Abstract
Peribunyaviruses are enveloped and possess three distinct, single-stranded, negative-sense RNA segments comprising 11.2–12.5 kb in total. The family includes globally distributed viruses in the genera Orthobunyavirus, Herbevirus, Pacuvirus and Shangavirus. Most viruses are maintained in geographically-restricted vertebrate–arthropod transmission cycles that can include transovarial transmission from arthropod dam to offspring. Others are arthropod-specific. Arthropods can be persistently infected. Human infection occurs through blood feeding by an infected vector arthropod. Infections can result in a diversity of human and veterinary clinical outcomes in a strain-specific manner. Segment reassortment is evident between some peribunyaviruses. This is a summary of the International Committee on Taxonomy of Viruses (ICTV) Report on the taxonomy of the family Peribunyaviridae, which is available at ictv.global/report/peribunyaviridae.
Keywords: Peribunyaviridae, bunyavirus, ICTV Report, Orthobunyavirus, Herbevirus, Pacuvirus, Shangavirus, taxonomy
Virion
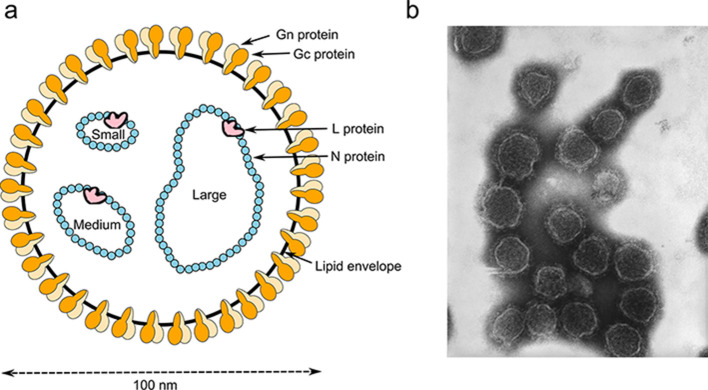
Peribunyavirus virions are spherical or pleomorphic, 80–120 nm in diameter [1] with glycoprotein surface projections (5–18 nm) embedded in a lipid bilayer envelope (about 5 nm) (Table 1, Fig. 1). The genome comprises three single-stranded negative-sense RNAs (designated S, small, M, medium and L, large) (Fig. 2), each with complementary terminal nucleotide sequences that base-pair to form non-covalently closed, circular RNAs [2] that are individually encapsidated.
Table 1.
Characteristics of members of the family Peribunyaviridae
|
Typical member |
Bunyamwera virus (S, D00353; M, M11852; L, X14383), species Bunyamwera orthobunyavirus, genus Orthobunyavirus |
|---|---|
|
Virion |
Enveloped, spherical or pleomorphic virions, 80–120 nm in diameter |
|
Genome |
Three single-stranded, negative-sense RNA molecules, S, M and L, each of about 1, 4 and 6.8 kb |
|
Replication |
Cytoplasmic; primary transcription is primed by ‘cap snatching’ of host RNAs |
|
Translation |
On ER-bound ribosomes for Gn and Gc and on free ribosomes in the cytoplasm for N and L |
|
Host range |
Vertebrates and invertebrates (including mammals, birds, mosquitoes, culicoids and psychodid sandflies) |
|
Taxonomy |
Phylum Negarnaviricota, subphylum Polyploviricotina, class Ellioviricetes, order Bunyavirales, several genera and >90 species |
Fig. 1.
Peribunyavirus virion structure. (a) representation of a virion in cross-section. The surface spikes comprise the Gn and Gc glycoproteins. The helical nucleocapsids are circular and comprise one each of the unique ssRNA segments (L, large; M, medium; S, small) encapsidated by N protein and associated with the L protein. (b) negative-stained transmission electron microscopy photograph of California encephalitis virus virions (image: CDC/Drs Frederick Murphy and Erskine Palmer).
Fig. 2.
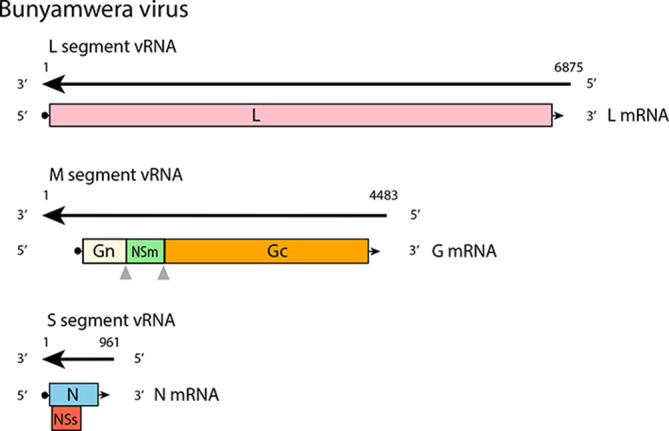
Coding strategy of Bunyamwera virus. Translation of NSs protein is initiated at an alternative start codon. The Gn, NSm and Gc proteins are generated by co-translational cleavage of M polyprotein.
Genome
The S segment encodes the nucleocapsid protein (N), which is abundant in infected cells; in some viruses an overlapping reading frame encodes the non-structural protein NSs [3–5]. The M segment encodes two structural glycoproteins (Gn and Gc). Some members also encode a non-structural protein (NSm) between the Gn and Gc coding regions [4]. The L segment encodes the L protein, which has RNA-directed RNA polymerase and endonuclease functions.
Replication
Virions attach via surface glycoproteins, entering the cell through clathrin-mediated endocytosis. Fusion of the viral Gc protein fusion peptide with endosomal membranes facilitates the release of ribonucleocapsids into the cytoplasm. The complementary 5′- and 3′-terminal ends serve as promoters for both mRNA and antigenome synthesis. Viral mRNAs are not polyadenylated and are truncated relative to the vRNA; a 5′-methylated cap is derived from host mRNA via ‘cap snatching’ mediated by the endonuclease function of the L protein. Proteins are translated on free ribosomes (S and L segment mRNAs) or membrane-bound ribosomes (M segment mRNA). The Gn and Gc proteins are generated by co-translational cleavage and targeted to and retained in the Golgi complex. Ribonucleoproteins are targeted near the Golgi complex. Genomes are packaged by signals from non-conserved sequences in the terminal untranslated regions. Virions bud into Golgi cisternae and are transported to the cell surface by the secretory pathway [6].
Taxonomy
Genera are monophyletic based on analysis of the virus L protein; members of a genus have similar genomic organizations and transmission cycles. Peribunyaviruses form a group in the phylum Negarnaviricota, subphylum Polyploviricotina, class Ellioviricetes, order Bunyavirales, being most closely related to viruses of the families Fimoviridae and Tospoviridae. Peribunyaviruses share some of the following characteristics: (i) enveloped spherical or pleomorphic virions; (ii) three segments of single-stranded, negative-sense RNA, with all proteins encoded in the same sense; (iii) capped but not polyadenylated viral mRNA; (iv) establish a persistent infection in an arthropod vector.
Resources
Current ICTV Report on the family Peribunyaviridae: ictv.global/report/peribunyaviridae.
Funding information
Production of this summary, the online chapter, and associated resources was funded by a grant from the Wellcome Trust (WT108418AIA). W. M. S. is supported by Fundação de Amparo à Pesquisa do Estado de São Paulo, Brazil (17/13981–0).
Acknowledgements
Members of the ICTV Report Consortium are Stuart G. Siddell, Andrew J. Davison, Elliot J. Lefkowitz, Peter Simmonds, Sead Sabanadzovic, Donald B. Smith, Richard J. Orton and Jens H. Kuhn. The findings and conclusions in this manuscript are those of the authors and do not necessarily represent the views of the CDC or other organizations.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: G, Glycoprotein; L, N and NS, large, nucleocapsid and non-structural proteins; S. M and L, Small, medium and large vRNA segments.
References
- 1.Martin ML, Lindsey-Regnery H, Sasso DR, McCormick JB, Palmer E. Distinction between Bunyaviridae genera by surface structure and comparison with Hantaan virus using negative stain electron microscopy. Arch Virol. 1985;86:17–28. doi: 10.1007/BF01314110. [DOI] [PubMed] [Google Scholar]
- 2.Raju R, Kolakofsky D. The ends of La Crosse virus genome and antigenome RNAs within nucleocapsids are base paired. J Virol. 1989;63:122–128. doi: 10.1128/jvi.63.1.122-128.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Melo AB, de Souza WM, Acrani GO, Carvalho VL, Romeiro MF, et al. Genomic characterization and evolution of Tacaiuma orthobunyavirus (Peribunyaviridae family) isolated in Brazil. Infect Genet Evol. 2018;60:71–76. doi: 10.1016/j.meegid.2018.02.027. [DOI] [PubMed] [Google Scholar]
- 4.Marklewitz M, Zirkel F, Rwego IB, Heidemann H, Trippner P, et al. Discovery of a unique novel clade of mosquito-associated bunyaviruses. J Virol. 2013;87:12850–12865. doi: 10.1128/JVI.01862-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodrigues DS, Medeiros DB, Rodrigues SG, Martins LC, de Lima CP, et al. Pacui virus, Rio Preto da Eva virus, and Tapirape virus, three distinct viruses within the family Bunyaviridae . Genome Announc. 2014;2:e00923–14. doi: 10.1128/genomeA.00923-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferron F, Weber F, de la Torre JC, Reguera J. Transcription and replication mechanisms of Bunyaviridae and Arenaviridae L proteins. Virus Res. 2017;234:118–134. doi: 10.1016/j.virusres.2017.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]