Abstract
Critical to managing the spread of COVID-19 is the ability to diagnose infection and define the acquired immune response across the population. While genomic tests for the novel Several Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) detect the presence of viral RNA for a limited time frame, when the virus is shed in the upper respiratory tract, tests able to define exposure and infection beyond this short window of detectable viral replication are urgently needed. Following infection, antibodies are generated within days, providing a durable read-out and archive of exposure and infection. Several antibody tests have emerged to diagnose SARS-CoV-2. Here we report on a qualified quantitative ELISA assay that displays all the necessary characteristics for high-throughput sample analysis. Collectively, this test offers a quantitative opportunity to define both exposure and levels of immunity to SARS-CoV-2.
1. Introduction
Coronaviruses (CoVs) represent a large family of RNA viruses that largely cause mild to moderate respiratory disease in humans (Vincent et al., 2020; Lia Van Der Hoek et al., 2004; Fielding, 2011; Fouchier et al., 2004). However, over the past 2 decades, several lethal CoVs have emerged including Severe Respiratory Acute Syndrome (SARS-CoV-1) and Middle Eastern Respiratory Syndrome (MERS-CoV), linked to 10% and 40% fatality rates, respectively (Chen, 2020; Petrosillo et al., 2020). Likewise, in December of 2019, a novel coronavirus, SARS-CoV-2, causing the Coronavirus Infection Disease 2019 (COVID-19) pandemic, represents the third coronavirus to jump the species barrier to cause severe fatal disease in humans, leading to an unprecedented devastating global pandemic that has left the world paralyzed.
Key to managing spread is the ability to diagnose infection and define “immunity”. While genomic tests can detect the presence of the virus at the time of infection, these tests only provide a measure of exposure at the time that virus is detectable in the upper respiratory tract (Wolfel et al., 2020). Conversely, the detection of pathogen-specific antibodies, that emerge within days of infection, represent a durable biomarker of previous exposure. Days after infection, IgM antibodies develop followed quickly by the evolution of more affinity matured class switched IgG/IgA. Recent studies from China and Germany show that although a small percentage of patients never seroconvert, over 40% of tested RNA-confirmed cases possess detectable levels of IgM and IgG within the first 7 days of illness, and that by day 15, all patients clearly possess detectable cross-isotype antibody levels (Zhao et al., 2020; Okba et al., 2020).
Large numbers of antibody-tests have emerged over the past few months (Adams et al., 2020; Amanat et al., 2020; Qian et al., 2020)with different performance characteristics, however, few provide quantitative results, not only defining infected/uninfected, but also providing precise quantitative metrics related to the level of humoral response found within a given individual. Moreover, scale up and access to these tests is a challenge. Thus, here we describe a robust ELISA test that can detect and quantify IgA, IgM and IgG against SARS-CoV-2 receptor binding domain (RBD), the target of the majority of neutralizing antibodies. This simple qualification plan provides a step-by-step approach to set up the assay and ensure robust assay performance. The ELISA can be rapidly scaled up and automated to test thousands of samples daily providing quantitative measures of exposure and immunity.
2. Material and method
2.1. Antigen production
The receptor binding domain (RBD) of the spike protein of SARS-CoV-2 (residues 319–529) (GenBank: MN975262.1), of SARS-CoV-1 (residues 306–515) (Urbani strain; AAP13441.1), and of CoV-HKU1 (residues 310–677) (Accession #: AY597011) were cloned into a pVRC vector with a C-terminal SBP-tag and expressed in HEK293F cells and purified as previously published Bajic et al., 2019.
2.2. CR3022 monoclonal construct and production
2.2.1. Plasmid design and construction
Five donor pUC plasmids encoding the variable heavy domain (DQ168569.1), the Fc domain, a furin P2A sequence, the variable light domain (DQ168570.1) of the antibody, and the kappa constant light domain were designed. The 5′ donor plasmids were combined with a destination vector in a single digestion-ligation reaction to generate a single expression plasmid encoding both the heavy chain and light chain of the monoclonal antibody. One alternate destination vector was additionally created for expression of only the light chain allowing optimization of the heavy chain to light chain ratio during antibody expression. Ligation products were transformed into Stellar competent cells (Clontech), and plated onto agar plates with kanamycin, and colonies were screened and sequenced to verify the identity of each target plasmid.
2.2.2. Production of antibodies in mammalian cells
Plasmids were expanded and transfected into 293F suspension cells grown in FreeStyle™ 293 Expression media (Gibco). 25 μg of total DNA was transfected onto cells using Polyethylenimine (PEI) (Polysciences) at 1 μg/μl in a ratio of 3 μg PEI to 1 μg DNA. Cell culture supernatants were harvested 5 days post transfection. CR3022 IgG1 was purified using Protein A/G magnetic beads (Millipore). IgA1/IgA2 cell culture supernatants were also harvested 5 days post transfection and antibodies were purified using Peptide M agarose (InvivoGen).
For production and purification of CR3022 IgM, 1.2 × 106 cells/ml were transfected with 25 μg of DNA in the following ratio: 10 μg CR3022 IgM expression construct, 10 μg CR3022 Light Chain expression construct, and 5 μg of a J-Chain expression construct that was previously designed in the lab. The plasmids were transfected with the same ratio of PEI to DNA as previously described. Cell culture supernatants were harvested 5 days post transfection and CR3022 IgM were purified with PierceTM Protein L magnetic beads (ThermoFisher Scientific).
For 500 ml production sizes, 250 μg of total DNA was transfected onto cells with PEI in the same ratio as the smaller production size. Purified antibodies were concentrated using Amicon Ultra-15 Centrifugal Filter Units with a 50 kDa molecular weight cut off.
2.3. Samples
Serum/plasma samples used in this study where obtained through the Massachusetts General Hospital Diagnostic Laboratories, the University of Washington, Brigham and Women Hospital, and the Ragon Institute Clinical Services. All subjects were collected after subjects provided signed informed consent or were collected as discarded samples under approved MGH institutional protocols. Four groups of subjects were enrolled: hospitalized individuals with a SARS-CoV-2 confirmed RNA test were included; hospitalized subjects with no presumption of SARS-CoV-2 infection were included; convalescents with a confirmed prior SARS-CoV-2 RNA+ and two repeat RNA-negative tests after 2 weeks of isolation; and a group of low risk community members. Subjects were included if they had a positive SARS-CoV-2 RNA test. Samples were heat inactivated at 60 °C for 1 h.
2.4. ELISA
384-well ELISA plates (Thermo Fisher #464718) were coated with 50 μl/well of 500 ng/ml SARS-CoV-2-RBD in coating buffer (1 capsule of carbonate-bicarbonate buffer (Sigma #C3041100CAP) per 100 ml Milli-Q H2O) for 30 min at room temperature. Plates were then washed 3 times with 100 μl/well of wash buffer (0.05% Tween-20, 400 mM NaCl, 50 mM Tris (pH 8.0) in Milli-Q H2O) using a Tecan automated plate washer. Plates were blocked by adding 100 μl/well of blocking buffer (1% BSA, 140 mM NaCl, 50 mM Tris (pH 8.0) in Milli-Q H2O) for 30 min at room temperature. Plates were then washed as described above. 50ul of diluted samples (in dilution buffer; 1% BSA, 0.05% Tween-20, 140 mM NaCl, 50 mM Tris (pH 8.0) in Milli-Q H2O) was added to the wells and incubated for 30 min at 37 °C. Plates were then washed 5 times as described above. 50 μl/well of diluted detection antibody solution (HRP-anti human IgG, IgA or IgM; Bethyl Laboratory #A80-104P, A80-100P, A80-102P) was added to the wells and incubated for 30 min at room temperature. Plates were then washed 5 times as described above. 40 μl/well of TMB peroxidase substrate (Thermo Fisher #34029) was then added to the wells and incubated at room temperature for 3 min (IgG), 5 min (IgA and IgM). The reaction was stopped by adding 40 μl/well of stop solution (1 M H2SO4 in Milli-Q H2O) to each well. OD were read at 450 nm and 570 nm on a Tecan infinite M1000pro plate reader.
For CR3022 dilution curves, the antibody was diluted to a concentration of 20 μg/ml in dilution buffer and a 12 two-fold serial dilution curve was generated and plated.
2.5. Statistics
Statistical calculations were done in Prism v8.2.1 and in R v3.6.1; packages Stars, pROC, ggplot2, ggpubr, caret, zetadiv, drc.
Receiver operating characteristic (ROC) curves: Logistic regression models for individual and all possible (Wolfel et al., 2020) combinations of IgG, IgA, IgM as predictors to discriminate SARS-CoV-2 positive and negative samples were trained for 100 repetitions of 10-fold cross-validation using the R function ‘glm.cons’ (package zetadiv). A negative parameter constraint for the intercept and positive parameter constraint for the coefficients were applied to enforce the direction of the model, i.e., higher values for the SARS-CoV-2 positives. As always. The minimal achievable area under the curve (AUC) was 0.5. ROC curves were calculated using the R function ‘roc’ (package pROC).
Youden index: The Youden index, J, combined the specificity and sensitivity as J = sensitivity + specificity - 1. The cut-off value for which the Youden index was maximal for the ROC curves generated without cross-validation was reported.
3. Results
3.1. Survey of SARS-CoV-2 positive samples
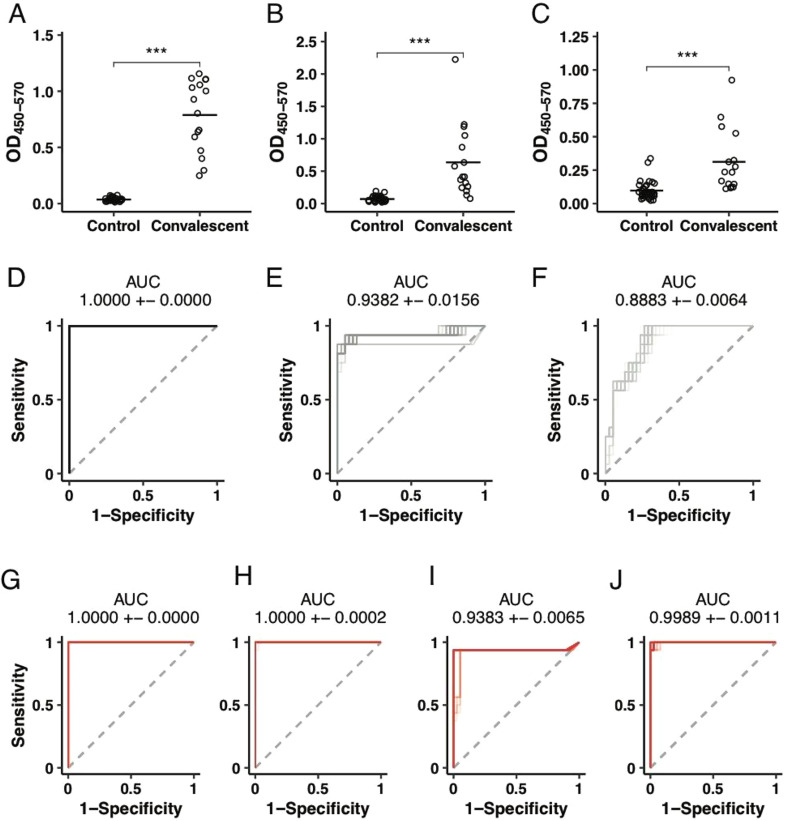
While nucleic acid testing provides a highly sensitive measure of the presence of the virus in the upper respiratory tract, high rates of false negativity are likely associated with transient and intermittent viral shedding, that may result in missed diagnoses (Wolfel et al., 2020; Paul Wikramaratna et al., 2020). Conversely, days after infection, pathogen-specific IgM rise early, followed by IgG and IgA antibodies, providing highly specific biomarkers of infection. Several ELISA assays have been described, including assays to the Coronavirus Spike (S) antigen (Adams et al., 2020), the main target for neutralizing antibodies, to the nucleocapsid protein (Liu et al., 2020) as well as to a small region of the S protein, the receptor binding domain (RBD), involved in viral attachment to the human angiotensin converting enzyme 2 (ACE2) receptor (Wang et al., 2020). Given that the RBD is likely the target of protective neutralizing antibodies against the virus (Poh et al., 2020; Quinlan et al., 2020), its highly stable structure, the ease to scale production, and previous ELISA development successes (Amanat et al., 2020) we focused on the development of an RBD-specific ELISA. To confirm that RBD-specific responses can differentiate individuals that have been previously exposed to SARS-CoV-2 from those that have not been exposed, RBD-specific IgM, IgG, and IgA responses were probed in a group of convalescent (individuals who were previously diagnosed with COVID-19 and later recovered, as confirmed by two negative RNA tests) and negative community controls. Significantly higher levels of SARS-CoV-2-RBD-specific IgG were observed across all convalescents compared to community controls (Fig. 1A). Conversely, more variable level of IgA and IgM were observed in convalescents, albeit levels of these two isotypes were significantly higher among convalescents compared to community controls (Fig. 1B and C). The ability of logistic regression models to discriminate between SARS-CoV-2 positive and negative samples based on IgG, IgA, IgM or all possible combinations of the three measurements was assessed. Receiver operating characteristic curves, used to define the diagnostic capacity of each isotype, revealed the robust ability of each isotype to classify individuals into convalescents and community controls (Fig. 1D-J).
Fig. 1.
Robust discrimination between convalescents and community controls by ELISA. (A-C) The dot plots show the differences in CoV2-receptor binding domain (RBD) IgG (A), IgA (B), or IgM (C) levels in convalescents and community controls. Groups were compared using a Mann-Whitney U test (***p < 0.001). (D-J) Receiver operator characteristic (ROC) curves. Area under the curve (AUC) was calculated in a 10-fold cross-validation framework for 100 repetitions for discriminating 38 SARS-CoV-2 negatives from 16 convalescent SARS-CoV-2 positives based on OD450–570 levels for (D) IgG (E) IgA (F) IgM (G) IgG and IgA, (H) IgG and IgM, (I) IgA and IgM, (J) IgG, IgA, IgM. AUCs are reported as mean + − SD.
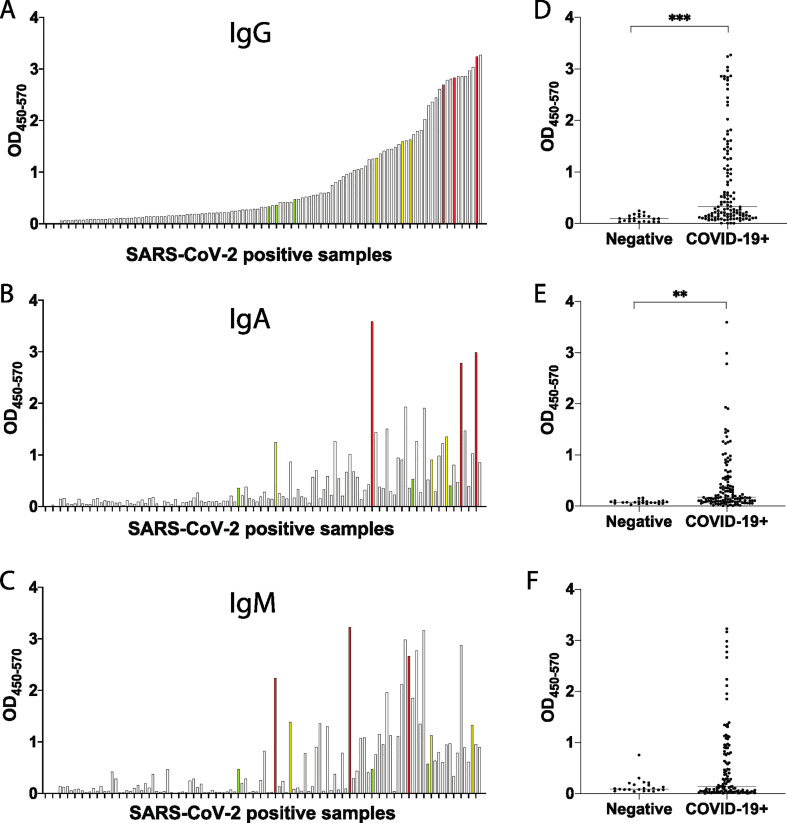
To further determine whether SARS-CoV-2-specific IgM, IgG, and IgA responses could also be detected in individuals with more variable levels of immunity, we next turned to early infection. We next profiled RBD-specific immunity in a cohort of recently SARS-CoV-2 infected and uninfected individuals. A total of 118 individuals, at variable days following symptom onset, were included in this analysis. A wide range of SARS-CoV-2-RBD-specific IgG, IgA and IgM responses were observed across the early infected samples (Fig. 2A-C), with significant levels of SARS-CoV-2-RBD-specific IgG, and IgA, expressed in SARS-CoV-2 RNA+ individuals compared to controls (Fig. 2 D-F). When individuals were stratified by IgG levels, individuals with the highest IgG levels exhibited variable levels of IgA (Fig. 2B) and low levels of IgM (Fig. 2C). Conversely, high levels of IgM (Fig. 2C) were observed in individuals that exhibited intermediate levels of IgG (Fig. 2A) and IgA (Fig. 2B).
Fig. 2.
CoV2-RBD-specific antibody levels in early CoV2 infection. 118 SARS-CoV-2 positive samples were screened for SARS-COV-2-RBD specific IgG (A), IgA (B) and IgM (C). Each graph shows the OD obtained for each sample (450 nm–570 nm). Selected high samples are shown in red, mediums in yellow and lows in green. The same set of SARS-CoV-2 positive samples was run against 22 negative samples. Data is shown for IgG (D), IgA (E), IgM (F). Groups were compared using a Mann-Whitney U test (**p < 0.01, ***p < 0.001). . (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
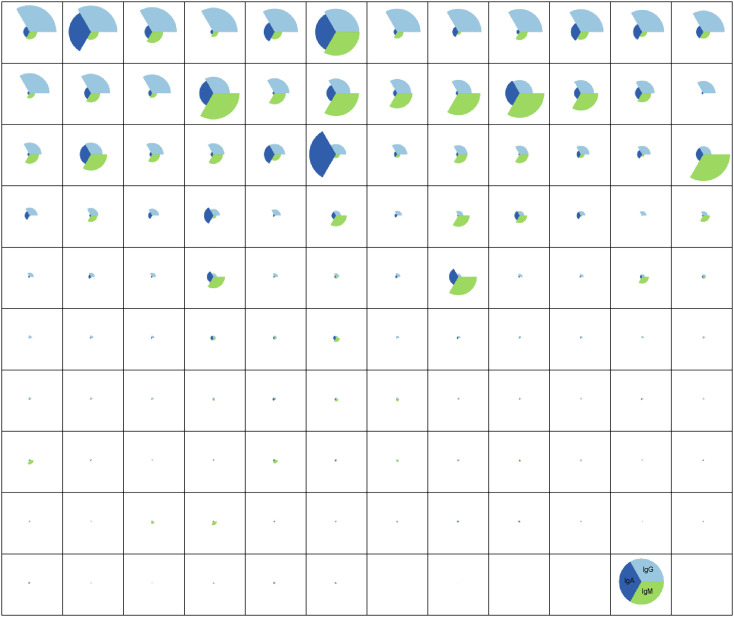
To gain a deeper sense of the relationships between isotypes across the population, pie charts were generated for each subject Fig. 3 , where the pie slice colour represented the isotype, and the size of the pie represented whether the individual belonged in the top, middle, or lowest tercile for that specific SARS-CoV-2-RBD-specific isotype. The top row points to a group of RNA+ individuals enriched for nearly only SARS-CoV-2-RBD-specific IgG responses. The second row consisted of a group of individuals with lower IgG responses, but stronger IgA responses. Finally, intermittent individuals exhibited levels of SARS-CoV-2-RBD-specific IgA responses. These data highlight the highly variable IgG, IgA, and IgM responses seen in hospitalized individuals. However, to go on to develop a robust assay, 3 subjects with high (yellow), medium (yellow), or low (green) IgG, IgA, and IgM (Fig. 2A-C) responses were selected for assay optimization.
Fig. 3.
CoV2-RBD-specific isotype profiles in early CoV2 infection. Relative amount of IgG, IgA and IgM against SARS-CoV-2-RBD are shown for the whole panel of 118 SARS-CoV-2 positive serum sample. For each sample, the relative amount of an isotype is reported as fold over background.
3.2. ELISA optimization
Despite the clear resolution between convalescents and community members, more variable detection was observed in the early infection cohort. These observations motivated further optimization to drive the development of an assay with top performance characteristics to ensure the collection of limited numbers of false negatives or positives. A systematic dissection of each step of the ELISA was undertaken to improve assay performance, including the identification of the optimal antigen-coating concentration, the definition of the optimal secondary antibody utilization, the analysis of specificity, and ultimately the final analysis of precision or performance accuracy.
3.2.1. Concentration of antigen
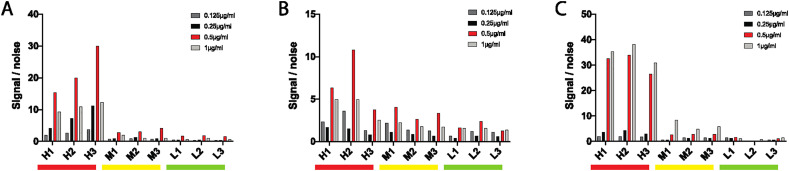
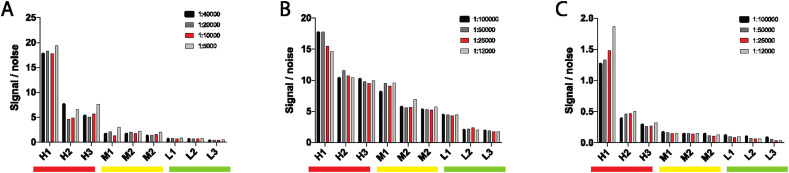
Four coating-antigen concentrations were tested to begin. Two-fold dilutions were tested beginning at 1 μg/ml. For each isotype, the selected 3 high, 3 medium and 3 low positive samples were all run in duplicate in addition to 2 known SARS-CoV-2-negative control samples. The 0.5μg/ml coating concentration was clearly superior for IgG and IgA detection (Fig. 4A and B). The 0.5μg/ml also showed similar performance for individuals with a high IgM response, although the 1μg/ml coating concentration showed some superiority in the medium-IgM responders (Fig. 4C). For simplicity and given the striking differential that was still observable between high/med/low at 0.5μg/ml, a single 0.5μg/ml coating concentration was selected for the assay.
Fig. 4.
Optimization of antigen amount. With high (H), medium (M) and low (L) samples, four different concentrations of SARS-CoV-2-RBD were tested for IgG (A), IgA (B) and IgM (C). Optimal concentrations are shown in red. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2.2. Concentration of detecting antibody
The following optimization step included the identification of the ideal concentration of secondary-horse radish peroxidase (HRP) conjugated antibodies for each isotype. Four dilutions were tested for each secondary antibody. Based on specifications, two-fold dilutions were generated for each isotype starting at 1:100000 for IgM and IgG and beginning at 1:40000 for IgA. The same RBD-specific 3 high, 3 medium and 3 low samples (Fig. 2A and B) were tested in duplicate in addition to 2 known SARS-CoV-2-RNA-negative control samples. Similar performance was observed across most dilutions for IgG (Fig. 5A). Similar patterns were observed for IgA and IgM, with the exception of individuals with high SARS-CoV-2-RBD specific titers (Fig. 5B and C). Ultimately, for simplicity an intermediate dilution was selected: 1:25000 for IgG and IgM and 1:10000 for IgA.
Fig. 5.
Optimization of detecting antibody dilution. With high (H), medium (M) and low (L) samples, four different dilutions of detecting antibody were tested for IgG (A), IgA (B) and IgM (C). Optimal dilutions are shown in red. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2.3. Binding specificity
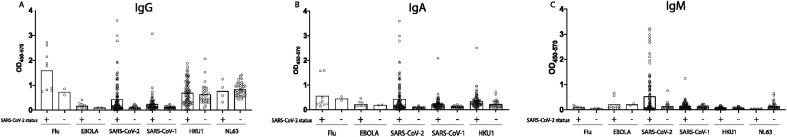
To assess the specificity of the assay, the 3 high, 3 medium and 3 low for IgG, IgA, and IgM responders were profiled against a pool of influenza hemagglutinin (HA) antigens as a positive control and against the Ebola virus glycoprotein as a negative control. In addition, responses were also tested against the related SARS-CoV-1 RBD, that shares above 70% homology with SARS-CoV-2 (Zhou et al., 2020). Robust HA-specific IgG responses were observed across all subjects, with more mixed IgM and IgA responses (Fig. 6A). No Ebola GP-specific responses were detected across subjects and isotypes (Fig. 6B).
Fig. 6.
Binding specificity. Binding specificity was assessed by running SARS-CoV-2 positive samples against Flu HA, EBOLA GP, CoV-HKU1-RBD, CoV-NL63-RBD, SARS-CoV-1-RBD and SARS-CoV-2-RBD for IgG (A), IGA (B) and IgM (C). Boxes show average result.
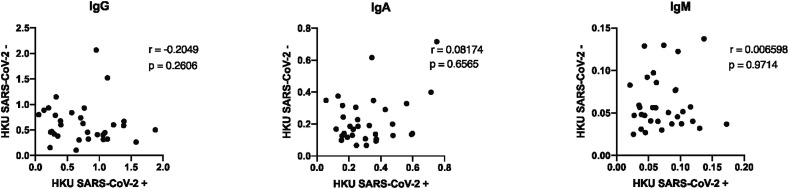
Several coronaviruses have entered the human population over the past few decades, including two lethal CoVs: SARS-CoV-1 and MERS that share 77.5% and 50% sequence identify with SARS-CoV-2 RBD (Kim et al., 2020a). Additionally, two additional common cold causing CoVs, HKU1 and NL63 circulate in the population, sharing 20% and 1.9% sequence identify to the SARS-CoV-2-RBD (Aaron Schmidt, personal communications, February 2020). While exposure to SARS-CoV-1 and MERS is likely to be negligible across most tested populations, an immune response to cold-causing CoVs is likely to be extensive. Thus, to gain a sense of the overall specificity, or cross-reactivity, in the assay, IgG, IgA and IgM responses were quantified across SARS-CoV-1-RBD, CoV-NL63 and HKU1-RBD, the 3 most related CoV RBDs. While no significant IgM reactivity was observed to SARS-CoV-1, significant numbers of SARS-CoV-2 positive samples showed cross-reactivity to SARS-CoV-1 (Fig. 6). Given the limited circulation of SARS-CoV-1, these data point to potential cross-reactivity across SARS-CoVs using RBD. Although we observed broad responses to CoV-NL63 and CoV-HKU1-RBD (Fig. 6), mostly for IgG and to some degree for IgA, there was no significant difference observed between the SARS-CoV-2 positive and the SARS-CoV-2 negative samples, clearly indicating that this response was due to a previous infection with HKU1 as opposed to any evidence of SARS-CoV-2 cross reactivity. The lack of cross-reactivity is further supported by the fact that SARS-CoV-2 responses do not correlate with antibody titers against CoV-HKU1 RBD (Fig. 7 ).
Fig. 7.
Absence of correlation between SARS-CoV-2 and HKU1 titers. Correlation analyses are plotted for HKU1 titers for SARS-CoV-2 positive and negative samples. Spearman rank correlation coefficient (r) and p value are shown.
3.3. ELISA qualification
Once optimized, the assay was qualified, per GCLP standards, which required that all assay performance characteristics fall within expected assay performance ranges defined through rigorous replicate testing by multiple operators. Again, samples with high, medium, and low IgG, IgA, and IgM response profiles were selected and utilized in parallel to a group of 48 known SARS-CoV-2-RNA-negative control samples.
3.3.1. Limit of detection
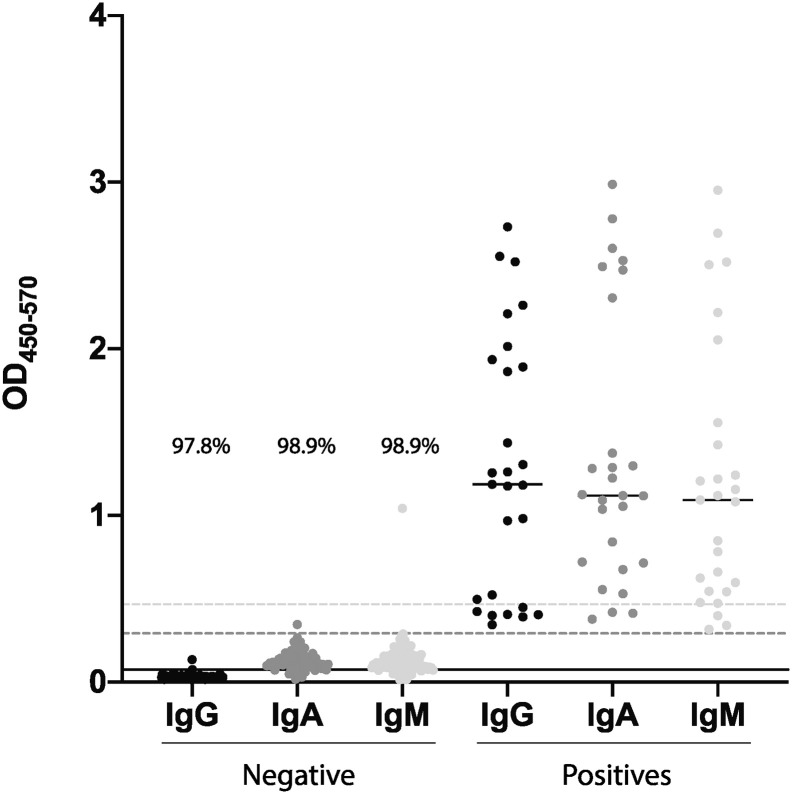
To determine the limit of detection of the assay, 90 known SARS-CoV-2-RNA-negative control samples were initially profiled across each isotype (Fig. 8 ). Low level reactivity was observed for IgG, with average background optical density (OD) value of 0.0295. In contrast, intermittent IgA and IgM responses were observed in the negative controls, resulting in a background average of 0.1274 and 0.1342 OD respectively. Thus, using three times the calculated SD, the limit of detection was established as 0.0736 for IgG, 0.2914 for IgA and 0.4662 for IgM, with 97.8% (88/90), 98.9% (89/90), and 98.9% (89/90) classification accuracy for each isotype, respectively.
Fig. 8.
Limit of detection. Known negative samples were run against SARS-CoV-2-RBD for IgG, IgA and IgM. The graph shows 90 negative samples for all three isotypes, the limit of detection (lines of corresponding colors) and the percentage of data point under the limit of detection. High, medium and low positive samples were also plotted to show the range in contrast to the limit of detection.
3.3.2. Quantitation
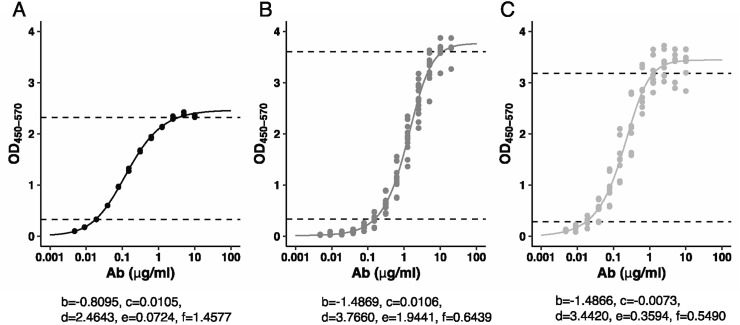
To estimate the concentration (μg/ml) of each isotype present in a sample, we developed standard curves with SARS-CoV-1-RBD specific monoclonal CR3022 that has been shown to cross-react with SARS-CoV-2 RBD (Yuan et al., 2020). In addition to the original IgG1 CR3022, we produced an IgA and IgM variants of the antibody with the same specificity. Each monoclonal was then serially diluted and run against SARS-CoV-2-RBD (Fig. 9A-C). The range of quantitation of the assay was established for each isotype as the range of the optical density (OD) where the sample dilution curve demonstrated linearity. As expected, the curves differed across the isotypes, with an optimal range of 0.3254–2.3177 for IgG, 0.3329–3.6051 for IgA, and 0.5296–3.1830 for IgM, thus establishing the lower and upper limit of quantitation (LLOQ and ULOQ) for each isotype.
Fig. 9.
Quantification. 12 points dilution curves were run for CR3022_IgG (A), CR3022_IgA (B) and CR3022_IgM (C) monoclonal. The dotted lines show linear range between ULOQ and LLOQ. A five-parameter log-logistic function (Y = c + (d - c)/((1 + exp.(b*(log(X) - log(e))))^f)) allows the conversion of OD in concentration of antibody (μg/ml).
We fitted a five-parameter log-logistic functions and the equation and estimated parameters are shown in Fig. 9. These equations allow the conversion of an OD value into a corresponding concentration of antibody (μg/ml). For example, to calculate the concentration of IgG in a sample with an OD of 1.5, we would use the following equation:
x(μg/ml) = e*exp.(log(((d-c)/(Y-c))^(1/f)-1)/b) = 0.0724*exp.(log(((2.4643–0.0105)/(1.5–0.0105))^(1/1.4577) - 1)/(−0.8095)) which would give us a concentration of 0.2187 μg/ml.
3.3.3. Sensitivity and specificity
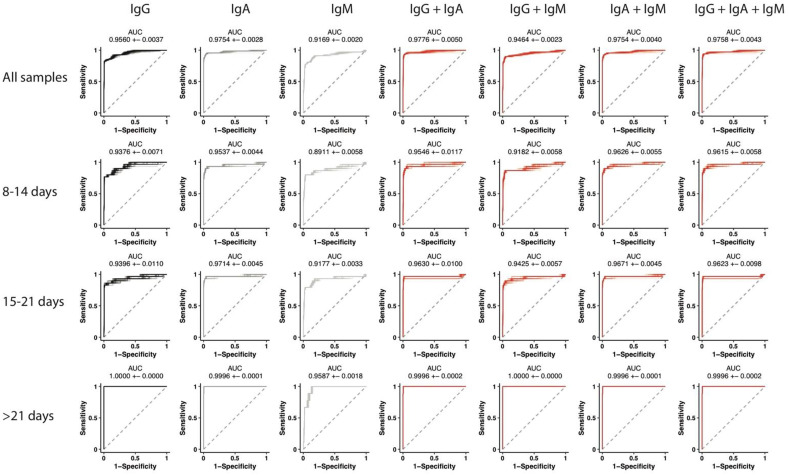
To assess the ability of our ELISA to accurately diagnose SARS-CoV-2 infection, 68 SARS-CoV-2 RNA positive samples and 232 negative samples were run against SARS-CoV-2-RBD for IgG, IgA and IgM. Each sample was run in duplicate. The ability of logistic regression models to discriminate between SARS-CoV-2 positive and negative samples based on IgG, IgA, IgM or all possible combinations of the three measurements was assessed. RBD-specific IgG, IgA and IgM levels overall were associated with an average AUC of 0.9560, 0.9754 and 0.9169, respectively (Fig. 10 ). Interestingly, IgM, our first antibody isotype, arose early following infection, persisting at variable levels depending on disease severity, likely contributing to more heterogeneity in IgM detection across populations. Combined, IgG and IgA displayed overall the highest predictive power with and AUC of 0.9776. Early in disease, IgA and IgM combined were the best predictors of SARS-CoV-2 infection with an AUC of 0.9626. However, within 15 to 21 days after symptom onset, IgA alone showed the highest predictive power with an AUC 0.9714. Finally, later in disease, AUCs similar to those observed originally in convalescent samples (Fig. 1) were observed (Fig. 10).
Fig. 10.
ROC curves. ROC curves were calculated based on OD450–570 levels for IgG, IgA, IgM, IgG and IgA, IgG and IgM, IgA and IgM, IgG, IgA and IgM. Area under the curve (AUC) were calculated to assess performance of the model in a 10-fold cross-validation framework for 100 repetitions and reported as mean + − SD. ROC curves are shown for all samples and for 8–14 days, 15–21 days or > 21 days from symptoms onset.
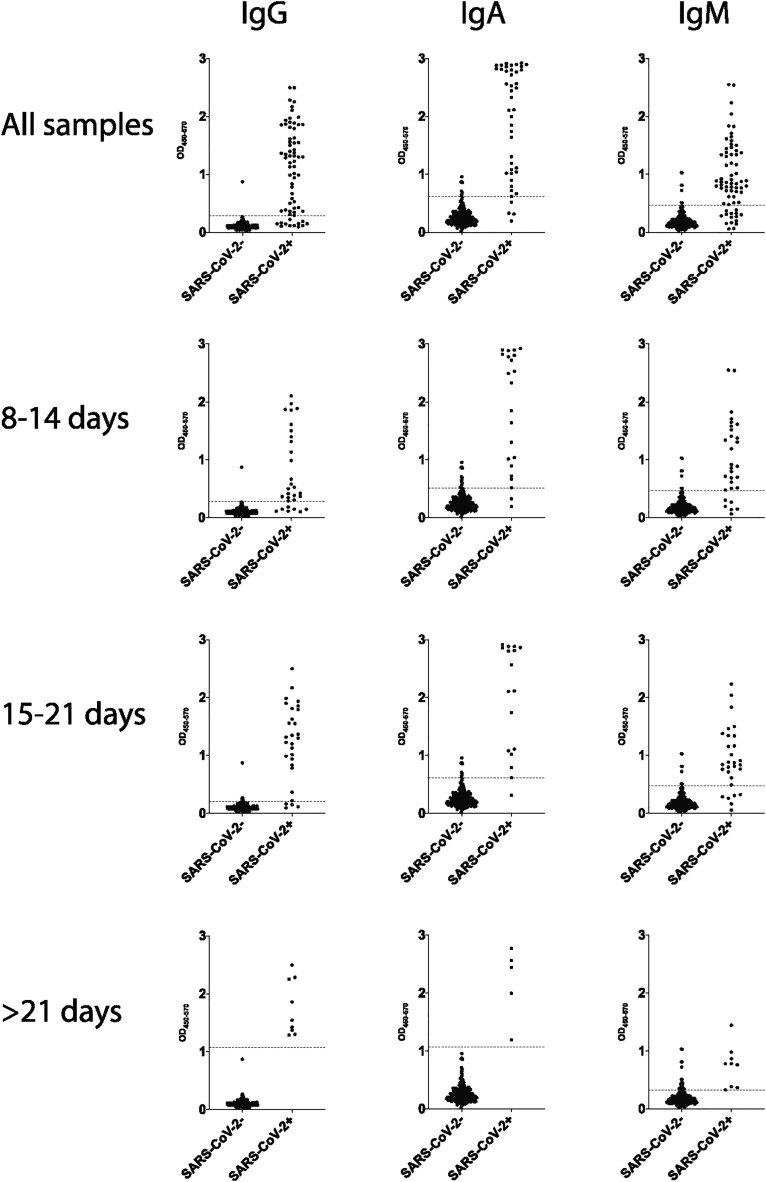
To determine a cutoff that would allow optimal separation of negative and positive samples for each isotype, Youden index were calculated for each ROC curves (Fig. 10) and a threshold was set at the OD value corresponding to the maximal Youden index (Fig. 11 ). As shown in Table 1 , robust specificity was obtained for IgG and IgA for every timepoint. Conversely, specificity was higher for IgM before 21 days from symptom onset. IgA demonstrated a high sensitivity throughout while IgG and IgM showed maximal sensitivity at later timepoints. These data confirm the robust predictive power of IgG, IgA and IgM responses both in convalescents (Fig. 1) as well as in newly and acutely infected individuals (Fig. 10).
Fig. 11.
Threshold at maximal Youden Index. Individual data points for SARS-CoV-2 positive and negative samples are plotted for IgG, IgA and IgM. Data is shown for all samples and for 8–14 days, 15–21 days or > 21 days from symptoms onset. Dashed lines show OD value at maximal Youden index as calculated from ROC curves shown in Fig. 9.
Table 1.
Specificity and sensitivity. The table shows specificity and sensitivity data for each isotype and for all samples and for 8–14 days, 15–21 days or > 21 days from symptoms onset.
| Days from symptoms onset | IgG |
IgA |
IgM |
|||
|---|---|---|---|---|---|---|
| Specificity | Sensitivity | Specificity | Sensitivity | Specificity | Sensitivity | |
| All | 99.56% | 82.35% | 96.55% | 94.12% | 96.98% | 77.94% |
| 8–14 | 99.57% | 76.67% | 94.83% | 93.33% | 96.98% | 80.00% |
| 15–21 | 97.41% | 86.21% | 96.55% | 96.55% | 96.98% | 79.31% |
| >21 | 100.00% | 100.00% | 100.00% | 100.00% | 88.36% | 100.00% |
3.3.4. Precision
Lastly, the overall precision of the ELISA was assessed as the repeatability across operators as well as across days (intermediate precision). Three different operators ran the same low, medium and high IgG, IgA and IgM samples and negative controls at two different dilutions in duplicate over 3 days. Precision was defined by calculating the average overall coefficient of variability (CV) between all the replicates for one sample for each operator. To determine intermediate precision, the CV was calculated for each operator day-to-day and between operators overall. The average %CV for intermediate precision was then calculated Table 2 .
Table 2.
Precision Results. The table lists the average precision (CV%) across operators (average repeatability) and across days and operators (intermediate precision) across isotypes and high, medium, and low samples. Per EMA guidelines, all results are below a coefficient of variation of 20% (25% for low samples), and often the results are below a 10%.
| Samples | IgG |
IgA |
IgM |
|||
|---|---|---|---|---|---|---|
| average repeatability | average intermediate precision | average repeatability | average intermediate precision | average repeatability | average intermediate precision | |
| High | 7.30% | 15.30% | 7.50% | 14.20% | 6.88% | 16.25% |
| Med | 10.00% | 15.70% | 8.48% | 14.29% | 9.96% | 18.27% |
| Low | 10.83% | 20.65% | 10.55% | 17.30% | 9.29% | 24.78% |
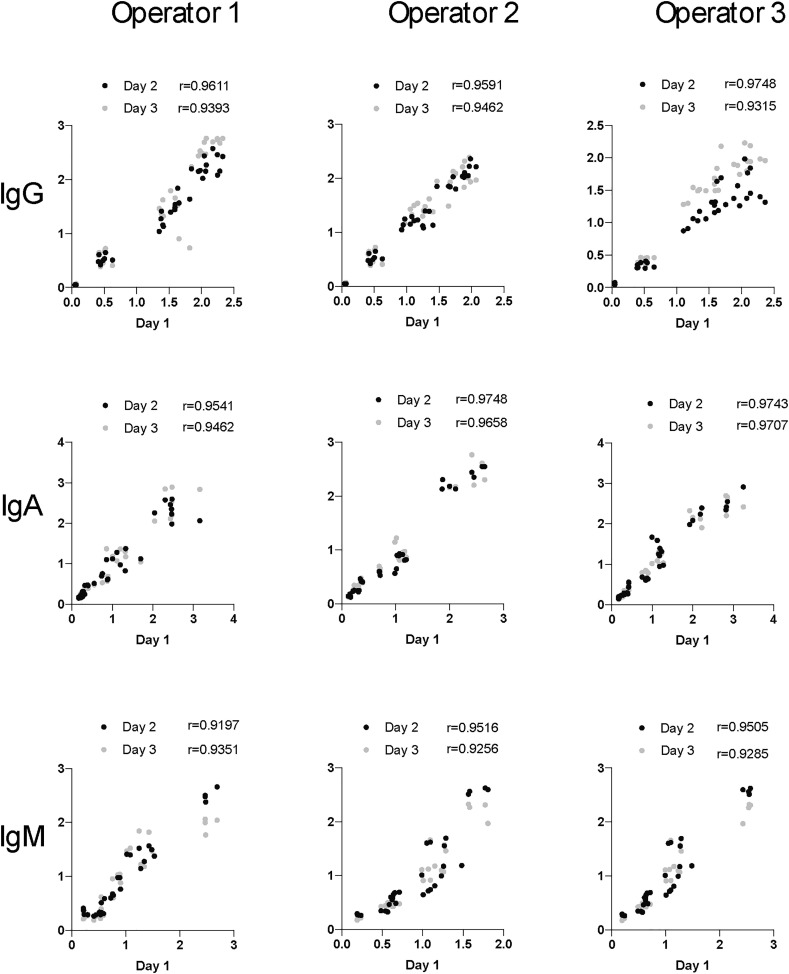
Robust correlations were observed between operators and days, across all isotypes (Fig. 12 ). Highly significant correlations were observed across all operators and days. Moreover, average Spearman rank correlation coefficients ranged from 0.9315–0.9748 for IgG, 0.9462–0.9748 for IgA, and 0.9197–0.9516 for IgM, highlighting the robustness of the assay day to day and operator to operator.
Fig. 12.
Precision Correlation. Correlation analyses are plotted for each operator across days. Spearman rank correlation coefficient are shown for each day.
According to the European Medicines Agency (EMA), a CV of 20% or lower is acceptable for repeatability and intermediate precision, although a 25% CV is acceptable for samples with antibody concentrations approaching the lower limit of quantitation. The IgG ELISA clearly met the required repeatability target CV with all values reaching levels lower than 11%. Additionally, IgG intermediate precision was below 16% for high and medium samples and just under 21% for low concentration samples, clearly meeting EMA regulations. The IgA and IgM ELISAs additionally met repeatability target CVs, with all CVs lower than 11% for IgA and below 10% for IgM. Finally, intermediate precision was below 15% for high and medium IgA samples, 16.25% for high IgM samples, 18.27% for medium IgM samples, and 17.30% and 24.78% for low IgA and IgM samples respectively, meeting the stringent requirements of the EMA. Thus, the assay met all the required performance criteria required for assay qualification. Given the simplicity of the assay, future automation may further reduce CVs and improve quantification and the precision of reported antibody concentration.
4. Discussion
Since entering the human population, SARS-CoV-2 has spread effortlessly across the globe, leading to an unprecedented global pandemic (Wolfel et al., 2020; Paul Wikramaratna et al., 2020). Social distancing and use of personal protective equipment (PPE), including face masks, have slowed the spread of the virus. However, in the absence of a vaccine, and in the setting of asymptomatic transmission, approaches are urgently needed to identify and quarantine infected individuals to slow down spread. The scale up of RNA testing has been instrumental for symptomatic testing in hospitals or health-care settings. However, the transient nature of viral shedding in the upper respiratory tract during the first few days of infection, may result in underreporting of the true burden or prevalence of infection at a global level. Thus, we currently have no concept of the actual number of individuals that have been exposed to SARS-CoV-2, which is readily achievable by seroprevalence studies. Understanding this “denominator” – or the number of individuals that have been exposed and whether they develop immunity- is absolutely key to managing the epidemic and to help bring an end to social isolation.
Serological tests, that capture pathogen-specific antibodies, represent a key tool used to define exposure/infection at a population level. Antibodies are generated days after infection, with an early rise in IgM, followed by class-switched IgG, then IgA, that collectively provide immunological evidence of exposure to pathogen components, irrespective of the level of viral replication at different sites within the respiratory tract, providing an immunological biomarker of exposure even after the virus is cleared. Dozens of tests have begun to emerge across the globe, most of which provide a qualitative means to define those that do or do not have antibodies. However, tests that offer a quantitative measure of the amount of antibody in the plasma are not currently available. Here we report a sensitive, quantitative IgG, IgA and IgM ELISA that provides a robust means to not only define those exposed to SARS-CoV-2, but also quantify the level of antibodies to guide the identification of antibody levels that may be key to defining “immunity” thresholds.
This current assay qualification builds on previously described ELISAs (Amanat et al., 2020), focused on capturing antibodies to the RBD, due to its critical role as the major target of neutralizing antibodies (Quinlan et al., 2020). Additionally, unlike the trimeric stabilized spike protein (S), which is more difficult to produce, RBD is a highly stable structure that can be produced and scaled for large scale ELISA testing. Given the immunogenic nature of RBD, it likely represents a reasonable target to both quantify population level exposure as well as to define antibody levels that may ultimately be associated with immunity. However, additional viral components including the membrane protein, the nucleocapsid, or envelope protein are generated in larger amounts during infection (Kim et al., 2020b; Hofmann and Pohlmann, 2004), and could provide enhanced diagnostic value, particularly in early infection, when antigen-concentration may influence the kinetics of antibody evolution. Moreover, antibodies that target alternate sites on the S antigen or with the ability to drive additional antibody effector functions, may also contribute to immunity. Thus, next generation assay development, using alternate antigens, may provide earlier diagnostic value, in the absence of RNA testing, as well as provide additional insights into antibody-levels that may track with immunity.
While several qualitative assays have been reported (Adams et al., 2020; Amanat et al., 2020; Qian et al., 2020), the presence or absence of antibody is unlikely to be key to defining whether a given individual is immune to SARS-CoV-2. Instead, emerging data has even suggested that high levels of neutralizing antibodies, that likely target the RBD, prior to infection, can lead to enhanced pathology in non-human primates (Liu et al., 2019). Whether similar results will also be observed in humans remains unclear. However, these data imply that deeper investigation will be required to fully define the nature of “protective” antibodies, studies that are emerging through sero-epidemiologic studies (Bendavid et al., 2020; Huang et al., 2020; Larremore et al., 2020), pre-clinical vaccine studies (Amanat and Krammer, 2020), as well as convalescent passive serum transfer studies (Shen et al., 2020; Zhang et al., 2020). However, what is clear, is that quantitative, rather than qualitative, assessments will be absolutely key in the context of immune correlates analyses to guide public policy.
A large number of antibody-diagnostics have recently emerged on the market, many of which have performed poorly (Panel, 2020), not only compromising patient care, but also in establishing the true number of individuals infected in the population, a number that is urgently needed to inform rapidly evolving public policies on opening up the world economies. The need for high quality, scalable, and cost-effective testing is therefore urgently needed to not only support testing in the developed world, but also for communities in all corners of the globe that are equally being ravaged by this viral infection. The qualification path reported here represents a simple process to establish a robust, inexpensive IgG, IgA, and IgM ELISA to capture antibody levels to SARS-CoV-2.
Declaration of Competing Interest
Galit Alter is a founder of SeromYx.
Acknowledgments
We thank Bruce Walker, Nancy Zimmerman, Mark and Lisa Schwartz, and Terry and Susan Ragon for their support. We would also like to Aaron Schmidt for protein production efforts and Alicja Piechocka-Trocha for sample collection. Funding support was provided by the Ragon Institute of MGH, MIT, and Harvard and MGH ECOR Scholars.
References
- Adams E.R. Evaluation of antibody testing for SARS-CoV-2 using ELISA and lateral flow immunoassay. medRxiv. 2020 doi: 10.1101/2020.04.15.20066407. [DOI] [Google Scholar]
- Amanat F., Krammer F. SARS-CoV-2 vaccines: status report. Immunity. 2020 doi: 10.1016/j.immuni.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanat F. A serological assay to detect SARS-CoV-2 seroconversion in humans. medRxiv. 2020 doi: 10.1101/2020.03.17.20037713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajic G. Influenza Antigen Engineering Focuses Immune Responses to a Subdominant but Broadly Protective Viral Epitope. Cell Host Microbe. 2019;25:827–835. doi: 10.1016/j.chom.2019.04.003. e826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendavid E. COVID-19 Antibody Seroprevalence in Santa Clara County, California. medRxiv. 2020 doi: 10.1101/2020.04.14.20062463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. Pathogenicity and transmissibility of 2019-nCoV-A quick overview and comparison with other emerging viruses. Microbes Infect. 2020;22:69–71. doi: 10.1016/j.micinf.2020.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielding B.C. Human coronavirus NL63: a clinically important virus? Future Medecine. 2011;6(2):153–159. doi: 10.2217/fmb.10.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouchier Ron A., Bestebroer Theo M., Berend Niemeyer, de Jong Jan C., Simon James H., Osterhaus Albert D. A previously undescribed coronavirus associated with respiratory disease in humans. PNAS. 2004;101:6212–6216. doi: 10.1073/pnas.0400762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann H., Pohlmann S. Cellular entry of the SARS coronavirus. Trends Microbiol. 2004;12:466–472. doi: 10.1016/j.tim.2004.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang A.T. 2020. A Systematic Review of Antibody Mediated Immunity to Coronaviruses: Antibody Kinetics, Correlates of Protection and Association of Antibody Responses with Severity of Disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.M. Identification of Coronavirus Isolated from a Patient in Korea with COVID-19. Osong Public Health Res Perspect. 2020;11:3–7. doi: 10.24171/j.phrp.2020.11.1.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. The architecture of SARS-CoV-2 transcriptome. bioRxiv. 2020 doi: 10.1101/2020.03.12.988865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larremore D.B. Estimating SARS-CoV-2 seroprevalence and epidemiological parameters with uncertainty from serological surveys. medRxiv. 2020 doi: 10.1101/2020.04.15.20067066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lia Van Der Hoek K.P., Jebbink Maarten F., Vermeulen-Oost Wilma, Berkhout Ron J.M., Wolthers Katja C., Pauline M.E., Kaandorp Jos, Spaargaren Joke, Berkhout Ben. Identification of a new human coronavirus. Nat. Med. 2004;10 doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L. Anti-spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight. 2019;4 doi: 10.1172/jci.insight.123158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W. Evaluation of Nucleocapsid and spike protein-based ELISAs for detecting antibodies against SARS-CoV-2. J. Clin. Microbiol. 2020 doi: 10.1128/JCM.00461-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okba N.M.A. SARS-CoV-2 specific antibody responses in COVID-19 patients. medRxiv. 2020 doi: 10.1101/2020.03.18.20038059. [DOI] [Google Scholar]
- Panel N.C.T.S. Evaluation of antibody testing for SARS-CoV-2 using ELISA and lateral flow immunoassays. medRxiv. 2020 doi: 10.1101/2020.04.15.20066407. [DOI] [Google Scholar]
- Paul Wikramaratna R.S.P., Ghafari Mahan, Lourenço José. Estimating false-negative detection rate of SARS-CoV-2 by RT-PCR. medRxiv. 2020 doi: 10.1101/2020.04.05.20053355.this. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrosillo N., Viceconte G., Ergonul O., Ippolito G., Petersen E. COVID-19, SARS and MERS: are they closely related? Clin. Microbiol. Infect. 2020 doi: 10.1016/j.cmi.2020.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poh C.M. Potent neutralizing antibodies in the sera of convalescent COVID-19 patients are directed against conserved linear epitopes on the SARS-CoV-2 spike protein. bioRxiv. 2020 doi: 10.1101/2020.03.30.015461. [DOI] [Google Scholar]
- Qian C. Development and Multicenter Performance Evaluation of The First Fully Automated SARS-CoV-2 IgM and IgG Immunoassays. medRxiv. 2020 doi: 10.1101/2020.04.16.20067231. [DOI] [PubMed] [Google Scholar]
- Quinlan B.D. The SARS-CoV-2 receptor-binding domain elicits a potent neutralizing response without antibody-dependent enhancement. bioRxiv. 2020 doi: 10.1101/2020.04.10.036418. [DOI] [Google Scholar]
- Shen C. Treatment of 5 critically ill patients with COVID-19 with convalescent plasma. JAMA. 2020 doi: 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent J., Munster P.D., Marion Koopmans D.V.M., van Doremalen Neeltje, Van Riel Debby, Emmie de Wit P.D. A Novel Coronavirus Emerging in China — Key Questions for Impact Assessment. N. Engl. J. Med. 2020;328(8) doi: 10.1056/NEJMp2000929. [DOI] [PubMed] [Google Scholar]
- Wang Q. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020 doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfel R. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020 doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Yuan M. A highly conserved cryptic epitope in the receptor-binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020 doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B. Treatment with convalescent plasma for critically ill patients with SARS-CoV-2 infection. Chest. 2020 doi: 10.1016/j.chest.2020.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J. Anyibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. medRxiv. 2020 doi: 10.1101/2020.03.02.20030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]