Abstract
Streptococcus anginosus is an often overlooked and understudied emerging pathogen inhabiting many areas of the human body. Through our sequencing of S. anginosus strains isolated from the female bladder microbiota, we detected numerous prophage sequences. Bioinformatic analysis of these sequences identified 17 distinct groups of S. anginosus prophages. The majority of these phages exhibit no sequence homology to previously characterized temperate or virulent phage sequences, indicating an unexplored diversity of Streptococcus phages. By culturing these bacterial isolates, we confirmed that the prophages of five of these groups are capable of induction. One of these putative phages was imaged, the first such evidence of an S . anginosus virus-like particle; it exhibits morphological characteristics of siphoviruses.
Keywords: Streptococcus anginosus, bacteriophage, urinary tract
Introduction
Streptococcus anginosus and other members of the S. anginosus group ( S. intermedius and S. constellatus ) are primarily commensal bacteria of the oral cavity, throat, gastrointestinal tract and vagina [1]. However, S. anginosus also has been found within the bladder [2], with strains from the vagina and urinary tract of the same individual belonging to essentially the same strain [3]. S. anginosus is now considered an emerging pathogen. It has been associated with infections of the respiratory tract [4–6], brain abscesses [7, 8], liver abscesses [9], and skin and soft tissue infections [10]. It also has been isolated from infections of the head and neck, central nervous system, gastrointestinal tract and blood [6, 9]. Furthermore, S. anginosus can cause infections of the genitourinary tract [6, 9, 11]. It has been associated with acute glomerulonephritis [12], bacterial vaginosis [13, 14], urge urinary incontinence [2] and urinary tract infections [15].
Previously, our group sequenced S. anginosus isolates from the urinary tract and vagina [3]. Our analysis of the genomes of these urinary isolates revealed that all harboured prophages. One strain, S. anginosus UMB0839, contained five prophage sequences [16]. At that time, no prophages had been described for the species. Subsequently, a bioinformatic study predicted prophage sequences within publicly available genomes of various Streptococcus species, including several S. anginosus strains [17]. Nevertheless, an in-depth analysis of S. anginosus prophages has yet to be conducted. Furthermore, the viability of these prophages remains unknown. We selected 14 S . anginosus strains from the urinary tract for complete genome sequencing and phage isolation, presenting here both novel prophage sequences and the first evidence of prophage induction for this species.
Streptococcus anginosus Isolates of the Bladder
Catheterized urine samples were collected from women as part of prior IRB-approved studies [2, 18–20]. Bacteria were isolated from these samples using the expanded quantitative urine culture (EQUC) method [20] and stored at −80 °C. Fourteen strains identified as S. anginosus by MALDI-TOF MS were selected for whole genome sequencing. Freezer stocks for each of the strains were first streaked on Columbia CNA agar with 5 % sheep blood plates (BD 221353) and incubated at 35 °C in 5 % CO2 for 24 h. A single colony was then selected and grown in BHI liquid medium at 35 °C in 5 % CO2 for 24 h. DNA was extracted with the Qiagen DNeasy Blood and Tissue Kit, and DNA libraries were constructed (Nextera XT Library Prep Kit) and sequenced using the MiSeq Reagent Kit v2, producing 250 bp paired-end reads. The raw reads were trimmed using sickle v1.33 (https://github.com/najoshi/sickle) and assembled with SPAdes v3.11.1 [21] (parameters: ‘only-assembler’ option for k=55, 77, 99 and 127). Genome coverage was calculated using bbmap v34 (https://sourceforge.net/projects/bbmap/); the scripts bbwrap.sh and pileup.sh were used to map trimmed reads to the assemblies and compute average genome coverage. Genome annotations were performed using NCBI’s Prokaryotic Genome Annotation Pipeline v4.8 [22]. Genome sequences as well as raw reads have been deposited in GenBank’s Assembly and SRA databases, respectively. Table 1 lists the genome assembly statistics for each strain. Genome sizes ranged between 1.87 and 2.26 Mbp. This is within the range of sizes for publicly available genomes (52 genomes: 1.79–2.31 Mbp). The average GC content was 38.72 %. Again, this in on par with genomes of other strains of S. anginosus .
Table 1.
Genome assembly statistics and the number of prophage sequences identified
|
Strain |
Genome size (bp) |
Number of contigs |
N50 |
Accession number |
Number of prophages |
|---|---|---|---|---|---|
|
UMB0248 |
2,261,348 |
91 |
53 274 |
5 |
|
|
UMB0567 |
1,948,733 |
56 |
78 484 |
2 |
|
|
UMB0595 |
1,870,792 |
39 |
85 075 |
2 |
|
|
UMB0619 |
1,927,957 |
41 |
71 633 |
2 |
|
|
UMB0622 |
1,969,304 |
49 |
118,764 |
3 |
|
|
UMB0633 |
1,943,307 |
44 |
116,838 |
2 |
|
|
UMB2128 |
2,204,898 |
105 |
43 663 |
6 |
|
|
UMB3444 |
1,923,186 |
42 |
88 335 |
2 |
|
|
UMB4683 |
2,066,950 |
32 |
100,828 |
3 |
|
|
UMB4708 |
2,106,325 |
54 |
104,455 |
6 |
|
|
UMB7052 |
1,956,946 |
73 |
61 834 |
4 |
|
|
UMB8390 |
2,155,037 |
87 |
51 935 |
2 |
|
|
UMB8616 |
2,005,385 |
34 |
111,649 |
7 |
|
|
UMB8710 |
2,026,560 |
43 |
118,775 |
1 |
Predicted S. anginosus prophage sequences
The assembled genome sequences were investigated for prophage sequences using PHASTER [23] and VirSorter [24]. The two tools predicted the same prophage regions. VirSorter predictions were selected for further analysis. Fifty-five prophage sequences were identified by VirSorter amongst the 14 S . anginosus strains. Four of the prophage sequences identified were >100 kbp in length. Eight prophages were removed from further consideration as they were incomplete; they were predicted as ‘incomplete’ or ‘questionable’ by PHASTER and predicted with low confidence by VirSorter. The number of prophages per genome is listed in Table 1. Prophage sequences were clustered based upon sequence similarity to identify related phages between strains. This clustering was performed using USEARCH v11.0.667 [25] with the cluster_fast parameter and an identity threshold of 80 %, and the results were manually inspected. The 47 prophage sequences were clustered into 10 groups (n=40). The remaining seven prophages did not resemble any other prophage sequence from our S. anginosus genomes and thus were not included in a group. Each predicted prophage sequence was annotated using PATRIC [26], and groups were aligned using MAFFT v7.388 through Geneious Prime 2019.1.1 (Biomatters) with default parameters [27].
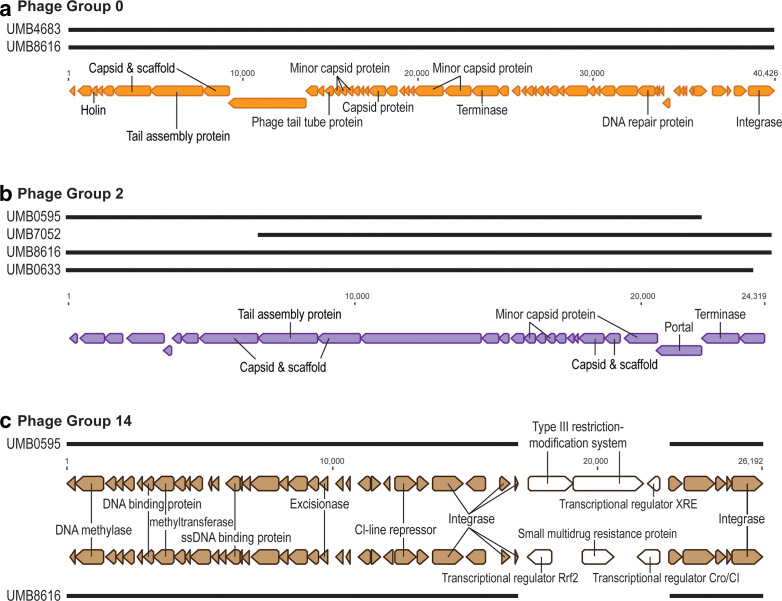
Insertion sequences, integrases and tRNAs are frequently found to be flanking the predicted prophage sequences within the bacterial host genomes. Annotation details for all 47 prophage sequences can be found in Table S1 (available in the online version of this article). Inspection of these gene annotations reveals phage terminases, tail proteins, lysins and holins, capsid and scaffolding proteins, and integrases, as well as a multitude of hypothetical proteins. Fig. 1 shows the gene annotations for prophages from three of these groups. For group 14 (Fig. 1c), two of the four members are shown to highlight the variation that can exist within even related predicted prophages. While the majority of the prophage sequences are identical for this group, one region is unique: the prophage from UMB0595 codes for a Type III restriction-modification system and the prophage from UMB8616 codes for a small multidrug resistance protein. The sequences of group 7 prophages (141_10, 162_2 and 165_16), group 174 prophages (174_1-c4 and 174_1-c5) and the singleton prophage 161_8 code for tetracycline resistance. Other Streptococcus -infecting phages, including the S. pyogenes bacteriophage Φm46.1 (which has sequence similarity to the group 3 and group 7 phages) [28], have been found to carry tetracycline resistance genes. The group 7, group 174 and singleton 161_8 prophage sequences also encode a partitioning system. Similar systems have been identified in temperate phages of other hosts, enabling these phages to replicate extra-chromosomally [29] (and citations therein), [30]. Additionally, prophage 161_8 codes for the epsilon/zeta toxin–antitoxin system. Further experimental work is required to ascertain if the group 7, group 174 and singleton prophage 161_8 are in fact phages or rather phage-like mobile genetic elements (MGEs).
Fig. 1.
Gene annotations for predicted prophage sequences for phage groups 0 (a), 2 (b) and 14 (c). Predicted prophage sequence alignments are shown. Only two of the representatives of group 14 are shown. Unlabelled coding regions are annotated as ‘hypothetical proteins'.
The 47 predicted prophage sequences were next compared to the NCBI nr/nt database of viruses and bacteria using megablast. All of the sequences most closely resembled annotated tailed Streptococcus prophages [17, 31], including prophages of S. anginosus , as well as S. agalactiae , S. constellatus , S. crispatus, S. equi , S. gallolyticus , S. intermedius , S. mitis , S. oralis , S. pneumoniae , S. porcinus , S. thermophilus , S. uberis and S. urinalis . Table 2 summarizes the blast similarities identified for the S. anginosus predicted prophage sequences. Full details of the blast analyses, including blast query results to the bacterial sequences of the nr/nt databases, can be found in Table S2. While most of the groups include prophage sequences with only modest sequence similarity to previously identified prophages, the prophages within groups 3 and 9 closely resemble the S. agalactiae prophage Javan 32 (GenBank: MK448906) and the S. anginosus prophage Javan 68 (GenBank: MK449004), respectively. Furthermore, the group 3 S . anginosus prophages and the S. agalactiae prophage Javan 32 also exhibit modest sequence similarity (43 % genome sequence coverage and 78.41 % sequence identity) to the characterized S. pyogenes phage Φm46.1 (GenBank: FM8642313). In a previous study, Φm46.1 was found to be capable of being transduced in vitro to strains of S. agalactiae , S. gordonii and S. suis [28]. This suggests that the group 3 phages may have a broad host range. With respect to the human urinary tract, this host range could include both the uropathogen S. agalactiae and S. gordonii [3, 32]. We have not found S. pyogenes or S. suis in urine samples of either men or women.
Table 2.
Phage group demographics and similarity to previously characterized phages
|
Phage group number |
Number of phages in group |
Urinary host strains |
Average prophage length (kbp) |
Average query coverage (%) |
Average sequence identity (%) |
Streptococcus phage/prophage hit |
|---|---|---|---|---|---|---|
|
0 |
2 |
UMB4683, UMB8616 |
37.4±1.8 |
31.00 |
86.85 |
Javan345 and Javan11 |
|
11 |
4 |
UMB0622, UMB2128, UMB7052, UMB7052 |
20.4±13.5 |
17.00 |
85.66 |
Abc2, Javan64, Javan648 and Javan355 |
|
14 |
4 |
UMB0595, UMB0633, UMB7052, UMB8616 |
19.4±9.3 |
19.75 |
94.83 |
IPP15 |
|
174 |
2 |
UMB8616, UMB8616 |
101.2±12.2 |
40.50 |
96.86 |
Javan618 |
|
2 |
4 |
UMB0595, UMB0633, UMB7052, UMB8616 |
21.6±2.9 |
24.25 |
92.23 |
Javan64 and Javan115 |
|
3 |
6 |
UMB0248, UMB2128, UMB4708, UMB4708, UMB8390, UMB8710 |
31.7±12.5 |
84.50 |
99.32 |
Javan32 and Javan618 |
|
4 |
8 |
UMB0248, UMB0619, UMB2128, UMB2128, UMB4683, UMB4708, UMB4708, UMB8616 |
31.0±23.5 |
24.50 |
92.87 |
Javan64, Javan68, Javan110 and Javan278 |
|
7 |
4 |
UMB0622, UMB2128, UMB4683, UMB4708 |
65.0±58.3 |
39.50 |
92.15 |
Javan426 and Javan638 |
|
8 |
3 |
UMB0248, UMB0567, UMB0619 |
60.7±44.5 |
3.00 |
86.31 |
Javan64 and Javan318 |
|
9 |
3 |
UMB0248, UMB0622, UMB8616 |
42.3±22.2 |
82.67 |
99.82 |
Javan68 |
|
Singleton 134_3 |
1 |
UMB0248 |
103.2 |
26.00 |
82.57 |
Javan112 |
|
Singleton 135_18 |
1 |
UMB0567 |
23.7 |
79.00 |
97.41 |
Javan64 |
|
Singleton 159_24 |
1 |
UMB2128 |
29.7 |
6.00 |
96.38 |
Javan104 |
|
Singleton 161_1 |
1 |
UMB3444 |
37.4 |
21.00 |
94.03 |
Javan226 |
|
Singleton 161_8 |
1 |
UMB3444 |
83.8 |
18.00 |
91.66 |
Javan638 |
|
Singleton 165_21 |
1 |
UMB4708 |
33.1 |
52.00 |
99.80 |
Javan38 |
|
Singleton 171_15 |
1 |
UMB8390 |
44.9 |
40.00 |
86.35 |
Javan191 |
Evidence of Spontaneous Induction
Using the predicted prophage sequences, we designed primers to recognize prophages in groups 0, 2, 3, 4 and 14 (Table S3) to test for lytic phages induced spontaneously in liquid culture under laboratory conditions. The S. anginosus strains were first grown in BHI liquid medium at 35 °C in 5 % CO2. After 24 h of growth, 1 ml of the liquid culture was centrifuged at 13 000g for 1 min, and the medium was filtered through a 0.2 µm cellulose acetate syringe filter. Lysates underwent a DNase treatment, using 1 U of OPTIZYME DNase I (Fisher BioReagents) following the manufacturer’s instructions. PCR was performed using 1 µl of the DNAsed lysates added to 1 µM of the forward and reverse primers, 25 µl PCR Master Mix (Taq DNA polymerase; Promega) and 22 µl nuclease-free water, for a final reaction volume of 50 µl. Amplification was conducted as follows: 95 °C denaturation temperature for 30 s (with an initial period of 5 min), 55 °C annealing temperature for 30 s, 72 °C elongation temperature for 30 s (with a final period of 10 min) and a total of 30 cycles. Additionally, the DNase-treated lysates underwent 16S rRNA gene PCR amplification, using primers 63f and 1387r [33] to check for bacterial DNA contamination. No 16S rRNA gene amplification was detected in any of the lysate PCRs. Following the above procedure, we conducted two trials of PCR-based detection (Table 3). Between these two trials, we detected the presence of all of the predicted prophages in groups 2, 4 and 14. Neither trial detected spontaneous induction for the group 3 phages in UMB8390 or UMB8710 or the group 0 phage in UMB4683. The PCR results for Trial 2 are shown in Fig. S1. While several of the prophages were detected in both trials, some were detected only in one of the trials, suggesting that spontaneous induction does not always occur or does not always occur at a detectable level. However, this is not surprising as the mechanisms triggering induction of some of the predicted prophages probably vary [34]. Nevertheless, detection of these predicted prophages in the medium suggests that prophages are capable of reproducing within the S. anginosus host.
Table 3.
PCR detection of lytic phages for five of the identified groups
+, Amplification; −, no amplification.
|
Phage group |
Strain |
Amplification |
|
|---|---|---|---|
|
Trial 1 |
Trial 2 |
||
|
4 |
UMB0248 |
− |
+ |
|
UMB0619 |
+ |
+ |
|
|
UMB2128 |
+ |
+ |
|
|
UMB4683 |
− |
+ |
|
|
UMB4708 |
+ |
− |
|
|
UMB8616 |
+ |
+ |
|
|
2 |
UMB0595 |
+ |
+ |
|
UMB0633 |
+ |
+ |
|
|
UMB7052 |
+ |
+ |
|
|
UMB8616 |
+ |
+ |
|
|
3 |
UMB0248 |
− |
+ |
|
UMB2128 |
− |
+ |
|
|
UMB4708 |
+ |
+ |
|
|
UMB8390 |
− |
− |
|
|
UMB8710 |
− |
− |
|
|
14 |
UMB0595 |
− |
+ |
|
UMB0633 |
− |
+ |
|
|
UMB7052 |
+ |
+ |
|
|
UMB8616 |
+ |
− |
|
|
0 |
UMB4683 |
− |
− |
|
UMB8616 |
+ |
+ |
|
In addition to PCR confirmation, lysates were prepared for microscopy. Lysate was applied to grids (Electron Microscopy Sciences) and negatively stained with 2 % uranyl acetate for visualization with a JEOL 1200 EX transmission electron microscope. Fig. 2 shows phages induced from the culture of S. anginosus UMB4708 from Trial 1 of the PCR screen. The observed morphology is representative of siphoviruses. While UMB4708 is predicted to contain six different prophage sequences (Table 1), the virus-like particles within the micrograph are quite homogeneous, having similar sized capsids and similar tail lengths. The transmission electron micrograph produced here shows similar morphology and size to the siphovirus Φm46.1 [35], which does exhibit sequence similarity to the group 3 prophage detected for UMB4708 (Table 3). As we also detected a group 4 prophage for this S. anginosus strain via PCR, we cannot definitively state which predicted prophage is imaged. Nevertheless, this is the first virus-like particle visualized for S. anginosus .
Fig. 2.
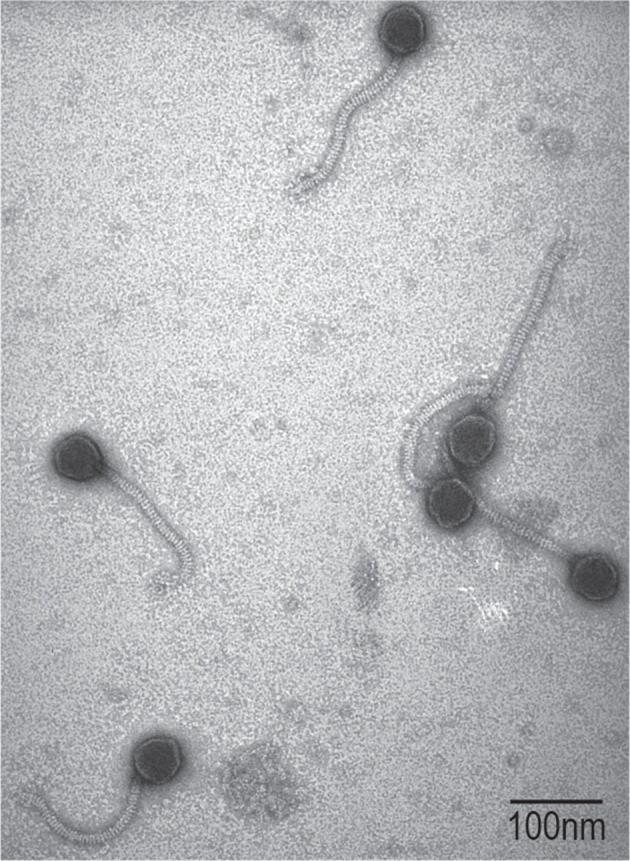
Transmission electron micrograph of virus-like particles induced from strain UMB4708. Bar, 100 nm.
While PCR amplification and transmission electron micropscopy confirmed the presence of phage particles, we were unable to identify plaques for any PCR-positive phage lysates. Each PCR-positive phage lysate was plated on each urinary S. anginosus strain in our collection listed in Table 1. Plaquing was assessed using the soft overlay method where 300 µl of overnight bacterial culture and 100 µl of lysate were mixed with 1 ml BHI soft agar (0.5 % agarose). Spotting 10 µl lysate on bacterial lawns was also tested, including different host densities (300 µl to 1 ml). Plates were incubated at 35 °C in 5 % CO2. While we also spotted lysates from cultures grown with the inducing agents mitomycin C and H2O2, again plaques were not observed. The lack of observable plaques, however, does not necessarily mean that these phages cannot propagate through the lytic cycle. The availability of a naïve, domesticated laboratory strain will greatly benefit future investigation of S. anginosus phages in the lytic cycle. To the best of our knowledge, only one phage has previously been documented to infect an S. anginosus strain – the temperate phage P10 from an S. oralis strain [36]. This is, however, the first analysis of S. anginosus prophage sequences and the first evidence of putative S. anginosus phages. Phages capable of infecting this emerging pathogen have potential therapeutic applications given the documented antibiotic resistance of many strains [37]. Thus, further characterization and isolation of S. anginosus is greatly needed.
Supplementary Data
Funding information
This work was funded by an Investigator Initiated grant from the Kimberly Clark Corporation (C.P. and A.J.W.) and by an NIH award (A.J.W., R01 DK104718).
Acknowledgements
We wish to acknowledge The Loyola Urinary Education and Research Collaborative for recruitment of study participants and attainment of urine samples, members of the Wolfe laboratory who isolated the S. anginosus strains from the samples, and Roberto Limeira of the Loyola Genomics Facility for sequencing of the bacterial and phage genomes.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
Catheterized urine samples were collected from women as part of prior studies approved by Loyola University Chicago’s IRB (numbers LU203986, LU206449 and LU207102).
Footnotes
Abbreviations: EQUC, Expanded Quantitative Urine Culture; MGE, Mobile Genetic Elements.
Three supplementary tables and one supplementary figure are available with the online version of this article.
Repositories: Streptococcus anginosus genome assemblies have been deposited under the following accession numbers: VYWV00000000, VYWU00000000, VYWR00000000, VYWQ00000000, VYWP00000000, VYWM00000000, VYVZ00000000, VYVX00000000, VYVW00000000, VYVU00000000, VYVS00000000, VYVP00000000, VYVM00000000 and VYVJ00000000.
References
- 1.Ruoff KL. Streptococcus anginosus (Streptococcus milleri): the unrecognized pathogen. Clin Microbiol Rev. 1988;1:102–108. doi: 10.1128/CMR.1.1.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pearce MM, Hilt EE, Rosenfeld AB, Zilliox MJ, Thomas-White K, et al. The female urinary microbiome: a comparison of women with and without urgency urinary incontinence. mBio. 2014;5:e01283–01214. doi: 10.1128/mBio.01283-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomas-White K, Forster SC, Kumar N, Van Kuiken M, Putonti C, et al. Culturing of female bladder bacteria reveals an interconnected urogenital microbiota. Nat Commun. 2018;9:1557. doi: 10.1038/s41467-018-03968-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parkins MD, Sibley CD, Surette MG, Rabin HR. The Streptococcus milleri group--an unrecognized cause of disease in cystic fibrosis: a case series and literature review. Pediatr Pulmonol. 2008;43:490–497. doi: 10.1002/ppul.20809. [DOI] [PubMed] [Google Scholar]
- 5.Chuang Y-M, Hsueh P-R, Lee P. Disseminated Streptococcus anginosus infection with empyema thoracis in a patient with sarcoma. J Formos Med Assoc. 2006;105:760–764. doi: 10.1016/S0929-6646(09)60205-9. [DOI] [PubMed] [Google Scholar]
- 6.Whiley RA, Beighton D, Winstanley TG, Fraser HY, Hardie JM. Streptococcus intermedius, Streptococcus constellatus, and Streptococcus anginosus (the Streptococcus milleri group): association with different body sites and clinical infections. J Clin Microbiol. 1992;30:243–244. doi: 10.1128/JCM.30.1.243-244.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moazzam AA, Rajagopal SM, Sedghizadeh PP, Zada G, Habibian M. Intracranial bacterial infections of oral origin. J Clin Neurosci. 2015;22:800–806. doi: 10.1016/j.jocn.2014.11.015. [DOI] [PubMed] [Google Scholar]
- 8.Esplin N, Stelzer JW, All S, Kumar S, Ghaffar E, et al. A case of Streptococcus anginosus brain abscess caused by contiguous spread from sinusitis in an immunocompetent patient. Cureus. 2017;9:e1745. doi: 10.7759/cureus.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suzuki H, Hase R, Otsuka Y, Hosokawa N. Bloodstream infections caused by Streptococcus anginosus group bacteria: A retrospective analysis of 78 cases at a Japanese tertiary hospital. J Infect Chemother. 2016;22:456–460. doi: 10.1016/j.jiac.2016.03.017. [DOI] [PubMed] [Google Scholar]
- 10.Fazili T, Riddell S, Kiska D, Endy T, Giurgea L, et al. Streptococcus anginosus group bacterial infections. Am J Med Sci. 2017;354:257–261. doi: 10.1016/j.amjms.2017.05.011. [DOI] [PubMed] [Google Scholar]
- 11.Furuichi M, Horikoshi Y. Sites of infection associated with Streptococcus anginosus group among children. J Infect Chemother. 2018;24:99–102. doi: 10.1016/j.jiac.2017.09.011. [DOI] [PubMed] [Google Scholar]
- 12.Maharaj S, Seegobin K, Chrzanowski S, Chang S. Acute glomerulonephritis secondary to Streptococcus anginosus . BMJ Case Rep. 2018;2018:bcr-2017-223314. doi: 10.1136/bcr-2017-223314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campisciano G, Zanotta N, Petix V, Corich L, De Seta F, et al. Vaginal microbiota dysmicrobism and role of biofilm-forming bacteria. Front Biosci. 2018;10:528–536. doi: 10.2741/E839. [DOI] [PubMed] [Google Scholar]
- 14.Zuñiga-Bahamon A, Tobar-Tosse F, Guillermo-Ortega J, Wibberg D, Tauch A. Draft genome sequence of Streptococcus anginosus BVI, a new vaginal pathogen candidate. Genome Announc. 2016;4:e01417–16. doi: 10.1128/genomeA.01417-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Price TK, Dune T, Hilt EE, Thomas-White KJ, Kliethermes S, et al. The clinical urine culture: enhanced techniques improve detection of clinically relevant microorganisms. J Clin Microbiol. 2016;54:1216–1222. doi: 10.1128/JCM.00044-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller-Ensminger T, Garretto A, Brenner J, Thomas-White K, Zambom A, et al. Bacteriophages of the urinary microbiome. J Bacteriol. 2018;200:e00738–17. doi: 10.1128/JB.00738-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rezaei Javan R, Ramos-Sevillano E, Akter A, Brown J, Brueggemann AB. Prophages and satellite prophages are widespread in Streptococcus and may play a role in pneumococcal pathogenesis. Nat Commun. 2019;10:4852. doi: 10.1038/s41467-019-12825-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomas-White KJ, Hilt EE, Fok C, Pearce MM, Mueller ER, et al. Incontinence medication response relates to the female urinary microbiota. Int Urogynecol J. 2016;27:723–733. doi: 10.1007/s00192-015-2847-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pearce MM, Zilliox MJ, Rosenfeld AB, Thomas-White KJ, Richter HE, et al. The female urinary microbiome in urgency urinary incontinence. Am J Obstet Gynecol. 2015;213:347.e1–34347. doi: 10.1016/j.ajog.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hilt EE, McKinley K, Pearce MM, Rosenfeld AB, Zilliox MJ, et al. Urine is not sterile: use of enhanced urine culture techniques to detect resident bacterial flora in the adult female bladder. J Clin Microbiol. 2014;52:871–876. doi: 10.1128/JCM.02876-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, et al. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016;44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arndt D, Grant JR, Marcu A, Sajed T, Pon A, et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44:W16–W21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roux S, Enault F, Hurwitz BL, Sullivan MB. VirSorter: mining viral signal from microbial genomic data. PeerJ. 2015;3:e985. doi: 10.7717/peerj.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 26.Wattam AR, Brettin T, Davis JJ, Gerdes S, Kenyon R, et al. Assembly, annotation, and comparative genomics in PATRIC, the all bacterial bioinformatics resource center. Methods Mol Biol. 2018;1704:79–101. doi: 10.1007/978-1-4939-7463-4_4. [DOI] [PubMed] [Google Scholar]
- 27.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giovanetti E, Brenciani A, Morroni G, Tiberi E, Pasquaroli S, et al. Transduction of the Streptococcus pyogenes bacteriophage Φm46.1, carrying resistance genes mef(A) and tet(O), to other Streptococcus species. Front Microbiol. 2014;5:746. doi: 10.3389/fmicb.2014.00746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dedrick RM, Mavrich TN, Ng WL, Cervantes Reyes JC, Olm MR, et al. Function, expression, specificity, diversity and incompatibility of actinobacteriophage parABS systems. Mol Microbiol. 2016;101:625–644. doi: 10.1111/mmi.13414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deutsch DR, Utter B, Verratti KJ, Sichtig H, Tallon LJ, et al. Extra-Chromosomal DNA Sequencing reveals episomal prophages capable of impacting virulence factor expression in Staphylococcus aureus . Front Microbiol. 2018;9:1406. doi: 10.3389/fmicb.2018.01406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brueggemann AB, Harrold CL, Rezaei Javan R, van Tonder AJ, McDonnell AJ, et al. Pneumococcal prophages are diverse, but not without structure or history. Sci Rep. 2017;7:42976. doi: 10.1038/srep42976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kline KA, Lewis AL. Gram-positive uropathogens, polymicrobial urinary tract infection, and the emerging microbiota of the urinary tract. Microbiol Spectr. 2016;4:UTI-0012-2012. doi: 10.1128/microbiolspec.UTI-0012-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marchesi JR, Sato T, Weightman AJ, Martin TA, Fry JC, et al. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl Environ Microbiol. 1998;64:795–799. doi: 10.1128/AEM.64.2.795-799.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nanda AM, Thormann K, Frunzke J. Impact of spontaneous prophage induction on the fitness of bacterial populations and host-microbe interactions. J Bacteriol. 2015;197:410–419. doi: 10.1128/JB.02230-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brenciani A, Bacciaglia A, Vignaroli C, Pugnaloni A, Varaldo PE, et al. Phim46.1, the main Streptococcus pyogenes element carrying mef(A) and tet(O) genes. Antimicrob Agents Chemother. 2010;54:221–229. doi: 10.1128/AAC.00499-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Szafrański SP, Winkel A, Stiesch M. The use of bacteriophages to biocontrol oral biofilms. J Biotechnol. 2017;250:29–44. doi: 10.1016/j.jbiotec.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 37.Babbar A, Kumar VN, Bergmann R, Barrantes I, Pieper DH, et al. Members of a new subgroup of Streptococcus anginosus harbor virulence related genes previously observed in Streptococcus pyogenes . Int J Med Microbiol. 2017;307:174–181. doi: 10.1016/j.ijmm.2017.02.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.