Abstract
Our previous research confirmed the repression of SMADs signaling pathway inhibits the invasion, migration, and EMT in breast cancer MCF-7 and SKBR-3 cell lines by DNMT1 up-regulating CLDN6, but the mechanism is unclear. Western blot was performed to detect the expression of SMAD2, SMAD3, P-SMAD2, and P-SMAD3. Then RT-PCR was carried out to examine the expression of tight junctions and cell adhesion molecule E-cadherin. According to the gene sequence of Claudin6, shRNA was linked with the green fluorescent protein-expressing eukaryotic expression vector pGC silencer TMΜ6/Neo/GFP, and it was transfected into breast cancer MCF-7 cells and SKBR-3 cells. RT-PCR and western blot were applied to verify the Claudin6 gene-silencing effect. We observed cellular morphology with inverted microscope, analyzed the capacity for clone formation, and detected transepithelial electrical resistance. The level of MMP2, and MMP9 in the cells treated with or without SB431542 and MCF-7-shGFP, MCF-7-shClaudin-6, SKBR-3-shGFP, and SKBR-3-shClaudin-6 cells pretreated with SB431542 were examined by RT-PCR and western blot. The expressions of Claudin-6, occludin, and cell adhesion molecule E-cadherin were enhanced by SB431542. SB431542 transformed mesenchymal cell morphology into epithelial cell morphology, inhibited capacity for clone formation, increased transepithelial electrical resistance, and downregulated the expression of MMP2 and MMP9. Knock down of Claudin6 can abolish SB431542 effects. We conclude that Claudin6 mediates the effects of SB431542 on the biologic phenotypes of the breast cancer cells we studied. We speculate Claudin6-mediated the SB431542 inhibition of invasion, migration, and EMT in breast cancer cells via MMP2/9.
Keywords: SMADs, Claudin6, TEERs, clone formation, morphology, MMPs
Introduction
Cancer cells escape from the primary sites to distant sites by local infiltration or intravasation into the blood or lymphatic vessels. The transportation is required during the formation of metastases [1,2]. This process facilitates changes in the morphology and movement of cancer cells and leads to changes in cell-cell adhesion molecules [1]. In epithelial tissues, the linker complex including adhesive junctions (AJs), tight junctions (TJs), desmosomes, and gap junctions is located between cells [3]. Epithelial cells are connected each other by adhesion molecules at the AJs that mediate stable cohesion between cells [4,5]. These junctional complexes include E-Cadherin (CDH1), which binds with β-catenin and α-catenin in the junctional complex [4-6]. The loss of cell-cell adhesion and cell junctions mediated by CDH1 homophilic binding promotes cells to dissociate from the primary tumor, invade neighboring tissues, and migrate to remote sites [7].
Occludin, Claudins (CLDNs) and junctional adhesion molecules (JAMs) constitute TJs [8]. CLDNs are a pivotal component of TJs [9]. CLDNs family consists 27 members [10]. As major transmembrane proteins, CLDNs play key roles in the formation and maintenance of tight junctions [11]. It is generally acknowledged that the disruption of tight junctions causes the loss of intercellular cohesion. It promotes the aggressiveness and lack of differentiation of cancer cells, which leads to metastasis. Previous studies have demonstrated that mRNA or membrane protein expression levels of CLDNs were strongly correlated with carcinogenesis in breast cancer [12-14].
Unlimited proliferation is a characteristic of cancer cell activity [12]. It is accompanied by morphologic manifestations both in vitro in cell culture and in vivo during tumor proliferation, invasion, and metastasis. During in vitro cell culture, the proliferation results in the formation of cell clones. The biologic activity of cancer cells can be reflected by the clone formation ability and morphologic features [16-18].
Cancer cells acquire invasive and migratory abilities by epithelial-to-mesenchymal transition (EMT). The capability facilitates tumor dissemination and metastasis [19]. Matrix metalloproteinases (MMPs) play a role in regulating EMT, invasion, and migration. Various MMPs play an important role in cancer progression and metastasis [20].
CLDNs can regulate a variety of cancers’ proliferation, invasion, and metastasis. The CLDNs possess different functions in different kinds of tumors. They play a key role during tumorigenesis [21]. Studies have shown that CLDN6 can induce the apoptosis, and inhibit the invasion, and migration of breast cancer cells MCF-7 [22]. Our previous studies showed that Smads signaling pathway can regulate the Claudin6 expression by DNA methyltransferase1 (DNMT1), and can regulate the invasion, migration, and EMT in breast cancer cells, but the mechanism is not clear [23]. Here, we show that CLDN6 mediates the Smads signaling pathway inducing morphologic change, enhances transepithelial resistance and inhibits clone formation in breast cancer. Knock down of CLDN6 can abolish downregulation of MMP2 and MMP9 expression by SB431542.
Materials and methods
Cell culture and reagents
We acquired the human breast cancer cell lines MCF-7 and SKBR-3 from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China) and cultured the cells in DMEM (Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (Hyclone, Logan, UT, USA) and 100 units/ml penicillin and streptomycin (Invitrogen, Carlsbad, CA, USA). All the cell lines were cultured in a humidified incubator (5% CO2, 37°C). SB431542 was obtained from Sigma (St. Louis, MO, USA).
Reverse transcription-polymerase chain reaction (RT-PCR)
Total RNA was extracted from cells using TRIzol (Invitrogen, USA) according to the manufacturer’s instructions. One microgram of total RNA was used for reverse transcription to synthesize cDNA using the MMuLVreverse transcriptase (TaKaRa, Japan) for 1 hour at 42°C, and 0.5 µg cDNA was used for PCR. ZO-2, ZO-1, Occludin, CLDN7, CLDN6, CDH1, MMP2 and MMP9 were amplified along with GAPDH as an endogenous control following the instructions of Premix LA Taq Kit (TaKaRa, Japan). The primer sequences and the PCR reaction conditions are listed in Table 1. After electrophoresis, the gel was imaged and analyzed by an imaging system (Syngene, Cambridge, UK).
Table 1.
Primers and information for RT-PCR
| Primer Name | Primer Sequence | Length (bp) | Annealing Temp (°C) | Cycles |
|---|---|---|---|---|
| GAPDH | 5’-TGTTGCCATCAATGACCCCTT-3’ | 178 | 56 | 25 |
| 5’-CTCCACGACGTACTCAGCG-3’ | ||||
| CLDN6 | 5’-TTCATCGGCAACAGCATCGT-3’ | 345 | 58 | 35 |
| 5’-GGTTATAGAAGTCCCGGATGA-3’ | ||||
| CLDN4 | 5’-TGCACTCTGCGAACGTTAAG-3’ | 141 | 56 | 35 |
| 5’-GCGATGGCCATTACCTGTAG-3’ | ||||
| CLDN7 | 5’-GGAGATCCCAGGTCACACAT-3’ | 140 | 50 | 30 |
| 5’-CAGGGTCTGCCCTAGTCATC-3’ | ||||
| Occludin | 5’-AAAATGTGTCTGCAGGCACACAGGACG-3’ | 101 | 55 | 35 |
| 5’-AGGCTGCCTGAAGTCATCCACAGGC-3’ | ||||
| ZO-2 | 5’-GCCAAAACCCAGAACAAAGA-3’ | 203 | 55 | 30 |
| 5’-AACACTGGCAAATATCACAGC-3’ | ||||
| ZO-1 | 5’-CACGATGCTCAGAGACGAAGG-3’ | 156 | 55 | 30 |
| 5’-TTCTACATATGGAAGTTGGGGATC-3’ | ||||
| CDH1 | 5’-ATTCTGGGGATTCTTGGAGG-3’ | 337 | 56 | 30 |
| 5’-GGTCAGTATCAGCCGCTTTC-3’ | ||||
| MMP2 | 5’-TCTCCGACATTGACCTTGGC-3’ | 302 | 60 | 30 |
| 5’-CAGGGTGCTGGCTGAGTAGATC-3’ | ||||
| MMP9 | 5’-TTGACAGCGACAAGAAGTGG-3’ | 180 | 58 | 30 |
| 5’-GCCATTCACGTTCGTCCTTAT-3’ |
Western blotting analysis
Western blotting experiments were carried out as described previously [24]. Anti-SMAD2, SMAD3, P-SMAD2, and P-SMAD3 antibodies were from Cell Signaling Technology (Beverly, MA, USA); Anti-MMP2, MMP9, CLDN6 antibodies were purchased from Abcam (Cambridge, UK). The anti-β-actin antibody was obtained from Santa Cruz (Santa Cruz, CA, USA). Primary and secondary antibodies were both diluted at 1:1000. The blots were imaged and analyzed applying an ECL western blotting system (GE, Fairfield, CT, USA).
Cell clone formation assay
Tumor cells were digested using 0.25% trypsin/0.02% EDTA solution at the logarithmic phase to make a single-cell suspension in culture medium. The cells were plated into six-well culture plates at 1,000 cells each well. The medium was changed every 3 days until cell clones could be observed by the unaided eye [25].
Transient transfection
Cells were transfected with short hairpin RNA (shRNA) by applying Lipofectamine 2000 (Invitrogen), according to the specification. ShRNA targeting of CLDN6 (50-GTGCAAGGTGTACGACTCA-30) and a negative control shRNA were both obtained from GeneChem Co. Ltd.
Transepithelial electrical resistance (TEER) measurement
The cells were cultured in Millicell chambers with 6-well culture plates at 5.0 × 105 cells per well; TEERs of cells were detected according to the Millicell-ERS instructions when cells were cultured until a monolayer of cell fused. The resistance of Millicell chambers’ unseeded cells was detected as blank control (Rblank). The resistance of each group of cells is the difference between the average of the resistance of each group of cells and the Rblank multiplied by the diameter of the chamber.
Statistical analysis
All analysis was performed using the SPSS version 22.0 for Windows (SPSS Inc., Chicago, IL, USA). Unpaired t-tests were performed to assess data for target mRNA and protein. The data are presented as means ± standard error from at least three independent experiments. P < 0.05 was considered significant.
Results
SMADs signaling pathway was inhibited by SB431542 in breast cancer cells
The breast cancer cells were incubated with SB431542, an inhibitor of SMADs signaling pathway, RT-PCR and western blot analysis showed that P-SMAD2 and P-SMAD3 expression was decreased in MCF-7 and SKBR-3 cells treated with SB431542 (Figure 1).
Figure 1.
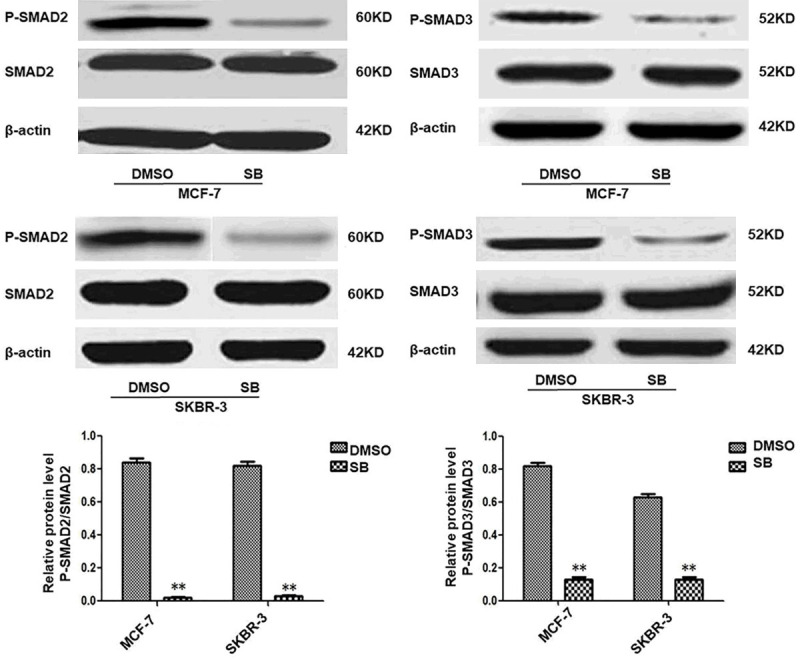
SB431542 inhibits SMADs signaling in MCF-7 and SKBR-3 cells. SB431542 downregulated P-SMAD2 and P-SMAD3 expression. MCF-7 and SKBR-3 cells were incubated with SB431542 for 24 h at 10 μM. Results of densitometry analysis of relative expression levels of P-SMAD2 and P-SMAD3 after normalization to SMAD2 and SMAD3 are presented (*P < 0.05 and **P < 0.01 are considered significant and highly significant, respectively). Bars represent mean ± SE (n = 3).
SMADs signaling pathway regulates the expression of tight junction protein and E-cadherin in breast cancer cells
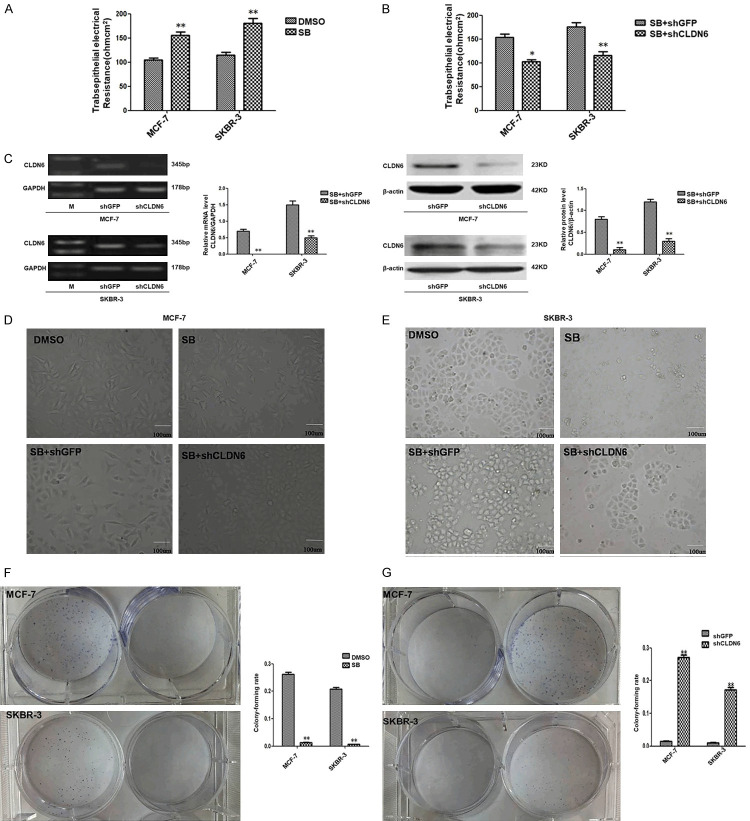
We have already reported that inhibition of the SMADs signal pathway can reverse the EMT process which is one of the key steps in the tumor metastasis and inhibits the migration and invasion of breast cancer cells. In order to study further the effects of CLDN6-mediated inhibition of the SMADs signaling pathway on the occurrence and development of breast cancer, MCF-7 and SKBR-3 cells were treated with 10 µM SB431542 for 24 h. RT-PCR demonstrated that inhibition of SMADs signaling pathway dysregulated the expression of tight junction protein CLDN6, occludin, and E-cadherin in MCF-7 and SKBR-3 cells (Figure 2). TEER is a generally acknowledged quantitative technique to measure the intactness of tight junction dynamics in cell culture models of endothelial and epithelial monolayers [21]. The TEER was detected. The result showed that SB431542 increased the TEER in MCF-7 and SKBR-3 cells (Table 2; Figure 3A) Thus, our results suggested that SB431542 enhanced TEER by regulating the expression of tight junction protein and E-cadherin in breast cancer cells. The changes in cell-cell connections are closely related to tumor formation and development. In order to confirm whether CLDN6 mediates the effect of SB431542 on TEER, CLDN6 in MCF-7 and SKBR-3 cells treated with SB431542 as knocked down by shRNA. The result showed that the TEER was decreased (Table 3; Figure 3B). RT-PCR and western blot analysis demonstrated that CLDN6 was significantly downregulated in the cells that had knocked down CLDN6, compared to the control group (Figure 3C). Therefore, we conclude that CLDN6 mediates the SB431542 effect on TEER in MCF-7 and SKBR-3 cells.
Figure 2.
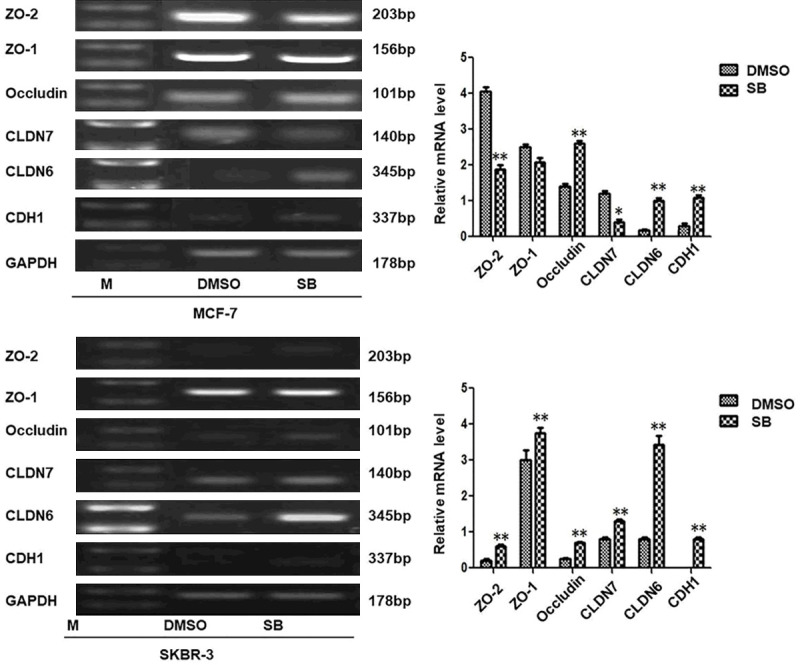
SMADs signaling regulates expression of tight junction protein and E-cadherin. Real-time polymerase chain reaction (RT-PCR) analysis of tight junction protein (ZO-2, ZO-1, occludin, CLDN7, and CLDN6) and E-cadherin; and densitometric analysis of relative gene expression levels after normalization to loading control GAPDH are presented. The lane “M” stands for marker; (*P < 0.05 and **P < 0.01 are considered significant and highly significant, respectively). Bars represent mean ± SE (n = 3).
Table 2.
Effect of SB431542 on TEER in MCF-7 and SKBR-3 cells
| Experimental Group | TEER (ohm·cm2) | |
|---|---|---|
|
| ||
| MCF-7 | SKBR-3 | |
| DMSO | 105 ± 7 | 114.3 ± 11.5 |
| SB431542 | 155.3 ± 12.5** | 180.3 ± 17.5** |
p<0.01 is considered highly significant.
Figure 3.
CLDN6 is required for the breast cancer cell TEER, morphology, and clone formation. A, B. CLDN6-mediated inactivation of SMADs signaling pathway regulated TEER (*P < 0.05 and **P < 0.01 are considered significant and highly significant, respectively). bars represent mean ± SE (n = 3). C. RT-PCR and immunoblot expression analyses of CLDN6 in MCF-7-shGFP, MCF-7-shCLDN6, SKBR-3-shGFP, and SKBR-3-shCLDN6 cells pretreated by SB431542, and densitometric analysis of relative expression levels after normalization to loading control GAPDH or β-actin are presented. The lane “M” stands for marker (*P < 0.05 and **P < 0.01 are significant and highly significant, respectively). Bars represent mean ± SE (n = 3). D, E. CLDN6-mediated inactivation of SMADs signaling pathway affected cell morphology (Bar, 100 μm). F, G. CLDN6 was required to inhibit cell clone formation (*P < 0.05 and **P < 0.01 are significant and highly significant, respectively). Bars represent mean ± SE (n = 3).
Table 3.
CLDN6 mediates the effect of SB431542 on TEER in MCF-7 and SKBR-3 cells
| Experimental Group | TEER (ohm·cm2) | |
|---|---|---|
|
| ||
| MCF-7 | SKBR-3 | |
| SB431542 + shGFP | 153.3 ± 12.5 | 176 ± 15 |
| SB431542 + shCLDN6 | 102.3 ± 7* | 116 ± 13** |
p<0.05 is considered significant and highly significant.
p<0.01 is considered significant and highly significant.
CLDN6 effects on breast cancer cellular morphology
MCF-7 and SKBR-3 cells were treated with 10 µM SB431542 for 24 h, and the cell morphology was observed. SB431542 caused a profound effect on the morphology of MCF-7 and SKBR-3 cells. MCF-7 cell morphology changed from long spindle shape to short spindle shape. SKBR-3 cells acquired a cobble-shaped epithelial phenotype as opposed to a polygon. MCF-7 and SKBR-3 cells treated with SB431542 were used to knock down CLDN6 through shRNA. The change in morphology caused by SB431542 was reversed (Figure 3D, 3E). Therefore, we conclude that knocking down CLDN6 blocks the effect of SB431542 on the morphology of MCF-7 and SKBR-3 cells.
CLDN6 effects on breast cancer cell clone formation
MCF-7 and SKBR-3 cells were treated with 10 µM SB431542 for one week. We observed the cell clone formation. SB431542 inhibited the cell clone formation (P < 0.05) (Figure 3F). Knocking down CLDN6 showed that the cell clone formation was increased significantly (Figure 3G).
CLDN6 effects on the expression of MMP2 and MMP9
In our previous experiment, it was confirmed CLDN6-mediated repression of SMADs signaling pathway inhibited the invasion, migration and EMT. CLDN6-mediated inhibition of Smad signaling pathway changed the breast cancer cell morphology, increased transepithelial resistance, and reduced clone formation which played a crucial role during the EMT, invasion, and migration progression in our current study. It is unclear that the molecular mechanism of CLDN6-mediated inhibition of SMADs signal pathway represses the invasion, migration, and EMT. To address this problem, MMP2 and MMP9 were detected by RT-PCR and Western blot. MMP2 and MMP9 were down-regulated following SB431542 treatment. CLDN6 was knocked down after the cells were treated with SB431542. The down-regulation of MMP2 and MMP9 expression by SB431542 was blocked in MCF-7 cells (Figure 4A, 4B). We got similar results in SKBR-3 cells (results not shown).
Figure 4.
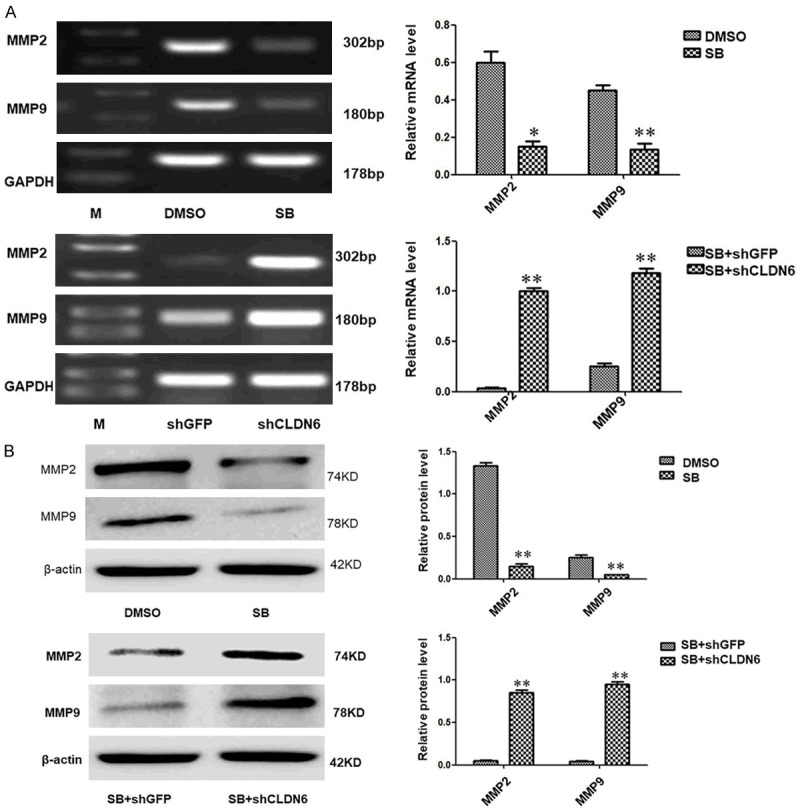
CLDN6-mediated Smads signaling pathway regulates the expression of MMP2 and MMP9. (A) RT-PCR and (B) immunoblot expression analyses of MMP2 and MMP9 in MCF-7 cells treated with or without SB431542, MCF-7-shGFP, and MCF-7-shCLDN6 pretreated with SB431542, and densitometric analysis of relative expression levels after normalization to loading control GAPDH or β-actin are presented (*P < 0.05 and **P < 0.01 are significant and highly significant, respectively). Bars represent mean ± SE (n = 3).
Discussion
Breast cancer is one of the most common tumors. It is derived from the ductal epithelium of the breast [27-29]. The metastasis and recurrence of breast cancer is the main cause of death [30]. The metastases require the cancer cells to break away from the original site. They infiltrate the surrounding tissue, invade into the vasculature, extravasate to distal sites, and grow [1,2]. The process accompanies morphologic change, movement of carcinoma cells, and repression of cell-cell adhesion molecule expression [1]. In an earlier study, we reported that CLDN6 can reverse EMT which is a step in tumor metastasis, and inhibit breast cancer cell migration and invasion by mediating the inhibition of the SMADs signaling pathway [23], In the current research, we aimed to further explore the effects of CLDN6 mediating the inhibition of SMADs signaling pathway on the development of the MCF-7 and SKBR-3 breast cancer cells.
RT-PCR was performed. The result manifested that inhibition of SMADs signaling pathway upregulated some of the expression of occludin and CLDN6 tight junction proteins and E-cadherin in MCF-7 and SKBR-3 cells. TEER is a quantitative technique to detect the integrity of tight junction dynamics in cell culture models of endothelial and epithelial monolayers [26]. The TEER was detected. The results showed that SB431542 increased the TEER in MCF-7 and SKBR-3 cells. Thus, our results suggested that SB431542 enhanced TEER by regulating the expression of tight junction protein and E-cadherin in breast cancer cells. Similar to our results, Krizbai [31] also found that SB431542 enhanced TEER, indicating disruption in rat brain endothelial cell monolayer integrity. The changes in cell-cell connections were closely related to tumor formation and development. CLDNs play crucial roles in the formation and maintenance of tight junctions [11]. It is generally accepted that the disruption of tight junctions leads to the loss of intercellular cohesion, which causes the invasiveness and de-differentiation of cancer cells and facilitates metastasis. Earlier studies have suggested that mRNA or membrane protein expression levels of CLDNs were strongly correlated with carcinogenesis in BC [12-14].
In order to confirm that CLDN6 mediates the SB431542 regulation of the TEER in MCF-7 and SKBR-3 cells, the cells were treated with SB431542. CLDN6 in the cells was knocked down. The result showed that the TEER was decreased. Therefore, we can conclude that CLDN6 mediates the SB431542 effect on TEER in MCF-7 and SKBR-3 cells.
Uncontrolled proliferation is a hallmark of cancer cells [15]. It accompanies morphologic manifestations in cells cultured in vitro and during tumor proliferation, invasion and metastasis in vivo. In cells cultured in vitro, cell proliferation results in the formation of cell clones. The biologic behavior of cancer cells can be reflected by the clone formation rate and morphologic characteristics [16-18]. CLDNs can regulate proliferation, invasion, and metastasis in a variety of cancer types [32-35]. In the current study, SB431542 inhibited the cell clone formation. Knocking down CLDN6 obstructed the inhibitory effects of SB431542 on clone formation.
In the present research, SB431542 resulted in a profound effect on the morphology of MCF-7 and SKBR-3 cells. MCF-7 cell morphology changed from a long spindle shape to short spindle shape. SKBR-3 cells acquired a cobble-shaped epithelial phenotype as opposed to a polygon. After knocking down CLDN6, the changes in morphology caused by SB431542 were abrogated. Therefore, we conclude that CLDN6 mediates the function of SB431542 on the clone formation and morphology of MCF-7 and SKBR-3 cells.
The CLDNs have different functions in different tumors. They play a key role during tumorigenesis [19]. Studies have shown that claudin6 can inhibit the apoptosis, invasion, and migration of breast cancer cells MCF-7 [22]. Our previous studies showed that Smads signaling pathway can regulate the CLDN6 expression by DNMT1, and can regulate the invasion, migration and EMT in breast cancer cells, but the mechanism is not clear [23]. Here, we show that Cloudin 6 mediates the inhibition of Smads signaling pathway regulating the breast cancer cell morphology, increasing transepithelial resistance, and lessening clone formation, which are closely involved in EMT [36,37]. The levels of MMP2 and MMP9 in cells were downregulated following SB431542 treatment. CLDN6 was knocked down after the cells treated with SB431542, and the down-regulation of MMP2 and MMP9 was blocked. We speculate that Claudin6-mediated effects of SB431542 inhibit the invasion, migration, and EMT in breast cancer cells through MMP2 and MMP9. We will further investigate the molecular mechanisms by which SB431542/CLDN6 regulate the expression of MMPs to inhibit EMT, migration, and invasion in breast cancer.
Despite recent prevention, diagnosis, and therapy advances, lomany patients still die due to metastasis and recurrence. Evidence reveals that the EMT process plays an essential role in metastasis and recurrence of cancer [38]. It reveals the mechanism responsible for EMT. Genes, especially CLDN6 and signaling associated with EMT should be given high priority in order to decrease cancer morbidity and mortality.
Acknowledgements
Manuscript preparation funded by Science and Technology Plan Project in Qiqihar City (Code: SFGG-201762) and Item of Scientific Research Fund For Doctor of Qiqihar Medical University (Code: QY2016B-25). The authors thank Dr William Orr, Department of Pathology, University of Manitoba, Canada, for help in this manuscript preparation.
Disclosure of conflict of interest
None.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Lambert AW, Pattabiraman DR, Weinberg RA. Emerging biological principles of metastasis. Cell. 2017;168:670–691. doi: 10.1016/j.cell.2016.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Franke WW. Discovering the molecular components of intercellular junctions--a historical view. Cold Spring Harb Perspect Biol. 2009;1:a003061. doi: 10.1101/cshperspect.a003061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wirtz-Peitz F, Zallen JA. Junctional trafficking and epithelial morphogenesis. Curr Opin Genet Dev. 2009;19:350–6. doi: 10.1016/j.gde.2009.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ragkousi K, Gibson MC. Cell division and the maintenance of epithelial order. J Cell Biol. 2014;207:181–8. doi: 10.1083/jcb.201408044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valenta T, Hausmann G, Basler K. The many faces and functions of beta-catenin. EMBO J. 2012;31:2714–36. doi: 10.1038/emboj.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petrova YI, Schecterson L, Gumbiner BM. Roles for E-cadherin cell surface regulation in cancer. Mol Biol Cell. 2016;27:3233–3244. doi: 10.1091/mbc.E16-01-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brennan K, Offiah G, McSherry EA, Hopkins AM. Tight junctions: a barrier to the initiation and progression of breast cancer? J Biomed Biotechnol. 2010;2010:460607. doi: 10.1155/2010/460607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakagawa S, Miyoshi N, Ishii H, Mimori K, Tanaka F, Sekimoto M, Doki Y, Mori M. Expression of CLDN1 in colorectal cancer: a novel marker for prognosis. Int J Oncol. 2011;39:791–6. doi: 10.3892/ijo.2011.1102. [DOI] [PubMed] [Google Scholar]
- 10.Mineta K, Yamamoto Y, Yamazaki Y, Tanaka H, Tada Y, Saito K, Tamura A, Igarashi M, Endo T, Takeuchi K, Tsukita S. Predicted expansion of the claudin multigene family. FEBS Lett. 2011;585:606–12. doi: 10.1016/j.febslet.2011.01.028. [DOI] [PubMed] [Google Scholar]
- 11.Hewitt KJ, Agarwal R, Morin PJ. The claudin gene family: expression in normal and neoplastic tissues. BMC Cancer. 2006;6:186. doi: 10.1186/1471-2407-6-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tokés AM, Kulka J, Paku S, Szik A, Páska C, Novák PK, Szilák L, Kiss A, Bögi K, Schaff Z. Claudin-1, -3 and -4 proteins and mRNA expression in benign and malignant breast lesions: a research study. Breast Cancer Res. 2005;7:R296–305. doi: 10.1186/bcr983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim TH, Huh JH, Lee S, Kang H, Kim GI, An HJ. Down-regulation of claudin-2 in breast carcinomas is associated with advanced disease. Histopathology. 2008;53:48–55. doi: 10.1111/j.1365-2559.2008.03052.x. [DOI] [PubMed] [Google Scholar]
- 14.Kominsky SL, Argani P, Korz D, Evron E, Raman V, Garrett E, Rein A, Sauter G, Kallioniemi OP, Sukumar S. Loss of the tight junction protein claudin-7 correlates with histological grade in both ductal carcinoma in situ and invasive ductal carcinoma of the breast. Oncogene. 2003;22:2021–33. doi: 10.1038/sj.onc.1206199. [DOI] [PubMed] [Google Scholar]
- 15.Dowsett M, Nielsen TO, A’Hern R, Bartlett J, Coombes RC, Cuzick J, Ellis M, Henry NL, Hugh JC, Lively T, McShane L, Paik S, Penault-Llorca F, Prudkin L, Regan M, Salter J, Sotiriou C, Smith IE, Viale G, Zujewski JA, Hayes DF International Ki-67 in Breast Cancer Working Group. Assessment of Ki67 in breast cancer: recommendations from the international Ki67 in breast cancer working group. J Natl Cancer Inst. 2011;103:1656–64. doi: 10.1093/jnci/djr393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soltysova A, Altanerova V, Altaner C. Cancer stem cells. Neoplasma. 2005;52:435–40. [PubMed] [Google Scholar]
- 17.Franken NA, Rodermond HM, Stap J, Haveman J, van Bree C. Clonogenic assay of cells in vitro. Nat Protoc. 2006;1:2315–9. doi: 10.1038/nprot.2006.339. [DOI] [PubMed] [Google Scholar]
- 18.Munshi A, Hobbs M, Meyn RE. Clonogenic cell survival assay. Methods Mol Med. 2005;110:21–8. doi: 10.1385/1-59259-869-2:021. [DOI] [PubMed] [Google Scholar]
- 19.Nieto MA, Huang RY, Jackson RA, Thiery JP. Emt: 2016. Cell. 2016;166:21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 20.Vos MC, Hollemans E, Ezendam N, Feijen H, Boll D, Pijlman B, van der Putten H, Klinkhamer P, van Kuppevelt TH, van der Wurff AA, Massuger LF. MMP-14 and CD44 in epithelial-to-mesenchymal transition (EMT) in ovarian cancer. J Ovarian Res. 2016;9:53. doi: 10.1186/s13048-016-0262-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwon MJ. Emerging roles of claudins in human cancer. Int J Mol Sci. 2013;14:18148–80. doi: 10.3390/ijms140918148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu Q, Liu X, Liu YF, Lu Y, Wang LP, Zhang XW, Li YL, Quan CS. Inhibition of p38 activity reverses claudin-6 induced cell apoptosis, invasion, and migration. Chin Med J (Engl) 2013;126:3539–44. [PubMed] [Google Scholar]
- 23.Lu Y, Wang L, Li H, Li Y, Ruan Y, Lin D, Yang M, Jin X, Guo Y, Zhang X, Quan C. SMAD2 inactivation inhibits CLDN6 methylation to suppress migration and invasion of breast cancer cells. Int J Mol Sci. 2017;18:1863. doi: 10.3390/ijms18091863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y, Jin X, Li Y, Ruan Y, Lu Y, Yang M, Lin D, Song P, Guo Y, Zhao S, Dong B, Xie Y, Dang Q, Quan C. DNA methylation of claudin-6 promotes breast cancer cell migration and invasion by recruiting MeCP2 and deacetylating H3Ac and H4Ac. J Exp Clin Cancer Res. 2016;35:120. doi: 10.1186/s13046-016-0396-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geng XF, Fang M, Liu SP, Li Y. Quantum dot-based molecular imaging of cancer cell growth using a clone formation assay. Mol Med Rep. 2016;14:3007–12. doi: 10.3892/mmr.2016.5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Srinivasan B, Kolli AR, Esch MB, Abaci HE, Shuler ML, Hickman JJ. TEER measurement techniques for in vitro barrier model systems. J Lab Autom. 2015;20:107–26. doi: 10.1177/2211068214561025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coleman MP, Quaresma M, Berrino F, Lutz JM, De Angelis R, Capocaccia R, Baili P, Rachet B, Gatta G, Hakulinen T, Micheli A, Sant M, Weir HK, Elwood JM, Tsukuma H, Koifman S, E Silva GA, Francisci S, Santaquilani M, Verdecchia A, Storm HH, Young JL CONCORD Working Group. Cancer survival in five continents: a worldwide population-based study (CONCORD) Lancet Oncol. 2008;9:730–56. doi: 10.1016/S1470-2045(08)70179-7. [DOI] [PubMed] [Google Scholar]
- 28.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 29.Akram M, Iqbal M, Daniyal M, Khan AU. Awareness and current knowledge of breast cancer. Biol Res. 2017;50:33. doi: 10.1186/s40659-017-0140-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wolfe AR, Debeb BG, Lacerda L, Larson R, Bambhroliya A, Huang X, Bertucci F, Finetti P, Birnbaum D, Van Laere S, Diagaradjan P, Ruffell B, Trenton NJ, Chu K, Hittelman W, Diehl M, Levental I, Ueno NT, Woodward WA. Simvastatin prevents triple-negative breast cancer metastasis in pre-clinical models through regulation of FOXO3a. Breast Cancer Res Treat. 2015;154:495–508. doi: 10.1007/s10549-015-3645-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krizbai IA, Gasparics Á, Nagyőszi P, Fazakas C, Molnár J, Wilhelm I, Bencs R, Rosivall L, Sebe A. Endothelial-mesenchymal transition of brain endothelial cells: possible role during metastatic extravasation. PLoS One. 2015;10:e0123845. doi: 10.1371/journal.pone.0119655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu Z, Ding L, Hong H, Hoggard J, Lu Q, Chen YH. Claudin-7 inhibits human lung cancer cell migration and invasion through ERK/MAPK signaling pathway. Exp Cell Res. 2011;317:1935–46. doi: 10.1016/j.yexcr.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dahiya N, Becker KG, Wood WH 3rd, Zhang Y, Morin PJ. Claudin-7 is frequently overexpressed in ovarian cancer and promotes invasion. PLoS One. 2011;6:e22119. doi: 10.1371/journal.pone.0022119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tabaries S, Dupuy F, Dong Z, Monast A, Annis MG, Spicer J, Ferri LE, Omeroglu A, Basik M, Amir E, Clemons M, Siegel PM. Claudin-2 promotes breast cancer liver metastasis by facilitating tumor cell interactions with hepatocytes. Mol Cell Biol. 2012;32:2979–91. doi: 10.1128/MCB.00299-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ashikari D, Takayama KI, Obinata D, Takahashi S, Inoue S. CLDN8, an androgen-regulated gene, promotes prostate cancer cell proliferation and migration. Cancer Sci. 2017;108:1386–1393. doi: 10.1111/cas.13269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–42. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- 37.Tomaskovic-Crook E, Thompson EW, Thiery JP. Epithelial to mesenchymal transition and breast cancer. Breast Cancer Res. 2009;11:213. doi: 10.1186/bcr2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. 2006;127:679–95. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]