Abstract
The aim of this study was to detect and characterize isolates of methicillin-/oxacillin-resistant Staphylococcus aureus (MRSA) carrying gene mecC (MRSA/mecC) and occurring in the Czech Republic within the period from 2002 to 2017. Altogether, 18 from 3,472 isolates of MRSA were mecC positive (0.52%). The first detection of MRSA/mecC in the Czech Republic is dated to 2004. MRSA/mecC isolates were susceptible to almost all tested antibiotics with few exceptions. Resistances to erythromycin (n = 2), clindamycin (n = 1), trimethoprim-sulfamethoxazole (n = 1), and rifampicin (n = 1) were found in the collection. Multilocus sequence typing and spa typing revealed a genetic heterogeneity of MRSA/mecC strains: three CCs (130, 425, and 2361), five STs (1245, 130, 2361, 425, and a new ST5480), and eight spa types (t843, t978, t1048, t1535, t1736, t6104, t8842, and t17153), which were detected in the study, with the highest prevalence of CC130/t843 lineage (n = 8, 44%). Except for two strains, none from 18 examined isolates harbored genes encoding any of S. aureus toxins: enterotoxins a–u, exfoliative toxins A, B, and D, toxic shock syndrome toxin-1, and the Panton-Valentine leukocidin.
Keywords: Staphylococcus aureus, MRSA; mecC; MLST; spa typing
Introduction
Staphylococcus aureus is the most important human Staphylococcus species associated with nosocomial and community-acquired infections.1 Since the first detection of diseases caused by methicillin-/oxacillin-resistant S. aureus (MRSA) (1961, UK), this pathogen has expanded into the whole world.2 MRSA isolates can exhibit resistance to a broad spectrum of antibiotics. Decreased susceptibility to methicillin/oxacillin as well as to other antibiotics is caused by acquired transpeptidase penicillin binding protein 2a (PBP2a) characterized by low affinity to β-lactams, with the exception of ceftaroline and ceftobiprole, so called antiMRSA cephalosporins. PBP2a is coded by mecA occurring in the staphylococcal chromosome cassette mec (SCCmec). SCCmec consists of variable and conserved genetic elements, including mec operon with mecA and regulatory (mecI and mecR1) genes, as well as recombinase genes (ccrA, ccrB, and ccrC) responsible for excision/integration of this genetic element within chromosome.3 Till now, 13 types of SCCmec (I–XIII) have been identified.4
In 2002 (Scotland) was detected a new variant of mecA gene in humans; mecALGA251 (later called mecC) producing an altered transpeptidase (PBP2c), also with a low affinity to β-lactam antibiotics.5 MecA and mecC homologs share similarity at the DNA level around 70%,3 and only 63% at amino acid level.5 Due to a nucleic acid divergence between mecA and mecC genes, all methods commonly used for mecA detection failed in mecC uncovering. One of the successful and simple methods used in the detection of MRSA isolates carrying gene mecC (MRSA/mecC) is PCR in combination with primers specific for gene mecC.5,6
Despite the fact that the presence of MRSA isolates carrying gene mecC was confirmed almost from all over Europe in humans,5,7 there is missing information about mecC detection in the Czech Republic. Therefore, the objective of this work was to examine the occurrence, an antibiotic susceptibility profile, virulence potential, as well as the genetic diversity of MRSA isolates carrying gene mecC in the long-term period, from 2002 to 2017 in the Czech Republic.
Materials and Methods
Bacterial isolates
Screening of MRSA/mecA isolates is provided as routinely in the National Reference Laboratory for Antibiotics (National Institute of Public Health, Prague, Czech Republic) as a part of European Antimicrobial Resistance Surveillance Network (EARS-Net). MRSA strains with an absence of mecA gene were further examined for the presence of mecC gene. Overall, 3,472 isolates of MRSA were isolated in the National Reference Laboratory for Antibiotics from 2002 to 2017. The majority of the clinical specimen, altogether 78.6% (n = 2,728), were of invasive origin (blood and cerebrospinal fluid), and 21.4% (n = 744) of MRSA strains were isolated from noninvasive materials (surgical wound, urine, sputum, and swab: nose, nasopharynx, and throat). All staphylococci were cultivated on nutrient agar (Oxoid) overnight at 36°C aerobically. Identification of S. aureus strains was performed by Matrix-assisted laser desorption ionization–time of flight mass spectrometry (Maldi-TOF; Microflex Brucker, Bremen, Germany).
Susceptibility testing
Screening for MRSA was performed by disk diffusion method using cefoxitin disc (30 μg); zone diameter <22 mm was interpreted as oxacillin resistance according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) recommendations.8 Minimal inhibition concentration of oxacillin (OXA), erythromycin (ERY), clindamycin (CLI), trimethoprim-sulfamethoxazole (SXT), rifampicin (RIF), ciprofloxacin (CIP), gentamicin (GEN), vancomycin (VAN), fusidic acid (FUS), chloramphenicol (CMP), linezolid (LNZ), and tigecycline (TGC) was determined by broth microdilution method according to ISO 20776-1. Susceptibility to tetracycline (TET) and tobramycin (TOB) was determined by disc diffusion method.9 Interpretation of susceptibility testing results was performed as recommended by the National Committee for Clinical Laboratory Standards (NCCLS) (later the Clinical & Laboratory Standards Institute: CLSI Guidelines)10 and EUCAST (version according to a corresponding year).11 Minimal inhibitory concentration of oxacillin was interpreted according to the NCCLS (later CLSI)10 and the EUCAST-ECOFF value (version according to a corresponding year).12
Molecular typing
PCR
DNA was isolated according to the procedure of isolation kit (GenElute™ Bacterial Genomic DNA Kits, Sigma Aldrich). The presence of mecA and mecC genes in MRSA isolates was identified by PCR. Gene mecA was amplified using primers F 5′ -GTA GAA ATG ACT GAA CGT CCG ATA A- 3′ and R 5′ -CCA ATT CCA CAT TGT TTC GGT CTA A- 3′. Conditions for PCR were 2 minutes at 94°C followed by 30 cycles of 30 seconds at 94°C, 1 minute at 55°C, 1 minute at 72°C, and finished by 10 minutes at 72°C (Bio-rad, DNA Engine Dyad® Dual–Bay Thermal Cycler). Gene mecC was detected according to the protocol of National Food Institute.6 PCR products were resolved in 1% agarose (Agarose SFR, VWR Life Science) in electrophoresis 5 V/cm for 50 minutes. S. aureus, NCTC13552 (Salisbury, UK), was used as positive control for mecC gene.
Spa and MLST typing
Spa typing: MRSA/mecC isolates were further analyzed using genotypic methods; repetitive sequences of protein A were amplified using the following primers: 1113f (5′- TAA AGA CGA TCC TTC GGT GAG C -3′) and 1514r (5′-CAG CAG TAG TGC CGT TTG CTT -3′), according to the procedure13 by analyzer (Applied Biosystems 3130xL). Data were analyzed using Bionumerics 7.6.2 (Applied Maths, Ghent, East Flanders, Belgium). The minimum spanning tree (MST) algorithm (gap creation cost 250%, gap extension cost 50%, duplicate creation cost 25%, and duplicate extension 25%, maximum duplication of three repetitions) used for cluster analysis of spa isolates was carried out also by Bionumerics 7.6.2.
Multilocus sequence typing (MLST) of mecC strains: seven housekeeping genes arcC, aroE, glpF, gmk, pta, tpi, and yqi were amplified according to the standard procedure described earlier.14 All strains were sequenced by analyzer (Applied Biosystems 3130xL). Data were analyzed by Bionumerics 7.6.2 and eBurst analysis was performed (database is no longer active).
Toxin detection
Isolates of S. aureus carrying mecC gene were also subjected to PCR analysis for confirmation or refutation of presence of genes encoding staphylococcus toxins: enterotoxins a-u (sea–seu), exfoliative toxins A, B, and D (eta, etb, and etd), staphylococci superantigens: toxic shock syndrome toxin-1 (tst), and the Panton–Valentine leukocidin (lukS/F-PVL; consisting of the LukS and LukF fragments). The list of primers and PCR conditions used for toxins amplification is in the Supplementary Table S1. S6 (sea, seb, seq, and sek), FRI137 (sec, seh, sel, sem, sen, seo, and seu), 01HMPL280 (sed, seg, sei, sej, sep, and ser), and FRI326 (see) were reference strains used for the detection of enterotoxins (all acquired from the French Food Safety Agency, Paris, France). Strains CCM7056 (eta and etb), CCM2331 (etd) (both acquired from Brno, Czech Republic), CNCTC5931 (Prague, Czech Republic, positive for tst,), and ATCC49775 (Virginia, positive for lukS/F-PVL,) were used for the detection of exfoliative toxins, tst, and the lukS/F-PVL.
Results
Antibiotic susceptibility of MRSA/mecC isolates
Altogether, 3,472 isolates of S. aureus resistant to CXT were tested for the presence of mecA/mecC gene in the period from 2002 to 2017. Eighteen of them harbored the mecC gene (0.52%). Resistance to methicillin/oxacillin in the rest of the MRSA samples (99.48%) was caused by the presence of mecA gene (data not shown). The magnitude of spreading differed from year to year, but the first detection of mecC in the Czech Republic was dated to the year 2004 (Table 1, isolate number 2132). MRSA/mecC isolates were detected in Ceske Budejovice, Pribram, Prostejov, Kyjov, Uherske Hradiste, Benesov, Prague, and Plzen. Positive strains were isolated especially from noninvasive material: swab (nose, nasopharynx, and throat) (n = 8), chirurgical wound (n = 5), and sputum (n = 1). Only four strains were acquired from blood.
Table 1.
The List of Methicillin-/Oxacillin-Resistant Staphylococcus aureus/mecC Isolates Involved in the Study
| Isolate | Age | Sex | Source | Year of isolation | Location | mm |
mg/L |
|
|
|
|
|
|
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CXT | TET | TOB | OXA | ERY | CLI | SXT | RIF | CIP | GEN | VAN | FUS | CMP | LNZ | TGC | spa | ST | CC | En | tst | Exf | lukS/F-PVL | ||||||
| 2132 | 59 | M | Blood | 2004 | Prague | 12 | 24 | 21 | >8 | 1 | 0.25 | ≤0.125 | ≤0.03 | 0.5 | 0.5 | 1 | 0.25 | 8 | 2 | 0.5 | t1736 | 5480 | 130 | − | − | − | − |
| 4747 | 67 | M | Sputum | 2004 | Ceske Budejovice | 14 | 28 | 22 | >8 | 1 | 0.25 | ≤0.125 | ≤0.03 | 0.25 | 0.5 | 1 | 0.125 | 8 | 2 | 0.5 | t843 | 130 | 130 | − | − | − | − |
| 4259 | 79 | M | Swab | 2010 | Ceske Budejovice | 12 | 27 | 22 | 8 | 0.5 | 0.125 | ≤0.125 | ≤0.03 | 0.5 | 0.25 | 0.5 | 0.125 | 8 | 2 | 0.125 | t843 | 1245 | 130 | − | − | − | − |
| 7468 | 79 | M | Blood | 2010 | Ceske Budejovice | 14 | 25 | 22 | >32 | 0.5 | 0.25 | ≤0.125 | ≤0.03 | 0.5 | 0.25 | 0.5 | 0.06 | 4 | 1 | 0.06 | t843 | 130 | 130 | − | − | − | − |
| 8223 | 79 | M | Blood | 2011 | Ceske Budejovice | 13 | 28 | 23 | 8 | 0.25 | 0.125 | ≤0.125 | ≤0.03 | 0.5 | 0.5 | 1 | 0.06 | 4 | 2 | 0.125 | t843 | 130 | 130 | − | − | − | − |
| 18063 | 73 | M | Chirurgical wound | 2013 | Benesov | 17 | 26 | 21 | 2 | >8 | 0.125 | ≤0.125 | ≤0.03 | 0.5 | 0.5 | 2 | 0.125 | 3 | 2 | 0.06 | t978 | 2361 | 2361 | # | + | − | − |
| 25255 | 65 | M | Swab | 2014 | Pribram | 18 | 25 | 22 | 1 | 0.5 | 0.125 | ≤0.125 | ≤0.03 | 0.5 | ≤0.125 | 1 | 0.125 | 8 | 2 | 0.06 | t843 | 1245 | 130 | − | − | − | − |
| 25994 | 6 | M | Swab | 2014 | Kyjov | 11 | 27 | 22 | 16 | 0.5 | 0.125 | ≤0.125 | ≤0.03 | 0.5 | 0.25 | 1 | 0.125 | 8 | 2 | 0.06 | t6104 | 425 | 425 | − | − | − | − |
| 26510 | 68 | F | Blood | 2014 | Plzen | 16 | 27 | 21 | 8 | 0.25 | 0.06 | ≤0.125 | ≤0.03 | 0.5 | ≤0.125 | 1 | 0.125 | 8 | 2 | 0.5 | t8842 | 1245 | 130 | − | − | − | − |
| 29732 | 62 | M | Chirurgical wound | 2015 | Prerov | 14 | 23 | 20 | 4 | 0.25 | 0.25 | ≤0.125 | ≤0.03 | 0.5 | 0.25 | 1 | ≤0.03 | 8 | 2 | 0.5 | t1048 | 130 | 130 | − | − | − | − |
| 30299 | 65 | M | Chirurgical wound | 2015 | Prerov | 17 | 27 | 21 | 4 | 0.25 | 0.125 | >4 | >2 | 0.5 | 0.25 | 1 | 0.125 | 4 | 2 | 0.06 | t1048 | 130 | 130 | − | − | − | − |
| 30720 | 37 | F | Swab | 2015 | Pribram | 16 | 25 | 21 | 8 | 0.25 | 0.125 | ≤0.125 | ≤0.03 | 0.5 | 0.5 | 1 | 0.06 | 8 | 2 | 0.06 | t843 | 1245 | 130 | − | − | − | − |
| 30721 | 5 | F | Swab | 2015 | Pribram | 16 | 28 | 23 | 8 | 0.5 | 0.25 | ≤0.125 | ≤0.03 | 0.5 | 0.25 | 1 | 0.125 | 8 | 2 | 0.125 | t843 | 1245 | 130 | − | − | − | − |
| 30722 | 5 | F | Swab | 2015 | Pribram | 15 | 27 | 21 | 4 | 0.5 | 0.25 | ≤0.125 | ≤0.03 | 0.5 | 0.5 | 1 | 0.125 | 8 | 2 | 0.125 | t843 | 1245 | 130 | − | − | − | − |
| 30730 | 84 | M | Chirurgical wound | 2015 | Uherske Hradiste | 16 | 27 | 22 | 4 | >8 | >4 | ≤0.125 | ≤0.03 | 0.5 | 0.5 | 2 | 0.06 | 8 | 2 | 0.125 | t1048 | 130 | 130 | − | − | − | − |
| 30792 | 57 | M | Swab | 2015 | Pribram | 15 | 26 | 22 | 8 | 0.5 | 0.25 | ≤0.125 | ≤0.03 | 0.5 | 0.5 | 1 | 0.06 | 8 | 2 | 0.125 | t1535 | 1245 | 130 | − | − | − | − |
| 35220 | 71 | F | Swab | 2016 | Prerov | 13 | 28 | 23 | 8 | 0.5 | 0.125 | ≤0.125 | ≤0.03 | 1 | 0.5 | 1 | 0.06 | 4 | 2 | 0.125 | t17153 | 130 | 130 | − | + | − | − |
| 37553 | 53 | M | Chirurgical wound | 2017 | Brno | 11 | 26 | 21 | 8 | 1 | 0.25 | 0.5 | ≤0.03 | 0.5 | ≤0.125 | 1 | 0.125 | 8 | 2 | 0.125 | t1048 | 130 | 130 | − | − | − | − |
CIP, ciprofloxacin; CLI, clindamycin; CMP, chloramphenicol; CXT, cefoxitin; ERY, erythromycin; FUS, fusidic acid; GEN, gentamicin; LNZ, linezolid; OXA, oxacillin; RIF, rifampicin; SXT, trimethoprim-sulfamethoxazole; TET, tetracycline; TGC, tigecycline; TOB, tobramycin; VAN, vancomycin.
#, genes for enterotoxins (sec, seg, sei, sel, and sem) detected in MRSA/mecC isolates; +, positive; −, negative; CC, clonal complex; En, enterotoxins; Exf, exfoliative toxins; F, female; lukS/F-PVL, Panton–Valentine leukocidin; M, man; spa, Staphylococcus protein A; ST, sequence type; tst, toxic shock syndrome toxin-1.
Antimicrobial susceptibility testing revealed that the majority of isolates was susceptible to most of tested antibiotics. Altogether, two isolates of MRSA/mecC (18063 and 25255) were resistant to CXT, but sensitive to OXA. One isolate showed resistance to ERY. The isolate 30730 was resistant to both ERY and CLI. The strain 30299 was resistant to SXT and RIF (Table 1).
Molecular typing
Among 18 isolates, eight different spa types were found in the collection. Spa type t843 (n = 8), followed by t1048 (n = 4) were the most frequent spa types detected in the study. The remaining spa types of MRSA/mecC isolates were as follows: t978, t1535, t1736, t6104, t8842, and t17153 (Table 1). The core of the group/the closest genetic relationship (one difference in repeat sequence) was observed between four spa types: t843, t1048, t1535, and t8842. The rest of isolates (t978, t1736, t6104, and t17153) differed from the core of the group by more than just one repeat sequence in the spa gene. The cluster analysis (MST) based on similarity of spa profiles is shown in Fig. 1.
FIG. 1.
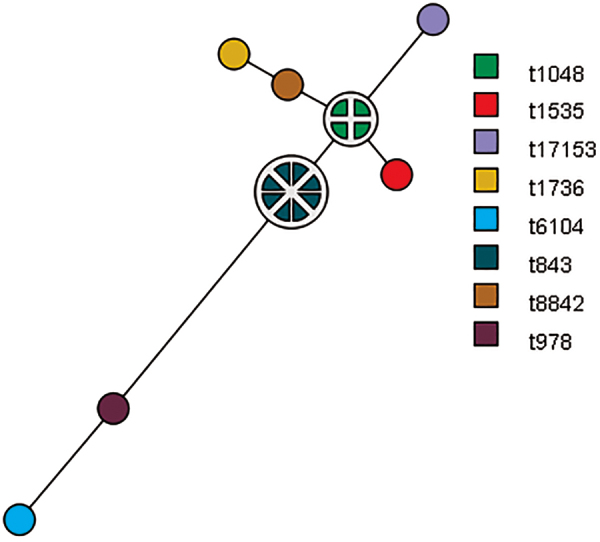
MST analysis of MRSA/mecC isolates (n = 18) illustrating the relationship within cluster. The nodes/one-colored circles consist of isolates with the same genotype, the size of node/circle correlates with the number of genetically identical isolates, and the distance between nodes represents the genetic relationship between nodes. MST, minimum spanning tree; MRSA, methicillin-/oxacillin-resistant Staphylococcus aureus. Color images are available online.
Altogether, there were five sequence types (STs) detected in MRSA/mecC isolates: ST130 (n = 8), ST425 (n = 1), ST1245 (n = 7), and ST2361 (n = 1). A new ST5480 (a new allele tpi 611) was found in the first MRSA-/mecC-positive isolate (number 2132) detected in the Czech Republic. MLST analysis revealed the prevalence of clonal complex (CC) 130 (89%, n = 16), including STs 1245, 130, and 5480. Other STs detected in the study, ST2361 and ST425, belonged to different CCs: CC2361 (ST2361) and CC425 (ST425) (Table 1).
Toxins
Strains were negative for virulence toxin genes encoding exfoliative toxins, and lukS/F-PVL (Table 1), with the exception of genes for enterotoxins sec, seg, sei, sel, sem (found in 18063), and tst that were detected in two isolates (18063 and 35220) (Table 1).
Discussion
Identification of MRSA isolates is key for controlling dissemination of this pathogen in human population and it is important for suitable management of colonized and infected patients. Except for mecA, there is also a new homologue of this gene, mecC, which has been already sporadically reported in MRSA strains all over Europe.15,16 Except for S. aureus, mecC has been already detected in other Staphylococcus species, namely Staphylococcus stepanovicii,17 Staphylococcus xylosus,18 and Staphylococcus sciuri.19
The most reliable tools used in the MRSA/mecC detection are PCR with specific primers for mecC gene and disc diffusion test with CXT. Two (18063 and 25255) from 18 strains were susceptible to OXA and resistant to CXT, along with a positive detection of mecC gene by PCR. The majority of mecC-positive MRSA was susceptible to the most of examined antibiotics, which was in line with results of other authors.7,20,21
Molecular typing is an important tool for control and prevention of infections. One of such methods used especially for analysis of genetic variability of S. aureus is spa typing. Spa analysis revealed the presence of eight spa types associated with mecC strains in the trial. The most prevalent spa type t843 (44%),22 altogether with t978, t1535, t8842,23 t1736,24 and t1048,7,25 has been already confirmed in mecC-positive strains in epidemiological studies all over Europe. Except for a susceptible S. aureus from France (Ridom SpaServer, Ridom GmbH, Würzburg, Germany), spa type t6104 was detected in this study too. MLST is also an epidemiology tool useful for understanding the molecular evolution of bacteria. Isolates of MRSA were assigned to three CCs: 130, 425, and 2361. CC130 has been previously reported in MRSA isolates of animal and human origin and it is a CC commonly associated with the occurrence of MRSA isolates carrying gene mecC in Europe.6,25,26 CC2361 and 425 were previously observed in MRSA/mecC isolates in Denmark and Spain.27,28
A virulence potential of mecC-positive MRSA strains has been also studied. A whole genome sequencing analysis has been pointed at an absence of genes coding exfoliative toxins as well as staphylococcal tst, and enterotoxins (sea–see and seg–sej).7,16 The absence of lukS/F-PVL in MRSA/mecC isolates was confirmed.25 On the other hand, it seems that ST profile could be associated with toxin production; an absence of toxin genes in the most samples of CC130 has been already described before.7,16 The only two toxins-positive isolates (enterotoxins sec, seg, sei, sel, sem, and tst) belonged to the CC130/t17153 and CC2361/t978, indicating that this toxin producing CCs/spa types might represent a risk to humans.
In line with our results, the spreading of MRSA/mecC isolates becomes more frequent in people older than 50 years.7,16,27 This part of human population is considered high risk due to the weakening of immune system and frequent consumption of antibiotics.27 Also, a hospitalization or contact with animals (zoonotic transmission) as well as travel activities belong to the risk factors for MRSA spreading.16
The limitation of this study lies in the kind of analyzed clinical specimen. The most of MRSA isolates was acquired from invasive material, preferentially sent to the National Reference Laboratory for Antibiotics. However, the majority of mecC-positive MRSA isolates, as showed this study, has been found in noninvasive clinical specimen. Most of isolates were acquired from swab (nose/throat).
The aim of this study was to detect and describe MRSA/mecC isolates occurring in the Czech Republic within a 15-year long period. And to our knowledge, this is the first report concerning the detection of these strains in the Czech Republic. The first detection of MRSA/mecC isolate was dated to 2004 (59 years, man, Prague), a new ST5480, a spa type t1763. The most of MRSA-positive mecC strains belonged to the susceptible lineage CC130/t843. A rare occurrence of MRSA/mecC isolates confirmed in this study (0.52%) was comparable with findings in Europe (2%, in Denmark).25
Due to a clinical relevance of MRSA strains and their spreading possibilities, there should be attention on a right detection as well as monitoring of abovementioned strains associated, especially with hospital-acquired infections.
Supplementary Material
Acknowledgments
We thank Iveta Vrbová and Markéta Čechová for their technical assistance.
Disclosure Statement
No competing financial interests exist.
Funding Information
This project was supported by Ministry of Health, Czech Republic—conceptual development of research organization (The National Institute of Public Health – NIPH, 75010330).
Supplementary Material
References
- 1. Donkor E.S. 2019. Nosocomial pathogens: an in-depth analysis of the vectorial potential of cockroaches. Trop Med Infect Dis. 4: E14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jevons M.P. 1961. Celbenin-resistant staphylococci. BMJ. 1:124–125 [Google Scholar]
- 3. Foster T.J. 2017. Antibiotic resistance in Staphylococcus aureus. Current status and future prospects. FEMS Microbiol Rev. 41:430–449 [DOI] [PubMed] [Google Scholar]
- 4. Baig S., Johannesen T.B, Overballe-Petersen S., Larsen J., Larsen A.R, and Stegger M.. 2018. Novel SCCmec type XIII (9A) identified in an ST152 methicillin-resistant Staphylococcus aureus. Infect Genet Evol. 61:74–76 [DOI] [PubMed] [Google Scholar]
- 5. García-Álvarez L., Holden M.T.G, Lindsay H., Webb C.R, Brown D.F, Curran M.D, Walpole E., Brooks K., Pickard D.J, Teale C., Parkhill J., Bentley S.D, Edwards G.F, Girvan E.K, Kearns A.M, Pichon B., Hill R.L, Larsen A.R, Skov R.L, Peacock S.J, Maskell D.J, and Holmes M.A.. 2011. Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: a descriptive study. Lancet Infect Dis. 11:595–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stegger M., Andersen P.S, Kearns A., Pichon B., Holmes M.A, Edwards G., Laurent F., Teale C., Skov R., and Larsen A.R.. 2012. Rapid detection, differentiation and typing of methicillin-resistant Staphylococcus aureus harbouring either mecA or the new mecA homologue mecALGA251. Clin Microbiol Infect. 18:395–400 [DOI] [PubMed] [Google Scholar]
- 7. Kerschner H., Harrison E.M, Hartl R., Holmes M.A, and Apfalter P.. 2014. First report of mecC MRSA in human samples from Austria: molecular characteristics and clinical data. New Microbes New Infect. 3:4–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. The European Committee on Antimicrobial Susceptibility Testing (EUCAST). 2017. EUCAST guidelines for detection of resistance mechanisms and specific resistances of clinical and/or epidemiological importance, Version 2.0. Available at www.eucast.org/resistance_mechanisms
- 9. The European Committee on Antimicrobial Susceptibility Testing (EUCAST). 2017. Antimicrobial susceptibility testing EUCAST disk diffusion method, Version 6.0. Available at www.eucast.org/ast_of_bacteria/disk_diffusion_methodology
- 10. The National Committee for Clinical Laboratory Standards (NCCLS). 2001. Performance standards for antimicrobial susceptibility testing, 11th edn. NCCLS, Wayne, PA. 19087-1898, USA
- 11. The European Committee on Antimicrobial Susceptibility Testing (EUCAST). 2010–2017. Clinical breakpoints – bacteria, version 2010–2017. Available at www.eucast.org/ast_of_bacteria/previous_versions_of_documents
- 12. The European Committee on Antimicrobial Susceptibility Testing (EUCAST). 2010–2017. The epidemiological cut-off value (ECOFF), version 2010–2017. Available at https://mic.eucast.org/Eucast2/SearchController/search.jsp?action=init
- 13. Ridom spaServer. 2005. DNA Sequencing of the spa Gene. Available at https://spa.ridom.de/background.shtml
- 14. Enright M.C., Day N.P, Davies C.E, Peacock S.J, and Spratt B.G.. 2000. Multilocus sequence typing for characterization of methicillin – resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 38:1008–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Diaz R., Ramalheira E., Afreixo V., and Gago B.. 2016. Methicillin-resistant Staphylococcus aureus carrying the new mecC gene—a meta-analysis. Diagn Microbiol Infect Dis. 84:135–140 [DOI] [PubMed] [Google Scholar]
- 16. García-Garrote F., Cercenado E., Marín M., Bal M., Trincado P., Corredoira J., Ballesteros C., Pita J., Alonso P., and Vindel A.. 2014. Methicillin-resistant Staphylococcus aureus carrying the mecC gene: emergence in Spain and report of a fatal case of bacteraemia. J Antimicrob Chemother. 69:45–50 [DOI] [PubMed] [Google Scholar]
- 17. Loncaric I. 2013. Characterization of methicillin-resistant Staphylococcus spp. carrying the mecC gene, isolated from wildlife. J. Antimicrob. Chemother.14:2222–2225 [DOI] [PubMed]
- 18. Harrison E.M. 2013. A Staphylococcus xylosus isolate with a new mecC allotype. Antimicrob. Agents Chemother. 57:1524–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Harrison E.M. 2014. A novel hybrid SCCmec–mecC region in Staphylococcus sciuri. J. Antimicrob. Chemother. 69:911–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Paterson G.K, Morgan F.J.E, Harrison E.M, Cartwright E.J.P, Török M.E, Zadoks R.N, Parkhill J., Peacock S.J and Holmes M.A.. 2014. Prevalence and characterization of human mecC methicillin-resistant Staphylococcus aureus isolates in England. J Antimicrob Chemother. 69:907–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Worthing K.A., Coombs G.W, Pang S., Abraham S., Saputra S., Trott D.J, Jordan D., Wong H.S, Abraham R.J, and Norris J.M.. 2016. Isolation of mecC MRSA in Australia. J Antimicrob Chemother. 71:2348–2349 [DOI] [PubMed] [Google Scholar]
- 22. Gómez P., Lozano C., Camacho M.C, Lima-Barbero J.F, Hernández J.M, Zarazaga M., Ú. Höfle, and Torres C.. 2016. Detection of MRSA ST3061-t843-mecC and ST398-t011-mecA in white stork nestlings exposed to human residues. J Antimicrob Chemother. 71:53–57 [DOI] [PubMed] [Google Scholar]
- 23. Kriegeskorte A., Idelevich E.A, Schlattmann A., Layer F., Strommenger B., Denis O., Paterson G.K, Holmes M.A, Werner G., and Becker K.. 2017. Comparison of different phenotypic approaches to screen and detect mecC-harboring 2 methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 56:pii: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Haenni M., Châtre P., Tasse J., Nowak N., Bes M., Madec J.Y, and Laurent F.. 2014. Geographical clustering of mecC-positive Staphylococcus aureus from bovine mastitis in France. J Antimicrob Chemother. 69:2292–2293 [DOI] [PubMed] [Google Scholar]
- 25. Paterson G.K., Harrison E.M, and Holmes M.A.. 2014. The emergence of mecC methicillin-resistant Staphylococcus aureus. Trends Microbiol. 22:42–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Benito D., Gómez P., Aspiroz C., Zarazaga M., Lozano C., and Torres C.. 2016. Molecular characterization of Staphylococcus aureus isolated from humans related to a livestock farm in Spain, with detection of MRSA-CC130 carrying mecC gene: a zoonotic case? Enferm Infecc Microbiol Clin. 34:280–285 [DOI] [PubMed] [Google Scholar]
- 27. Petersen A., Stegger M., Heltberg O., Christensen J., Zeuthen A., Knudsen L.K, Urth T., Sorum M., Schouls L., Larsen J., Skov R., and Larsen A.R.. 2013. Epidemiology of methicillin-resistant Staphylococcus aureus carrying the novel mecC gene in Denmark corroborates a zoonotic reservoir with transmission to humans. Clin Microbiol Infect. 19:16–22 [DOI] [PubMed] [Google Scholar]
- 28. Porrero M.C., Valverde A., Fernández-Llario P., Díez-Guerrier A., Mateos A., Lavín S., Cantón R., Fernández-Garayzabal J.F, and Domínguez L.. 2014. Staphylococcus aureus carrying mecC gene in animals and urban wastewater, Spain. Emerg Infect Dis. 20:899–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


