Abstract
The paramyxoviruses Hendra virus (HeV) and parainfluenza virus 5 (PIV5) require the fusion (F) protein to efficiently infect cells. For fusion to occur, F undergoes dramatic, essentially irreversible conformational changes to merge the viral and cell membranes into a continuous bilayer. Recently, a transmembrane (TM) domain leucine/isoleucine (L/I) zipper was shown to be critical in maintaining the expression, stability and pre-fusion conformation of HeV F, allowing for fine-tuned timing of membrane fusion. To analyse the effect of the TM domain L/I zipper in another paramyxovirus, we created alanine mutations to the TM domain of PIV5 F, a paramyxovirus model system. Our data show that while the PIV5 F TM L/I zipper does not significantly affect total expression and only modestly affects surface expression and pre-fusion stability, it is critical for fusogenic activity. These results suggest that the roles of TM L/I zipper motifs differ among members of the family Paramyxoviridae.
Keywords: pre-fusion stability, fusogenic activity, leucine/isoleucine zipper, transmembrane domain
Observation
Measles virus (MeV), parainfluenza virus 5 (PIV5) and the zoonotic Hendra virus (HeV), are enveloped viruses that belong to the family Paramyxoviridae [1]. MeV and HeV are both highly pathogenic viruses of worldwide significance [2], while PIV5 serves as an important Paramyxoviridae viral model [3]. For these enveloped viruses, successful infection requires fusion of their membranes with target cell membranes to allow for content mixing [4, 5]. Since membrane fusion is energetically costly [6], the fusion protein (F) and attachment proteins (G, HN or H) serve as critical viral surface proteins that lower the kinetic barrier to drive the fusion and entry process. For fusion to occur, the attachment protein tethers the viral particle to the host cell via interactions with cellular receptors; subsequently, the F protein drives the fusion process by undergoing large-scale, essentially irreversible conformational changes from a metastable pre-fusion structure to a highly stable post-fusion conformation that results in the merging of the viral and target cell membranes [7–9].
Like other class I fusion proteins, paramyxovirus F proteins are synthesized in a metastable pre-fusion state and folded in the endoplasmic reticulum (ER) into a homotrimer that must be proteolytically cleaved to become fusogenically active [6]. The cleavage unveils a fusion peptide (FP) that interacts with the target membrane to facilitate fusion [10, 11]. Upon synthesis, HeV F is trafficked through the secretory pathway to be expressed on the cell surface in its fusogenically inactive form (F0). Subsequently, HeV F0 is endocytosed and cleaved by the protease cathepsin L within endosomes and retrafficked to the surface in a disulfide-linked fusogenically active form (F1 +F2) [4, 12]. Conversely, PIV5 F trafficking is more straightforward: it is similarly synthesized in the secretory pathway, but undergoes cleavage within the trans-Golgi network by furin during transport to the cell surface to be expressed as F1 + F2 [4, 7, 9, 12, 13]. Importantly, throughout the trafficking process, paramyxovirus F proteins must be maintained in a metastable pre-fusion state, as premature triggering renders the protein fusion dead [14].
Although certain paramyxovirus F proteins such as Sendai virus (SeV) F can trigger in the absence of their homotypic attachment proteins [15], most paramyxoviruses engage in complex interactions with their attachment proteins to begin refolding from the pre-fusion to the post-fusion state [9, 16]. A number of studies have focused on how external domains and cytoplasmic tails of F proteins impact fusion [5, 17, 18]. However, the transmembrane (TM) domain, which was initially thought to mainly serve as a membrane anchor, has recently been shown to play critical roles in the pre-fusion stability of the paramyxovirus F proteins [4, 14, 19–23]. In isolation, the TM domains of HeV F, PIV5 F and the closely related pneumovirus human metapneumovirus (HMPV) F self-associate in trimers. For HeV F, an AXXXG motif, similar to the GXXXG motif known to support association of hydrophobic residues, was found to be important for maintaining surface levels of the cleaved pre-fusion form [4]. On further investigation, another important association motif, the leucine/isoleucine (L/I) zipper was identified in the TM domain of HeV F and similar β-branched residues in heptad repeats were found for 140 other paramyxoviruses, including PIV5 F. Studies on HeV F showed that not only is the L/I zipper important for the self-association of the TM domains in isolation, but it is also important in the pre-fusion stability of the full HeV F protein. These studies showed a severe reduction in surface and total expression for the HeV L/I zipper mutants, termed LIZ. Not surprisingly, HeV F LIZ is also deficient in forming syncytia, a consequence of not being stably present on the cell surface [14, 19, 20].
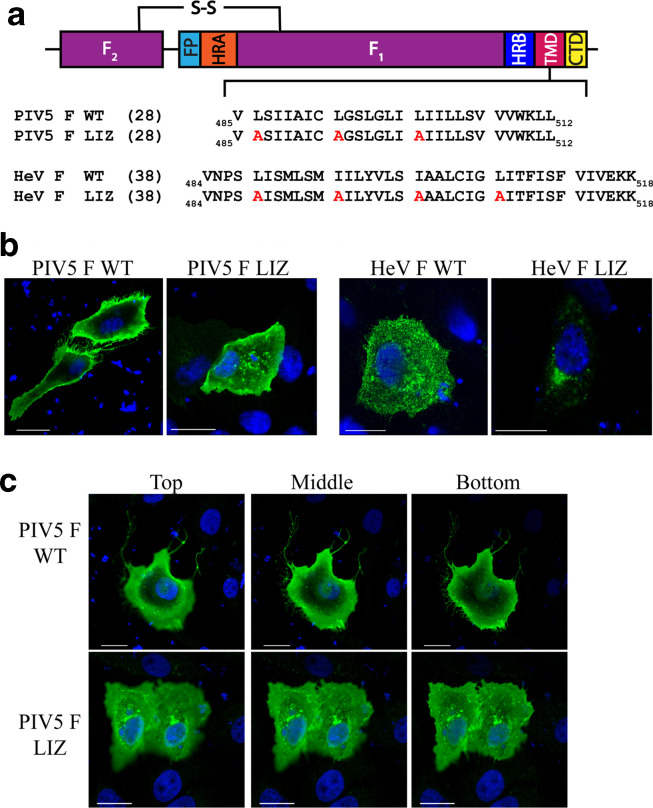
To test the role of the TM L/I zipper motif in another paramyxovirus F, we introduced TM L/I zipper alanine mutations to create PIV5 F LIZ (Fig. 1a). With this mutant, we performed immunofluorescence [20] on cells transfected with 0.75 µg of DNA to analyse the intracellular localization compared to PIV5 F WT. For PIV5, we used a monoclonal pre-fusion-specific antibody PIV5 F1a (kindly provided by Dr Richard Randall, University of St Andrews). HeV F WT and HeV F LIZ localization were also analysed using HeV F mAb 5G7 (kindly provided by Dr Chris Broder, Uniformed Services University of Health Sciences). Our results demonstrate that while HeV F WT displays a global cellular distribution, HeV F LIZ is mostly confined in pockets around the nucleus consistent with the endoplasmic reticulum (Fig. 1b), corroborating previous data that showed that surface and total expression of HeV F LIZ was considerably lower than for HeV F WT [14]. Interestingly, we found that PIV5 F WT primarily localizes at the surface of the cell and is present within membrane ruffles, while PIV5 F LIZ shows a higher level of intracellular distribution. We further examined multiple focal planes of cells transfected with PIV5 F WT and LIZ using Z-stacks (Fig. 1c ). We found that PIV5 F WT is mostly absent from the immediate perinuclear region in different optical slices, but PIV5 F LIZ is distributed throughout the cells and is present in puncta close to the nucleus. These results suggest a more subtle yet significant effect of the PIV5 F L/I zipper in trafficking and intracellular localization of the protein. Likewise, these data indicate a more modest effect of the TM L/I zipper in protein folding for PIV5 F than for HeV F.
Fig. 1.
Mutations to the L/I zipper of HeV F and PIV5 F have variable effects. (a). Schematic of the paramyxovirus fusion protein highlighting the TM domain L/I zipper of HeV F and PIV5 F, and the mutant constructs. FP, fusion peptide; HRA, heptad repeat A; HRB, heptad repeat B; TMD, transmembrane domain; CT, cytoplasmic tail); S–S, disulfide bond. (b). Immunofluorescence to visualize localization of HeV and PIV5 F proteins. Vero cells were seeded in eight-well chamber plates and transfected with 0.75 µg PIV5 F WT or LIZ mutant (left), and HeV F WT or LIZ mutant (right). Localization of HeV F was analysed with anti-F 5G7 antibodies, and PIV5 F analysed with mAb F1a (green). Images were taken with a Nikon 1A confocal microscope. Images are representative. Scale bars represent 10 µm. (c). Z-stack images from (b) were collected in 0.3 µm sections, and images corresponding to top, bottom and middle slices are shown. Images are representative of two independent experiments carried out in triplicate. Scale bars represent 10 µm.
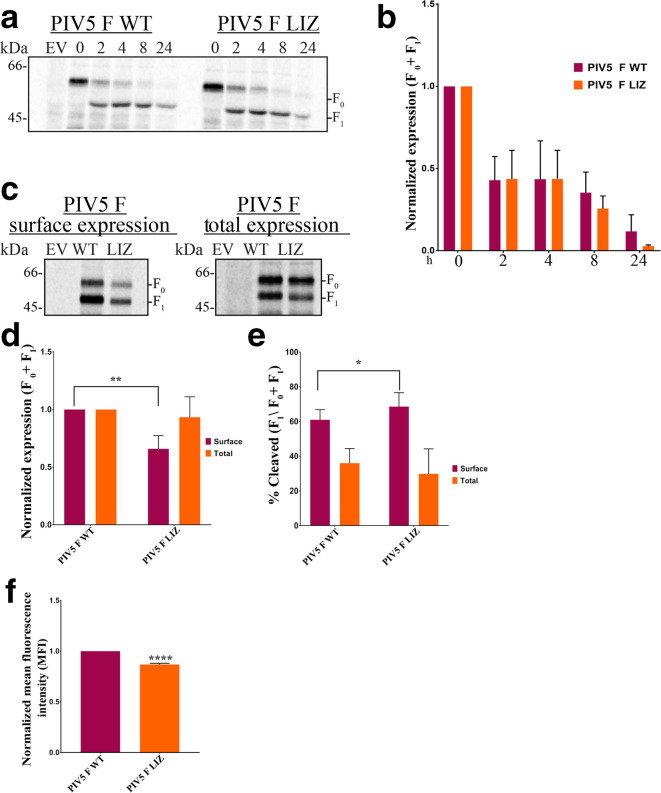
To biochemically assess the effect of the L/I zipper on PIV5 F synthesis and stability, we performed a pulse-chase time course assay as previously described [14] with PIV5 cytoplasmic tail antibody 516–529 to examine PIV5 F WT and PIV5 F LIZ stability (Fig. 2a). Previous studies showed that total expression over time for HeV F LIZ was significantly decreased compared to WT [14]. Surprisingly, between 0 and 8 h, PIV5 F LIZ expression was comparable to WT (Fig. 2b), suggesting that the PIV5 F L/I zipper is not critical for stability, unlike the L/I zipper of HeV F [14].
Fig. 2.
Expression and stability of PIV5 F WT and LIZ are comparable. (a). Assessment of stability of PIV5 F WT or LIZ over time course. A pulse-chase experiment was carried out 18 h after cells were transfected with 2.5 µg of indicated DNA for Vero cells in six-well plates. Following a 30 minute S35 metabolic radiolabel, samples were chased for indicated times. (b). Quantitation of PIV5 F and LIZ expression shown in (a). Expression levels of total F protein (F0+F1) were determined by band densitometry normalized to WT levels. (c). Percentage of cleaved F compared to total F (F1/F0+F1) normalized to WT. The averages represent three independent experiments, each carried out in duplicate. (d). Surface and total expression of PIV5 F protein. Quantitation of expression levels (e) and percentage cleavage (f) of surface and total PIV5 F protein. (g). Flow cytometry to quantify expression of pre-fusion PIV5 F only present at the surface of cells. The averages represent three independent experiments, each carried out in duplicate. The LIZ mutant was compared to WT using Student’s t-test. *, P<0.05; **, P<0.005; ****P<0.0001
Since the presence of F at the membrane is crucial for biological activity, we probed the surface expression of PIV5 F WT and LIZ in transfected cells using a surface biotinylation assay [11]. We found that after radiolabelling newly synthesized F protein for 3 h with S35 at 18–24 h post-transfection, PIV5 F LIZ surface expression is decreased by more than 30 % when compared to WT, but total amounts of protein between PIV5 F WT and LIZ remain comparable (Fig. 2c–e). Therefore, at the surface of cells, the total of pre-fusion and post-fusion forms of PIV5 F WT is slightly higher than for PIV5 F LIZ. To build on these findings, we selectively quantified the population of potentially fusogenically active protein at the surface by performing flow cytometry, as outlined by Bose et al. [18], using the pre-fusionspecific PIV5 F mAb F1a. These data also show that the pre-fusion form of PIV5 F LIZ is only slightly lower than for PIV5 F WT (Fig. 2f), showing a significant presence of potentially fusogenically active PIV5 at the surface of cells in the absence of the L/I zipper.
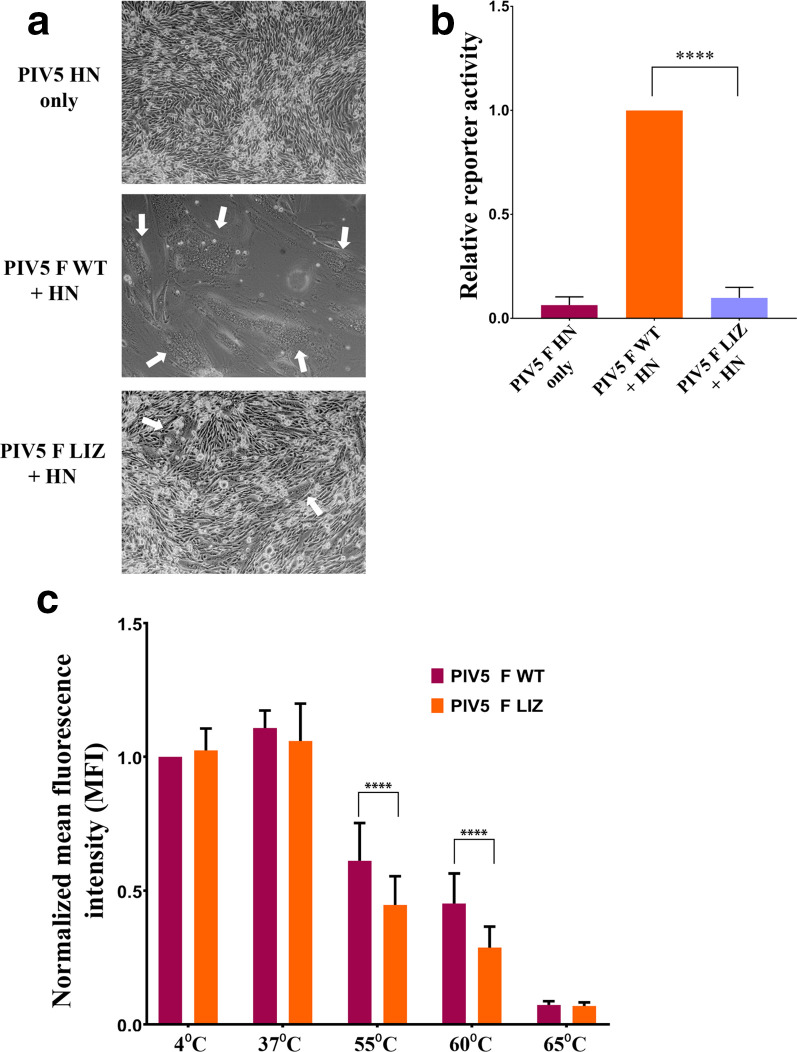
Having established that pre-fusion PIV5 F LIZ can still be trafficked to the surface of cells, we utilized a syncytia assay, as detailed by Webb et al. [14], to test the fusogenic activity of PIV5 F LIZ in comparison to WT. We observed that PIV5 F WT is highly fusogenic, as the expression of PIV5 F WT with HN resulted in BHK cells fusing into a few large syncytia. Remarkably, syncytial activity was abolished in the PIV5 F LIZ mutant (Fig. 3a). Further characterization of fusogenic activity using a luciferase reporter system (described by Barrett et al. [20]) also showed quantitatively that the PIV5 F TM L/I zipper is critical for fusion (Fig. 3b). Notably, a leucine residue at position 468 (L486) has been reported to be important for fusogenic activity. L486 was shown to be critical for both membrane mixing and content mixing, thus identifying L486 as essential in the events leading up to the merge of lipid bilayers driven by F [21]. L486 is present in the proposed L/I zipper of PIV5 F, corroborating this finding in the context of the L/I zipper.
Fig. 3.
Mutations to the L/I zipper of PIV5 reduce F-mediated fusion activity. (a). Syncytia assay. BHK cells plated in six-well plates were transfected with 2.5 µg of total DNA with the PIV5 HN attachment protein alone, PIV5 WT F and HN or PIV5 LIZ F and HN. Syncytia formation was analysed 24 h post-transfection. Images were taken with a Nikon TS100 microscope. White arrows indicate syncytia. Images are representative of two independent experiments, each carried out in triplicate. (b). Luciferase reporter gene assay to quantify F fusogenic activity. Vero cells in 24-well plates were transfected with 1.0 µg total DNA with a T7 promoter plasmid and PIV5 F WT+HN or pIV5 F LIZ+HN. The following day, Vero cells were overlaid with BSR cells and incubated for 3 h to allow for luciferase production. Luciferase activity was measured using a luciferase assay system. The average represents three independent experiments, each performed in duplicate. (c). Thermal triggering assay to observe PIV5 F WT and LIZ pre-fusion thermostability. Cells expressing surface PIV5 F or WT were exposed to 4, 37, 55, 60 or 65 °C for 15 min. Cells were immediately placed on ice for 15 min and prepared for flow cytometry using PIV5 mAb F1a. The average represents two independent experiments, each performed in triplicate. The LIZ mutant was compared to WT using a using Student’s t-test. ****P<0.0001
The comparable PIV5 F WT and LIZ pre-fusion surface expression levels, in contrast to the dramatic decrease in HeV F LIZ when compared to HeV F WT, show that L/I zippers in the TM domains of HeV F and PIV5 F play distinct but critical roles in maintaining biological activity. While these studies suggest that fusion, rather than surface or total expression, is significantly affected by residues within the PIV5 F TM L/I zipper, the exact mechanism by which these LIZ mutations abrogate fusion is currently unknown. To understand whether PIV5 F LIZ is capable of being triggered from its pre-fusion form to undergo the conformational changes that are critical for membrane fusion, we transfected cells with either PIV5 F WT or PIV5 F LIZ for a thermal triggering assay [18]. Previous studies show that PIV5 F can be triggered in the absence of its cognate attachment protein when exposed to heat [18, 24]. Thus PIV5 F WT- and LIZ-expressing cells were exposed to increasing temperatures (Fig. 3c), and flow cytometry using the pre-fusion-specific mAb PIV5 F1a was utilized to quantitate the levels of pre-fusion F. Triggering of conformational changes in response to heat would lead to loss of F1a binding. As the temperature increased, the detected levels of pre-fusion F decreased for both the WT and LIZ F proteins (Fig. 3c). Interestingly, at 55 and 60 °C, a statistically significant increase in the triggering of PIV5 F LIZ compared to WT (Fig. 3c) was observed, potentially indicating a role for the TM L/I zipper in stabilizing PIV5 F in the pre-fusion conformation. This stabilization is less dramatic than was observed for HeV F [14], but does suggest that a role for the LIZ in pre-fusion stability may be a property across the viral family. However, it is unlikely that this small decline in the thermostability of pre-fusion PIV5 F LIZ would fully account for the drastic loss of fusogenic activity shown in the syncytia and reporter gene assays (Fig. 3a and b).
Reports show that for class I fusion proteins such as Ebola virus GP2, influenza virus HA and PIV5 F, the FP and TM domains interact in the post-fusion conformation [11, 22, 25, 26]. It is possible that for PIV5 F, the L/I zipper within the TM domain contributes to making essential contacts with the fusion peptide to hold the post-fusion conformation in place and merge viral and target membranes. Additionally, studies demonstrate that the TMs of class I fusion proteins induce local membrane changes that decrease the energy barrier needed for fusion [10, 22, 26, 27] – as such, the L/I zipper of PIV5 F may contribute in this local disruption. Finally, it is important to note that PIV5 F is known to make contact with HN through an Ig-like domain at the ectodomain [18]. The L/I zipper may be involved in transmitting conformational changes that result from this initial contact, and thus in refolding. Alternatively, F could have important interactions with HN through contacts with the TM domain L/I zipper, which are disrupted by the LIZ mutations.
Funding information
This work was supported by NIAID grant R01AI051517 and NIH grant 2P20 RR02017 to R. E. D.
Acknowledgements
We gratefully acknowledge Dr Richard Randall (University of St Andrews) for kindly providing PIV5 mAb F1a and Dr Chris Broder (Uniformed Services University of Health Sciences) for kindly providing HeV F Mab 5G7. We also thank the Dutch laboratory for input on the manuscript.
Author contributions
Conceptualization, J. M. B and R. E. D.; methodology, J. M. B and R. E. D; investigation, J. M. B. and R. E. D.; writing J. M. B. and R. E. D.; funding acquisition, R. E. D.; resources, R. E. D.; supervision, R. E. D.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: CT, cytoplasmic tail; F, Fusion protein; FP, fusion peptide; G, Glycoprotein; HeV, Hendra virus; H/HA, Hemagglutinin; HN, Hemagglutinin neuramninidase attachment protein; HRA, heptad repeat A; HRB, heptad repeat B; LIZ, Leucine/isoleucine zipper-alanine mutant; L/I zipper, leucine/isoleucine zipper; MeV, Measles virus; PIV5, parainfluenza virus 5; S-S, disulfide bond; TM, transmembrane; TMD, transmembrane domain.
References
- 1.Rima B, Collins P, Easton A, Fouchier R, Kurath G, et al. Problems of classification in the family Paramyxoviridae. Arch Virol. 2018;163:1395–1404. doi: 10.1007/s00705-018-3720-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cox RM, Plemper RK. Structure and organization of paramyxovirus particles. Curr Opin Virol. 2017;24:105–114. doi: 10.1016/j.coviro.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harrison MS, Sakaguchi T, Schmitt AP. Paramyxovirus assembly and budding: building particles that transmit infections. Int J Biochem Cell Biol. 2010;42:1416–1429. doi: 10.1016/j.biocel.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith EC, Smith SE, Carter JR, Webb SR, Gibson KM, et al. Trimeric transmembrane domain interactions in paramyxovirus fusion proteins: roles in protein folding, stability, and function. J Biol Chem. 2013;288:35726–35735. doi: 10.1074/jbc.M113.514554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aguilar HC, Henderson BA, Zamora JL, Johnston GP. Paramyxovirus glycoproteins and the membrane fusion process. Curr Clin Microbiol Rep. 2016;3:142–154. doi: 10.1007/s40588-016-0040-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martens S, McMahon HT. Mechanisms of membrane fusion: disparate players and common principles. Nat Rev Mol Cell Biol. 2008;9:543–556. doi: 10.1038/nrm2417. [DOI] [PubMed] [Google Scholar]
- 7.El Najjar F, Schmitt AP, Dutch RE. Paramyxovirus glycoprotein incorporation, assembly and budding: a three way dance for infectious particle production. Viruses. 2014;6:3019–3054. doi: 10.3390/v6083019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim YH, Donald JE, Grigoryan G, Leser GP, Fadeev AY, et al. Capture and imaging of a prehairpin fusion intermediate of the paramyxovirus PIV5. Proc Natl Acad Sci USA. 2011;108:20992–20997. doi: 10.1073/pnas.1116034108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang A, Dutch RE. Paramyxovirus fusion and entry: multiple paths to a common end. Viruses. 2012;4:613–636. doi: 10.3390/v4040613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yao H, Hong M. Membrane-Dependent conformation, dynamics, and lipid interactions of the fusion peptide of the paramyxovirus PIV5 from solid-state NMR. J Mol Biol. 2013;425:563–576. doi: 10.1016/j.jmb.2012.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith EC, Gregory SM, Tamm LK, Creamer TP, Dutch RE. Role of sequence and structure of the Hendra fusion protein fusion peptide in membrane fusion. J Biol Chem. 2012;287:30035–30048. doi: 10.1074/jbc.M112.367862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pager CT, Craft WW, Patch J, Dutch RE. A mature and fusogenic form of the Nipah virus fusion protein requires proteolytic processing by cathepsin L. Virology. 2006;346:251–257. doi: 10.1016/j.virol.2006.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Welch BD, Liu Y, Kors CA, Leser GP, Jardetzky TS, et al. Structure of the cleavage-activated prefusion form of the parainfluenza virus 5 fusion protein. Proc Natl Acad Sci USA. 2012;109:16672–16677. doi: 10.1073/pnas.1213802109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Webb S, Nagy T, Moseley H, Fried M, Dutch R. Hendra virus fusion protein transmembrane domain contributes to pre-fusion protein stability. J Biol Chem. 2017;292:5685–5694. doi: 10.1074/jbc.M117.777235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leyrer S, Bitzer M, Lauer U, Kramer J, Neubert WJ, et al. Sendai virus-like particles devoid of haemagglutinin-neuraminidase protein infect cells via the human asialoglycoprotein receptor. J Gen Virol. 1998;79 (Pt 4:683–687. doi: 10.1099/0022-1317-79-4-683. [DOI] [PubMed] [Google Scholar]
- 16.Smith EC, Popa A, Chang A, Masante C, Dutch RE. Viral entry mechanisms: the increasing diversity of paramyxovirus entry. Febs J. 2009;276:7217–7227. doi: 10.1111/j.1742-4658.2009.07401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee JK, Prussia A, Paal T, White LK, Snyder JP, et al. Functional interaction between paramyxovirus fusion and attachment proteins. J Biol Chem. 2008;283:16561–16572. doi: 10.1074/jbc.M801018200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bose S, Heath CM, Shah PA, Alayyoubi M, Jardetzky TS, et al. Mutations in the parainfluenza virus 5 fusion protein reveal domains important for fusion triggering and metastability. J Virol. 2013;87:13520–13531. doi: 10.1128/JVI.02123-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Webb SR, Smith SE, Fried MG, Dutch RE. Transmembrane domains of highly pathogenic viral fusion proteins exhibit trimeric association in vitro . mSphere. 2018;3:e00047-18. doi: 10.1128/mSphere.00047-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barrett CT, Webb SR, Dutch RE. A hydrophobic target: using the paramyxovirus fusion protein transmembrane domain to modulate fusion protein stability. J Virol. 2019;93 doi: 10.1128/JVI.00863-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bissonnette MLZ, Donald JE, DeGrado WF, Jardetzky TS, Lamb RA. Functional analysis of the transmembrane domain in paramyxovirus F protein-mediated membrane fusion. J Mol Biol. 2009;386:14–36. doi: 10.1016/j.jmb.2008.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Donald JE, Zhang Y, Fiorin G, Carnevale V, Slochower DR, et al. Transmembrane orientation and possible role of the fusogenic peptide from parainfluenza virus 5 (PIV5) in promoting fusion. Proc Natl Acad Sci USA. 2011;108:3958–3963. doi: 10.1073/pnas.1019668108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Porotto M, Yokoyama CC, Palermo LM, Mungall B, Aljofan M, et al. Viral entry inhibitors targeted to the membrane site of action. J Virol. 2010;84:6760–6768. doi: 10.1128/JVI.00135-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poor TA, Jones LM, Sood A, Leser GP, Plasencia MD, et al. Probing the paramyxovirus fusion (F) protein-refolding event from pre- to postfusion by oxidative footprinting. Proc Natl Acad Sci USA. 2014;111:E2596–E2605. doi: 10.1073/pnas.1408983111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lai AL, Freed JH. The interaction between influenza HA fusion peptide and transmembrane domain affects membrane structure. Biophys J. 2015;109:2523–2536. doi: 10.1016/j.bpj.2015.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee J, Nyenhuis DA, Nelson EA, Cafiso DS, White JM, et al. Structure of the Ebola virus envelope protein MPER/TM domain and its interaction with the fusion loop explains their fusion activity. Proc Natl Acad Sci U S A. 2017;114:E7987–E7996. doi: 10.1073/pnas.1708052114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwon B, Lee M, Waring AJ, Hong M. Oligomeric structure and three-dimensional fold of the HIV gp41 membrane-proximal external region and transmembrane domain in phospholipid bilayers. J Am Chem Soc. 2018;140:8246–8259. doi: 10.1021/jacs.8b04010. [DOI] [PMC free article] [PubMed] [Google Scholar]