Abstract
The family Hepadnaviridae comprises small enveloped viruses with a partially double-stranded DNA genome of 3.0–3.4 kb. All family members express three sets of proteins (preC/C, polymerase and preS/S) and replication involves reverse transcription within nucleocapsids in the cytoplasm of hepatocytes. Hepadnaviruses are hepatotropic and infections may be transient or persistent. There are five genera: Parahepadnavirus, Metahepadnavirus, Herpetohepadnavirus, Avihepadnavirus and Orthohepadnavirus. This is a summary of the International Committee on Taxonomy of Viruses (ICTV) Report on the family Hepadnaviridae, which is available at ictv.global/report/hepadnaviridae.
Keywords: Hepadnaviridae, ICTV Report, taxonomy, hepatitis B virus
Virion
Hepadnaviruses are spherical and occasionally pleomorphic. The envelope contains two or three surface proteins, which suffice to induce protective immunity (Table 1). The icosahedral nucleocapsid encloses a partially double-stranded DNA genome, the viral DNA polymerase, and a cell-derived protein kinase and chaperones [1]. Hepadnaviruses induce overproduction of surface proteins secreted into the blood as pleomorphic particles. For hepatitis B virus, these are 17–22 nm spherical particles and filaments (Fig. 1). Virions and empty particles contain two or three surface proteins, each in more than one isoform due to alternative glycosylation.
Table 1.
Characteristics of members of the family Hepadnaviridae
|
Typical member: |
hepatitis B virus, genotype D (V01460), species Hepatitis B virus, genus Orthohepadnavirus |
|---|---|
|
Virion |
Envelope of 42–50 nm diameter surrounding a nucleocapsid usually composed of 240 protein subunits |
|
Genome |
3.0–3.4 kb partially double-stranded DNA |
|
Replication |
Pre-genomic RNA transcripts from covalently closed circular DNA in the nucleus are encapsidated and reverse-transcribed in the cytoplasm |
|
Translation |
Five or six mRNA transcripts, depending on genus, with different 5′-ends and a common 3′-end linked to a polyadenylation site |
|
Host range |
Teleost fish (Parahepadnavirus and Metahepadnavirus), reptiles and frogs (Herpetohepadnavirus), birds (Avihepadnavirus) and mammals (Orthohepadnavirus) |
|
Taxonomy |
Realm Riboviria, kingdom Pararnavirae, phylum Artverviricota, class Revtraviricetes, order Blubervirales, several genera and > 15 species |
Fig. 1.
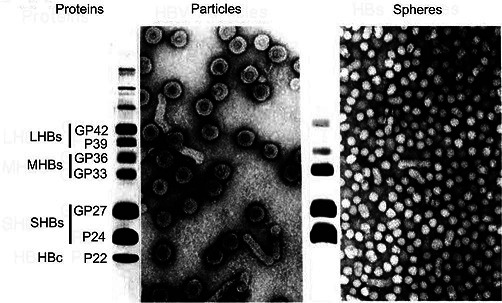
Negative-contrast electron micrographs of hepatitis B virus virions (left) and virus-associated particles (right; spheres are 17–22 nm in diameter), together with their SDS-PAGE protein profile. LHBs, MHBs and SHBs refer to large, middle and small hepatitis B virus surface proteins, respectively. HBc, hepatitis B virus core proteins. GP, glycoprotein; P, protein. (Courtesy of W. Gerlich).
Genome
The genome is kept in circular conformation by base pairing of cohesive overlaps between the 5′-ends of the two DNA strands (Fig. 2). In the nucleocapsid, the minus-sense DNA strand is full length and has an 8–9 nt terminal redundancy, whereas the positive-sense DNA strand varies in length, since its 3′-end terminates at variable positions, creating a single-stranded gap accounting for up to 60 % of the genome. The genome has three open reading frames (ORFs); precore/core (preC/C), polymerase (P), env or surface (preS/S) and, regarding orthohepadnaviruses, an additional X ORF (Fig. 2). Apart from HBcAg, the PreC/C ORF in avi- and orthohepdnaviruses encodes HBeAg, a protein with immunomodulatory function secreted into serum. Genomic and sub-genomic RNAs are transcribed by RNA polymerase II into mRNAs with varying 5ʹ-ends but a common 3ʹ-end [2]. The genome is compact, with regulatory elements located within overlapping reading frames.
Fig. 2.
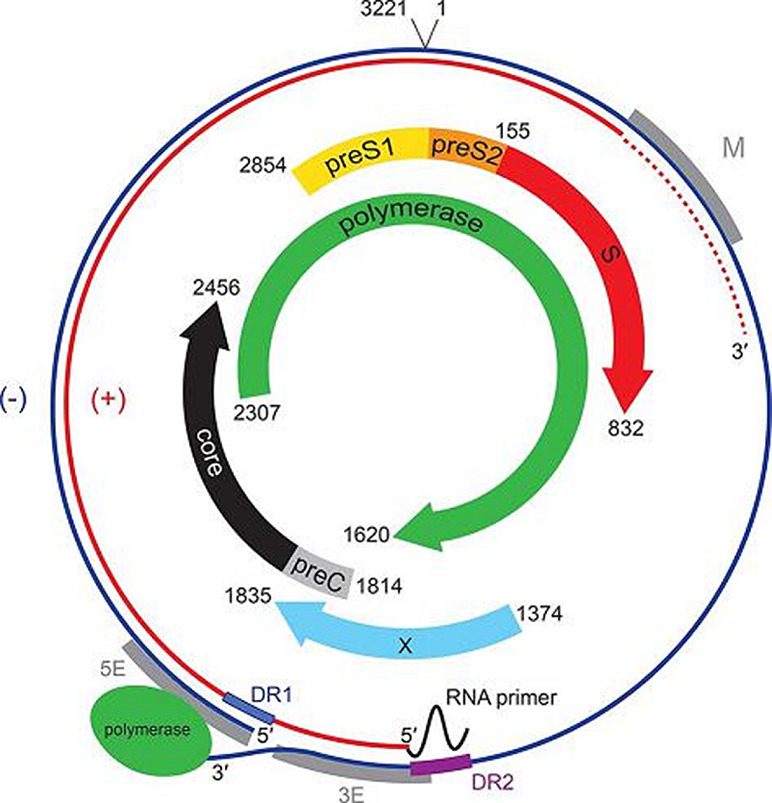
Genome organization of hepatitis B virus genotype A (X02763). The outer circle represents the relaxed circular negative-sense viral DNA found within virions that has a terminal redundancy. Coloured arrows indicate the position of ORFs together with their start and termination coordinates numbered from a unique EcoR1 restriction site. M, 5E and 3E are regulatory elements required for genome circularization; DR1 and DR2 are direct repeats; plus-strand DNA synthesis is initiated at DR2.
Replication
The virion of the hepatitis B virus binds first to heparan sulfate proteoglycans on the cell surface via its major/small surface protein, followed by a specific interaction of the preS1 domain of the large surface protein to a hepatocyte receptor, sodium taurocholate co-transporting polypeptide [3]. Following uptake, the nucleocapsid is transported to the nuclear periphery. The capsids pass the nuclear pore and release the genome to the nucleoplasm, where repair of the single-stranded DNA gap occurs [4]. The DNA ends are ligated to form covalently closed circular DNA (cccDNA), forming a histone-associated minichromosome, which provides the template for transcription of viral mRNAs. The unspliced pregenomic mRNA transcript is encapsidated and reverse-transcribed within core particles [1]. Some nucleocapsids are transported back to the nucleus, thereby increasing the pool of cccDNA (as shown for avihepadnaviruses). Other nucleocapsids bud through the endoplasmic reticulum enriched for co-translationally inserted envelope proteins and are secreted as virions.
Taxonomy
Species are delimited by about 20 % nucleotide divergence. Regarding para-, meta- and herpetoviruses see Table 1 of [5]. The genus Avihepadnavirus includes the species Duck hepatitis B virus, Heron hepatitis B virus and Parrot hepatitis B virus, members of which infect duck and crane, heron and stork, and parrot, respectively. Orthohepadnaviruses infect mammals, with a narrow host range for members of each viral species. Members of the species Hepatitis B virus infect humans and apes, members of the species Woolly monkey hepatitis B virus infect woolly monkey and members of the species Capuchin monkey hepatitis B virus infect capuchin monkey, while members of other species infect sciurid rodents (woodchuck and ground squirrel), bats (four species), one shrew, one artiodactyl and one carnivore species. Endogenous hepadnaviral elements (eHBVs) are present in the genomes of sauropsid amniotes, for example in zebra finches and chicken, snakes, turtles and crocodilians.
Resources
Current ICTV Report on the family Hepadnaviridae: ictv.global/report/hepadnaviridae
Funding information
Production of this summary, the online chapter, and associated resources was funded by a grant from the Wellcome Trust (WT108418AIA).
Acknowledgements
Members of the ICTV Report Consortium are Stuart G. Siddell, Andrew J. Davison, Elliot J. Lefkowitz, Peter Simmonds, Sead Sabanadzovic, Donald B. Smith, Richard J. Orton and Balázs Harrach.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: cccDNA, covalently closed circular DNA; P, polymerase; preC/C, precore/core; preS/S, env or surface.
References
- 1.Hu J, Seeger C. Hepadnavirus genome replication and persistence. Cold Spring Harb Perspect Med. 2015;5:a021386. doi: 10.1101/cshperspect.a021386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rall LB, Standring DN, Laub O, Rutter WJ. Transcription of hepatitis B virus by RNA polymerase II. Mol Cell Biol. 1983;3:1766–1773. doi: 10.1128/MCB.3.10.1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yan H, Peng B, He W, Zhong G, Qi Y, et al. Molecular determinants of hepatitis B and D virus entry restriction in mouse sodium taurocholate cotransporting polypeptide. J Virol. 2013;87:7977–7991. doi: 10.1128/JVI.03540-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gallucci L, Kann M. Nuclear import of hepatitis B virus capsids and genome. Viruses. 2017;9:21. doi: 10.3390/v9010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lauber C, Seitz S, Mattei S, Suh A, Beck J, et al. Deciphering the origin and evolution of hepatitis B viruses by means of a family of non-enveloped fish viruses. Cell Host Microbe. 2017;22:387–399. doi: 10.1016/j.chom.2017.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]


