Abstract
Objectives
We aimed to develop and validate a risk score to predict severe respiratory failure (SRF) among patients hospitalized with coronavirus disease-2019 (COVID-19).
Methods
We performed a multicentre cohort study among hospitalized (>24 hours) patients diagnosed with COVID-19 from 22 February to 3 April 2020, at 11 Italian hospitals. Patients were divided into derivation and validation cohorts according to random sorting of hospitals. SRF was assessed from admission to hospital discharge and was defined as: Spo2 <93% with 100% Fio2, respiratory rate >30 breaths/min or respiratory distress. Multivariable logistic regression models were built to identify predictors of SRF, β-coefficients were used to develop a risk score. Trial Registration NCT04316949.
Results
We analysed 1113 patients (644 derivation, 469 validation cohort). Mean (±SD) age was 65.7 (±15) years, 704 (63.3%) were male. SRF occurred in 189/644 (29%) and 187/469 (40%) patients in the derivation and validation cohorts, respectively. At multivariate analysis, risk factors for SRF in the derivation cohort assessed at hospitalization were age ≥70 years (OR 2.74; 95% CI 1.66–4.50), obesity (OR 4.62; 95% CI 2.78–7.70), body temperature ≥38°C (OR 1.73; 95% CI 1.30–2.29), respiratory rate ≥22 breaths/min (OR 3.75; 95% CI 2.01–7.01), lymphocytes ≤900 cells/mm3 (OR 2.69; 95% CI 1.60–4.51), creatinine ≥1 mg/dL (OR 2.38; 95% CI 1.59–3.56), C-reactive protein ≥10 mg/dL (OR 5.91; 95% CI 4.88–7.17) and lactate dehydrogenase ≥350 IU/L (OR 2.39; 95% CI 1.11–5.11). Assigning points to each variable, an individual risk score (PREDI-CO score) was obtained. Area under the receiver-operator curve was 0.89 (0.86–0.92). At a score of >3, sensitivity, specificity, and positive and negative predictive values were 71.6% (65%–79%), 89.1% (86%–92%), 74% (67%–80%) and 89% (85%–91%), respectively. PREDI-CO score showed similar prognostic ability in the validation cohort: area under the receiver-operator curve 0.85 (0.81–0.88). At a score of >3, sensitivity, specificity, and positive and negative predictive values were 80% (73%–85%), 76% (70%–81%), 69% (60%–74%) and 85% (80%–89%), respectively.
Conclusion
PREDI-CO score can be useful to allocate resources and prioritize treatments during the COVID-19 pandemic.
Keywords: Age, Coronavirus disease 2019, C-reactive proteine, Lactate dehydrogenase, Obesity, Prognostic tool, Severe acute respiratory syndrome coronavirus 2, Severe respiratory failure
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) -associated coronavirus disease 2019 (COVID-19) has gripped the world in a pandemic, challenging its culture, economy and healthcare system. The virus was first reported in China in December 2019 and has subsequently spread worldwide.
The clinical spectrum of COVID-19 is broad with the majority of infected individuals experiencing only mild or subclinical illness, especially in the early phase of disease [1]. However, approximately 14%–30% of hospitalized patients diagnosed with COVID-19 develop a severe respiratory failure (SRF) requiring intensive care [[2], [3], [4]].
To date, no therapy has proven effective, so supportive care aimed to protect multi-organ function represents the main resource to reduce mortality [5]. The capacity of the system is limited, prompting the need of rationing decisions [6], but a number of promising innovative drugs and treatment strategies are under investigation [7]. We deemed that an early identification of patients at risk of developing SRF could support the planning of resources and help to set up organizational and clinical interventions, including early pharmacological treatment to prevent admission to the intensive care unit.
The objectives of the study were therefore (a) develop a risk model to identify individuals at high risk of developing SRF on hospital admission using a cohort of hospitalized patients with microbiologically confirmed diagnosis of COVID-19; and (b) to validate this risk model in an external multicentre cohort.
Methods
Design and setting
We performed a retrospective multicentre cohort study of prospectively collected data from patients with laboratory-confirmed SARS-CoV-2 infection, hospitalized from 22 February through 3 April, 2020. Last follow-up date was 23 April 2020.
Eleven hospitals from four Italian regions, including four tertiary teaching hospitals, five non-teaching tertiary hospitals and two secondary hospitals, participated in the study (see Supplementary material, Fig. S1).
Diagnostic testing for COVID-19 and hospitalization were performed according to local policy and clinical judgement, and were not dictated by a study protocol. The local microbiology laboratory information and management systems were used to identify patients. Clinical charts and hospital electronic records were used as data sources. De-identified data were collected and managed using REDCap electronic data capture tools, Alma Mater University of Bologna [8,9].
The study was approved by the Ethics Committee of the promoting centre (Comitato Etico Indipendente di Area Vasta Emilia Centro, n.283/2020/Oss/AOUBo). A waiver of informed consent was granted by the Ethics Committee due to safety risk. The study protocol was registered on clinicaltrials.gov with the number NCT04316949.
Participants
All consecutive adults (≥18 years) diagnosed with SARS-CoV-2 infection during the study period were included.
Exclusion criteria were hospital discharge within 24 hours of admission to Emergency Department and occurrence of SRF within 24 hours of hospitalization.
Participants were divided into two cohorts: the derivation cohort consisted of patients admitted to hospitals C, D(a, c) and I, the validation cohort consisted of patients admitted to hospitals A, B, D(b), E, F, G and H (see Supplementary material, Fig. S1). Hospitals were sorted randomly and assigned initially to the derivation cohort. Once 50% of participants with a new assignation was reached, the remaining centres were assigned to the validation cohort.
Variables and definitions
Microbiological diagnosis of SARS-CoV-2 infection was defined as a positive RT-PCR test on nasopharyngeal swabs.
The end-point variable was occurrence of SRF. Occurrence SRF was assessed through review of the collected data from admission to hospital discharge by a blinded investigator (ST). SRF was defined according to WHO criteria as: Spo 2 <93% with 100% Fio 2 (reservoir mask or continuous positive airway pressure ventilation or other non-invasive ventilation), respiratory rate >30 breaths/minute, or respiratory distress [10].
Exposure variables were assessed at hospital admission and included: age, older age (>70 years), sex, body mass index, being obese (body mass index >30 kg/m2). Underlying conditions were recorded according to the Charlson co-morbidity index [11]. Hypertension was defined as history of permanent increase of systolic blood pressure over 140 mmHg, and a diastolic increase to more than 90 mmHg. Immunosuppression included neutropenia (neutrophil count <500/mm3), solid organ transplantation, haematopoietic stem cell transplantation, corticosteroid therapy at a dosage higher then or equivalent to prednisone 16 mg/day ≥15 days, uncontrolled human immunodeficiency virus infection (<200 CD4/mm3). Regarding the SARS-CoV-2 infection, symptoms at onset and hospitalization, vital signs and laboratory tests were collected. Severity of illness at hospitalization was recorded according to sequential organ failure assessment (SOFA) score, quickSOFA (qSOFA), CURB-65 score and Modified Early Warning Score (MEWS).
End-point variables were assessed from hospital admission to discharge. In addition to SRF, we collected in-hospital all-cause mortality and date of hospital discharge.
Microbiological testing
The presence of SARS-CoV-2 was detected by RT-PCR assay. Briefly, UTM-RT swab specimens (Copan, Brescia, Italy) were immediately tested or stored at 4°C until processed, no more than 48 hours. Total genomic DNA/RNA was extracted from 280 μL of the clinical sample by Nuclisens EasyMag (BioMérieux, Marcy l’Étoile, France) following the manufacturer's instructions. Detection of SARS-CoV-2 was performed by real time RT-PCR following the WHO and/or CDC protocol in a QuantStudio S5 Real-time PCR system (ThermoFisher, Waltham, MA, USA). Microbiological analysis was not performed in a centralized laboratory.
Study size
For the sample size calculation we followed recent recommendations from Riley et al. [12]. We aimed to enroll at least 370 patients in the derivation cohort, with an expected number of events of 148 (an expected 40% rate, based on preliminary raw observations) and a maximum eight binary variables in the model, using the pmsampsize procedure in Stata 10 [12]. For the validation cohort, we aimed for a similar sample size.
Statistical analysis
For descriptive analysis, categorical variables are presented as counts and percentages. Continuous variables as mean and standard deviation if normally distributed or as median and interquartile range (IQR) if non-normally distributed.
For group comparison, Student's t test, Mann–Whitney U test and analysis of variance, or Kruskal–Wallis test were used for normally distributed quantitative variables, skewed distributed quantitative variables and more than two groups, respectively. Pearson's χ2 test (Fisher exact test where appropriate) for categorical variables. Shapiro Wilk's and Kolmogorov–Smirnov test, as well as visual methods, were applied to test for normality.
To develop and validate the score, analyses were initially performed on the derivation cohort and repeated identically in the validation cohort.
Univariate and multivariate mixed logistic regression models were performed to investigate risk factors for SRF. Variables were included in the multivariable model according the following strategy: clinically relevant variables, significance at the univariable analysis (p < 0.10), lack of co-linearity (in case of co-linearity, the model with lower Akaike Information Criterion was chosen), missing data in <10% of cases (i.e. we performed a complete case analysis). Overall goodness of fit was analysed by Akaike's Information Criteria and Nagelkerke's R2. Discrimination of the model was assessed by receiver-operator characteristics (ROC) curve of the predicted probability, Brier score and Somers' D. Calibration of the model was assessed by comparing predicted probability with actual probability of SRF in deciles of risk. Cluster-robust variance was used, to take into account within-hospital correlation.
To develop the risk score (PREDI-CO score), variables in the multivariate logistic regression model regardless of their significance were assigned a point value corresponding to the β-coefficient (fixed effects) rounded to the nearest integer; the total score was obtained by summation of the individual variables scores.
The discrimination of PREDI-CO score towards SRF was then analysed by non-parametric analysis of ROC curve under covariates, using bootstrap (1000 replications), with clustering per hospital. An optimal cut-point was then assigned using the Youden's J statistic, and performance characteristics at the cut-point (sensitivity, specificity, positive and negative likelihood, diagnostic accuracy, positive and negative predictive values) were calculated with the corresponding 95% CI.
In the validation cohort, the slope and intercept of the linear predictor were also assessed. The results of multivariable analysis in the validation cohort were not used to change the model obtained in the derivation cohort.
All statistical tests were two-sided. Stata computer software version 16.0 (Stata Corp., College Station, TX, USA) was used for statistical analysis.
Results
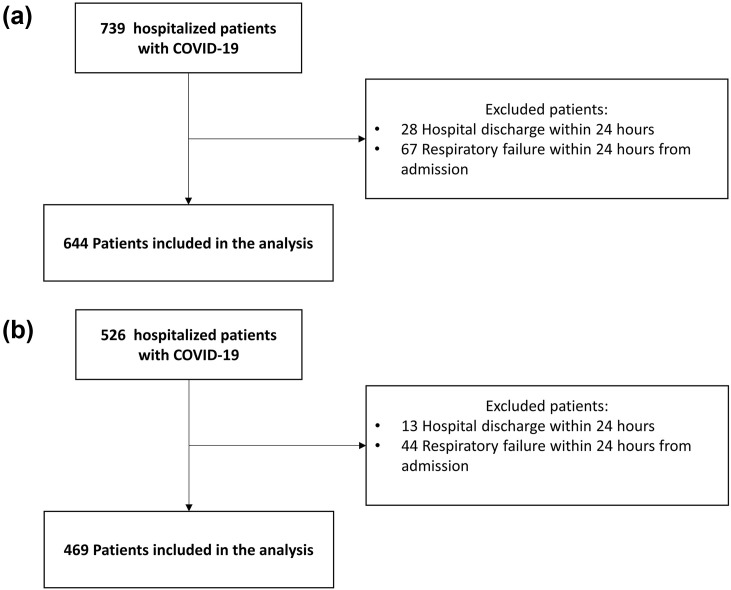
The initial population consisted of 1265 individuals: 739 in the derivation cohort and 526 in the validation cohort. One-hundred and fifty-two individuals were excluded according to eligibility criteria. Of the 1113 participants analysed: 644 were in the derivation cohort and 469 in the validation cohort (Fig. 1 ). The median number of patients included per hospital was 40 (IQR 11–84, range 4–384).
Fig. 1.
Study flow-chart: derivation cohort (a) and validation cohort (b).
The mean age of participants was 65.7 ± 15 years, and 704 (63.3%) were male. The median time from onset of symptoms to hospital admission was 6 (IQR 3–9) days. The two cohorts were different in several patient characteristics (Table 1 ).
Table 1.
Comparison of patients in derivation and validation cohort
| Overall cohort ( n = 1113) | Derivation cohort (n = 644) | Validation (n = 469) | p | |
|---|---|---|---|---|
| Demographics | ||||
| Age (years), mean (±SD) | 65.7 (±15.2) | 63.7 (±15.6) | 68.5 (±14.1) | <0.001 |
| Sex, male | 704 (63.3) | 376 (58.4) | 328 (69.9) | <0.001 |
| Underlying diseases | ||||
| Obesity | 196 (17.6) | 122 (18.9) | 74 (15.8) | 0.003 |
| BMI (kg/m2), median (IQR) | 26 (24–29) | 25 (23–29) | 26.1 (24–29) | 0.03 |
| Hypertension | 579 (52) | 321(49.8) | 258 (55) | 0.20 |
| Diabetes mellitus | 60 (5.4) | 37 (5.7) | 23 (4.9) | 0.04 |
| Coronary disease | 83 (7.5) | 56 (8.7) | 27 (5.8) | 0.08 |
| Congestive heart failure | 73 (6.6) | 32 (5) | 41 (8.7) | 0.014 |
| Cerebrovascular disease | 93 (8.4) | 44 (6.8) | 49 (10.5) | 0.04 |
| Peripheral vascular disease | 114 (10.2) | 38 (5.9) | 76 (16.2) | <0.001 |
| Chronic kidney disease | 115 (10.3) | 61 (9.5) | 54 (11.5) | 0.3 |
| COPD | 113 (10.2) | 58 (9) | 55 (11.7) | 0.16 |
| ESLD | 25 (2.3) | 11 (1.7) | 14 (3) | 0.22 |
| Immunosuppression | 42 (3.8) | 21 (3.3) | 21 (4.5) | <0.001 |
| Charlson index, median (IQR) | 3.3 (1–5) | 3.1 (1–5) | 3.7 (2–5) | <0.001 |
| Symptoms at onset | ||||
| Fever ≥38°C | 597 (53.6) | 332 (51.6) | 265 (56.5) | 0.03 |
| Cough | 635 (57.1) | 380 (59) | 255 (54.4) | 0.06 |
| Dyspnoea | 381 (34.2) | 241 (37.4) | 140 (29.9) | 0.007 |
| Symptoms at hospitalization | ||||
| Fever ≥38°C | 435 (39.1) | 248 (38.5) | 187 (39.9) | 0.47 |
| Cough | 609 (54.3) | 376 (58.4) | 233 (49.7) | <0.001 |
| Dyspnoea | 470 (42.2) | 256 (39.8) | 214 (45.6) | 0.03 |
| Vital signs at hospitalization | ||||
| GCS, median (IQR) | 15 (15–15) | 15 (15–15) | 15 (15–15) | 0.54 |
| MAP, median (IQR) | 90 (83–98) | 90 (83–97) | 90 (83–98) | 0.59 |
| PR, median (IQR) | 85 (75–95) | 85 (75–95) | 86 (76–95) | 0.31 |
| RR, median (IQR) | 20 (16–24) | 20 (16–24) | 20 (18–24) | 0.002 |
| Sato2 on ambient air, median (IQR) | 95.4 (93–97) | 96.5 (94–98) | 94 (92–96) | <0.001 |
| Laboratory tests at hospitalization | ||||
| Lymphocytes (109/L) median (IQR) | 0.97 (0.7–1.3) | 1.06 (0.79–1.4) | 0.89 (0.63–1.2) | <0.001 |
| CRP (mg/dL), median (IQR) | 5.2 (2.2–10.6) | 5 (2.1–9.8) | 5.6 (2.4–11) | 0.03 |
| LDH (IU/L), median (IQR) | 287 (224–391) | 271 (214–356) | 316 (245–414) | <0.001 |
| Treatments | ||||
| Hydroxychloroquine | 896 (80) | 477 (74) | 419 (89) | <0.001 |
| Lopinavir/ritonavir | 341 (31) | 154 (24) | 187 (40) | <0.001 |
| Darunavir/ritonavir | 251 (22) | 9 (1) | 242 (52) | <0.001 |
| Darunavir/cobicistat | 31 (3) | 14 (2) | 17 (4) | 0.87 |
| LMWH | 357 (32) | 231 (36) | 126 (27) | <0.001 |
| Tociluzumab | 129 (12) | 87 (13) | 42 (9) | 0.23 |
| Outcome | ||||
| ICU admission | 139 (12) | 71 (11) | 68 (15) | <0.001 |
| In-hospital mortality | 218 (19) | 102 (15) | 116 (25) | <0.001 |
Abbreviations: BMI, body mass index; COPD, chronic obstructive pulmonary disease; CRP, C-reactive protein; ESLD, end-stage liver disease; GCS, Glasgow coma scale; HRCT, high-resolution computed tomography; IQR, interquartile range; IU, international units; LDH, lactate dehydrogenase; MAP, mean arterial pressure; PR, pulse rate.
All values given are n (%) unless otherwise stated.
Three-hundred and seventy-six individuals (33%) developed SRF after ≥24 hours of admission. Median time to SRF in this group was 4 (IQR 2–7) days from hospital admission and 10 (7–13) days from onset of symptoms. The rates of SRF were 29% (189/644) and 40% (187/469) in the derivation and validation cohorts, respectively.
There were several differences between individuals with and without SRF in the derivation (Table 2 ) and validation (Table 3 ) cohorts.
Table 2.
Univariate analysis for severe respiratory failure among patients with SARS-CoV-2 pneumonia: derivation cohort
| Cases with available data | Severe respiratory failure (n = 189) | No severe respiratory failure (n = 455) | OR (95% CI) | |
|---|---|---|---|---|
| Demographics | ||||
| Age (years), mean (±SD) | 644 | 72.2 (±13.9) | 60.1 (±14.8) | 1.06 (1.045–1.073)a |
| Sex, male | 644 | 108 (57) | 268 (59) | 0.93 (0.66–1.31) |
| Underlying diseases | ||||
| Obesity | 633 | 76 (40) | 46 (10) | 6.09 (3.99–9.3) |
| BMI (kg/m2), median (IQR) | 393 | 28.3 (25–31) | 25.9 (23–27) | 1.14 (1.085–1.21)a |
| Hypertension | 636 | 126 (67) | 195 (42) | 2.75 (1.92–3.93) |
| Diabetes mellitus | 643 | 18 (9) | 19 (4) | 2.11 (1.04–4.3) |
| Coronary artery disease | 644 | 25 (13) | 31 (6) | 2.09 (1.2–3.64) |
| Congestive heart failure | 644 | 16 (8) | 16 (3) | 2.54 (1.2–5.2) |
| Cerebrovascular disease | 644 | 30 (18) | 14 (3) | 5.94 (3.07–11.5) |
| Peripheral vascular disease | 644 | 19 (10) | 16 (3) | 2.57 (1.33–4.96) |
| Chronic kidney disease (moderate to severe) | 644 | 20 (11) | 41 (9) | 1.2 (0.68–2.1) |
| COPD | 644 | 32 (16) | 26 (6) | 3.36 (1.94–5.8) |
| Immunosuppression | 618 | 9 (5) | 12 (3) | 1.98 (0.82–4.79) |
| Charlson index (median, IQR) | 588 | 4.4 (2–6) | 2.5 (1–4) | 1.32 (1.23–1.42)a |
| Symptoms at onset | ||||
| Fever ≥38°C | 626 | 96 (51) | 236 (51) | 0.96 (0.57–1.62) |
| Cough | 629 | 98 (52) | 282 (62) | 0.69 (0.49–0.99) |
| Dyspnoea | 630 | 93 (49) | 148 (32) | 2.09 (1.47–2.96) |
| Time to hospital admission (days), median (IQR) | 560 | 6 (3–9) | 6 (3–8) | 0.95 (0.93–0.97)a |
| Symptoms at hospitalization | ||||
| Fever ≥38°C | 637 | 98 (52) | 150 (33) | 2.23 (1.58–3.17) |
| Cough | 635 | 93 (49) | 283 (62) | 0.59 (0.42–0.83) |
| Dyspnoea | 636 | 108 (57) | 148 (32) | 2.83 (1.99–4.02) |
| Vital signs at hospitalization | ||||
| GCS (median, IQR) | 597 | 15 (15–15) | 15 (15–15) | 0.68 (0.53–0.87)a |
| MAP (median, IQR) | 598 | 90.7 (83–96) | 91.4 (83–96) | 0.99 (0.98–1.01)a |
| PR (median, IQR) | 585 | 85 (76–94) | 85 (75–95) | 1.00 (0.99–1.01)a |
| RR (median, IQR) | 623 | 24 (20–27) | 18 (16–21) | 1.14 (1.1–1.18)a |
| Sato2 on ambient air (%), (median, IQR) | 580 | 95 (93–97) | 97 (95–98) | 0.98 (0.96–1.00)a |
| Laboratory tests at hospitalization | ||||
| Lymphocytes (109/L), median (IQR) | 595 | 0.84 (0.60–1.06) | 1.17 (0.88–1.51) | 0.16 (0.10–0.28)a |
| CRP (mg/dL), median (IQR) | 601 | 11.0 (5.3–16.0) | 3.3 (1.6–6.99) | 1.2 (1.16–1.25)a |
| LDH (IU/L), median (IQR) | 569 | 350 (255–491) | 255 (201–313) | 1.0 (1.003–1.006)a |
| Glucose (mg/dL), median (IQR) | 487 | 116 (102–137) | 107 (94–123) | 1.01 (1.003–1.01)a |
| Creatinine (mg/dL), median (IQR) | 623 | 1.06 (0.86–1.36) | 0.86 (0.71–1.03) | 1.44 (1.15–1.81)a |
| Sodium (mmol/L), median (IQR) | 525 | 137 (135–141) | 137 (135–140) | 1.02 (0.98–1.06)a |
| Potassium (mmo/L), median (IQR) | 513 | 4 (3.7–4.4) | 4 (3.7–4.3) | 0.96 (0.82–1.14)a |
| Bilirubin (mg/dL), median (IQR) | 502 | 0.65 (0.45–0.85) | 0.60 (0.46–0.80) | 1.57 (1.03–2.34)a |
| Aspartate aminotransferase (IU/L), median (IQR) | 531 | 35 (27–45) | 31 (23–42) | 1.00 (1.00–1.01)a |
| Alanine aminotransferase (IU/L) median (IQR) | 566 | 22 (16–32) | 27 (18–40) | 1.00 (0.99–1.00)a |
All values given are n (%) unless otherwise stated.
Abbreviations: BMI, body mass index; COPD, chronic obstructive pulmonary disease; CRP, C-reactive protein; ESLD, end-stage liver disease; GCS, Glasgow coma scale; HRCT, high-resolution computed tomography; IQR interquartile range; LDH, lactate dehydrogenase; MAP, mean arterial pressure; PR, pulse rate.
For each year, point or unit increase.
Table 3.
Univariate analysis for severe respiratory failure among patients with SARS-CoV-2 pneumonia: validation cohort
| Cases with available data | Severe respiratory failure (n = 187) | No severe respiratory failure (n = 282) | OR (95% CI) | |
|---|---|---|---|---|
| Demographics | ||||
| Age (years), mean (±SD) | 469 | 72.4 (±12.3) | 65.8 (±14.6) | 1.04 (1.02–1.05)a |
| Sex, male | 469 | 145 (77) | 183 (64) | 1.87 (1.23–2.85) |
| Underlying diseases | ||||
| Obesity | 469 | 42 (22) | 32 (11) | 2.26 (1.37–3.74) |
| BMI (kg/m2), median (IQR) | 195 | 28 (25–31) | 25 (24–28) | 1.13 (1.04–1.23)a |
| Hypertension | 469 | 114 (61) | 144 (51) | 1.51 (1.04–2.23) |
| Diabetes mellitus | 469 | 17 (9) | 5 (2) | 4.1 (1.27–13.3) |
| Coronary artery disease | 469 | 17 (9) | 10 (3) | 2.72 (1.22–6.08) |
| Congestive heart failure | 469 | 24 (13) | 17 (6) | 2.3 (1.19–4.4) |
| Cerebrovascular disease | 469 | 22 (12) | 27 (10) | 1.26 (0.69–2.29) |
| Peripheral vascular disease | 469 | 46 (25) | 30 (11) | 2.74 (1.66–4.54) |
| Chronic kidney disease (moderate to severe) | 469 | 30 (16) | 24 (9) | 2.05 (1.16–3.64) |
| COPD | 469 | 29 (16) | 26 (9) | 1.81 (1.03–3.2) |
| Immunosuppression | 469 | 14 (7) | 7 (2) | 3.18 (1.26–8.03) |
| Charlson index (median, IQR) | 461 | 5 (3–7) | 3 (1–5) | 1.25 (1.16–1.35)a |
| Symptoms at onset | ||||
| Fever ≥38°C | 469 | 115 (61) | 150 (53) | 0.99 (0.5–1.95) |
| Cough | 469 | 98 (52) | 157 (98) | 0.93 (0.64–1.35) |
| Dyspnoea | 469 | 77 (41) | 63 (122) | 2.55 (1.7–3.8) |
| Time to hospital admission (days), median (IQR) | 451 | 6 (2–9) | 6 (2–9) | 0.94 (0.90–1.09)a |
| Symptoms at hospitalization | ||||
| Fever ≥38°C | 469 | 91 (48) | 96 (34) | 1.85 (1.26–2.7) |
| Cough | 469 | 91 (48) | 142 (59) | 0.94 (0.65–1.35) |
| Dyspnoea | 469 | 108 (57) | 142 (50) | 2.26 (1.57–3.29) |
| Vital signs at hospitalization | ||||
| GCS (median, IQR) | 446 | 15 (15–15) | 15 (15–15) | 0.56 (0.32–0.98)a |
| MAP (median, IQR) | 461 | 90.7 (83–96) | 91.4 (83–96) | 0.97 (0.31–3.00)a |
| PR (median, IQR) | 468 | 87 (79–99) | 85 (75–93) | 1.02 (1.00–1.03)a |
| RR (median, IQR) | 459 | 22 (16–22) | 20 (16–22) | 1.12 (1.07–1.16)a |
| Sato2 on ambient air (%), (median, IQR) | 416 | 95 (93–97) | 97 (95–98) | 0.91 (0.86–0.96)a |
| Laboratory tests at hospitalization | ||||
| Lymphocytes (10ˆ9/L), median (IQR) | 468 | 0.72 (0.51–0.98) | 0.96 (0.73–1.34) | 0.25 (0.15–0.41)a |
| CRP (mg/dL), median (IQR) | 454 | 11.2 (6.19–15.8) | 3.5 (1.8–6.5) | 1.27 (1.21–1.33)a |
| LDH (IU/L), median (IQR) | 406 | 398 (309–476) | 278(228–355) | 1.01 (1.00–1.01)a |
| Glucose (mg/dL), median (IQR) | 412 | 124 (110–155) | 112 (101–129) | 1.00 (1.00–1.01)a |
| Creatinine (mg/dL), median (IQR) | 460 | 1.12 (0.89–1.59) | 0.99 (0.82–1.15) | 2.46 (1.63–3.71)a |
| Sodium (mmol/L), median (IQR) | 403 | 136 (133–139) | 137 (134–139) | 1.00 (0.98–1.02)a |
| Potassium (mmo/L), median (IQR) | 381 | 3.9 (3.5–4.3) | 3.9 (3.7–4.2) | 1.18 (0.8–1.73)a |
| Bilirubin (mg/dL), median (IQR) | 174 | 0.55 (0.38–0.80) | 0.50 (0.34–0.74) | 1.88 (0.89–3.97)a |
| Aspartate aminotransferase (IU/L), median (IQR) | 206 | 44 (21–66) | 28 (23–34) | 1.04 (1.01–1.06)a |
| Alanine aminotransferase (IU/L) median (IQR) | 566 | 26 (16–42) | 24 (17–35) | 1.01 (1–1.02)a |
All values given are n (%) unless otherwise stated.
Abbreviations: BMI, body mass index; COPD, chronic obstructive pulmonary disease; CRP, C-reactive protein; ESLD, end-stage liver disease; GCS, Glasgow coma scale; HRCT, high-resolution computed tomography; IQR interquartile range; LDH, lactate dehydrogenase; MAP, mean arterial pressure; PR, pulse rate.
For each year/day, point or unit increase.
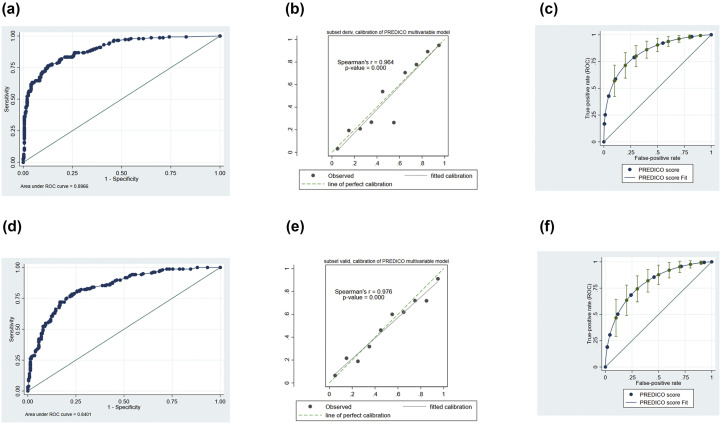
In the derivation cohort, multivariate analysis showed that age ≥70 years, obesity, fever at hospitalization (body temperature ≥38°C), respiratory rate ≥22 breaths/minute, lymphocytes ≤900 cells/mm3, creatinine ≥1 mg/dL, C-reactive protein ≥10 mg/dL and lactate dehydrogenase ≥350 UI/L were independent risk factors for developing SRF (Table 4 ). The model was highly discriminant: area under the ROC 0.90 (Fig. 2 a), Brier score 0.11, Somers' D 0.79 (95% CI 0.73–0.85). Calibration (Fig. 2b) and fitting (Fig. 2c) of the model were also good. In the validation cohort the model performed similarly in terms of discrimination, calibration (Fig. 2d,e, respectively), fitting (Fig. 2f) and distribution (see Supplementary material, Fig. S2b). Area under the ROC curve was 0.84 with Brier score 0.16 and Somers' D 0.68 (95% CI 0.60–0.76). Linear prediction coefficient in the validation cohort was 0.79 (95% CI 0.73–0.95).
Table 4.
Multivariate analysis of risk factors for respiratory failure in derivation and validation cohort, and score development
| Derivation cohort |
Validation cohort |
|||||||
|---|---|---|---|---|---|---|---|---|
| OR | 95% CI | p | β-coefficient | Points | OR | 95% CI | p | |
| Age ≥70 years | 2.74 | 1.66–4.50 | <0.001 | 1.01 | 1 | 2.25 | 1.45–3.49 | <0.001 |
| Obesity | 4.62 | 2.78–7.70 | <0.001 | 1.53 | 1 | 1.07 | 0.72–1.60 | 0.73 |
| Fever ≥38°C at hospitalization | 1.73 | 1.30–2.29 | <0.001 | 0.55 | 1 | 1.87 | 0.99–3.52 | 0.05 |
| RR ≥ 22 breaths/min | 3.75 | 2.01–7.01 | <0.001 | 1.32 | 1 | 2.44 | 1.41–4.21 | 0.001 |
| Lymphocytes ≤0.9 × 109/L | 2.69 | 1.60–4.51 | <0.001 | 0.99 | 1 | 1.94 | 1.15–3.27 | 0.01 |
| CRP ≥10 mg/dL | 5.91 | 4.88–7.17 | <0.001 | 1.78 | 2 | 8.44 | 4.72–15.07 | <0.001 |
| LDH ≥350 IU/L | 2.39 | 1.11–5.11 | 0.025 | 0.87 | 1 | 3.34 | 2.51–4.44 | <0.001 |
| Creatinine ≥1 mg/dL | 2.38 | 1.59.–3.56 | <0.001 | 0.87 | 1 | 1.35 | 1.16–1.57 | <0.001 |
Abbreviations: CRP, C-reactive protein; LDH, lactate dehydrogenase; OR, odds ratio; RR, respiratory rate.
Fig. 2.
Discrimination (a) and calibration (b) of the multivariable model and discrimination (c) of the PREDI-CO score in the derivation cohort. Discrimination (d), calibration (e) and discrimination (f) of the PREDICO score in the validation cohort.
Assignment of points on the basis of the β coefficient for these eight independent variables generated an individual risk score for each patient ranging from 0 to 9 (Table 4). Median PREDI-CO score was 4 (IQR 2–7) (see Supplementary material, Fig. S3a).
In the derivation cohort, the area under the ROC curve of the PREDI-CO score was 0.89 (95% CI 0.86–0.92). At a risk score of >3, the sensitivity, specificity, positive predictive value and negative predictive value were 72% (65%–79%), 86% (89%–92%), 74% (67%–80%) and 89% (85%–91%), respectively. The positive and negative likelihood ratios associated with a >3 score cut-off were 6.73 (95% CI 5.1–8.9) and 0.31 (95% CI 0.25–0.39), respectively (see Supplementary material, Table S1).
In the validation cohort, the PREDI-CO score showed an area under the ROC curve of 0.85 (95% CI 0.81–0.88). At risk score of >3, the sensitivity, specificity, positive predictive value and negative predictive value, and postive likelihood ratio 3.30 (2.65–4.11), negative likelihood ratio 0.27 (0.20–0.36) (Supplementary material, Table S1).
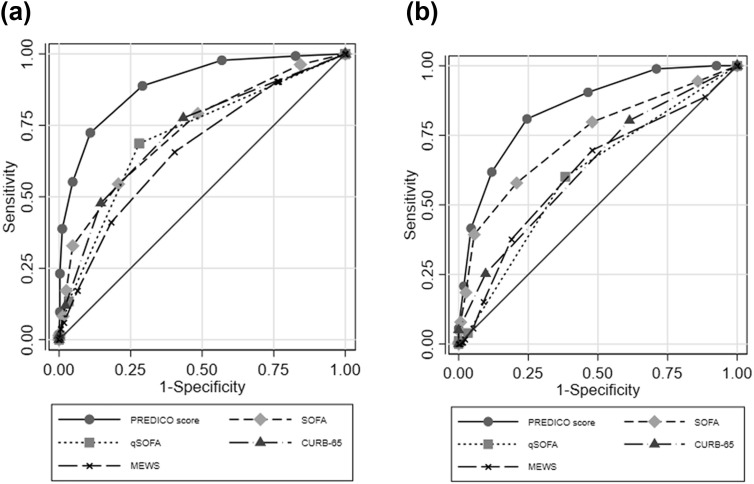
Finally, according to the ROC curve analysis the prediction ability for SRF of our score was higher than that of SOFA, qSOFA, CURB-65 and MEWS scores in both the derivation (Fig. 3 a) and validation (Fig. 3b) cohorts.
| Derivation cohort |
Validation cohort |
|||||
|---|---|---|---|---|---|---|
| AUC | Lower 95% CI | Upper 95% CI | AUC | Lower 95% CI | Upper 95% CI | |
| PREDI-CO score | 0.89 | 0.86 | 0.92 | 0.85 | 0.81 | 0.88 |
| SOFA | 0.73 | 0.68 | 0.78 | 0.74 | 0.69 | 0.79 |
| qSOFA | 0.71 | 0.66 | 0.76 | 0.61 | 0.56 | 0.65 |
| CURB-65 | 0.72 | 0.67 | 0.77 | 0.64 | 0.59 | 0.68 |
| MEWS | 0.66 | 0.61 | 0.72 | 0.62 | 0.56 | 0.67 |
Abbreviations: AUC area under the curve MEWS Modified Early Warning Score, SOFA Sequential Organ Failure Assessment.
Fig. 3.
Comparison of prediction ability for severe respiratory failure in hospitalized individuals with a diagnosis of COVID-19 of the PREDICO score with qSOFA, SOFA, CURB-65 and MEWS scores. (a) Derivation cohort; (b) validation cohort.
All the models and overall score performance were revaluated after the inclusion of covariates that are supposed to change the natural history of the disease including hydroxychloroquine, tocilizumab and corticosteroids without any significant change in the overall performance (data not shown).
Discussion
We developed and independently validated a simple individual risk score (the PREDI-CO score) to identify at the time of hospitalization individuals with COVID-19 who were at high risk of developing SRF during hospitalization. We found that of the individuals hospitalized with COVID-19 on the wards for at least 24 hours, a high percentage (33%) developed worsening of symptoms with SRF after this initial period. A predictive model was built and validated, using age >70 years, obesity, fever at hospitalization, respiratory rate ≥22 breaths/minute, lymphocyte count ≤900 cells/mm3, creatinine ≥1 mg/dL, C-reactive protein ≥10 mg/dL and lactate dehydrogenase ≥350 IU/L. Our model and risk score performed similarly even in different cohorts, as defined by different hospitals, providing independent validation.
The rate of SRF in our cohort of hospitalized patients with COVID-19 was higher than that in initial reports [4,13], but was in line with more recent findings [14,15]. Demographic characteristics of population, socio-cultural issues and local strategies for diagnostic testing have been appointed among the factors contributing to the different severity of COVID-19 across countries [14]. Indeed, the mean age of our patients was 65.7 years, compared with 47 and 49 years in the cohorts from Singapore and China, respectively [4,13].
It is worth mentioning that in most of the published prognostic studies on COVID-19, demographic characteristics (older age and male sex), underlying co-morbidities and altered laboratory tests (e.g. C-reactive protein, lactate dehydrogenase and lymphocyte counts) correlated with poor outcome, as in our study [16,17]. The strongest underlying condition influencing outcome in our analysis was obesity, as observed for other severe viral pneumonia, like H1N1 flu [18]. Recently, a similar score was developed and validated in Chinese hospitals [19]. This score compared with ours requires an online calculator so it could be less applicable in emergency situations and some of the included variables like haemoptysis were rarely reported in our cohort. This may represent differences between populations and settings.
Our study has a number of limitations. First, being a retrospective study, several variables were not systematically collected across all centres, especially in these times of increased clinical duties and stresses of the health-care system. This might introduce bias if patients with more severe clinical conditions had a higher chance of missed information. For example, interleukin-6 and D-dimer previously showed a significant correlation with disease progression [20], but were not available in this study. However, the strict correlation between interleukin-6 and all acute-phase proteins, including C-reactive protein is well known [21]. Additionally, interleukin-6 is not available in most laboratory chemistry panels of emergency rooms or wards of non-tertiary hospitals. The inclusion of such parameters in our score could reduce its applicability. Second, we included only individuals with SARS-CoV-2-positive nasopharyngeal swabs; this could contribute to a selection bias. In fact, the testing algorithm may have been affected by local policies [14]. Additionally, some patients could have been excluded from the study considering the suboptimal sensitivity of nasopharyngeal swabs [22]. Third, individuals with SRF within the first 24 hours after admission were excluded; we made this choice because we aimed to identify patients at risk of unfavourable clinical evolution, rather than discriminating between those already in severe clinical condition at admission. Fourth, our score has been developed and validated in Italian hospitals; even if restricted to single-country analysis, local care practices might have a strong impact on SRF rates. However, the PREDI-CO score performed similarly in different cohorts, providing external validation. Lastly, one risk factor for SRF (respiratory rate) may overlap with its definition. Being aware that this may constitute a bias we preferred to maintain this parameter as is commonly used in other clinical scores (qSOFA and CURB-65) to increase the applicability of our model.
To conclude, we developed and validated an individual risk score including eight strong predictors of SRF to identify at hospital admission those individuals with a COVID-19 diagnosis deserving a high level of care and prompt medical treatment. In particular, in our setting with a high frequency of respiratory failure (as was seen in the first phases of the pandemic in Italy) the negative predictive values were good, so our score might be useful to identify those patients who might not need intensive or high intensity care. If further validated in a prospective study our score might serve for both rationing decisions at health-care levels and selecting patients to include in randomized controlled trials on new treatment options.
Authors' contribution
PV, MB, MG, LS, CM, ST, MT, VMR and TT contributed to conceptualization; MB, LS, MG, MR, MT and TT to methodology; LB, GF, RP, LP, ZP, FT, LB, CC, LA, MMer, MMen, MMes, AL, SR and PG to investigations; and MB, MG and LS to the formal analysis. MG and MB wrote the original draft and LS, PV, TT, FB and VMR contributed to reviewing and editing. FB, MC, MP; CM, FC and PV supervised the work.
Transparency declaration
The authors declare that they have no conflicts of interest. No external funding was received for the present study.
Acknowledgement
We would like to acknowledge Prof. Russell Lewis (Department of Medical and Surgical Sciences, Alma Mater Studiorum, University of Bologna) for his advice on methodology. No funding was available for this study.
Editor: J. Rodriguez-Baño
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cmi.2020.08.003.
Contributor Information
PREDICO study group:
Luigi Raumer, Luca Guerra, Fabio Tumietto, Alessandra Cascavilla, Eleonora Zamparini, Gabriella Verucchi, Simona Coladonato, Arianna Rubin, Stefano Ianniruberto, Eugenia Francalanci, Francesca Volpato, Giulio Virgili, Nicolò Rossi, Elena Rosselli Del Turco, Viola Guardigni, Giovanni Fasulo, Nicola Dentale, Ciro Fulgaro, Giorgio Legnani, Emanuele Campaci, Cristina Basso, Alberto Zuppiroli, Amalia Sanna Passino, Giulia Tesini, Lucia Angelelli, Adriana Badeanu, Agostino Rossi, Giulia Santangelo, Flovia Dauti, Vidak Koprivika, Nicholas Roncagli, Ioannis Tzimas, Guido Maria Liuzzi, Irid Baxhaku, Letizia Pasinelli, Mattia Neri, Tommaso Zanaboni, Francesco Dell'Omo, Oana Vatamanu, Alice Gori, Idina Zavatta, Stefano Antonini, Chiara Pironi, Elena Piccini, Luca Esposito, Alessandro Zuccotti, Giacomo Urbinati, Agnese Pratelli, Alberto Sarti, Michela Semprini, Enrico Evangelisti, Mara D'Onofrio, Giuseppe Sasdelli, Giacinto Pizzilli, Elisabetta Pierucci, Giada Rossini, Caterina Vocale, Lorenzo Marconi, Maria Cristina Leoni, Elisa Fronti, Giovanni Guaraldi, Davide Bavaro, and Paola Laghetti
Appendix.
PREDICO study group
Luigi Raumer, Luca Guerra, Fabio Tumietto, Alessandra Cascavilla, Eleonora Zamparini, Gabriella Verucchi, Simona Coladonato, Arianna Rubin, Stefano Ianniruberto, Eugenia Francalanci, Francesca Volpato, Giulio Virgili, Nicolò Rossi, Elena Rosselli Del Turco, Viola Guardigni, Giovanni Fasulo, Nicola Dentale, Ciro Fulgaro, Giorgio Legnani, Emanuele Campaci, Cristina Basso, Alberto Zuppiroli, Amalia Sanna Passino, Giulia Tesini, Lucia Angelelli, Adriana Badeanu, Agostino Rossi, Giulia Santangelo, Flovia Dauti, Vidak Koprivika, Nicholas Roncagli, Ioannis Tzimas, Guido Maria Liuzzi, Irid Baxhaku, Letizia Pasinelli, Mattia Neri, Tommaso Zanaboni, Francesco Dell'Omo, Oana Vatamanu, Alice Gori, Idina Zavatta, Stefano Antonini, Chiara Pironi, Elena Piccini, Luca Esposito, Alessandro Zuccotti, Giacomo Urbinati, Agnese Pratelli, Alberto Sarti, Michela Semprini, Enrico Evangelisti, Mara D'Onofrio, Giuseppe Sasdelli, University of Bologna, Bologna, Italy.
Giacinto Pizzilli, Elisabetta Pierucci; Intensive Care Unit, Department of Medical and Surgical Sciences, Policlinico Sant’Orsola, Bologna, Italy.
Giada Rossini, Caterina Vocale; Centro di riferimento regionale per le emergenze microbiologiche (CRREM), Clinical Microbiology Unit, Department of Experimental, Diagnostic and Specialty Medicine, Policlinico Sant’Orsola, Bologna, Italy.
Lorenzo Marconi; Infectious Diseases Unit, Rimini-Forlì-Cesena Hospitals, Rimini, Italy.
Maria Cristina Leoni, Elisa Fronti; Infectious Diseases Unit, “Guglielmo da Saliceto” Hospital, Piacenza, Italy.
Giovanni Guaraldi; Infectious Diseases Unit, Policlinico di Modena, Università degli Studi di Modena e Reggio Emilia, Modena, Italy.
Davide Bavaro, Paola Laghetti, Lucia Diella Infectious Disease Unit - Department of Biomedical Sciences and Human Oncology, University of Bari, Policlinico di Bari, Italy.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72314 cases from the Chinese center for disease control and prevention. JAMA. 2020 doi: 10.1001/jama.2020.2648. epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 2.Richardson S., Hirsch J.S., Narasimhan M., Crawford J.M., McGinn T., Davidson K.W., et al. Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York City Area. JAMA. 2020 doi: 10.1001/jama.2020.6775. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grasselli G., Pesenti A., Cecconi M. Critical Care utilization for the COVID-19 outbreak in Lombardy, Italy: early experience and forecast during an emergency response. JAMA. 2020 doi: 10.1001/jama.2020.4031. epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 4.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alhazzani W., Moller M.H., Arabi Y.M., Loeb M., Gong M.N., Fan E., et al. Surviving sepsis campaign: guidelines on the management of critically ill adults with coronavirus disease 2019 (COVID-19) Crit Care Med. 2020 doi: 10.1097/CCM.0000000000004363. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.White D.B., Lo B. A framework for rationing ventilators and critical care beds during the COVID-19 pandemic. JAMA. 2020 doi: 10.1001/jama.2020.5046. epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 7.Sanders J.M., Monogue M.L., Jodlowski T.Z., Cutrell J.B. Pharmacologic treatments for coronavirus disease 2019 (COVID-19): a review. JAMA. 2020 doi: 10.1001/jama.2020.6019. epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 8.Harris P.A., Taylor R., Thielke R., Payne J., Gonzalez N., Conde J.G. Research electronic data capture (REDCap)—a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42:377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harris P.A., Taylor R., Minor B.L., Elliott V., Fernandez M., O'Neal L., et al. The REDCap consortium: building an international community of software platform partners. J Biomed Inform. 2019;95:103208. doi: 10.1016/j.jbi.2019.103208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.WHO Clinical management of COVID-19. 2020. https://www.who.int/publications-detail/clinical-management-of-severe-acute-respiratory-infection-when-novel-coronavirus-(ncov)-infection-is-suspected Available at:
- 11.Charlson M.E., Pompei P., Ales K.L., MacKenzie C.R. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis. 1987;40:373–383. doi: 10.1016/0021-9681(87)90171-8. [DOI] [PubMed] [Google Scholar]
- 12.Riley R.D., Ensor J., Snell K.I.E., Harrell F.E., Jr., Martin G.P., Reitsma J.B., et al. Calculating the sample size required for developing a clinical prediction model. BMJ. 2020;368:m441. doi: 10.1136/bmj.m441. [DOI] [PubMed] [Google Scholar]
- 13.Young B.E., Ong S.W.X., Kalimuddin S., Low J.G., Tan S.Y., Loh J., et al. Epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. JAMA. 2020 doi: 10.1001/jama.2020.3204. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Onder G., Rezza G., Brusaferro S. Case-fatality rate and characteristics of patients dying in relation to COVID-19 in Italy. JAMA. 2020 doi: 10.1001/jama.2020.4683. epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 15.Grasselli G., Zangrillo A., Zanella A., Antonelli N., Cabrini L., Castelli A., et al. Baseline characteristics and outcomes of 1591 patients infected with SARS-CoV-2 admitted to ICUs of the Lombardy Region. Italy JAMA. 2020 doi: 10.1001/jama.2020.5394. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wynants L., Van Calster B., Bonten M.J., Collins G.S., Debray T., De Vos M., et al. Systematic review and critical appraisal of prediction models for diagnosis and prognosis of COVID-19 infection. BMJ. 2020 doi: 10.1136/bmj.m1328. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ji D., Zhang D., Xu J., Chen Z., Yang T., Zhao P., et al. Prediction for progression risk in patients with COVID-19 pneumonia: the CALL Score. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa414. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rubin R. Obesity and influenza A shedding. JAMA. 2018;320:1230. doi: 10.1001/jama.2018.13078. [DOI] [PubMed] [Google Scholar]
- 19.Liang W., Liang H., Ou L., Chen B., Chen A., Li C., et al. He, Development and validation of a clinical risk score to predict the occurrence of critical illness in hospitalized patients with COVID-19. JAMA Intern Med. 2020 doi: 10.1001/jamainternmed.2020.2033. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao W., Zhong Z., Xie X., Yu Q., Liu J. Relation between chest CT findings and clinical conditions of coronavirus disease (COVID-19) pneumonia: a multicenter study. Am J Roentgenol. 2020:1–6. doi: 10.2214/AJR.20.22976. [DOI] [PubMed] [Google Scholar]
- 21.Gabay C., Kushner I. Acute-phase proteins and other systemic responses to inflammation. N Engl J Med. 1999;340:448–454. doi: 10.1056/NEJM199902113400607. [DOI] [PubMed] [Google Scholar]
- 22.Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020 doi: 10.1001/jama.2020.3786. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.