Abstract
The family Botourmiaviridae includes viruses infecting plants and filamentous fungi containing a positive-sense, ssRNA genome that can be mono- or multi-segmented. Genera in the family include: Ourmiavirus (plant viruses), and Botoulivirus, Magoulivirus and Scleroulivirus (fungal viruses). This is a summary of the International Committee on Taxonomy of Viruses (ICTV) Report on the taxonomy of the family Botourmiaviridae, which is available at ictv.global/report/botourmiaviridae.
Keywords: Botourmiaviridae, ICTV Report, taxonomy
Virion
Members of the genus Ourmiavirus are plant viruses with non-enveloped bacilliform virions that are composed of a single 23.8 kDa coat protein. Electron microscopy reveals particles with conical ends (apparently hemi-icosahedral) and cylindrical bodies that are 18 nm in diameter (Table 1, Fig. 1). Most particles consist of two discs (giving a particle length of 30 nm), with other particles having three (37 nm) or, more rarely, either four (45.5 nm) or six discs (62 nm). Members of other genera infect fungi and are not encapsidated.
Table 1.
Characteristics of members of the family Botourmiaviridae
|
Typical member: |
Ourmia melon virus VE9 (RNA1: EU770623; RNA2: EU770624; RNA3: EU770625), species Ourmia melon virus, genus Ourmiavirus |
|---|---|
|
Virion |
Bacilliform (18×30–62 nm) with a 23.8 kDa coat protein (Ourmiavirus) or unencapsidated (members of other genera) |
|
Genome |
Positive-sense RNA of 2–3 kb (Botoulivirus, Magoulivirus and Scleroulivirus) or three segments of 2.8, 1.1 and 0.97 kb (Ourmiavirus) |
|
Replication |
Cytoplasmic; virion assembly is coupled to active replication |
|
Translation |
From genomic RNA; each genomic segment is monocistronic |
|
Host range |
Plants and fungi |
|
Taxonomy |
Realm Riboviria, kingdom Orthornavira, phylum Lenarviricota, class Miaviricetes, order Ourlivirales, family Botourmiaviridae, several genera each with multiple species |
Fig. 1.
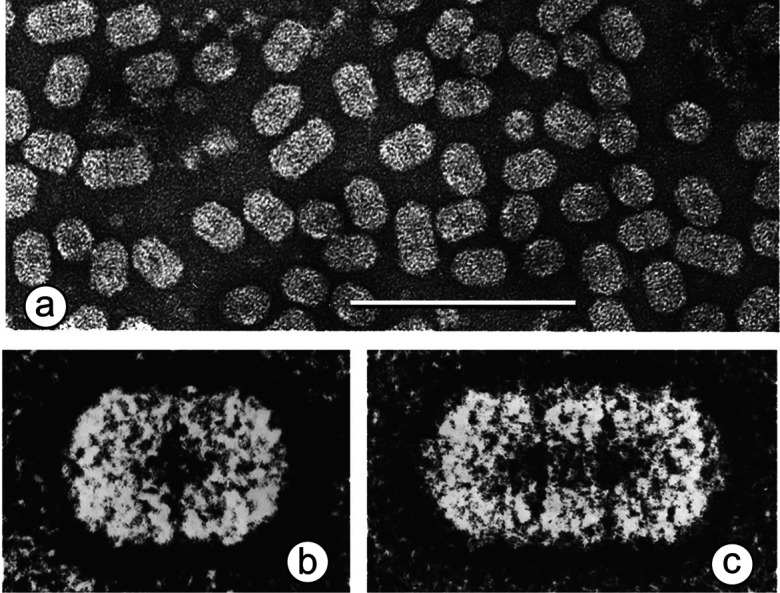
Virion morphology: (a) negative-contrast electron micrographs (uranyl acetate) of purified particles of Ourmia melon virus (bar, 100 nm); (b, c) features of the two commonest particle types (two- and three-disc), enhanced by photographic superimposition.
Genome
The genome of ourmiaviruses consists of three segments of positive-sense, ssRNA (2814, 1064 and 974 nt for Ourmia melon virus) [1] encoding an RNA-directed RNA polymerase (97.5 kDa; RNA1), a movement protein (31.6 kDa; RNA2) and a coat protein (23.8 kDa; RNA3) (Fig. 2) [2]. Members of other genera have a genome with a single segment of 2000–3200 nt encoding an RNA-directed RNA polymerase [3–6]. Unusually for the family, the genome of Magnaporthe oryzae ourmia-like virus 1 (species Magnaporthe magoulivirus 1, genus Magoulivirus) is polyadenylated at the 3′-end (4).
Fig. 2.
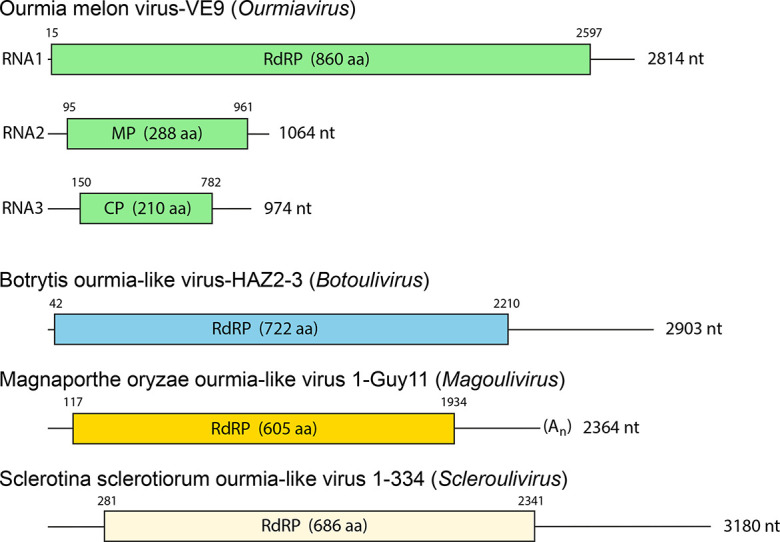
Genome organization of representative isolates of the family Botourmiaviridae. Boxes indicate the position and size of the ORFs encoding the coat protein (CP), movement protein (MP) and RNA-directed RNA polymerase (RdRP).
Replication
Ourmiavirus replication is dependent on the virus RNA-directed RNA polymerase. Synthesis of the ourmiavirus coat protein from actively replicating RNA3 is necessary for both virion assembly and systemic infection of the host [2]. The ourmiavirus movement protein determines symptoms and forms tubular structures involved in cell-to-cell movement [7] and may undergo post-translational modification. Botoulivirus, magoulivirus and scleroulivirus replication is strictly dependent on the virus RNA-directed RNA polymerase.
Pathogenicity
Members of the genera Botoulivirus, Magoulivirus and Scleroulivirus infect fungi from the genera Botrytis, Magnaporthe or Sclerotinia, respectively [3–5]. Members of the genus Ourmiavirus infect plants; Ourmia melon virus infects melon, producing chlorotic spots and irregular ringspots [8], Epirus cherry virus produces rasp-leaf symptoms in cherry, and cassava virus C induces severe stunting and a yellow mosaic pattern in cassava.
Taxonomy
Botourmiaviruses are more closely related to members of the genus Narnavirus (family Narnaviridae) than to members of the genus Mitovirus (family Narnaviridae). Members of different botourmiavirus genera differ by >70 % in RNA-directed RNA polymerase amino acid sequence.
Resources
Current ICTV Report on the family Botourmiaviridae: ictv.global/report/botourmiaviridae
Funding information
Production of this summary, the online chapter and associated resources was funded by a grant from the Wellcome Trust (WT108418AIA).
Acknowledgements
Members of the ICTV Report Consortium are Stuart G. Siddell, Andrew J. Davison, Elliot J. Lefkowitz, Sead Sabanadzovic, Peter Simmonds, Donald B. Smith, Richard J. Orton and F. Murilo Zerbini.
Conflicts of interest
The authors declare that there are no conflicts of interest.
References
- 1.Rastgou M, Habibi MK, Izadpanah K, Masenga V, Milne RG, et al. Molecular characterization of the plant virus genus Ourmiavirus and evidence of inter-kingdom reassortment of viral genome segments as its possible route of origin. J Gen Virol. 2009;90:2525–2535. doi: 10.1099/vir.0.013086-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crivelli G, Ciuffo M, Genre A, Masenga V, Turina M. Reverse genetic analysis of Ourmiaviruses reveals the nucleolar localization of the coat protein in Nicotiana benthamiana and unusual requirements for virion formation. J Virol. 2011;85:5091–5104. doi: 10.1128/JVI.02565-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Donaire L, Rozas J, Ayllón MA. Molecular characterization of Botrytis ourmia-like virus, a mycovirus close to the plant pathogenic genus Ourmiavirus . Virology. 2016;489:158–164. doi: 10.1016/j.virol.2015.11.027. [DOI] [PubMed] [Google Scholar]
- 4.Marzano S-YL, Nelson BD, Ajayi-Oyetunde O, Bradley CA, Hughes TJ, et al. Identification of diverse mycoviruses through metatranscriptomics characterization of the viromes of five major fungal plant pathogens. J Virol. 2016;90:6846–6863. doi: 10.1128/JVI.00357-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marzano S-YL, Domier LL. Novel mycoviruses discovered from metatranscriptomics survey of soybean phyllosphere phytobiomes. Virus Res. 2016;213:332–342. doi: 10.1016/j.virusres.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 6.Illana A, Marconi M, Rodríguez-Romero J, Xu P, Dalmay T, et al. Molecular characterization of a novel ssRNA ourmia-like virus from the rice blast fungus Magnaporthe oryzae . Arch Virol. 2017;162:891–895. doi: 10.1007/s00705-016-3144-9. [DOI] [PubMed] [Google Scholar]
- 7.Margaria P, Anderson CT, Turina M, Rosa C. Identification of Ourmiavirus 30K movement protein amino acid residues involved in symptomatology, viral movement, subcellular localization and tubule formation. Mol Plant Pathol. 2016;17:1063–1079. doi: 10.1111/mpp.12348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lisa V, Milne RG, Accotto GP, Boccardo G, Caciagli P, et al. Ourmia melon virus, a virus from Iran with novel properties. Ann Appl Biol. 1988;112:291–302. doi: 10.1111/j.1744-7348.1988.tb02065.x. [DOI] [Google Scholar]


