Abstract
Fish bacterial pathogens cause diseases which result in a considerable economic impact on the aquaculture industry, necessitating the use of antimicrobials for their control. However, intensive and indiscriminate use of antimicrobials has led to increased occurrence of drug resistance in pathogenic bacteria, as well as normal flora. The aim of the current study was to determine the susceptibility patterns of bacteria isolated from fish, with respect to some commonly used antibiotics and disinfectants. Bacteria were isolated between December 2017 and April 2018 from farmed Nile tilapia, African catfish, goldfish, and koi carp in Kirinyaga County, Kenya. Antibiotic and disinfectant susceptibility patterns of 48 isolates belonging to the genera Aeromonas, Proteus, Klebsiella, Citrobacter, Salmonella, Streptococcus, Pseudomonas, Escherichia, Serratia, and Micrococcus were established using the Kirby–Bauer disc diffusion method and agar well diffusion technique, respectively. The antibiotics evaluated included ampicillin, tetracycline, co-trimoxazole, streptomycin, kanamycin, gentamicin, co-trimoxazole, and chloramphenicol, while the disinfectants tested were quaternary ammonium compound, formalin, hydrogen peroxide, sodium hypochlorite, and iodine. All the bacteria except Micrococcus, Escherichia, and Salmonella species showed multiple drug resistance patterns. Streptococcus showed resistance to six antibiotics, while Proteus, Pseudomonas, and Serratia were resistant to five antibiotics. The multiple antibiotic resistance index ranged from 0.1 to 0.8, with Streptococcus spp. having the highest score value. All the organisms were sensitive to gentamicin, while co-trimoxazole and ampicillin showed the highest resistance at 73% (n = 34) and 62% (n = 31), respectively. Most of the disinfectants showed antibacterial activity with varying magnitudes. The isolates were 100% sensitive to hydrogen peroxide and formalin, but were resistant to sodium hypochlorite at recommended user-dilution. The study has shown that some of the bacterial isolates were resistant to common antibiotics and disinfectants; thus, it is recommended to include an antibiogram whenever making any therapeutic decision. The resistant bacteria may transmit resistance genes to other fish bacteria and also to human bacteria, thus making it difficult to treat the resultant disease(s); thus, there is a possibility that these resistant bacteria may be transmitted to humans who consume or handle the carrier fish. It is, therefore, advisable that fish are cooked properly before consumption, so as to kill bacteria that may be present.
1. Introduction
A wide range of fish bacteria have the potential to cause diseases, making them one of the most important concerns for the aquaculture sector. So far, many pathogenic microorganisms, such as Aeromonas hydrophila, Pseudomonas spp., Citrobacter freundii, Streptococcus iniae, Flavobacterium columnare, and Edwardsiella tarda, among others, have been reported as aetiological agents of fish diseases [1]. Disease outbreaks can, in turn, bear negative implications on the quality and rate of production, worse being fish mortality. Thus, based on this aspect and the increased disease rates, there has been increased reliance on antimicrobials within aquaculture worldwide, especially in countries where regulatory limits lack clear definition and monitoring.
Antimicrobials refer to substances which include antibiotics, food additives, antiseptics, disinfectants, and other substances that act against microorganisms [2], disinfectants being substances that inhibit multiplication of microbes on inanimate/nonliving objects or surfaces. Substances such as iodophors, formalin, metal hydroxides and oxides, hydrogen peroxide, and potassium permanganate are used extensively in aquaculture facilities for control and prevention of fish diseases [1, 3, 4]. According to Manyi-Loh et al. [5], aiming to respond to the proliferation of pathogen and disease rates in aquaculture systems, farmers are quickly turning to the use of antimicrobials. Antimicrobials are used in fish farming to control bacterial diseases, ectoparasites, fungal outbreaks, as well as to improve poor water quality, sterilize eggs and equipment, and control aquatic weeds and free-living molluscs [6]. According to Austin and Austin [1], antimicrobials may be administered in fish through oral (via medicated feed), bath, dip, flush, topical application, or injection.
However, a seemingly gray area exists in the use and utilization of antimicrobial in aquaculture, tending to contradict what is observed on the ground. Available studies tend to suggest that there is low usage of antimicrobials in pisciculture in most developed countries, while in some countries, the usage appeared to decline with time. This has lead Manyi-Loh et al. [5] to postulate that the apparent scarcity of information on the amount of antibiotic used in aquaculture stems from the fact that only a few countries monitor the quantity of antibiotics utilized, since large quantities of antimicrobials are used in aquaculture in some countries often without professional consultation or supervision. In addition, as they are being used in land animal husbandry and human medicine, some antimicrobials may end up in water bodies with fish. These two factors may potentiate development of drug resistance exhibited by microorganisms. Manyi-Loh et al. [5] elaborate that antimicrobial resistance (AMR) is a phenomenon that predates antibiotic clinic usage, as it is an ancient process. Antimicrobial resistance in bacterial pathogens is a worldwide public health concern. This is of concern since, albeit the vast advancement being made in the aquaculture sector in Kenya, AMR continues to remain a gray area, due to there being no precise and concrete valuation on the quantity and existence of AMR in the Kenya aquaculture sector. This gap is a detrimental aspect for the fish veterinarian, pisciculturists, fisheries managers, policy planners, and fish farmers. There is, therefore, avid need to bridge this gap through effective and elaborate surveys and studies on AMR, so as to generate sufficient data for intervention purposes. This study was carried out to determine antibiotic and disinfectant susceptibility patterns of bacteria isolated from farmed fish in Kirinyaga County, Kenya.
2. Materials and Methods
2.1. Study Area and Design
The isolates used in the present study were recovered from apparently healthy farmed fishes (Oreochromis niloticus, Clarias gariepinus, Carassius auratus, and Cyprinus carpio carpio) and source pond water in Kirinyaga County, Kenya. The study was cross-sectional where bacteria were isolated and identified by conventional bacteriological methods following Markey et al. [7] and Austin and Austin [1] and by use of Analytic Profile Index 20E (API 20E) kits. Antibiotic and disinfectant susceptibility profiles of 48 bacterial isolates (belonging to the genera Aeromonas, Proteus, Klebsiella, Citrobacter, Salmonella, Streptococcus, Pseudomonas, Escherichia, Serratia, and Micrococcus) were established using the Kirby–Bauer disc diffusion method and agar well diffusion technique, respectively. Members of genera Klebsiella, Salmonella, Escherichia, and Streptococcus were tested due to zoonotic potential; members of the genus Serratia, Micrococcus, Pseudomonas, and Aeromonas were tested owing to their high pathogenicity potential in fish, as well as some being of public health concern, while members of the genus Proteus were tested due to their high prevalence. Escherichia coli (25922), Pseudomonas aeruginosa (27853), and Staphylococcus aureus (25923) from the American type culture collection (ATCC) were used as quality control organisms.
2.2. Antibiotic Susceptibility Testing
The antibiotic susceptibility patterns for the 48 bacterial pathogens were established in vitro, following the Kirby–Bauer disc diffusion method on Mueller–Hinton agar (Oxoid®), as described by Hudzicki [8] with certain modifications. There are no registered antibiotic formulations for use in aquaculture in Kenya, and therefore, the choice was guided by different classes of drugs reported elsewhere to treat diseases in aquaculture facilities [1]. Moreover, these drugs are equally used in veterinary and human medicine therapy. Eight commercially available antibiotic disks (Himedia® OD 2/4) were used in the following concentrations: ampicillin (25 μg); tetracycline (25 μg); co-trimoxazole (25 μg); streptomycin (10 μg); kanamycin (30 μg); gentamicin (10 μg); sulphamethoxazole (200 μg); and chloramphenicol (30 μg).
The isolates and reference strains were inoculated on nutrient broth separately and incubated aerobically at 37°C. After overnight incubation, the bacterial suspension was vortexed and diluted to turbidity equivalent to that of 0.5 McFarland standards. The bacterial suspension was, then, spread onto the surface of the Mueller–Hinton agar (Oxoid) to make confluent growth. Antibiotic discs were immediately placed on the surface of the agar plate using forceps and incubated aerobically at 37°C for 16 hours. Inhibition zones for various isolates were measured and interpreted as sensitive, intermediate, or resistant according to the Clinical Laboratory Standards Institute (CLSI) [9–11].
2.3. Multiple Antibiotic Resistance Indices
The Multiple Antibiotic Resistance (MAR) indices of the study isolates against tested antibiotic were calculated using the formula MAR=a/bc , where “a” is the number of antibiotics to which the bacterial isolate was resistant, “b” is number of antibiotics tested, and “c” is the total number of isolates [12].
2.4. Disinfectant Susceptibility Testing
In order to determine the antibacterial profile of a disinfectant, the antibacterial susceptibility was performed by the agar well diffusion method on Mueller–Hinton agar as described by Schillinger and Lucke [14] and Njagi et al. [15], with certain modifications. Five (5) disinfectants available in the market were used at the concentration shown in Table 1.
Table 1.
Disinfectants and their concentrations used in the disinfectants efficacy assay for the study bacteria.
| Trade name | Chemical name | Active ingredient | Concentration |
|---|---|---|---|
| [1] Noro-cleanse® (Norbrook, Kenya) | Quaternary ammonium compound | 15% glutaraldehyde and 10% coco-benzyl-dimethyl-ammonium chloride | 1% |
| 1.5%∗ | |||
|
| |||
| [3] Formalin | Formalin | 40% formaldehyde | 1% |
| 2%∗∗ | |||
|
| |||
| [5] Hydrogen peroxide (Impact Chemicals, Kenya) | Hydrogen peroxide | 6% hydrogen peroxide | 1% |
| 3%∗∗ | |||
|
| |||
| [7] Sodium hypochlorite solution (Diarim, Kenya) | Sodium hypochlorite | 5% sodium hypochlorite | 1% |
| 2%∗ | |||
|
| |||
| [9] Novasept®(Navketan Research and Laboratories Pvt, India) | Iodine | 1% available iodine | 0.5% |
| 1%∗ | |||
Key: ∗User-recommended concentration by the manufacturer; ∗∗the concentrations are as recommended by Noga [16].
Owing to differences observed in phenotypic antibiotic susceptibility of isolates of the same genera, disinfectant analysis was carried out for all the 48 isolates against the 5 disinfectants. The bacteria suspensions were spread in the same way as for the antibiotic susceptibility testing. After spreading out, wells were punched using a sterile 6 mm diameter agar well-puncher. Dilutions of all the disinfectants used were prepared using sterile distilled water. Each disinfectant was dispensed separately, using a sterile micropipette, into different wells, using one Petri-dish per disinfectant. Each well was laden with 50 μl of the pertinent disinfectant concentration. The Petri-dishes were then incubated in an upright position at 37°C. After 16 hours of incubation, each inhibition zone was measured and recorded in millimetres (mm). The results were construed as follows: a radius of 0–5 mm from the edge of the well to the inhibition front was taken as no inhibition; a radius of 6–9 mm was taken as a moderate inhibition; a radius of 10–14 mm was considered a strong inhibition, and beyond 15 mm was taken as a very strong inhibition [17].
2.5. Data Analysis
The collected data were validated, entered, and stored in a Microsoft Excel® spreadsheet, which was also used to calculate means and proportions. Descriptive and inferential statistics were performed using Statistical Package for the Social Sciences (SPSS®), version 22.0. The overall antibiotic response of each antibiotic was calculated as the number of bacteria resistant or sensitive to antibiotics over total number of bacteria isolates tested. One-way ANOVA was used to compute antibiotic and disinfectant susceptibility. All the tests were carried out at a significance level of 0.05.
3. Results
3.1. Antibiotic Susceptibility/Resistance Testing
Results of susceptibility testing for the 8 tested antibiotics on the 48 bacterial isolates are tabulated in Table 2. From the table, the overall susceptibility rates for each antibiotic for all the bacterial isolates were the highest in gentamicin (100%, n = 48) and kanamycin (92%, n = 44). Also, the bacterial isolates showed highest resistance rates to co-trimoxazole (73%, n = 34) and ampicillin (65%, n = 31). The overall antibiotic response for ampicillin, co-trimoxazole, streptomycin, kanamycin, sulphamethoxazole, and chloramphenicol was statistically significant (p < 0.05). However overall antibiotic response for tetracycline was insignificant (p=0.086).
Table 2.
Antibiotic susceptibility patterns and overall antibiotic responses for the bacterial isolates, pegged on CLSI's inhibition zones interpretive criterion.
| Bacterial isolates | Pattern | Number of isolates susceptible or resistant, n (%), to the antibiotic agents tested | |||||||
|---|---|---|---|---|---|---|---|---|---|
| AMP | TET | COT | S | K | GEN | SX | C | ||
| A. hydrophila (N = 6) | S | 0 (0) | 5 (83) | 4 (67) | 6 (100) | 6 (100) | 6 (100) | 6 (100) | 6 (100) |
| R | 6 (100) | 1 (17) | 2 (33) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
|
| |||||||||
| Proteus spp. (N = 6) | S | 0 (0) | 2 (33) | 0 (0) | 6 (100) | 6 (100) | 6 (100) | 3 (50) | 3 (50) |
| R | 6 (100) | 4 (67) | 6 (100) | 0 (0) | 0 (0) | 0 (0) | 3 (50) | 3 (50) | |
|
| |||||||||
| K. pneuzmoniae (N = 7) | S | 6 (86) | 5 (71) | 5 (71) | 7 (100) | 7 (100) | 7 (100) | 7 (100) | 7 (100) |
| R | 1 (14) | 2 (29) | 2 (29) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
|
| |||||||||
| C. freundii (N = 6) | S | 5 (83) | 6 (100) | 5 (83) | 5 (83) | 6 (100) | 6 (100) | 6 (100) | 6 (100) |
| R | 1 (17) | 0 (0) | 1 (17) | 1 (17) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
|
| |||||||||
| Salmonella enteritidis (N = 4) | S | 2 (50) | 4 (100) | 0 (0) | 4 (100) | 4 (100) | 4 (100) | 4 (100) | 4 (100) |
| R | 2 (50) | 0 (0) | 4 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
|
| |||||||||
| Streptococcus spp. (N = 3) | S | 0 (0) | 2 (67) | 0 (0) | 0 (0) | 1 (33) | 3 (100) | 1 (33) | 3 (100) |
| R | 3 (100) | 1 (33) | 3 (100) | 3 (100) | 2 (67) | 0 (0) | 2 (67) | 0 (0) | |
|
| |||||||||
| P. aeruginosa (N = 5) | S | 0 (0) | 4 (80) | 0 (0) | 5 (100) | 5 (100) | 5 (100) | 0 (0) | 0 (0) |
| R | 5 (100) | 1 (20) | 5 (100) | 0 (0) | 0 (0) | 0 (0) | 5 (100) | 5 (100) | |
|
| |||||||||
| E. coli (N = 3) | S | 2 (67) | 3 (100) | 0 (0) | 3 (100) | 3 (100) | 3 (100) | 3 (100) | 3 (100) |
| R | 1 (33) | 0 (0) | 3 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
|
| |||||||||
| S. marcescens (N = 6) | S | 0 (0) | 6 (100) | 0 (0) | 4 (67) | 4 (67) | 6 (100) | 4 (67) | 6 (100) |
| R | 6 (100) | 0 (0) | 6 (100) | 2 (33) | 2 (33) | 0 (0) | 2 (33) | 0 (0) | |
|
| |||||||||
| Micrococcus spp. (N = 2) | S | 2 (100) | 2 (100) | 0 (0) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) |
| R | 0 (0) | 0 (0) | 2 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
|
| |||||||||
| Overall antibiotic response (N = 48) | S | 17 (35) | 39 (81) | 14 (27) | 42 (88) | 44 (92) | 48 (100) | 36 (75) | 40 (83) |
| R | 31 (65) | 9 (19) | 34 (73) | 6 (12) | 4 (8) | 0 (0) | 12 (25) | 8 (17) | |
S: susceptible, R: resistance, n: number of isolates, AMP: ampicillin, TET: tetracycline, COT: co-trimoxazole, S: streptomycin, K: kanamycin, GEN: gentamicin, SX: sulphamethoxazole, C: chloramphenicol.
Figure 1 shows the antibiotic susceptibility pattern of one of the isolates. From the figure, there is a clear distinction between resistant (no inhibition) and susceptible (inhibition/clear zones).
Figure 1.
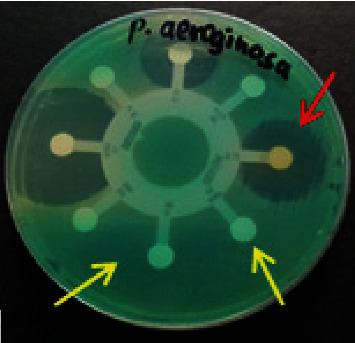
Antimicrobial susceptibility test results on the Mueller–Hinton agar culture plate showing susceptible (red arrow) and resistant (yellow arrow) zones of one of the isolates.
3.2. Multiple Drug Resistance and Multiple Antibiotic Resistance Indices of the Isolates
Table 3 shows that the MAR indices of the bacterial isolates ranged from 0.1 to 0.8. The MAR index values of all the bacterial isolates were higher than 0.2 except Micrococcus spp. which had an MAR index of 0.1. This implies that most of the bacterial isolates were from high-risk source pollution such as manure-fertilized ponds. From the table, it can be seen that all the isolates showed multidrug resistant patterns to at least three antibiotics except Micrococcus spp., Salmonella enteritidis, and Escherichia coli. Genera Proteus, Pseudomonas, Serratia, and Streptococcus showed the highest MDR pattern, in that order.
Table 3.
Multiple antibiotic resistance indices and multiple drug resistance patterns of bacterial isolates.
| Bacterial strains | Antibiotic(s) to which the bacterial isolate was resistant to | MDR pattern | MAR index |
|---|---|---|---|
| Micrococcus spp. | COT | No | 0.1 |
| Salmonella enteritidis | AMP and COT | No | 0.3 |
| Escherichia coli | AMP and COT | No | 0.3 |
| Aeromonas hydrophila | AMP, TET, and COT | Yes | 0.4 |
| Klebsiella pneumoniae | AMP, TET, and COT | Yes | 0.4 |
| Citrobacter freundii | AMP, COT, and S | Yes | 0.4 |
| Proteus spp. | AMP, TET, COT, SX, and C | Yes | 0.6 |
| Pseudomonas aeruginosa | AMP, TET, COT, SX, and C | Yes | 0.6 |
| Serratia marcescens | AMP, COT, S, K, and SX | Yes | 0.6 |
| Streptococcus spp. | AMP, TET, COT, S, K, and SX | Yes | 0.8 |
AMP: ampicillin, TET: tetracycline, COT: co-trimoxazole, S: streptomycin, K: kanamycin, GEN: gentamicin, SX: sulphamethoxazole, C: chloramphenicol, MDR: multiple drug resistance, MAR: multiple antibiotic resistance.
3.3. Disinfectant Susceptibility Profiles of Bacterial Isolates
Table 4 gives the mean radius of the inhibition zone from the edge of the well to the inhibition zone front of disinfectants tested against selected bacteria. From Table 4, it can be gleamed that 3% and 1% hydrogen peroxide demonstrated the largest inhibition zone among the study bacteria at 23 mm to 20 mm zones, respectively, showing the broadest spectrum of antibacterial activity utilized by the study bacteria. Other disinfectants at different concentration(s) produced inhibition zones among the study bacteria as follows: by 2% formalin (19 mm), 1% iodine (15 mm), 1% formalin (14 mm), 0.5% iodine (12 mm), and 1.5% quaternary ammonium compound (10 mm).
Table 4.
Mean radius ± SD of the inhibition zone from the edge of the well to the inhibition zone front of disinfectants tested against the study bacteria.
| Bacterial isolates | Sodium hypochlorite | Iodine | Hydrogen peroxide | Formalin | QAC | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1% | 2%∗ | 0.5% | 1%∗ | 1% | 3%∗∗ | 1% | 2%∗∗ | 1% | 1.5%∗ | |
| Aer. hydrophila | 1 ± 1.0 | 2 ± 0.5 | 5 ± 0.5 | 8 ± 0.5 | 17 ± 0.5 | 21 ± 0.5 | 10 ± 0.5 | 14 ± 1.0 | 3 ± 0.5 | 5 ± 0.0 |
| Proteus spp. | 0 ± 0.0 | 0 ± 0.0 | 5 ± 0.0 | 8 ± 2.0 | 19 ± 0.5 | 23 ± 0.5 | 11 ± 0.5 | 15 ± 0.5 | 3 ± 1.0 | 5 ± 1.0 |
| Kleb. pneumoniae | 0 ± 0.0 | 0 ± 0.0 | 3 ± 0.0 | 6 ± 2.0 | 15 ± 0.0 | 18 ± 0.0 | 9 ± 1.0 | 14 ± 1.0 | 4 ± 0.5 | 5 ± 0.5 |
| Citr. freundii | 1 ± 0.0 | 3 ± 0.0 | 5 ± 1.5 | 9 ± 3.0 | 16 ± 1.5 | 19 ± 2.0 | 12 ± 0.5 | 15 ± 0.0 | 4 ± 0.5 | 6 ± 0.5 |
| Salm. enteritidis | 0 ± 0.0 | 0 ± 0.0 | 5 ± 0.5 | 10 ± 0.5 | 15 ± 1.0 | 20 ± 1.0 | 12 ± 0.5 | 14 ± 0.5 | 3 ± 0.0 | 5 ± 0.5 |
| Streptococcus spp. | 1 ± 0.5 | 2 ± 1.5 | 5 ± 1.0 | 7 ± 1.5 | 17 ± 3.0 | 21 ± 1.0 | 12 ± 2.0 | 15 ± 2.0 | 4 ± 2.0 | 6 ± 1.5 |
| Escherichia coli | 1 ± 0.0 | 2.0 ± 0 | 3 ± 0.0 | 5 ± 0.5 | 15 ± 0.5 | 20 ± 0.5 | 14 ± 0.5 | 19 ± 0.5 | 3 ± 0.5 | 6 ± 0.5 |
| Ser. marcescens | 1 ± 0.5 | 2 ± 1.5 | 4 ± 0.5 | 7 ± 0.5 | 12 ± 0.0 | 18 ± 1.0 | 11 ± 0.5 | 14 ± 1.5 | 2 ± 0.0 | 4 ± 0.0 |
| Ps. aeruginosa. | 0 ± 0.0 | 2 ± 0.5 | 3 ± 0.0 | 6 ± 0.5 | 13 ± 0.5 | 15 ± 1.0 | 9 ± 0.5 | 11 ± 0.5 | 4 ± 0.5 | 7 ± 0.5 |
| Micrococcus spp. | 0 ± 0.0 | 0.0 ± 0 | 10 ± 0.5 | 12 ± 1.0 | 11 ± 0.5 | 17 ± 0.5 | 14 ± 0.5 | 17 ± 0.5 | 9 ± 0.0 | 10 ± 0 |
| Range | 0–2 | 0–4 | 3–12 | 4–15 | 10–20 | 14–23 | 8–14 | 10–19 | 2–9 | 4–10 |
| Mean | 1 ± 0.2 | 1 ± 0.3 | 4.9 ± 0.6 | 7.9 ± 0.7 | 15 ± 0.6 | 19 ± 0.5 | 11 ± 0.4 | 15 ± 0.5 | 4 ± 0.4 | 6 ± 0.4 |
∗User-recommended concentration by the manufacturer; ∗∗the concentrations are as recommended by Noga [16]; QAC- quaternary ammonium compound; Aer.- Aeromonas; Kleb.- Klebsiella; Citr.- Citrobacter; Salm.- Salmonella; Ser.- Serratia; Ps. -Pseudomonas.
Figure 2 shows disinfectant susceptibility patterns of study bacterial isolates against some disinfectants on Mueller–Hinton agar plates. The inhibition zone of the given disinfectant is shown by the clear areas.
Figure 2.
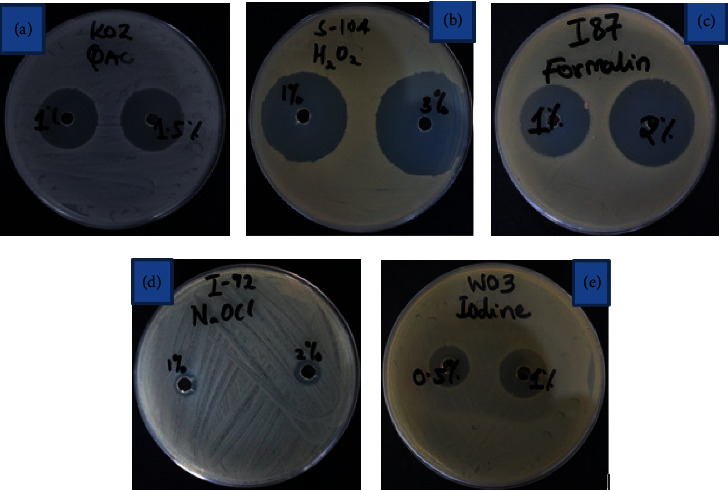
Antibacterial activity (clear areas) of various disinfectants to bacteria isolates (QAC (a), hydrogen peroxide (b), formalin (c), sodium hypochlorite (d), and iodine (e)).
Table 5 shows the extent of the inhibitory effect of the disinfectants tested on growth of test bacterial. Generally, most of the disinfectants showed an action of varying magnitudes against the bacterial isolates. The inhibitory activity of formalin and hydrogen peroxide at all concentrations was highly visible. The isolates were 100% sensitive to hydrogen peroxide and formalin at the recommended user-dilution. Iodine at user-recommended concentration showed moderate antibacterial activity. However, sodium hypochlorite at all concentrations showed little to no growth inhibition.
Table 5.
Inhibitory activity of disinfectants on bacterial isolates.
| Bacterial strains | Sodium hypochlorite | Iodine | Hydrogen peroxide | Formalin | QAC | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1% | 2%∗ | 0.5% | 1%∗ | 1% | 3%∗∗ | 1% | 2%∗∗ | 1% | 1.5%∗ | |
| Aer. hydrophila | _ | _ | _ | + | +++ | +++ | ++ | ++ | _ | _ |
| Proteus spp. | _ | _ | + | + | +++ | +++ | ++ | +++ | _ | _ |
| Kleb. pneumoniae | _ | _ | _ | + | +++ | +++ | + | ++ | _ | _ |
| Citrobacter freundii | _ | _ | _ | + | +++ | +++ | ++ | +++ | _ | + |
| Salm. enteritidis | _ | _ | _ | ++ | +++ | +++ | ++ | ++ | _ | _ |
| Streptococcus spp. | _ | _ | _ | + | +++ | +++ | ++ | +++ | _ | + |
| Escherichia coli | _ | _ | _ | _ | +++ | +++ | ++ | +++ | _ | + |
| Ser. marcescens | _ | _ | _ | + | ++ | +++ | ++ | ++ | _ | _ |
| Ps. aeruginosa | _ | _ | _ | _ | ++ | +++ | + | ++ | _ | ++ |
| Micrococcus spp. | _ | _ | + | ++ | ++ | +++ | ++ | +++ | + | ++ |
| Mean inhibition | _ | _ | _ | + | ++ | +++ | ++ | +++ | _ | + |
∗User-recommended concentration by the manufacturer; ∗∗the concentrations are as recommended by Noga [16]; QAC- quaternary ammonium compound; Aer.- Aeromonas; Kleb.- Klebsiella; Salm.- Salmonella; Ser.- Serratia; Ps.- Pseudomonas.
Inhibition zone interpretive criterion: - = No clear zone (0–5 mm); + = Moderate clear zone (6–9 mm); ++ = Strong clear zone (10–14 mm); and +++ = Very strong clear zone (≥ 15 mm).
4. Discussion
4.1. Antibiotic Resistance and Multiple Antibiotic Resistance Indexing
Antimicrobial resistance (AMR) is a worldwide public health concern that has drawn attention in the recent time. Existence of antibiotic resistance amongst these bacterial strains may have public health concern in fish consumers and handlers (fish harvesters and retailers). This is so especially if these resistant strains are transferred to humans. While some of these bacteria are ubiquitous to aquatic environments, some are exogenous or shed from the terrestrial environment into water bodies. Pond fertilization using livestock manure may also facilitate transfer of antibiotic-resistant bacteria from animals into aquatic life via soil and/or water [5]. This, therefore, calls for a holistic one health approach in curbing AMR scourge.
As corroborated by several other sources, AMR requires proper evaluation, monitoring, and assessment to articulate necessary legislative and regulatory measures [18]. As a potentially growing fishery nation, Kenya is yet to embark on formulating relevant regulations and legislations on the use of antibiotics aquaculture. It will be prudent to do so, so as to set ground for greater development in the Kenyan aquaculture sector similar to its international counterparts who are already on this path.
All the bacterial isolates showed multiple resistance to various drugs tested except for Micrococcus spp., Salmonella enteritidis, and Escherichia coli. However, all bacterial isolates were susceptible to gentamicin, and this is in agreement with the findings of Gufe et al. [13] and Karimi [19]. The resistance levels to kanamycin, streptomycin, chloramphenicol, and tetracycline were relatively low (below 20%). Indeed, cot-trimoxazole had highest levels of resistance; this in contrast with the findings of Newaj‐Fyzul et al. [20] who reported a relatively low resistance of 6.4%. Resistance against ampicillin was markedly high (65%); it is comparable to studies by Karimi [19] and Newaj‐Fyzul et al. [20].
Multiple antibiotic resistance indexing has been an effective and efficient tool in tracing bacteria exposure [21]. The MAR index values of all the bacterial isolates were higher than 0.2 except for the Micrococcus spp. which had an MAR index of 0.1. When the use of antibiotics is rare or underdosed for an animal treatment, the MAR value is equal to or less than 0.2. However, the MAR index value is greater than 0.2 in cases of high risk exposure with frequent therapeutic use of antibiotics in an animal. These findings further suggest the need to explore the role of pond fertilization using animal manure as a probable high-risk contaminant. Furthermore, the increased indiscriminate use and misuse of antibiotics (without veterinary supervision or consultation) in the livestock [22] and aquaculture sector is a major contributor to AMR and may have given rise to resistant bacteria. Since diseased fish are often treated as groups and not as individuals, this tends to expose both the healthy and sick fish in that population to the antimicrobials used.
4.2. Disinfectant Susceptibility Testing
Disinfection is applied as a common health management tool in aquaculture facilities [4], and the applications range from egg disinfection [23] to system disinfection [24]. Disinfectants are often used to inhibit fungal and bacterial growth and parasitic load in systems where preventive biosecurity measures are insufficient [25, 26], or it may be used for disease eradication (stamping out) efforts [4].
Precaution is crucial in the usage and dealing of disinfectants. The choice of disinfectant to use in an aquaculture establishment depends on the efficacy, volume required, cost, toxicity, and potential effluent concerns. The efficacy of liquid disinfectants is affected by the temperature, contact time, pH, concentration and presence of suspended solids, and organic and inorganic constituents [27].
Despite all the disinfectants tested being not registered and approved for use in aquaculture establishments in Kenya, results revealed the antibacterial potential of all 5 disinfectants tested. Among the disinfectants evaluated in this study, hydrogen peroxide and formalin were the most effective, with respect to the study bacteria, followed by iodine and quaternary ammonium compound (QAC). Hydrogen peroxide and formalin were effective at dilutions lower than what Noga [16] recommended while iodine and quaternary ammonium compound were only effective at the user-recommended concentration. Sodium hypochlorite showed slight to no effect at all the concentrations tested. This finding is in agreement with that of a study conducted by Njagi et al. [15] on sensitivity of the Listeria spp. recovered from indigenous chickens. Hydrogen peroxide (3%) showed the largest zone of inhibition, while sodium hypochlorite showed the smallest zone, with some isolates showing no inhibition (i.e., were resistant). Hydrogen peroxide does not affect the fish negatively and appears to improve water quality [28]. The use of hydrogen peroxide, especially in pond water disinfection, may be advantageous as its decomposition by-products (water and oxygen) are harmless and, therefore, environmental-friendly.
Formalin is one of the most used disinfectants in aquaculture to eliminate infectious agents; however, it may be responsible for adverse effects on fish and water quality [29]. Moreover, its carcinogenic potential to humans limits its use in fish intended for human consumption [30].
Quaternary ammonium compounds are promptly deactivated by organic matter and are usually ineffective against organisms such as Pseudomonas, Proteus, and other Gram-negative bacteria [27]. Chlorine compounds call for a contact time of 10–30 minutes to be potent and are highly corrosive and toxic [27].
5. Conclusions
Concomitant to other issues highlighted in this paper, the authors have demonstrated the existence of resistance among the fish bacterial isolates against antibiotics which might pose a challenge in disease control, as well as being hazardous to health of consumers and fish handlers. The authors have also demonstrated that all bacterial isolates were susceptible to most disinfectants tested. The potency of these disinfectants gives a reprieve in the wake of failing antibiotics.
5.1. Recommendations
To the fish veterinarians, managers, and farmers, it is recommended that, before making any therapeutic decision, antibiotic/disinfectant susceptibility testing is carried out, so as to use the effective antibiotic/disinfectant. There is also a need to strengthen government and industry institutions for aquatic animal health through development and enforcement of regulations on the use of antimicrobials. In addition, the institutions may consider registering and approving some of the chemical disinfectants for use in aquaculture establishments for the control of bacterial pathogens.
Acknowledgments
This research work was financed by the Norwegian Agency for Development Cooperation (NORAD) through the Capacity Building for Training and Research in Aquatic and Environmental Health in Eastern and Southern Africa (TRAHESA) project (Grant TAN 13/0027).
Data Availability
Data associated with this research article are available on request from the corresponding author.
Conflicts of Interest
There are no conflicts of interest.
References
- 1.Austin B., Austin D. A. Bacterial Fish Pathogens: Disease of Farmed and Wild Fish. 6. New York, NY, USA: Springer; 2016. p. p. 732. [Google Scholar]
- 2.MacDonnell G., Russell D. Antiseptics and disinfectants: activity, action and resistance. Clinical Microbiology Reviews. 1999;12(1):147–179. doi: 10.1128/cmr.12.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Torgersen Y., Håstein T. Disinfection in aquaculture. Revue Scientifique et Technique de l’OIE. 1995;14(2):419–434. doi: 10.20506/rst.14.2.845. [DOI] [PubMed] [Google Scholar]
- 4.World Organisation for Animal Health (OIE) Manual of Diagnostic Tests for Aquatic Animals. Paris, France: Office international des epizooties; 2009. Methods for disinfection of aquaculture establishments; p. p. 366. [Google Scholar]
- 5.Manyi-Loh C., Mamphweli S., Meyer E., Okoh A. Antibiotic use in agriculture and its consequential resistance in environmental sources: potential public health implications. Molecules. 2018;23(4):p. 795. doi: 10.3390/molecules23040795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodgers C. J., Furones M. D. Antimicrobial agents in aquaculture: practice, needs and issues. Options Méditerranéennes. 2009;86:41–59. [Google Scholar]
- 7.Markey B., Leonard F., Archambault M., Cullinane A., Maguire D. Clinical Veterinary Microbiology. 2nd. New York, NY, USA: Oxford University Press; 2013. [Google Scholar]
- 8.Hudzicki J. Kirby-Bauer Disk Diffusion Susceptibility Test Protocol. Washington, DC, USA: American Society for Microbiology (ASM); 2009. p. 23. [Google Scholar]
- 9.CLSI. Methods for Dilution of Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically: Approved Standard M7-A7. Wayne, PA, USA: Clinical and Laboratory Standards Institute; 2006. p. p. 88pp. [Google Scholar]
- 10.CLSI. Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria: CLSI Guideline M45. Wayne, PA, USA: Clinical and Laboratory Standards Institute; 2015. p. p. 19pp. [Google Scholar]
- 11.CLSI. Performance Standards for Antimicrobial Susceptibility Testing: CLSI Supplement M100S. Wayne, PA, USA: Clinical and Laboratory Standards Institute; 2016. p. p. 256pp. [Google Scholar]
- 12.Krumperman P. H. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Applied and Environmental Microbiology. 1983;46(1):165–170. doi: 10.1128/aem.46.1.165-170.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gufe C., Canaan Hodobo T., Mbonjani B., Majonga O., et al. Antimicrobial profiling of bacteria isolated from fish sold at informal market in Mufakose, Zimbabwe. International Journal of Microbiology. 2019;2019:7. doi: 10.1155/2019/8759636.8759636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schillinger U., Lücke F. K. Antibacterial activity of Lactobacillus sake isolated from meat. Applied and Environmental Microbiology. 1989;55(8):1901–1906. doi: 10.1128/aem.55.8.1901-1906.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Njagi L. W., Mbuthia P. G., Bebora L. C., Nyaga P. N., Minga U., Olsen J. E. Sensitivity of Listeria species, recovered from indigenous chickens to antibiotics and disinfectants. East African Medical Journal. 2004;81(10):534–537. doi: 10.4314/eamj.v81i10.9237. [DOI] [PubMed] [Google Scholar]
- 16.Noga E. J. Fish Disease: Diagnosis and Treatment. 2nd. Hoboken, NJ, USA: Wiley Blackwell; 2010. [Google Scholar]
- 17.Liasi S. A., Azmi T. I., Hassan M. D., Shuhaimi M., Rosfarizan M., Ariff A. B. Antimicrobial activity and antibiotic sensitivity of three isolates of lactic acid bacteria from fermented fish product. Malaysian Journal of Microbiology. 2009;5(1):33–37. [Google Scholar]
- 18.World Health Organization. Monitoring and evaluation of the global action plan on antimicrobial resistance: framework and recommended indicators. Vol. 43. Geneva, Switzerland: World Health Organization; 2019. [Google Scholar]
- 19.Karimi R. D. Nairobi, Kenya: Kenyatta University; 2015. The bacterial flora of tilapia (oreochromis niloticus) and catfish (clarias gariepinus) from earthen ponds in sagana fish farm and masinga dam. [Google Scholar]
- 20.Newaj‐Fyzul A., Mutani A., Ramsubhag A., Adesiyun A. Prevalence of bacterial pathogens and their anti‐microbial resistance in tilapia and their pond water in Trinidad. Zoonoses and Public Health. 2008;55(4):206–213. doi: 10.1111/j.1863-2378.2007.01098.x. [DOI] [PubMed] [Google Scholar]
- 21.Osundiya O. O., Oladele R. O., Oduyebo O. O. Multiple antibiotic resistance (MAR) indices of Pseudomonas and Klebsiella species isolates in Lagos University Teaching Hospital. African Journal of Clinical and Experimental Microbiology. 2013;14(3):164–168. doi: 10.4314/ajcem.v14i3.8. [DOI] [Google Scholar]
- 22.Katakweba A. A. S., Mtambo M. M. A., Olsen J. E., Muhairwa A. P. Awareness of human health risks associated with the use of antibiotics among livestock keepers and factors that contribute to selection of antibiotic resistance bacteria within livestock in Tanzania. Livestock Research for Rural Development. 2012;24(10):p. 170. [Google Scholar]
- 23.Wagner E. J., Arndt R. E., Billman E. J., Forest A., Cavender W. Comparison of the efficacy of iodine, formalin, salt, and hydrogen peroxide for control of external bacteria on rainbow trout eggs. North American Journal of Aquaculture. 2008;70(2):118–127. doi: 10.1577/a06-068.1. [DOI] [Google Scholar]
- 24.Waldrop T., Gearheart M., Good C. Disinfecting recirculating aquaculture systems: post-harvest cleaning. Hatchery International. 2009;3:38–39. [Google Scholar]
- 25.Schmidt L. J., Gaikowski M. P., Gingerich W. H. Environmental assessment for the use of hydrogen peroxide in aquaculture for treating external fungal and bacterial diseases of cultured fish and fish eggs. 2006:p. 180. USGS Report.
- 26.Jørgensen T. R., Larsen T. B., Buchmann K. Parasite infections in recirculated rainbow trout (Oncorhynchus mykiss) farms. Aquaculture. 2009;289(1-2):91–94. [Google Scholar]
- 27.Mainous M. E., Smith S. A. Efficacy of common disinfectants againstMycobacterium marinum. Journal of Aquatic Animal Health. 2005;17(3):284–288. doi: 10.1577/h04-051.1. [DOI] [Google Scholar]
- 28.Pedersen L.-F., Pedersen P. B. Hydrogen peroxide application to a commercial recirculating aquaculture system. Aquacultural Engineering. 2012;46:40–46. doi: 10.1016/j.aquaeng.2011.11.001. [DOI] [Google Scholar]
- 29.Leal J. F., Neves M. G. P. M. S., Santos E. B. H., Esteves V. I. Use of formalin in intensive aquaculture: properties, application and effects on fish and water quality. Reviews in Aquaculture. 2016;10(2):281–295. doi: 10.1111/raq.12160. [DOI] [Google Scholar]
- 30.Duong A., Steinmaus C., McHale C. M., Vaughan C. P., Zhang L. Reproductive and developmental toxicity of formaldehyde: a systematic review. Mutation Research/Reviews in Mutation Research. 2011;728(3):118–138. doi: 10.1016/j.mrrev.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data associated with this research article are available on request from the corresponding author.


