Abstract
Background: Malignant melanoma is a skin cancer with a high rate of metastasis. Numerous circular RNAs (circRNAs) have been shown to play vital roles in melanoma. This research aimed to investigate the role and molecular basis of circ_0016418 in melanoma progression. Methods: The abundanced of circ_0016418, miR-605-5p and glutaminase (GLS) were measured using quantitative real-time polymerase chain reaction or western blot analysis. Cell proliferation was evaluated using Cell Counting Kit-8 (CCK-8) assay and colony formation assay. Cell migration and invasion were assessed by transwell assay. Cell cycle and apoptosis were monitored by flow cytometry. The levels of glutamine consumption and glutamate were examined using commercial kits. The interaction among circ_0016418, miR-605-5p and GLS was verified with the dual-luciferase reporter assay. A xenograft model was used to analyze tumor growth in vivo. Results: Circ_0016418 and GLS were up-regulated, while miR-605-5p was down-regulated in melanoma tissues and cells. Circ_0016418 silencing hindered cell proliferation, metastasis, and glutamine catabolism and promoted cell cycle arrest and apoptosis in A375 and A875 cells. Circ_0016418 modulated melanoma progression and glutamine catabolism through sponging miR-605-5p. Also, miR-605-5p inhibited melanoma progression and glutamine catabolism by targeting GLS. Moreover, circ_0016418 depletion blocked tumor growth in vivo. Conclusion: Knockdown of circ_0016418 suppressed melanoma development and glutamine catabolism by modulating the miR-605-5p/GLS pathway.
Keywords: Circ_0016418, miR-605-5p, GLS, melanoma, glutamine catabolism
Introduction
Malignant melanoma derived from melanocytes is the leading cause of skin cancer-related death, and its incidence is increasing year by year [22]. Melanoma is the most aggressive form of skin cancer and can quickly become life-threatening if it spreads [17]. Metastatic cutaneous melanoma has no radical treatment, and its prognosis is unsatisfactory with a 5-year overall survival rate of only 5% [23]. The pathogenesis of melanoma is not completely clear. Hence, identifying novel molecular pathways in melanoma is critical to improving poor prognosis.
Circular RNAs (circRNAs) are novel non-coding RNAs with covalently closed loops formed by back-splicing [21]. A growing number of studies have manifested that circRNAs exert a crucial function in tumorigenesis and development [12]. Also, aberrantly expressed circRNAs participate in the progression of melanoma [1]. For example, circ_0017247 facilitated melanoma metastasis by interacting with miR-145 [4]. In addition, circRNA_0084043 contributed to cell growth and metastasis in malignant melanoma by directly binding to miR-153-3p and up-regulating Snail [15]. Previous research unveiled that circ_0016418 derived from Vasohibin-2 (VASH2) was overtly up-regulated in skin melanoma [25]. Nevertheless, the mechanism of circ_0016418 in melanoma is still insufficient.
MicroRNAs (miRNAs) are non-coding RNAs consisting of ~22 nucleotides that modulate post-transcriptional silencing of target genes [7]. Mounting evidence has validated that miRNA dysregulation is closely associated with human diseases, including cancer [14]. For instance, miR-139-5p ameliorated the malignancy of melanoma through targeting IGF1R to weaken the PI3K/AKT pathway [26]. Additionally, miR-140-5p suppressed melanoma progression by binding to SOX4 and inhibiting the activation of the Wnt/β-catenin and NF-κB pathways [30]. Moreover, miR-145-5p impeded the development of malignant melanoma by repressing the TLR4-mediated NF-κB pathway [9]. Herein, bioinformatics analysis discovered that circ_0016418 might bind to miR-605-5p. Therefore, we speculated that circ_0016418 regulated melanoma progression by mediating miR-605-5p.
Furthermore, we first demonstrated that circ_0016418 facilitated glutamine catabolism. Abnormal metabolism is a hallmark of tumor cells, for example, increased glutamine catabolism [8]. Glutamine has multiple effects in cancer cells, including maintaining mitochondrial metabolism, activating cell signaling, and promoting cell replication [27]. Glutaminase (GLS) is a rate-limiting enzyme for glutamine catabolism, catalyzing the hydrolysis of glutamine to glutamate [11]. In addition, recent research revealed that GLS up-regulation expedited glutamine catabolism and malignant melanoma development [16].
In this research, we elucidated the function of circ_0016418 in melanoma progression and glutamine catabolism. We also investigated the association between circ_0016418 and the miR-605-5p/GLS pathway. Thus, our research provided a potential therapeutic target for melanoma.
Materials and methods
Specimen collection
Thirty patients diagnosed with melanoma were recruited from the First Affiliated Hospital of Northwest Minzu University. All patients did not receive preoperative treatment and signed written informed consent. Melanoma tissues and adjacent normal tissues were collected by resection. This research was ratified by the Ethics Committee of the First Affiliated Hospital of Northwest Minzu University.
Cell culture
Human epidermal melanocytes (HEMn-LP) were purchased from Invitrogen (Carlsbad, CA, USA) and incubated in medium 254 (Gibco, Carlsbad, CA, USA) supplemented with Human Melanocyte Growth Supplement (HMGS; Gibco). Human malignant melanoma cell lines (A375 and A875) were purchased from Shanghai Fusheng Industrial Co., Ltd. (Shanghai, China) and maintained in Dulbecco’s Modified Eagle Medium (DMEM; Gibco) supplemented with 10% fetal bovine serum (FBS; Gibco). All cells were cultured in an atmosphere of 5% CO2 at 37°C.
Cell transfection
Small interfering RNA (siRNA) against circ_0016418 (si-circ_0016418-1 and si-circ_0016418-2), the siRNA control (si-NC), miR-605-5p mimic, the mimic control (miRNA NC), miR-605-5p inhibitor, the inhibitor control (inhibitor NC), GLS overexpression vector (pc-GLS), and the negative control (pc-NC) were synthesized with the use of Ribobio (Guangzhou, China). Lipofectamine 3000 (Invitrogen) was utilized to transfect oligonucleotides and vectors into melanoma cells.
Quantitative real-time polymerase chain reaction (qRT-PCR)
RNA extracted by Trizol reagent (Invitrogen) was reverse-transcribed into complementary DNA using a specific RT-PCR kit (Takara, Dalian, China). Then, SYBR Green PCR Master Mix (LMAI Bio, Shanghai, China) was used to perform quantitative PCR. The expression abundance was calculated using the 2-ΔΔCt method. For RNase R digestion assay, 2 μg RNA was incubated with or without RNase R (Solarbio, Beijing, China) for 30 min. Next, circ_0016418 and VASH2 levels were measured using qRT-PCR. The primers included: circ_0016418-F: 5’-CTCCGACCCAAGTGAGAAGC-3’, circ_0016418-R: 5’-CAGCCTGTAGTTTGGGACC-3’; VASH2-F: 5’-CTCTTCCAGCCTTCCTTCCT-3’, VASH2-R: 5’-AGCACTGTGTTGGCGTACAG-3’; miR-605-5p-F: 5’-TAAATCCCATGGTGCCT-3’, miR-605-5p-R: 5’-TGGTGTCGTGGAGTCG-3’; GAPDH-F: 5’-GGGAAACTGTGGCGTGAT-3’, GAPDH-R: 5’-GAGTGGGTGTCGCTGTTGA-3’; U6-F: 5’-CTCGCTTCGGCAGCACA-3’, U6-R: 5’-AACGCTTCACGAATTTGCGT-3’. GAPDH or U6 was regarded as an internal reference.
Cell viability assay
5×103 transfected melanoma cells were injected into 96-well plates. After incubation for 0 h, 24 h, 48 h or 72 h, the medium was discarded, and the fresh medium containing 10 μL Cell Counting Kit-8 (CCK-8) solution (Beyotime, Shanghai, China) was mixed with melanoma cells. After reaction for 4 h, the absorbance was measured using a Microplate Reader (BioTek, Burlington, VT, USA).
Transwell assay
Unlike s cell migration test, the upper chamber in cell invasion test is covered with Matrigel (BD Biosciences, San Diego, CA, USA). In brief, the medium with 10% FBS was injected into the lower chamber. After 24 h in culture, the invaded and migrated cells were stained with 0.5% crystal violet for 15 min and counted in three random fields using a microscope.
Colony formation assay
Transfected A375 and A875 cells were maintained in six-well plates and incubated in medium containing 10% FBS at 37°C for 14 days. After staining with 0.5% crystal violet, the colonies were photographed and counted in at least three independent replications.
Flow cytometry
For cell cycle analysis, the transfected cells were digested with trypsin and suspended in PBS buffer after transfection for 48 h. Subsequently, the precipitate obtained by centrifugation was re-suspended in PBS buffer. After digestion with RNAase, the cells were stained with 1% propidium iodide (PI; ab14083, Abcam, Cambridge, UK) for 25 min. Then, the cell cycle distribution was monitored by a flow cytometer (Beckman Coulter, Miami, FL, USA).
For cell apoptosis analysis, A375 and A875 cells were washed with PBS after digestion with trypsin. Then, the cells were reacted with AnnexinV-fluorescein isothiocyanate and PI for 15 min in the dark. Next, the apoptotic rate was examined using a flow cytometer (Beckman Coulter).
Detection of glutamine consumption and glutamate
Transfected melanoma cells were cultured in DMEM harboring 2 mM glutamine. After incubation for 24 h, glutamine consumption and glutamate were examined using corresponding kits (Biovision, Milpitas, CA, USA) according to the manufacturer’s requirements.
Western blot assay
Protein was extracted using RIPA buffer (Solarbio). Next, the equal amount of protein was separated by polyacrylamide gel electrophoresis and transferred onto polyvinylidene difluoride membranes (Millipore, Billerica, MA, USA). After blocking with 5% non-fat milk, the membranes were incubated with primary antibodies against GLS (1:1000; ab260047, Abcam) or GAPDH (1:2500; ab9485, Abcam). Subsequently, the membranes were placed with the secondary antibody (1:20000; ab205718, Abcam). Finally, the signal intensity was measured using an ECL reagent (Solarbio).
Dual-luciferase reporter assay
The wild-type luciferase reporter vectors (WT-circ_0016418 or WT-GLS 3’UTR) were constructed using cloning circ_0016418 or GLS 3’UTR containing miR-605-5p binding sequence into pmirGLO vectors (Promega, Madison, WI, USA). The mutant luciferase reporter vectors (MUT-circ_0016418 or MUT-GLS 3’UTR) were formed through mutating the binding site. Then, the constructed luciferase reporter and miR-605-5p mimic or miRNA NC were incubated into A375 and A875 cells. The luciferase activity was examined using Dual-Lucy Assay Kit (Solarbio).
Xenograft tumor experiment
The xenograft assay was ratified by the Animal Research Committee of the First Affiliated Hospital of Northwest Minzu University. BALB/c nude mice (5-week-old) were randomly divided into two groups (n=6). Lentivirus containing circ_0016418 short hairpin RNA (sh-circ_0016418) or negative control (sh-NC) constructed by Genechem (Shanghai, China) was transfected into A875 cells. Then, 5×106 stably transfected cells were subcutaneously injected into the right abdomen of mice. Tumor volume was measured every 7 days. After 28 days, the mice were sacrificed, and the xenograft tumors were weighed.
Statistical analysis
Data were shown as mean ± standard deviation. The differences were evaluated by Student’s t-test or one-way analysis of variance using GraphPad Prism 7 software (GraphPad Inc., La Jolla, CA, USA). P<0.05 was considered significant.
Results
Circ_0016418 is up-regulated in melanoma tissues and cells
We first tested circ_0016418 level in 30 pairs of primary malignant melanoma tissues and adjacent normal tissues. As shown in Figure 1A, circ_0016418 expression was markedly increased relative to normal tissues. We also examined circ_0016418 level in HEMn-LP and melanoma cells (A375 and A875) and revealed that circ_0016418 expression was distinctly elevated in melanoma cells (Figure 1B). In addition, RNase R digestion analysis exhibited that circ_0016418 level had no change, while the mRNA level of its linear isoform VASH2 was significantly reduced after treatment with RNase R (Figure 1C). These data evidenced that circ_0016418 was highly expressed and stable in melanoma.
Figure 1.
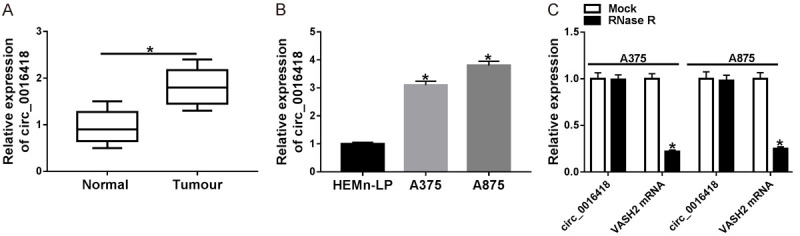
Circ_0016418 is up-regulated in melanoma tissues and cells. A. Circ_0016418 level was examined in melanoma tissues (n=30) and adjacent normal tissues (n=30). B. Circ_0016418 level was measured in human epidermal melanocytes (HEMn-LP) and melanoma cells (A375 and A875). C. After treatment of A375 and A875 cells with RNase R, the expression of circ_0016418 and VASH2 mRNA was tested. *P<0.05.
Circ_0016418 silencing hinders melanoma development and glutamine catabolism in melanoma cells
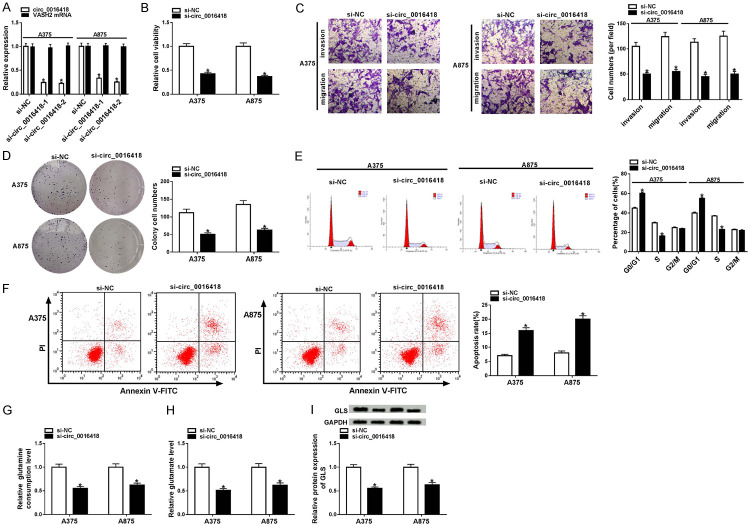
To investigate the biologic function of circ_0016418 in melanoma cells, A375 and A875 cells were incubated with si-NC, si-circ_0016418-1 or si-circ_0016418-2. qRT-PCR analysis suggested that si-circ_0016418-2 had the most significant knockdown efficiency, so si-circ_0016418-2 (si-circ_0016418) was selected for subsequent experiments (Figure 2A). Next, CCK-8 and colony formation assays unveiled that down-regulation of circ_0016418 suppressed the proliferative ability of A375 and A875 cells (Figure 2B and 2D). Meanwhile, transwell analysis showed that circ_0016418 silencing inhibited the invasion and migration of melanoma cells (Figure 2C). Flow cytometry displayed that knockdown of circ_0016418 markedly triggered cell cycle arrest and apoptosis in A375 and A875 cells (Figure 2E and 2F). Additionally, the levels of glutamine consumption and glutamate in the si-circ_0016418 group were significantly decreased in comparison with the si-NC group (Figure 2G and 2H). Also, GLS is a rate-limiting enzyme in glutamine catabolism. Western blot analysis suggested that transfection with si-circ_0016418 dramatically reduced GLS protein level (Figure 2I). Collectively, these data indicated that circ_0016418 silencing alleviated the malignancy of melanoma and suppressed glutamine catabolism in melanoma cells.
Figure 2.
Circ_0016418 silencing hinders melanoma development and glutamine catabolism in melanoma cells. (A) A375 and A875 cells were incubated with si-NC, si-circ_0016418-1 or si-circ_0016418-2, and the levels of circ_0016418 and VASH2 mRNA were examined using qRT-PCR. (B-I) A375 and A875 cells were transfected with si-NC or si-circ_0016418-2 (si-circ_0016418). Cell viability (B), invasion and migration (C), and colony numbers (D) were tested using CCK-8 assay, transwell assay (100×) and colony formation assay. (E and F) Flow cytometry was utilized to assess cell cycle and apoptosis. (G and H) The levels of glutamine consumption and glutamate were detected using commercial kits. (I) GLS level was measured using western blot. *P<0.05.
Circ_0016418 modulates melanoma progression and glutamine catabolism by regulating miR-605-5p
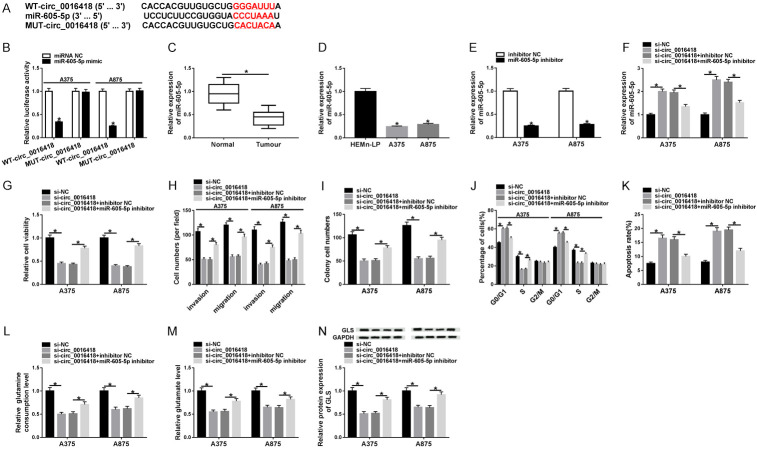
Next, we used Circular RNA Interactome database to predict possible targets for circ_0016418. As exhibited in Figure 3A, miR-605-5p contained potential binding sites for circ_0016418. Next, dual-luciferase reporter assay displayed that miR-605-5p mimic strikingly reduced the luciferase activity of WT-circ_0016418 reporter (Figure 3B). In addition, miR-605-5p levels in melanoma tissues and cells were markedly lower than those in the normal group (Figure 3C and 3D). Besides, the inhibition efficiency of miR-605-5p was confirmed using qRT-PCR (Figure 3E). Furthermore, to explore whether circ_0016418 played a role in melanoma by modulating miR-605-5p, A375 and A875 cells were incubated with si-NC, si-circ_0016418, si-circ_0016418+inhibitor NC, or si-circ_0016418+miR-605-5p inhibitor, respectively. First of all, transfection with miR-605-5p inhibitor attenuated the increase of miR-605-5p level caused by circ_0016418 depletion (Figure 3F). Meanwhile, circ_0016418 silencing blocked melanoma cell proliferation, invasion, and migration, and accelerated melanoma cell cycle arrest and apoptosis, while these impacts were abolished by down-regulating miR-605-5p (Figure 3G-K). Moreover, circ_0016418 depletion reduced the levels of glutamine consumption, glutamate, and GLS, whereas introduction of miR-605-5p inhibitor abrogated these effects (Figure 3L-N). Altogether, these data evidenced that circ_0016418 regulated melanoma progression and glutamine catabolism by sponging miR-605-5p.
Figure 3.
Circ_0016418 modulates melanoma progression and glutamine catabolism by regulating miR-605-5p. (A) The putative binding sequence between circ_0016418 and miR-605-5p is exhibited. (B) Luciferase activity was determined in A375 and A875 cells co-transfected with WT-circ_0016418 or MUT-circ_0016418 and miRNA NC or miR-605-5p mimic. (C and D) The expression of miR-605-5p was measured in melanoma tissues and cells. (E) MiR-605-5p level was determined in A375 and A875 cells transfected with inhibitor NC or miR-605-5p inhibitor. MiR-605-5p level (F), cell viability (G), invasion and migration (H), colony numbers (I), cell cycle and apoptosis (J and K), glutamine consumption (L), glutamate level (M) and GLS protein level (N) were examined in A375 and A875 cells incubated with si-NC, si-circ_0016418, si-circ_0016418+inhibitor NC or si-circ_0016418+miR-605-5p inhibitor. *P<0.05.
MiR-605-5p inhibits melanoma progression and glutamine catabolism through targeting GLS
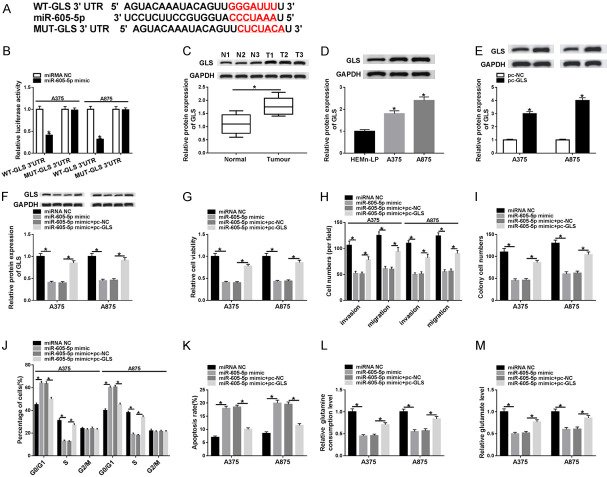
TargetScan online database displayed that GLS was a putative target gene for miR-605-5p (Figure 4A). Then, dual-luciferase reporter analysis showed that mature miR-605-5p overtly decreased the luciferase activity of WT-GLS 3’UTR reporter (Figure 4B). Compared with the normal group, GLS protein level was remarkably increased in melanoma tissues and cells (Figure 4C and 4D). Additionally, the protein expression of GLS in pc-GLS group was significantly elevated relative to pc-NC group, indicating an efficient overexpression efficiency (Figure 4E). In order to verify whether miR-605-5p modulated melanoma growth by targeting GLS, a series of rescue experiments were performed in A375 and A875 cells incubated with miRNA NC, miR-605-5p mimic, miR-605-5p mimic+pc-NC or miR-605-5p mimic+pc-GLS. The results revealed that transfection with pc-GLS abated the reduction of GLS level induced by miR-605-5p mimic (Figure 4F). Next, up-regulation of miR-605-5p impeded melanoma cell proliferation, invasion, and migration and promoted cell cycle arrest and apoptosis, while these effects were reversed following overexpression of GLS (Figure 4G-K). Furthermore, the levels of glutamine consumption and glutamate were strikingly reduced after introduction of miR-605-5p mimic, whereas the changes were undermined by up-regulating GLS (Figure 4L, 4M). To sum up, these data demonstrated that miR-605-5p suppressed melanoma development and glutamine catabolism by modulating GLS.
Figure 4.
MiR-605-5p inhibits melanoma progression and glutamine catabolism by targeting GLS. A. The predicted binding sites of miR-605-5p on GLS 3’UTR and its mutant sites are shown. B. Luciferase activity was examined in A375 and A875 cells co-transfected with WT-GLS 3’UTR or MUT-GLS 3’UTR and miRNA NC or miR-605-5p mimic. C and D. The protein level of GLS was detected in melanoma tissues and cells. E. GLS protein expression was examined in A375 and A875 cells transfected with pc-NC or pc-GLS. F-M. After A375 and A875 cells were incubated with miRNA NC, miR-605-5p mimic, miR-605-5p mimic+pc-NC or miR-605-5p mimic+pc-GLS, GLS expression, cell viability, invasion and migration, colony numbers, cell cycle and apoptosis, glutamine consumption, and glutamate level were measured using corresponding methods. *P<0.05.
Circ_0016418 regulates GLS expression by sponging miR-605-5p
Firstly, co-transfection of si-circ_0016418 and pc-GLS alleviated the decrease in GLS protein level caused by circ_0016418 silencing (Figure 5A). To explore the regulatory effect of circ_0016418 on GLS expression, A375 and A875 cells were transduced with si-NC, si-circ_0016418, si-circ_0016418+inhibitor NC or si-circ_0016418+miR-605-5p inhibitor. As depicted in Figure 5B, circ_0016418 depletion significantly reduced GLS protein level, while the impact was abated after transfection with miR-605-5p inhibitor. These data indicated that circ_0016418 sponged miR-605-5p to regulate GLS expression.
Figure 5.

Circ_0016418 regulates GLS expression by sponging miR-605-5p. A. GLS protein level was measured in A375 and A875 cells transfected with si-NC, si-circ_0016418, si-circ_0016418+ pc-NC or si-circ_0016418+pc-GLS. B. GLS protein level was examined in A375 and A875 cells incubated with si-NC, si-circ_0016418, si-circ_0016418+inhibitor NC or si-circ_0016418+miR-605-5p inhibitor. *P<0.05.
Knockdown of circ_0016418 blocks tumor growth in vivo
To further elucidate the role of circ_0016418 in tumorigenesis in vivo, we established a melanoma xenograft mouse model. As exhibited in Figure 6A, circ_0016418 silencing markedly reduced tumor volume. After 28 days, the nude mice were killed, and the tumors removed. The results illustrated that circ_0016418 knockdown remarkably decreased tumor weight (Figure 6B). Meanwhile, the levels of circ_0016418 and GLS were overtly decreased, and miR-605-5p level was strikingly elevated in the sh-circ_0016418 group compared with the sh-NC group (Figure 6C-E). These results evidenced that circ_0016418 depletion inhibited melanoma growth in vivo.
Figure 6.

Knockdown of circ_0016418 blocks tumor growth in vivo. A875 cells transfected with sh-NC or sh-circ_0016418 were subcutaneously injected into nude mice. A. Tumor volume was measured every 7 days. B. After the mice were killed, the xenograft tumors were weighed. C-E. The levels of circ_0016418, miR-605-5p, and GLS were examined using qRT-PCR or western blot. *P<0.05.
Discussion
Accumulating evidence has illuminated that altered glutamine catabolism in tumor cells exerts a critical effect on cell proliferation [10]. On the other hand, the energy requirements of glutamine in tumors are generally highly heterogeneous [5]. Recently, several studies have corroborated that circRNAs play a regulatory role in tumor progression by regulating aspects of cellular metabolism, including glycolysis, glutamine catabolism, and fat metabolism [28]. Nevertheless, the function of circ_0016418 in glutamine catabolism remains unknown.
circRNAs regulate a range of biological functions, including cellular metabolism, since they function as miRNA sponges or directly interacting with proteins [31]. For example, Bian et al. demonstrated that circ_0025039 expedited melanoma progression and glucose metabolism by sponging miR-198 to elevate CDK4 [2]. Lin et al. disclosed that circRNA ITCH up-regulation restrained glucose uptake by reducing GLUT1 expression, thereby preventing melanoma cell proliferation [13]. Moreover, Han et al. suggested that circ-FOXM1 facilitated melanoma development and glycolysis by inhibiting miR-143-3p and activating FLOT2 [24]. In this research, we confirm that circ_0016418 level was remarkably increased in melanoma. Besides, previous research indicated that circ_0016418 contributed to cell proliferation and metastasis in skin melanoma by sponging miR-625 to activate YY1 [34]. Similarly, we show that circ_0016418 depletion impeded melanoma progression and glutamine catabolism.
Furthermore, we used prediction software to mine possible targets for circ_0016418 to elucidate the potential mechanism of circ_0016418 in melanoma. Due to the novelty of the circ_0016418/miR-605-5p pathway, miR-605-5p was chosen as a candidate for further research. Several studies have revealed that miR-605 has a tumor-suppressive effect in a variety of tumors. In prostate cancer, knockdown of miR-605 strengthened oncogenicity by activating EN2 [33]. In non-small-cell lung cancer, miR-605 overexpression hindered cell proliferation and metastasis through down-regulation of FOXP1 [32]. In melanoma, miR-605 suppressed tumor progression through inhibition of INPP4B-induced SGK3 activation [3]. In the current research, miR-605-5p level was prominently reduced in melanoma. More importantly, we evidenced that circ_0016418 was a decoy for miR-605-5p, and miR-605-5p down-regulation eliminated the inhibition of circ_0016418 silencing on melanoma development and glutamine catabolism.
Increasing evidence has manifested that miRNAs modulate gene expression by base-pairing with cytoplasmic mRNA 3’UTR [6]. Herein, we first verified that miR-605-5p directly bound to GLS 3’UTR and negatively regulated GLS expression. Hence, we hypothesized that circ_0016418 might serve as a competing endogenous RNA (ceRNA) for miR-605-5p to mediate GLS expression. Also, GLS is a key enzyme in glutamine metabolism and a tumor-promoting factor in various cancers [19,20]. In prostate cancer, GLS silencing repressed cell proliferation and accelerated apoptosis through suppression of Wnt/β-catenin pathway [29]. In glioma, GLS down-regulation blocked cell proliferation through modulation of oxidative stress [18]. In the present research, we discovered that GLS up-regulation mitigated the inhibition of miR-605-5p overexpression on melanoma progression and glutamine catabolism. Mechanistically, we revealed that circ_0016418 elevated GLS level by sponging miR-605-5p.
In conclusion, our research unveiled that circ_0016418 contributed to melanoma development and glutamine catabolism, acting as a ceRNA for miR-605-5p to up-regulate GLS. Furthermore, circ_0016418 depletion impeded tumor growth in vivo. These findings hinted that circ_0016418/miR-605-5p/GLS pathway might provide new treatment strategies for melanoma.
Acknowledgements
This work was approved by the Basic Research Project of the Central University of Northwest Minzu University (NO. 31920190199).
Disclosure of conflict of interest
None.
References
- 1.Abi A, Farahani N, Molavi G, Gheibi Hayat SM. Circular RNAs: epigenetic regulators in cancerous and noncancerous skin diseases. Cancer Gene Ther. 2020;27:280–293. doi: 10.1038/s41417-019-0130-x. [DOI] [PubMed] [Google Scholar]
- 2.Bian D, Wu Y, Song G. Novel circular RNA, hsa_circ_0025039 promotes cell growth, invasion and glucose metabolism in malignant melanoma via the miR-198/CDK4 axis. Biomed Pharmacother. 2018;108:165–176. doi: 10.1016/j.biopha.2018.08.152. [DOI] [PubMed] [Google Scholar]
- 3.Chen L, Cao Y, Rong D, Wang Y, Cao Y. MicroRNA-605 functions as a tumor suppressor by targeting INPP4B in melanoma. Oncol Rep. 2017;38:1276–1286. doi: 10.3892/or.2017.5740. [DOI] [PubMed] [Google Scholar]
- 4.Chen Z, Kang K, Chen S, Wang S, Zhang J, Zhang XY, Chen Z. Circular RNA circ_0017247 promotes melanoma migration and invasion via targeting miR-145. Eur Rev Med Pharmacol Sci. 2020;24:1932–1938. doi: 10.26355/eurrev_202002_20371. [DOI] [PubMed] [Google Scholar]
- 5.Cluntun AA, Lukey MJ, Cerione RA, Locasale JW. Glutamine metabolism in cancer: understanding the heterogeneity. Trends Cancer. 2017;3:169–180. doi: 10.1016/j.trecan.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 7.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 8.Hensley CT, Wasti AT, DeBerardinis RJ. Glutamine and cancer: cell biology, physiology, and clinical opportunities. J Clin Invest. 2013;123:3678–3684. doi: 10.1172/JCI69600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin C, Wang A, Liu L, Wang G, Li G, Han Z. miR-145-5p inhibits tumor occurrence and metastasis through the NF-kappaB signaling pathway by targeting TLR4 in malignant melanoma. J Cell Biochem. 2019 doi: 10.1002/jcb.28388. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 10.Jin L, Alesi GN, Kang S. Glutaminolysis as a target for cancer therapy. Oncogene. 2016;35:3619–3625. doi: 10.1038/onc.2015.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katt WP, Cerione RA. Glutaminase regulation in cancer cells: a druggable chain of events. Drug Discov Today. 2014;19:450–457. doi: 10.1016/j.drudis.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lei M, Zheng G, Ning Q, Zheng J, Dong D. Translation and functional roles of circular RNAs in human cancer. Mol Cancer. 2020;19:30. doi: 10.1186/s12943-020-1135-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin Q, Jiang H, Lin D. Circular RNA ITCH downregulates GLUT1 and suppresses glucose uptake in melanoma to inhibit cancer cell proliferation. J Dermatolog Treat. 2019:1–5. doi: 10.1080/09546634.2019.1654069. [DOI] [PubMed] [Google Scholar]
- 14.Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15:321–333. doi: 10.1038/nrc3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luan W, Shi Y, Zhou Z, Xia Y, Wang J. circRNA_0084043 promote malignant melanoma progression via miR-153-3p/Snail axis. Biochem Biophys Res Commun. 2018;502:22–29. doi: 10.1016/j.bbrc.2018.05.114. [DOI] [PubMed] [Google Scholar]
- 16.Luan W, Zhou Z, Zhu Y, Xia Y, Wang J, Xu B. miR-137 inhibits glutamine catabolism and growth of malignant melanoma by targeting glutaminase. Biochem Biophys Res Commun. 2018;495:46–52. doi: 10.1016/j.bbrc.2017.10.152. [DOI] [PubMed] [Google Scholar]
- 17.Luo C, Shen J. Research progress in advanced melanoma. Cancer Lett. 2017;397:120–126. doi: 10.1016/j.canlet.2017.03.037. [DOI] [PubMed] [Google Scholar]
- 18.Martin-Rufian M, Nascimento-Gomes R, Higuero A, Crisma AR, Campos-Sandoval JA, Gomez-Garcia MC, Cardona C, Cheng T, Lobo C, Segura JA, Alonso FJ, Szeliga M, Albrecht J, Curi R, Marquez J, Colquhoun A, Deberardinis RJ, Mates JM. Both GLS silencing and GLS2 overexpression synergize with oxidative stress against proliferation of glioma cells. J Mol Med (Berl) 2014;92:277–290. doi: 10.1007/s00109-013-1105-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masisi BK, El Ansari R, Alfarsi L, Rakha EA, Green AR, Craze ML. The role of glutaminase in cancer. Histopathology. 2020;76:498–508. doi: 10.1111/his.14014. [DOI] [PubMed] [Google Scholar]
- 20.Mates JM, Segura JA, Martin-Rufian M, Campos-Sandoval JA, Alonso FJ, Marquez J. Glutaminase isoenzymes as key regulators in metabolic and oxidative stress against cancer. Curr Mol Med. 2013;13:514–534. doi: 10.2174/1566524011313040005. [DOI] [PubMed] [Google Scholar]
- 21.Qian L, Yu S, Chen Z, Meng Z, Huang S, Wang P. The emerging role of circRNAs and their clinical significance in human cancers. Biochim Biophys Acta Rev Cancer. 2018;1870:247–260. doi: 10.1016/j.bbcan.2018.06.002. [DOI] [PubMed] [Google Scholar]
- 22.Schadendorf D, van Akkooi ACJ, Berking C, Griewank KG, Gutzmer R, Hauschild A, Stang A, Roesch A, Ugurel S. Melanoma. Lancet. 2018;392:971–984. doi: 10.1016/S0140-6736(18)31559-9. [DOI] [PubMed] [Google Scholar]
- 23.Strub T, Ballotti R, Bertolotto C. The “ART” of epigenetics in melanoma: from histone “alterations, to resistance and therapies”. Theranostics. 2020;10:1777–1797. doi: 10.7150/thno.36218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tian S, Han G, Lu L, Meng X. Circ-FOXM1 contributes to cell proliferation, invasion, and glycolysis and represses apoptosis in melanoma by regulating miR-143-3p/FLOT2 axis. World J Surg Oncol. 2020;18:56. doi: 10.1186/s12957-020-01832-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Q, Chen J, Wang A, Sun L, Qian L, Zhou X, Liu Y, Tang S, Chen X, Cheng Y, Cao K, Zhou J. Differentially expressed circRNAs in melanocytes and melanoma cells and their effect on cell proliferation and invasion. Oncol Rep. 2018;39:1813–1824. doi: 10.3892/or.2018.6263. [DOI] [PubMed] [Google Scholar]
- 26.Yang C, Xia Z, Zhu L, Li Y, Zheng Z, Liang J, Wu L. MicroRNA-139-5p modulates the growth and metastasis of malignant melanoma cells via the PI3K/AKT signaling pathway by binding to IGF1R. Cell Cycle. 2019;18:3513–3524. doi: 10.1080/15384101.2019.1690881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang L, Venneti S, Nagrath D. Glutaminolysis: a hallmark of cancer metabolism. Annu Rev Biomed Eng. 2017;19:163–194. doi: 10.1146/annurev-bioeng-071516-044546. [DOI] [PubMed] [Google Scholar]
- 28.Yu T, Wang Y, Fan Y, Fang N, Wang T, Xu T, Shu Y. CircRNAs in cancer metabolism: a review. J Hematol Oncol. 2019;12:90. doi: 10.1186/s13045-019-0776-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang J, Mao S, Guo Y, Wu Y, Yao X, Huang Y. Inhibition of GLS suppresses proliferation and promotes apoptosis in prostate cancer. Biosci Rep. 2019;39 doi: 10.1042/BSR20181826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao G, Yin Y, Zhao B. miR-140-5p is negatively correlated with proliferation, invasion, and tumorigenesis in malignant melanoma by targeting SOX4 via the Wnt/beta-catenin and NF-kappaB cascades. J Cell Physiol. 2020;235:2161–2170. doi: 10.1002/jcp.29122. [DOI] [PubMed] [Google Scholar]
- 31.Zhong Y, Du Y, Yang X, Mo Y, Fan C, Xiong F, Ren D, Ye X, Li C, Wang Y, Wei F, Guo C, Wu X, Li X, Li Y, Li G, Zeng Z, Xiong W. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17:79. doi: 10.1186/s12943-018-0827-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou W, Li R. microRNA-605 inhibits the oncogenicity of non-small-cell lung cancer by directly targeting Forkhead Box P1. Onco Targets Ther. 2019;12:3765–3777. doi: 10.2147/OTT.S193675. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Zhou YJ, Yang HQ, Xia W, Cui L, Xu RF, Lu H, Xue Z, Zhang B, Tian ZN, Cao YJ, Xing ZY, Yin S, He XZ. Down-regulation of miR-605 promotes the proliferation and invasion of prostate cancer cells by up-regulating EN2. Life Sci. 2017;190:7–14. doi: 10.1016/j.lfs.2017.09.028. [DOI] [PubMed] [Google Scholar]
- 34.Zou Y, Wang SS, Wang J, Su HL, Xu JH. CircRNA_0016418 expedites the progression of human skin melanoma via miR-625/YY1 axis. Eur Rev Med Pharmacol Sci. 2019;23:10918–10930. doi: 10.26355/eurrev_201912_19795. [DOI] [PubMed] [Google Scholar]