatan GIF Quantum K2 direct detector. Fourier transform of the drift-corrected and summed movies represented a clear indexing of the first-order reciprocal lattice of graphene (SI Appendix, Fig. S2 D and E), indicating that the raw signal in the micrograph has been recorded and well-maintained up to 2.14 Å in frequency.
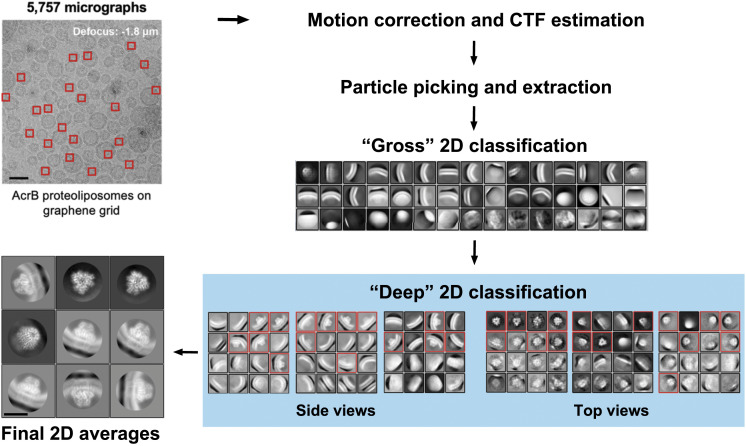
Fig. 2.
Deep 2D classification for selection of good protein particles embedded in liposomes. Protein particles (indicated by red squares) were extracted from 5,757 motion-corrected micrographs. (Scale bar: 50 nm.) After a gross 2D classification for all extracted particles, a “deep” 2D classification strategy was applied to the 2D classes containing over 1,000 particles. Particles in deep 2D classes with clear protein signal were collected (red boxes). Final 2D averages from the selected particles display clear secondary structure features. (Scale bar: 10 nm.) More details are provided in SI Appendix, Fig. S3.
Picking liposome-embedded protein particles was not straightforward, as only very few protein particles were discernible on the micrograph. The reported “signal subtraction” method removes the liposome signal to facilitate particle picking (28). However, it involves a separate reprocessing of all of the raw micrographs and requires the liposomes to be perfect spheres. Furthermore, high accuracy for per-liposome size fitting and signal scaling is required to avoid artifact during signal subtraction. These complications make this method inconvenient for SPA analysis.
We attempted to develop a more straightforward method that is compatible with the prevalent image-processing programs. Template matching-based auto-picking was performed in Relion (40), using two different types of reference: 1) 2D class averages generated from a few hundred manually picked particles; 2) random 2D projections generated from a 20 Å low-pass filtered three-dimensional (3D) map of AcrB (SI Appendix, Fig. S3). We intentionally reduced the picking threshold to include more potential particles. After reaching ∼4.6 million, the particles were divided into three subsets for more efficient processing and multiround 2D classification was applied to exclude junk particles.
A “deep 2D classification” strategy proved to be useful in efficient enrichment of good particles. One round of gross 2D classification for each subset was followed by further 2D classification of individual classes containing over 1,000 particles, a procedure defined as the deep 2D classification (Fig. 2). With this procedure, particles with distinctive structural features, including the lipid bilayer signal, were successfully isolated (Fig. 2, Final 2D averages). Particularly, in our case, this deep 2D classification approach can separate useful particles from those seemingly “lipid-only” classes and uncover the top-view particles that were embedded deeply in the gross 2D classification stage (Fig. 2). With efficient exclusion of junk particles, this procedure also downscaled the size of the dataset dramatically.
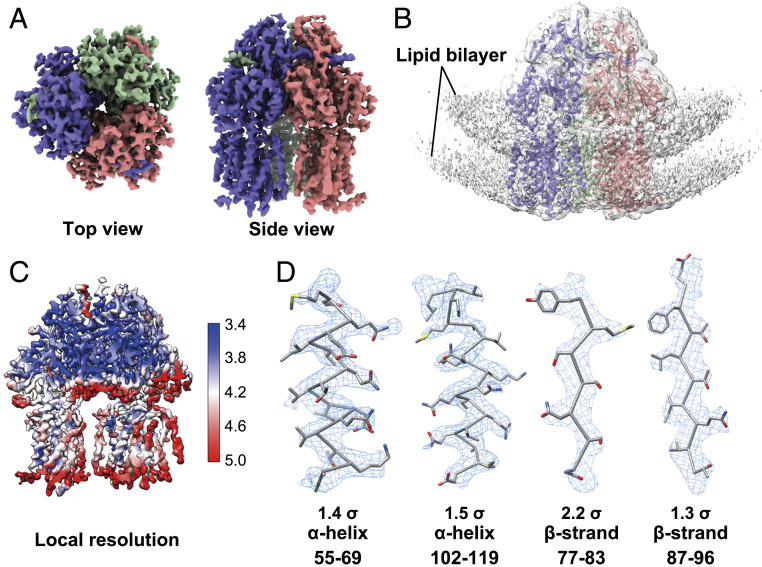
Followed with a few rounds of regular global and local angular search 3D classifications, we successfully obtained a 3D reconstruction of AcrB at an overall resolution of 3.9 Å according to the gold standard Fourier shell correlation (FSC) (Fig. 3 and SI Appendix, Fig. S4). The proteoliposomal AcrB did not suffer from obvious preferred orientation as assessed by the angular distribution and 3D-FSC results (SI Appendix, Fig. S4). Local resolution map and local densities further validate the quality of the reconstruction (Fig. 3 C and D). Density modification was performed to further improve the map quality (SI Appendix, Fig. S5A).
Fig. 3.
Single-particle reconstruction of AcrB embedded in proteoliposomes. (A) Top view and side view of the single-particle reconstruction of AcrB at 3.9 Å resolution. (B) EM map with low display threshold reveals lipid bilayer signal. The AcrB structure (PDB code: 1IWG) is docked into the density (before B-factor sharpening) contoured at 1.0σ. (C) Local resolution map of the AcrB reconstruction. The unit for resolutions shown on the right is Å. (D) Representative densities of two alpha helices and two beta strands of AcrB, shown as blue mesh with indicated contour level.
Previous study demonstrated that the trimeric AcrB in different conditions could adopt conformations with either C1 or C3 symmetry (20, 32). We performed 3D reconstructions with selected particles in either C1 or C3 symmetry and low-pass filtered both maps to 6 Å for structural comparison (SI Appendix, Fig. S5 B and C). No significant conformational difference was observed between these two reconstructions, indicating that the three protomers of liposome-embedded AcrB exhibit identical conformations at the current resolution. We further compared our reconstruction with published ones for apo AcrB with C3 (PDB: 1IWG) (41) or C1 AcrB (PDB: 6BAJ) (42) symmetry. Our reconstruction is nearly identical with that of the C3 apo-state, but deviates from the C1 in the soluble domain (SI Appendix, Fig. S5D).
Proteoliposomal AcrB with Membrane Feature.
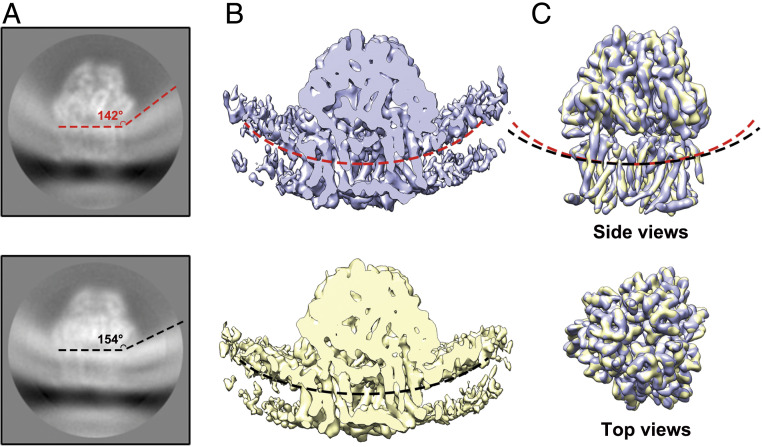
An important advantage of our protocol is to preserve the signal for the lipid bilayer during data processing. At a low display threshold, the reconstruction signal for the surrounding membrane could also be clearly recognized (Fig. 3B). According to the membrane curvature, roughly 98% of AcrB were found to have the periplasmic domains facing the lumen of the liposome (defined as inside-facing) (SI Appendix, Fig. S6). A slight difference in the curvature of the surrounding lipid bilayer membrane was observed when comparing the lipid signal of different side view classes in the final 2D class averages. (Fig. 4A and Movie S1). To investigate whether the difference in membrane curvature would influence the conformation of embedded AcrB, we selected the particles from each individual side-view class and performed 3D reconstruction independently.
Fig. 4.
AcrB surrounded by different membrane curvatures exhibits identical structure. (A) 2D class averages shown different membrane curvatures around AcrB. Two representative classes stand out. (B) Reconstruction of the membrane fraction. The 3D reconstructions of the particles in the two 2D classes shown in A are low-pass filtered to 7 Å. Membrane curvatures are indicated by red and black, dashed line. (C) AcrB from the two classes are nearly identical. The two 3D reconstructions are overlaid, with the respective membrane curvatures indicated by red and black dashed lines.
Although limited by the particle number and orientation distribution in one class, we successfully obtained the reconstructions with correct structure features. For a better comparison, these reconstructions were both low-pass filtered to 7 Å (Fig. 4 B and C). Indeed, different membrane curvatures were clearly seen in these two reconstructions, consistent with the 2D class averages (Fig. 4 A and B and Movie S1). Nevertheless, the structures of AcrB in different membrane curvatures did not exhibit an obvious conformational change at 7 Å (Fig. 4C).
Discussion
Here we present a complete workflow for high-resolution cryo-EM structural analysis of membrane proteins embedded in liposomes by cryo-EM, combining an optimized proteoliposome preparation protocol, high-quality graphene grids, and a deep 2D classification (SI Appendix, Fig. S7). A SEC-based preparation approach can effectively control the size of the proteoliposomes. Graphene grids can dramatically improve the density and distribution of the proteoliposomes in the field of view, which supports a rationalized data collection strategy based on the patterns in low magnification images. The deep 2D classification procedure enables convenient selection of the liposome-embedded protein particles for regular single particle analysis, avoiding complex and demanding preprocessing.
It is noteworthy that the signals from the soluble domain of the embedded AcrB have made major contributions to particle selection in deep 2D classification. Without a soluble domain with distinctive contour, it would be impractical to select transmembrane domain from the membrane. Therefore, our method is mainly applicable to membrane proteins with distinctive-shaped soluble domain(s) of reasonable size. On the other hand, an initial model has been applied as a template to facilitate particle picking. However, we reasoned that if a membrane protein could be successfully reconstituted into the liposomes, it should be simply easier to obtain at least a low-resolution 3D reconstruction in detergent micelles or nanodiscs by regular single particle cryo-EM analysis. Therefore, this prerequisite does not represent a roadblock for the application of our methods.
Interestingly, we also found that 98% of particles in all AcrB side views have their soluble domain inside the liposomes in our case (SI Appendix, Fig. S6). The orientation analysis could be extended to correct the calculations of transport efficiencies in liposome functional assays, which usually have an assumption of 50/50 distribution (43). It might be caused by the lipid or detergent composition (44), but an advanced understanding of the preferred orientation in liposomes requires further investigation.
We envision that our method can be applied to a spectrum of structure-function correlative studies. One of our purposes is to investigate the impact of the transmembrane gradient of ions or other substrates on the structure and function of the target proteins, such as the voltage-gated ion channels and proton-driven transporters. The biggest technical hurdle would be to prepare proteoliposome cryo-samples with controllable transmembrane gradient, because liposome leakage is a serious issue particularly during plunge freezing. Nonetheless, our method can be immediately applied to the analysis of the interdependence between the membrane curvature and the proteins which respond to membrane shaping, fusion, and fission, and mechanical sensitive channels, such as Piezo, whose gating depends on the membrane curvature (45). Our investigation reported here thus lays the ground for a broader application of cryo-EM in the investigation of membrane proteins and membrane.
Materials and Methods
Overexpression and Purification of E. coli AcrB.
Full-length AcrB was subcloned into the pET15b vector with an amino terminal His6 tag. Overexpression of AcrB was induced in E. coli BL21 (DE3) by 0.5 mM isopropyl-β-D-thiogalactoside (IPTG, Sigma-Aldrich) when optical density (OD600) reached 0.8 to 1.0. After growth at 20 °C for 16 h, the cells were harvested and resuspended in a buffer containing 20 mM Tris⋅HCl, pH 8.0, 500 mM NaCl, and 2 mM MgCl2, and disrupted by sonication (1.5 min/L). Cell debris was removed by high-speed centrifugation at 27,000 g for 10 min. The membrane-containing supernatant was collected and applied to ultra-speed centrifugation at 250,000 g for 1 h. The membrane-containing pellet was resuspended in the extraction buffer containing 20 mM Tris⋅HCl, pH 7.5, 150 mM NaCl, 10 mM imidazole, and 10% glycerol and incubated with 1.0% (wt/vol) n-dodecyl-β-D-maltopyranoside (DDM, Anatrace) for 2 h at 4 °C. After additional ultracentrifugation at 250,000 g for 30 min, the supernatant was loaded onto a Ni2+-nitrilotriacetate affinity resin (Qiagen), which was subsequently rinsed three times, each with 10 mL buffer containing 20 mM Tris⋅HCl, pH 7.5, 150 mM NaCl, 20–40 mM imidazole, and 0.03% DDM. The protein was eluted from the affinity resin with 10 mL wash buffer plus 220 mM imidazole and concentrated to ∼1 mL before applying to size-exclusion chromatography (Superose 6 Increase 10/300 GL, GE Healthcare) pre-equilibrated with the buffer containing 20 mM Tris⋅HCl, pH 7.5, 150 mM NaCl, and 0.03% DDM. The protein concentration of the pooled peak fractions was quantified using Quick Start Bradford dye (Bio-Rad) and measured with biophotometer (Eppendorf) at the wavelength of 595 nm.
AcrB Reconstitution into Liposomes.
POPC (Avanti) dissolved in 80 μL chloroform at 25 mg/mL was dried under a nitrogen stream and resuspended in 400 μL reconstitution buffer containing 20 mM Hepes-NaOH, pH 7.4, and 150 mM KCl. After sonication for 15 min, the lipid solution was frozen with liquid nitrogen and thawed in water bath for 10 times. Twenty microliters of 10% (wt/vol) DM (Anatrace) was added and incubated with the lipid at 4 °C for 30 min. Then approximately purified AcrB protein was added to make the protein-to-lipid molar ratio of 1:1,000. After incubation at 4 °C for 2 h, the mixture was loaded to manually packed Sephadex G-50 column (10 mm inner diameter and 300 mm height, Bio-Rad; Sephadex G-50 Superfine, Sigma-Aldrich), which was pre-equilibrated with the reconstitution buffer. Fractions that could be stained by Bradford dye (Bio-Rad) were collected and concentrated using Amicon Ultra centrifugal filter (0.5 mL, 100 kDa cutoff) to a final concentration of ∼0.7 mg/mL The protein concentration after reconstitution was measured in the same way as described above. Protein recovery rate was calculated by dividing the residual amount of protein in the collected fractions by the input (300 μg for each preparation).
Other trials for the choice of detergent and lipid compositions prepared proteoliposome in the same manner. For E. coli polar lipid and DM combination, E. coli polar lipid extract (Avanti), was dissolved in 80 μL chloroform at 25 mg/mL, hydrated with 400 μL reconstitution buffer after getting dried by nitrogen stream. After sonication and freeze-thaw, 20 μL of 10% (wt/vol) DM was added and incubated with the lipid at 4 °C for 30 min. For another combination, POPC and OG, 10 μL of 20% (wt/vol) OG (Anatrace) was added into 400 μL POPC suspension before the addition of AcrB.
Cryo-Sample Preparation on Homemade Graphene Grids.
Graphene grids were prepared following our published protocol (39). Briefly, monolayer graphene on copper foil (purchased from graphene supermarket) was first protected by methyl methacrylate (MMA). After removing backside graphene by glow discharge, the copper foil was etched by 0.5 M ammonium persulfate (Sigma). Then the floated graphene flakes were washed on water surface and transferred to Quantifoil Au R1.2/1.3 grids (300 mesh). The graphene grids were further cleaned by acetone and isopropanol. Ultraviolet (UV)/ozone (ProCleaner) was applied to functionalize the graphene grid surface. Freshly ozonated graphene grids were used for cryo-sample preparation.
Cryo-samples were prepared in Vitrobot Mark IV (Thermo Fisher), which was set to 10 °C with 100% humidity. Four microliters proteoliposome solution was applied to the graphene surface of the grid or regular Quantifoil grids as indicated. After waiting for 30 s, grids were blotted for 4 s with a blot force of 0 and rapidly plunged into precooled liquid ethane for vitrification. For a multiapplication approach, the applied sample solution will be taken away by pipetting after 30 s waiting on grid in Vitrobot. The sample application-waiting-removing cycle was repeated for two to four times before the blotting. Sample preparation with Quantifoil grids or graphene grids was performed more than three times, respectively. The liposome distributions in different conditions are repeatable in our case.
Data Acquisition.
A total number of 5,757 raw movie stacks were automatically collected from two individual sessions by SerialEM 3.7 on a 300 kV Cs-corrected Titan Krios using a K2 Summit detector (with GIF Bio-Quantum Energy Filters, Gatan). The acquisition areas were selected based on the liposome distribution pattern in view mode (magnification of 4,800). Raw movies were collected in K2 superresolution mode at a magnification of 105,000 (in energy-filtered transmission electron microscopy [EFTEM] mode, slit width 20 eV, spot size 6, C2 aperture 70 μm) with a superresolution pixel size of 0.557 Å. The total exposure time was set to 5.6 s with 0.175 s per frame to generate 32-frame gain normalized mrc stacks. The total dose for a stack is 50 e−/Å2.
Data Processing.
Movie stacks were drift corrected using Relion 3.1-beta’s own interpretation with a 5 × 5 patch and a twofold binning (46). Contrast transfer function (CTF) information was estimated from nondose weighted images by CTFFind4.1 in Relion 3.1-beta with exhaustive searching (47). Particles were picked by Relion Auto-picking from two different strategies: 1) with 2D reference generated from manual picked particles and 2) projections from a 3D reference. The picked particles were merged in later processes. After particle extraction, a regular gross 2D classification was performed. Classes with over 1,000 particles from the gross 2D classification were selected to perform further 2D classification individually. This strategy, named deep 2D classification was automatically run by a script developed by Chuangye Yan at Tsinghua University and Qiang Zhou at Westlake University, who have kindly authorized us to use and share the script. After collecting good particles from deep 2D classification, multiple rounds of 3D classification, 3D auto-refine, and Bayesian polishing were performed in Relion 3.1 with standard procedure (46). Particles outside the proteoliposome area were manually excluded before the final refinement. Individual 2D class-based reconstruction was done by relion_reconstruct program. Density modification was done in phenix (1.18rc1-3777) by ResolveCryoEM program (48). Local resolution map was calculated by Blocres (49). Directional FSC was calculated by 3DFSC program (50). Density maps were prepared using UCSF Chimera (51) and ChimeraX (52).
Supplementary Material
Acknowledgments
We thank Chuangye Yan in Tsinghua University and Qiang Zhou in Westlake University for developing and sharing the script for deep 2D classification. We thank the support by Paul Shao and Nan Yao for data collection at the cryo-EM facility at Princeton Imaging and Analysis Center, which is partially supported by the Princeton Center for Complex Materials, an NSF Materials Research Science and Engineering Center (MRSEC) program (DMR-1420541). We thank Xuelan Wu for helping manual particle exclusion and Collin McManus for suggestions during manuscript revision. This work is supported by grants from the Ara Parseghian Medical Research Foundation and the NIH (5R01GM130762), the Dean for Research Innovation Fund for New Ideas in the Natural Sciences, and a School of Engineering and Applied Science (SEAS) Innovation Research Grant from Princeton University. N.Y. is supported by the Shirley M. Tilghman endowed professorship from Princeton University. X.F. is supported by the HFSP long-term fellowship (LT000754) from the International Human Frontier Science Program Organization (HFSPO).
Footnotes
The authors declare no competing interest.
Data deposition: Data supporting the findings of this manuscript are available in public repositories. The EM map of AcrB reported in this paper is available in the Electron Microscopy Data Bank (accession no. EMD-22050). The raw data were deposited in the Electron Microscopy Public Image Archive at EMPIAR-10426.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2009385117/-/DCSupplemental.
Data Availability.
Data supporting the findings of this manuscript are available in public repositories. The accession number for the EM map of AcrB reported in this paper is EMD-22050 (53). The raw data were deposited at EMPIAR-10426 (54).
References
- 1.Watson H., Biological membranes. Essays Biochem. 59, 43–69 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sundelacruz S., Levin M., Kaplan D. L., Role of membrane potential in the regulation of cell proliferation and differentiation. Stem Cell Rev. Rep. 5, 231–246 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maloney P. C., Energy coupling to ATP synthesis by the proton-translocating ATPase. J. Membr. Biol. 67, 1–12 (1982). [DOI] [PubMed] [Google Scholar]
- 4.Bezanilla F., How membrane proteins sense voltage. Nat. Rev. Mol. Cell Biol. 9, 323–332 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Phillips R., Ursell T., Wiggins P., Sens P., Emerging roles for lipids in shaping membrane-protein function. Nature 459, 379–385 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frost A., Unger V. M., De Camilli P., The BAR domain superfamily: Membrane-molding macromolecules. Cell 137, 191–196 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shibata Y., Hu J., Kozlov M. M., Rapoport T. A., Mechanisms shaping the membranes of cellular organelles. Annu. Rev. Cell Dev. Biol. 25, 329–354 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Davies K. M., Anselmi C., Wittig I., Faraldo-Gómez J. D., Kühlbrandt W., Structure of the yeast F1Fo-ATP synthase dimer and its role in shaping the mitochondrial cristae. Proc. Natl. Acad. Sci. U.S.A. 109, 13602–13607 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perozo E., Kloda A., Cortes D. M., Martinac B., Physical principles underlying the transduction of bilayer deformation forces during mechanosensitive channel gating. Nat. Struct. Biol. 9, 696–703 (2002). [DOI] [PubMed] [Google Scholar]
- 10.Cheng Y., Membrane protein structural biology in the era of single particle cryo-EM. Curr. Opin. Struct. Biol. 52, 58–63 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liao M., Cao E., Julius D., Cheng Y., Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature 504, 107–112 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Q. et al., Structure of mammalian endolysosomal TRPML1 channel in nanodiscs. Nature 550, 415–418 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao Y., Cao E., Julius D., Cheng Y., TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature 534, 347–351 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katayama H. et al., Three-dimensional structure of the anthrax toxin pore inserted into lipid nanodiscs and lipid vesicles. Proc. Natl. Acad. Sci. U.S.A. 107, 3453–3457 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taylor N. M. I. et al., Structure of the human multidrug transporter ABCG2. Nature 546, 504–509 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Dang S. et al., Cryo-EM structures of the TMEM16A calcium-activated chloride channel. Nature 552, 426–429 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fan G. et al., Gating machinery of InsP3R channels revealed by electron cryomicroscopy. Nature 527, 336–341 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hite R. K. et al., Cryo-electron microscopy structure of the Slo2.2 Na(+)-activated K(+) channel. Nature 527, 198–203 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan Z. et al., Structure of the rabbit ryanodine receptor RyR1 at near-atomic resolution. Nature 517, 50–55 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qiu W. et al., Structure and activity of lipid bilayer within a membrane-protein transporter. Proc. Natl. Acad. Sci. U.S.A. 115, 12985–12990 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen W., Kudryashev M., Structure of RyR1 in native membranes. EMBO Rep. 21, e49891 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lucić V., Leis A., Baumeister W., Cryo-electron tomography of cells: Connecting structure and function. Histochem. Cell Biol. 130, 185–196 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan Y.-H. M., Boxer S. G., Model membrane systems and their applications. Curr. Opin. Chem. Biol. 11, 581–587 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Racker E., Stoeckenius W., Reconstitution of purple membrane vesicles catalyzing light-driven proton uptake and adenosine triphosphate formation. J. Biol. Chem. 249, 662–663 (1974). [PubMed] [Google Scholar]
- 25.Rigaud J.-L., Membrane proteins: Functional and structural studies using reconstituted proteoliposomes and 2-D crystals. Braz. J. Med. Biol. Res. 35, 753–766 (2002). [DOI] [PubMed] [Google Scholar]
- 26.Viitanen P., Newman M. J., Foster D. L., Wilson T. H., Kaback H. R., “Purification, reconstitution, and characterization of the lac permease of Escherichia coli” in Methods in Enzymology, (Elsevier, 1986), Vol. 125, pp. 429–452. [DOI] [PubMed] [Google Scholar]
- 27.Wang L., Sigworth F. J., Structure of the BK potassium channel in a lipid membrane from electron cryomicroscopy. Nature 461, 292–295 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tonggu L., Wang L., Broken symmetry in the human BK channel. bioRxiv:494385 (2018).
- 29.Nikaido H., Takatsuka Y., Mechanisms of RND multidrug efflux pumps. Biochim. Biophys. Acta 1794, 769–781 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eicher T. et al., Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop. Proc. Natl. Acad. Sci. U.S.A. 109, 5687–5692 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsunaga Y. et al., Energetics and conformational pathways of functional rotation in the multidrug transporter AcrB. eLife 7, e31715 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murakami S., Nakashima R., Yamashita E., Yamaguchi A., Crystal structure of bacterial multidrug efflux transporter AcrB. Nature 419, 587–593 (2002). [DOI] [PubMed] [Google Scholar]
- 33.Seeger M. A. et al., Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science 313, 1295–1298 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Rigaud J.-L., Lévy D., “Reconstitution of membrane proteins into liposomes” in Methods in Enzymology, (Elsevier, 2003), Vol. 372, pp. 65–86. [DOI] [PubMed] [Google Scholar]
- 35.Lichtenberg D., Ahyayauch H., Alonso A., Goñi F. M., Detergent solubilization of lipid bilayers: A balance of driving forces. Trends Biochem. Sci. 38, 85–93 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Rigaud J.-L., Levy D., Mosser G., Lambert O., Detergent removal by non-polar polystyrene beads. Eur. Biophys. J. 27, 305–319 (1998). [Google Scholar]
- 37.Brunner J., Skrabal P., Hauser H., Single bilayer vesicles prepared without sonication. Physico-chemical properties. Biochim. Biophys. Acta 455, 322–331 (1976). [DOI] [PubMed] [Google Scholar]
- 38.Snijder J. et al., Vitrification after multiple rounds of sample application and blotting improves particle density on cryo-electron microscopy grids. J. Struct. Biol. 198, 38–42 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han Y. et al., High-yield monolayer graphene grids for near-atomic resolution cryoelectron microscopy. Proc. Natl. Acad. Sci. U.S.A. 117, 1009–1014 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zivanov J. et al., New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murakami S., Nakashima R., Yamashita E., Yamaguchi A., Data from “Crystal structure of Bacterial Multidrug Efflux transporter AcrB.” Protein Data Bank. https://www.rcsb.org/structure/1IWG. Accessed 1 April 2020.
- 42.Qiu W., Fu Z., Guo Y., Data from “Cryo-EM structure of lipid bilayer in the native cell membrane nanoparticles of AcrB.” Protein Data Bank. https://www.rcsb.org/structure/6BAJ. Deposited 1 April 2020.
- 43.Scalise M., Pochini L., Giangregorio N., Tonazzi A., Indiveri C., Proteoliposomes as tool for assaying membrane transporter functions and interactions with xenobiotics. Pharmaceutics 5, 472–497 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tunuguntla R. et al., Lipid bilayer composition can influence the orientation of proteorhodopsin in artificial membranes. Biophys. J. 105, 1388–1396 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin Y.-C. et al., Force-induced conformational changes in PIEZO1. Nature 573, 230–234 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zivanov J., Nakane T., Scheres S. H. W., Estimation of high-order aberrations and anisotropic magnification from cryo-EM data sets in RELION-3.1. IUCrJ 7, 253–267 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Terwilliger T. C., Ludtke S. J., Read R. J., Adams P. D., Afonine P. V., Improvement of cryo-EM maps by density modification. bioRxiv:845032 (2019). [DOI] [PMC free article] [PubMed]
- 49.Heymann J. B., Guidelines for using Bsoft for high resolution reconstruction and validation of biomolecular structures from electron micrographs. Protein Sci. 27, 159–171 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tan Y. Z. et al., Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nat. Methods 14, 793–796 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pettersen E. F. et al., UCSF Chimera–A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 52.Goddard T. D. et al., UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yao X., Fan X., Yan N., 3.9 Angstrom reconstruction of E. coli AcrB embedded in the liposome. Electron Microscopy Data Bank. https://www.ebi.ac.uk/pdbe/entry/emdb/EMD-22050. Deposited 25 May 2020.
- 54.Yao X., Fan X., Yan N., 3.9 Angstrom reconstruction of E. coli AcrB embedded in the liposome. Electron Microscopy Public Image Archive. https://www.ebi.ac.uk/pdbe/emdb/empiar/entry/10426. Deposited 28 May 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting the findings of this manuscript are available in public repositories. The accession number for the EM map of AcrB reported in this paper is EMD-22050 (53). The raw data were deposited at EMPIAR-10426 (54).