Abstract
Purpose
The purpose of this study was to establish a deep learning model for automated sub-basal corneal nerve fiber (CNF) segmentation and evaluation with in vivo confocal microscopy (IVCM).
Methods
A corneal nerve segmentation network (CNS-Net) was established with convolutional neural networks based on a deep learning algorithm for sub-basal corneal nerve segmentation and evaluation. CNS-Net was trained with 552 and tested on 139 labeled IVCM images as supervision information collected from July 2017 to December 2018 in Peking University Third Hospital. These images were labeled by three senior ophthalmologists with ImageJ software and then considered ground truth. The areas under the receiver operating characteristic curves (AUCs), mean average precision (mAP), sensitivity, and specificity were applied to evaluate the efficiency of corneal nerve segmentation. The relative deviation ratio (RDR) was leveraged to evaluate the accuracy of the corneal nerve fiber length (CNFL) evaluation task.
Results
The model achieved an AUC of 0.96 (95% confidence interval [CI] = 0.935–0.983) and an mAP of 94% with minimum dice coefficient loss at 0.12. For our dataset, the sensitivity was 96% and specificity was 75% in the CNF segmentation task, and an RDR of 16% was reported in the CNFL evaluation task. Moreover, the model was able to segment and evaluate as many as 32 images per second, much faster than skilled ophthalmologists.
Conclusions
We established a deep learning model, CNS-Net, which demonstrated a high accuracy and fast speed in sub-basal corneal nerve segmentation with IVCM. The results highlight the potential of the system in assisting clinical practice for corneal nerves segmentation and evaluation.
Translational Relevance
The deep learning model for IVCM images may enable rapid segmentation and evaluation of the corneal nerve and may provide the basis for the diagnosis and treatment of ocular surface diseases associated with corneal nerves.
Keywords: corneal nerve fiber segmentation, deep learning, convolutional neural network, in vivo confocal microscopy
Introduction
Sub-basal corneal nerves penetrate the peripheral cornea radially and form the nerve plexus between Bowman's layer and the basal epithelium, playing an important role in the sensation of touch, pain, and temperature1,2 and mediating wound healing, blink reflex, and tear film stability.3
In vivo confocal microscopy (IVCM) is a noninvasive imaging modality that enables corneal visualization and evaluation in vivo. Conditions such as diabetic peripheral neuropathy (DPN),4–6 dry eye disease (DED),7–9 inflammation keratitis,10 contact lens wearing,11 and refractive surgery12,13 have demonstrated corneal nerve abnormities by IVCM. The corneal nerve fiber length (CNFL) is significantly reduced in individuals with diabetic mellitus (DM), and studies have suggested IVCM to be a more sensitive detection method for DPN than other established tests.5,6 Studies have also revealed significantly reduced CNFL in patients with DED.14 Moreover, IVCM research has documented a lower nerve density of the central cornea after laser-assisted in situ in keratomileusis (LASIK) and photorefractive keratectomy (PRK).15 Thus, IVCM is a critical method in ocular surface disease evaluation. Rapid segmentation and evaluation of sub-basal corneal nerves are essential in diagnosis and assessments of various diseases that may have an impact on clinical decision making and prediction.
Traditional corneal nerve fiber (CNF) segmentation and evaluation rely on time-consuming manual or semi-automated methods that require further improvements in speed and accuracy.16 To solve this problem, software for automatic CNF segmentation and evaluation has been developed. Scarpa et al. identified CNFs in IVCM based on seed points, and a tracking module detected the nerve direction and point movement through some successive segments perpendicular to the nerve.17 Another software package was used to detect CNFs based on ridge map calculation.18 Dabbah et al. first used a 2D Gabor filter to detect CNFs and then extended this filter to a dual-model detector.19,20 Nevertheless, software associated with CNFs, as examined by IVCM, was determined to be impractical in the clinic.
Deep learning-based approaches have been revolutionized over the past few years and are widely used in numerous fields, including image recognition, detection, and segmentation.21 In combining the ability to extract and represent features and layer-by-layer abstraction, the convolutional neural network (CNN) has demonstrated amazing performance on computers vision tasks because of its natural consistency with a two-dimensional structure of image data. CNNs based on deep learning models have been applied in various fields of ophthalmology, including glaucoma, retinopathy of prematurity (ROP), and age-related macular degeneration (AMD).22 Previous research has demonstrated the efficacy of deep learning models in the identification of diabetic retinopathy (DR) and DPN. Abramoff et al. showed a deep learning system that was able to achieve an area under the receiver operating characteristic curve (AUC) of 0.980, with a sensitivity of 96.8%, and a specificity of 87.0% in DR detection,23,24 which achieved a better performance than ophthalmologists. The present research aimed to train a CNN model named corneal nerve segmentation network (CNS-Net) for CNF segmentation and evaluation with IVCM images. The model was intended to directly and rapidly provide us with information on sub-basal corneal nerve segmentation and evaluation that may assist the decision making process.
Methods
Dataset
A total of 5221 images from 104 patients were collected at Peking University Third Hospital from July 2017 to December 2018. The present study was conducted following the Helsinki Declaration and was approved by the local review board. Informed consent was not obtained for each participant because it was a retrospective study, but we pledged that the images enrolled in the present research would not be linked to individual patients.
The images were taken with the Heidelberg Retina Tomograph Rostock Corneal Module (Heidelberg Engineering GmbH, Heidelberg, Germany). The images were captured by IVCM with a setting of 484 by 384 pixels over an area of 400 µm by 400 µm and a lateral spatial resolution of 0.5 µm and a depth resolution of 1 to 2 µm. A total of 40 to 50 pictures were captured for each eye from the corneal epithelium to the endothelium.
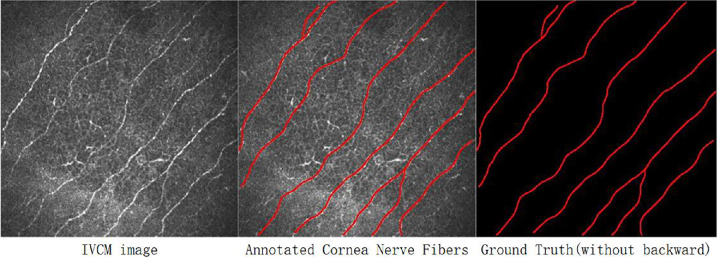
The flow chart of images obtaining, screening, and labeling is shown in Figure 1. All images were cropped first with a valid area of 384 pixels by 384 pixels. Then, some basic principles were followed in quality control. Images should reveal at least one clearly visible corneal nerve, and images with strong artifacts were excluded. Afterward, CNFs were drawn manually by the same three ophthalmologists independently (as shown in Fig. 1) as ground truth applying the Java-based software ImageJ (National Institute of Health, Bethesda, MD) with the NeuronJ (Biomedical Imaging Group, Lausanne, Switzerland) plugin, and the CNFL was calculated automatically with ImageJ software based on manual CNF annotation. The discrepancy of the image screening and labeling results among the 3 ophthalmologists was 65 resolved by a simple majority vote to obtain the final ground truth.
Figure 1.
Data obtaining, screening and labeling.
A dataset with 691 samples was proposed following quality control processes, which had an average of 5 to 6 images for each participant. Each of the samples consisted of an original IVCM image and its binary CNF annotation image, as shown in Figure 2. For building a reliable benchmark and ensuring the generalization and stability of artificial intelligence models, the dataset was divided into 3 parts in a ratio of 7:1:2, namely, a training set (483 samples, 70%), validation set (69 samples, 10%), and test set (139 samples, 20%), which are widely applied parameters in computer vision and artificial intelligence.
Figure 2.
Data example.
CNS-Net
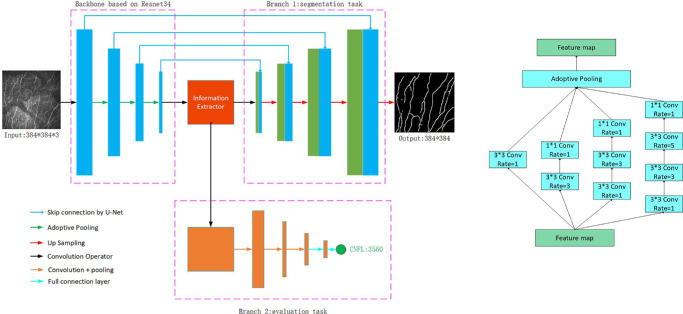
A CNN framework was established to output the CNF segmentation and CNFL from IVCM images. The CNS-Net model architecture is shown in Figure 3. The CNN model consisted of an encoder, an information extractor, and a decoder.
Figure 3.
CNS-Net architecture.
The encoder used a ResNet34 architecture25 without the last output layers, which was transferred from the ImageNet dataset with a powerful capability for image feature extraction. The information extractor consisted of a series of densely connected dilated convolution and concatenation operators with a final adoptive pooling layer. Finally, the generated feature map was analyzed by a decoder for the CNF segmentation task and the CNFL evaluation task.
In the CNF segmentation branch, the transposed convolution operator with a bilinear interpolation algorithm was used to upscale the feature map from an encoder and information extractor. During this process, the size of the feature map was amplified to the same size as that of the input IVCM image. Inspired by residual connections in the ResNet model and U-Net architecture, skip connections from encoder to decoder segmentation task branch were leveraged to improve the information flow from shadow features to high-level semantic features. This helped to restore the feature map with more detailed information intuitively. Finally, a softmax layer was introduced to classify every pixel of the feature map as the CNF or background along the channel axis.
In the CNFL evaluation branch, two convolution layers with a pooling operator and two fully connected layers were used to act on the feature map from the information extractor, and the CNFL was obtained.
Loss Function
Traditional loss functions are designed for a single image segmentation task or numerical calculation task, but our CNS-Net model combined two types of computer vision calculation tasks. For faster and better convergence of the end-to-end training process for the present CNS-Net, a novel loss function was introduced by a combination of different loss forms to optimize our model.
The dice coefficient (DC), a statistic used to gauge the similarity of two sets X and Y, is defined as . Thus, the loss of CNF segmentation task can be calculated when the ground truth is available, as in Equation 1:
| (1) |
where N is the number of pixels in one image and K is the class number. Here, p(k, i) ∈ [0, 1] and g(k, i) ∈ {0, 1}, respectively, denote the predicted probability and the ground truth for the labeled class k for the ith pixel. For the CNFL evaluation task, the common mean square error (MSE) is used, as in Equation 2:
| (2) |
where n is the number of samples. In addition, yi and , respectively, denoted the predicted CNFL and its actual value. Moreover, a weight decay (also called the regularization loss) Lreg was added to avoid overfitting because of our small dataset size.
The final loss function was formuled as Equation 3:
| (3) |
Experiment
Data Augmentation
To reduce the risk of overfitting in the model training phase, some data augmentation methods were used. First, image normalization of the red, green, and blue (RGB) channels, horizontal flip, vertical flip, and diagonal flip were applied. With these direct and effective methods, each image in the training set was augmented to eight different copies. Subsequently, random data augmentation methods were used in the model training procedure, including image shifting randomly in the range [—30°, 30°] and color jittering in the hue, saturation, and value (HSV) color space, image scaling from 1.1 to 1.2. We then cropped the input size randomly and added principal component analysis (PCA) noise with a coefficient sampled from a normal distribution with parameters .
Experiment Details
The training process was implemented in Python version 3.7.6 based on the open-source deep learning platform MxNet. A fixed ResNet34 model with pretrained parameters on ImageNet was applied on the encoder, whereas other parameters of CNS-Net were initialized with the Xavier algorithm.26 Nesterov accelerated gradient (NAG) descent was used for training with batch size 32, momentum 0.9, and weight decay 0.0001. Learning rate warmup27 and cosine learning rate decay policy25 were also used. The maximum epoch was 300 with an automatic stop mechanism. In artificial intelligence research, iteration refers to the process of updating model parameters with mini-batch data, whereas epoch refers to the process of making use of the entire training data set by many iterations. We can formulate the relation of the two terms as:
In our paper, one epoch is 16 iterations.
Evaluation Metrics
All of the metrics were calculated against the ground truth. For the accuracy of CNF segmentation, the AUCs, mean average precision (mAP), sensitivity, and specificity were reported. The mAP was calculated when the model was trained on the training set with the supervision of the validation set. The mAP was defined as:
| (4) |
where CNFseg is the segmentation result of our model for IVCM images and CNFgt is the manual ground truth by ophthalmologists. N is the size of the dataset, is the overlapped area between the model segmented result and manual ground truth for the ith IVCM image, and is the union area between them. The accuracy of CNFL task was evaluated by the relative deviation ratio, which was defined as the ratio of the absolute value of the difference between the output CNFL value and the input CNFL value to the input CNFL value, as
| (5) |
Results
As shown in Figure 4, the training loss and the validation loss quickly dropped to approximately 0.2 after the first 5 epochs of training and ultimately decreased to 0.12 and 0.13 as the training process continued. When the train loss did not decrease with 20 consecutive epochs, the procedure of model training stopped automatically, and we obtained a 92% validation mAP. The synchronous decline of train loss and validation loss verified the stability of the training process and ensured the generalization ability of the model.
Figure 4.
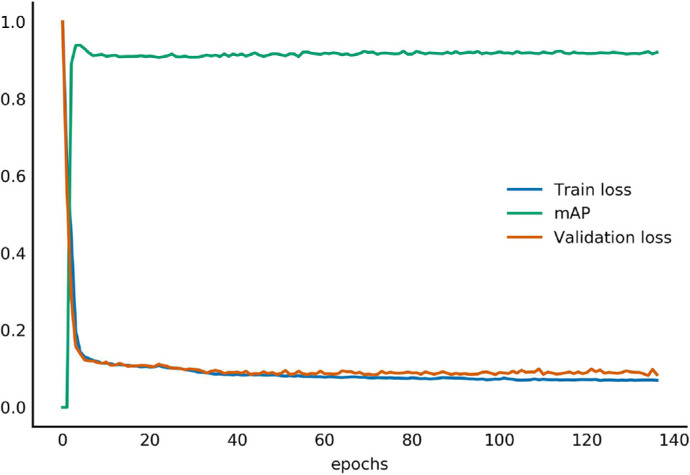
Training and validation loss curve.
The receiver operating characteristic (ROC) curve of the model's segmentation result on the test set is illustrated in Figure 5. The model achieved an AUC of 0.96 (95% confidence interval [CI] = 0.935-0.983) and an mAP of 94% with minimum dice coefficient loss 0.12, sensitivity 96%, and specificity 75%. The model also achieved a speed of 32 fps (32 images per second), an efficiency much higher than that of ophthalmologists.
Figure 5.

Receiver operating characteristic (ROC) curve.
An example of CNF segmentation with manual ground truth by an ophthalmologist is shown in Figure 6. Despite the fact that some tiny nerve endings were missed, the model was able to identify most of the CNFs.
Figure 6.
Example of CNFL segmentation.
On the CNFL evaluation task, a 0.91 (95% CI = 0.892–0.923) Pearson correlation coefficient and a 16% relative deviation ratio were reported by our method. We performed a Bland-Altman analysis to determine the consistency between manual work and CNS-Net, and the results are shown in Figure 7 (all 139 images used here were all from a fixed test set). The 95% limits of agreement between the ground truth and CNS-Net analysis were between -749.0 and 650.7 (concordance correlation coefficient [CCC] = 0.912).
Figure 7.
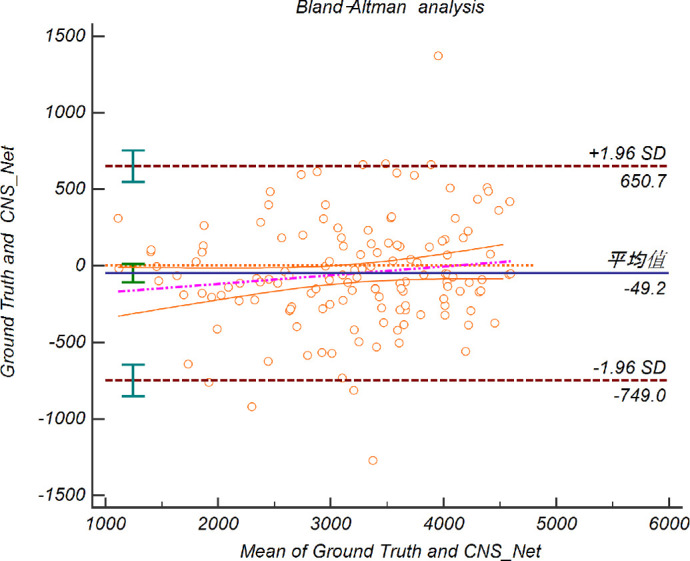
Bland-Altman Plots comparing pixels between manual work and CNS-Net. The middle dotted line represents the bias for the comparison between manual work and CNS-Net. The upper dotted and lower dotted lines show the upper and lower limits of agreement between the two methods.
Discussion
IVCM imaging has become an important method for the diagnosis of many ocular surfaces diseases, both in clinical practice and in scientific studies. Therefore, a concrete quantitative evaluation of CNF loss in IVCM remains to be explored. One ophthalmologist may exhibit different performances in different situations, and it is unlikely that manual operation can conduct favorable repeatability corneal nerve tracing. They can analyze these images several times to assure a relatively accurate result for scientific research. In clinical practice, however, this repetition is unlikely to happen due to limited time and other manual factors. In comparison, once informed of the identification processes, a mathematical model can perform nerve tracing and evaluation repeatedly, achieving relatively stable and consistent results.
Therefore, after obtaining the dataset as accurately as possible, we addressed these problems by proposing a fully automated model to identify and segment corneal nerve fibers from IVCM images. Unlike so-called feature engineering methods, CNNs can extract features automatically through a large quantity of data. Following the construction of the CNF segmentation dataset, feed-forward and back-propagation approaches were applied to train the CNS-Net model. Then, the model can segment and evaluate CNFs from IVCM images accurately, efficiently, and automatically. We collected a widely and sufficiently representative dataset, then the dataset was randomly divided into a training set, validation set, and test set. The trainer randomly obtains minibatch data from the training set and carries out data augmentation operations to optimize model parameters in each iteration of model training. Hence, our model can cover most of the IVCM images with accurate results, and has excellent generalization and stability. The results of the present research highlight the potential for the system to be applied in automatic CNF segmentation for ocular surface disease evaluations in the future.
In our study, the ophthalmologist (C.Y.L.) reviewed those results with a dice coefficient of lower than 85%. When further comparing the predicted CNF image with the ground truth, we found that the manual annotation missed some of the nerves that were identified by the algorithm. Additionally, the speed of the model identifying the image was 32 fps (32 images per second). The results demonstrated that our algorithm is orders of magnitude faster than manual annotation and achieves a much better performance in segmenting corneal nerves.
The main difference between CNS-Net and other automatic methods for corneal segmentation is the use of deep learning. ACCMetrics is a software applied in IVCM analysis, based on traditional computer vision algorithms, and CNS-Net has a higher accuracy and speed than ACCMetrics.28 Scarpa's algorithm was found to detect corneal nerves with a sensitivity of 80% and could calculate sub-basal corneal nerve length.18 Traditional machine learning based on the dual-model filter has also been applied in detecting corneal nerves in DPN according to curvilinear features.16 However, the features in certain kinds of corneal neuropathy could not be generalized to other diseases using IVCM. Therefore, we proposed a new model based on popular U-Net construction, and the model was trained with CNF images of all kinds. The model can extract features and classify each pixel into a corneal nerve or background efficiently.
However, there are several limitations of this automatic system for clinical application by ophthalmologists. First, the dataset in our study was based on only a small group of patients at Peking University Third Hospital and cannot represent the entire population. Therefore, data from a larger patient cohort should be applied to validate the system. Second, the current CNS-Net model cannot account for all of the parameters in IVCM images, such as the CNF width and tortuosity, which limits its generalizability. Hence, the present model was not able to replace the manual image annotation completely. Further studies are required to assess the performance of the model for CNF segmentation and quantitation in different populations and different types of IVCM equipment. With future research, the model is expected to further improve the accuracy of diagnosis and can be applied in clinics more efficiently.
Acknowledgments
The research was supported by the National Science and Technology Major Project 2018ZX10101004003003.
Disclosure: S. Wei, None; F. Shi, None; Y. Wang, None; Y. Chou, None; X. Li, None
References
- 1. Hamrah P, Cruzat A, Dastjerdi MH, et al.. Corneal sensation and subbasal nerve alterations in patients with herpes simplex keratitis: an in vivo confocal microscopy study. Ophthalmology. 2010; 117: 1930–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shaheen BS, Bakir M, Jain S. Corneal nerves in health and disease. Surv Ophthalmol. 2014; 59: 263–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nishida T, Chikama T, Sawa M, Miyata K, Matsui T, Shigeta K. Differential contributions of impaired corneal sensitivity and reduced tear secretion to corneal epithelial disorders. Jpn J Ophthalmol. 2012; 56: 20–25. [DOI] [PubMed] [Google Scholar]
- 4. Li Q, Zhong Y, Zhang T, et al.. Quantitative analysis of corneal nerve fibers in type 2 diabetics with and without diabetic peripheral neuropathy: comparison of manual and automated assessments. Diabetes Res Clin Pr. 2019; 151: 33–38. [DOI] [PubMed] [Google Scholar]
- 5. Callaghan BC, Cheng HT, Stables CL, Smith AL, Feldman EL. Diabetic neuropathy: clinical manifestations and current treatments. Lancet Neurol. 2012; 11: 521–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cruzat A, Qazi Y, Hamrah P. In vivo confocal microscopy of corneal nerves in health and disease. Ocul Surf. 2017; 15: 15–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Villani E, Magnani F, Viola F, et al.. In vivo confocal evaluation of the ocular surface morpho-functional unit in dry eye. Optom Vis Sci. 2013; 90: 576–586. [DOI] [PubMed] [Google Scholar]
- 8. Giannaccare G, Pellegrini M, Sebastiani S, Moscardelli F, Versura P, Campos EC. In vivo confocal microscopy morphometric analysis of corneal subbasal nerve plexus in dry eye disease using newly developed fully automated system. Graefe's Arch Clin Exp Ophthalmol. 2019; 257: 583–589. [DOI] [PubMed] [Google Scholar]
- 9. Kheirkhah A, Dohlman TH, Amparo F, et al.. Effects of corneal nerve density on the response to treatment in dry eye disease. Ophthalmology. 2015; 122: 662–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Müller RT, Abedi F, Cruzat A, et al.. Degeneration and regeneration of subbasal corneal nerves after infectious keratitis: a longitudinal in vivo confocal microscopy study. Ophthalmology. 2015; 122: 2200–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Patel DV, Zhang J, McGhee CNJ. In vivo confocal microscopy of the inflamed anterior segment: A review of clinical and research applications. Clin Experiment Ophthalmol. 2019; 47: 334–345. [DOI] [PubMed] [Google Scholar]
- 12. Zhao J, Yu J, Yang L, Liu Y, Zhao S. Changes in the anterior cornea during the early stages of severe myopia prior to and following LASIK, as detected by confocal microscopy. Exp Ther Med. 2017; 14: 2869–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pahuja NK, Shetty R, Deshmukh R, et al.. In vivo confocal microscopy and tear cytokine analysis in post-LASIK ectasia. Br J Ophthalmol. 2017; 101: 1604–1610. [DOI] [PubMed] [Google Scholar]
- 14. Labbé A, Alalwani H, Van Went C, Brasnu E, Georgescu D, Baudouin C. The relationship between subbasal nerve morphology and corneal sensation in ocular surface disease. Invest Ophthalmol Vis Sci. 2012; 53: 4926–4931. [DOI] [PubMed] [Google Scholar]
- 15. Benítez-del-Castillo JM, Acosta MC, Wassfi MA, et al.. Relation between corneal innervation with confocal microscopy and corneal sensitivity with noncontact esthesiometry in patients with dry eye. Invest Ophthalmol Vis Sci. 2007; 48: 173–181. [DOI] [PubMed] [Google Scholar]
- 16. Chen X, Graham J, Dabbah MA, Petropoulos IN, Tavakoli M, Malik RA. An automatic tool for quantification of nerve fibers in corneal confocal microscopy images. IEEE Trans Biomed Eng. 2016; 64: 786–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Calvillo MP, McLaren JW, Hodge DO, Bourne WM. Corneal reinnervation after LASIK: prospective 3-year longitudinal study. Invest Ophthalmol Vis Sci. 2004; 45: 3991–3996. [DOI] [PubMed] [Google Scholar]
- 18. Scarpa F, Grisan E, Ruggeri A. Automatic recognition of corneal nerve structures in images from confocal microscopy. Invest Ophthalmol Vis Sci. 2008; 49: 4801–4807. [DOI] [PubMed] [Google Scholar]
- 19. Holmes TJ, Pellegrini M, Miller C, et al.. Automated software analysis of corneal micrographs for peripheral neuropathy. Invest Ophthalmol Vis Sci. 2010; 51: 4480–4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dabbah MA, Graham J, Petropoulos I, Tavakoli M, Malik RA. Dual-model automatic detection of nerve-fibres in corneal confocal microscopy images. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. 2010: 300–307. [DOI] [PMC free article] [PubMed]
- 21. Chilamkurthy S, Ghosh R, Tanamala S, et al.. Deep learning algorithms for detection of critical findings in head CT scans: a retrospective study. Lancet. 2018; 392: 2388–2396. [DOI] [PubMed] [Google Scholar]
- 22. Ting DSW, Pasquale LR, Peng L, et al.. Artificial intelligence and deep learning in ophthalmology. Br J Ophthalmol. 2019; 103: 167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Abràmoff MD, Lou Y, Erginay A, et al.. Improved automated detection of diabetic retinopathy on a publicly available dataset through integration of deep learning. Invest Ophthalmol Vis Sci. 2016; 57: 5200–5206. [DOI] [PubMed] [Google Scholar]
- 24. Williams BM, Borroni D, Liu R, et al.. An artificial intelligence-based deep learning algorithm for the diagnosis of diabetic neuropathy using corneal confocal microscopy: a development and validation study. Diabetologia. 2020; 63: 419–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016: 770–778.
- 26. Glorot X, Bengio Y. Understanding the difficulty of training deep feedforward neural networks. In: Teh YW, Titterington M, eds. Proceedings of the Thirteenth International Conference on Artificial Intelligence and Statistics. Vol. 9 Proceedings of Machine Learning Research. PMLR; 2010: 249–256. [Google Scholar]
- 27. Goyal P, Dollár P, Girshick R, et al.. Accurate, large minibatch sgd: training imagenet in 1 hour. arXiv Prepr arXiv170602677 2017. Available at: https://arxiv.org/abs/1706.02677. [Google Scholar]
- 28. Dabbah MA, Graham J, Petropoulos IN, Tavakoli M, Malik RA. Automatic analysis of diabetic peripheral neuropathy using multi-scale quantitative morphology of nerve fibres in corneal confocal microscopy imaging. Med Image Anal. 2011; 15: 738–747. [DOI] [PubMed] [Google Scholar]