Abstract
The ability of amphipathic polymers to self-assemble with lipids and form nanodiscs has been a boon for the field of functional reconstitution of membrane proteins. In a field dominated by detergent micelles, a unique feature of polymer nanodiscs is their much-desired ability to align in the presence of an external magnetic field. Magnetic alignment facilitates the application of solid-state nuclear magnetic resonance (NMR) spectroscopy and aids in the measurement of residual dipolar couplings via well-established solution NMR spectroscopy. In this study, we comprehensively investigate the magnetic alignment properties of styrene maleimide quaternary ammonium (SMA-QA) polymer-based nanodiscs by using 31P and 14N solid-state NMR experiments under static conditions. The results reported herein demonstrate the spontaneous magnetic alignment of large-sized (≥20 nm diameter) SMA-QA nanodiscs (also called as macro—nanodiscs) with the lipid bilayer normal perpendicular to the magnetic field direction. Consequently, the orientation of macro—nanodiscs is further shown to flip the alignment axis parallel to the magnetic field direction upon the addition of a paramagnetic lanthanide salt. These results demonstrate the use of SMA-QA polymer nanodiscs for solid-state NMR applications including structural studies on membrane proteins.
Graphical Abstract
INTRODUCTION
Membrane proteins play a central and intricate part to many necessary cellular functions. However, the study of membrane proteins is formidable because of the common loss of solubility, function, and structure when removed from a native-like membrane environment essential for membrane protein stability.1–4 To overcome this major hindrance in the membrane protein field, a major active area of research has been the development of new membrane mimetic systems.1,5–15 Although several solubilization systems have been introduced for the study of membrane proteins (detergent micelles, bicelles, and nanodiscs),5,16 nanodiscs have been proven to have advantages over other solubilization/membrane mimetics systems.17–20 Nanodiscs are lipid bilayer discs surrounded by amphiphilic macromolecules comprising a protein18, peptide,21–23 or polymer24. Compared to other systems, polymer-based nanodiscs have the unique capability of being able to extract membrane proteins directly from their native environment without the membrane proteins ever leaving the lipid bilayer.25,26 Recent developments in the polymer nanodisc field expanded the applications of nanodiscs by using a wide variety of biophysical techniques to study membrane proteins.17,19,27–39
For the accommodation of different sizes of membrane proteins and membrane-associated protein-protein complexes and to achieve magnetic alignment, the size of nanodiscs should be variable. This is shown to be achievable with bicelles,40,41 peptide-containing lipid bilayers,42–46 and with different polymer-to-lipid ratios.47,48 The large nanodiscs, called as macro–nanodiscs, (>20 nm diameter) that align in an external magnetic field47,49,50 have been shown to be useful for structural studies using static solid-state nuclear magnetic resonance (NMR) experiments47,51 as well as an alignment medium to measure residual dipolar couplings via solution NMR experiments.52 In spite of such unique magnetic alignment properties of nanodiscs, a comprehensive study on understanding the factors affecting the alignment of nanodiscs is lacking. In this study, we undertook a comprehensive list of experiments to study the magnetic alignment of polymer-based nanodiscs made from a positively charged polymer styrene maleimide quaternary ammonium (SMA-QA)50 and 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) lipids. Magnetic alignment of SMA-QA + DMPC nanodiscs was studied using 31P and 14N solid-state NMR experiments. SMA-QA nanodiscs spontaneously align with the lipid bilayer normal perpendicular to the applied magnetic field direction. We also show that the nanodisc alignment direction can be changed (or flipped) by the addition of lanthanide salts.
MATERIALS AND METHODS
Poly(styrene-co-maleic anhydride) cumene terminated (SMA) with a 1.3:1 molar ratio of styrene/maleic anhydride and an average molecular weight (Mn) of 1600 g/mol, N,N-dimethylformamide (DMF), triethylamine (Et3N), 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, acetic acid (HOAc), hydrochloric acid (HCl), (2-aminoethyl)trimethylammonium chloride hydrochloride, and sodium hydroxide (NaOH) were purchased from Sigma-Aldrich. DMPC was purchased from Avanti Lipids Polar, Inc.
Synthesis and Characterization of SMA-QA Polymer.
SMA-QA polymer was synthesized as reported previously.50 Briefly, 2 g of SMA and 2.86 g of (2-aminoethyl)trimethylammonium chloride hydrochloride (15 equiv) was dissolved in 50 mL of DMF, followed by the addition of 4 mL of Et3N (30 equiv). The reaction mixture was stirred for 3 h at 80 °C. After 3 h, the reaction mixture was cooled to room temperature and precipitated with ice-cooled diethyl ether. The resulting precipitate was washed three times with ether and dried under a vacuum to give a white powder. The resulting powder was added to 2.25 g of sodium acetate, 50 mL of acetic anhydride, and 6 mL of Et3N. The reaction mixture was stirred for 12 h at 80 °C, and acetonitrile (50 mL) was added after the reaction. The white powder present at the bottom of the reaction mixture was discarded by carefully taking the supernatant with a pipette. The polymer was precipitated by the addition of diethyl ether. The resulting product was washed three times with ether and dried under a vacuum to give a brown powder. The resulting crude polymer product was purified using Sephadex LH-20 to remove the salts and lyophilized to produce 1.8 g brown powder of SMA-QA polymer.
Preparation and Characterization of Polymer Nanodiscs.
In this study, we used two different approaches to prepare the nanodisc samples. In the first approach, nanodiscs were formed by directly adding SMA-QA to DMPC vesicles (LUVs or MLVs) at a polymer/lipid ratio of 0.5:1 w/w, and the resultant sample was directly used in experiments. In the second approach, the nanodisc sample system was prepared using the same first approach; however, we included a size exclusion chromatography (SEC) purification step at the end to remove any non-nanodiscs forming free polymer from the solution, as explained below (Figure 1). Both types of nanodisc samples prepared were characterized by dynamic light scattering and transmission electron microscopy (TEM) experiments and were investigated by solid-state NMR experiments under static conditions, as explained in the Results and Discussion section below.
Figure 1.
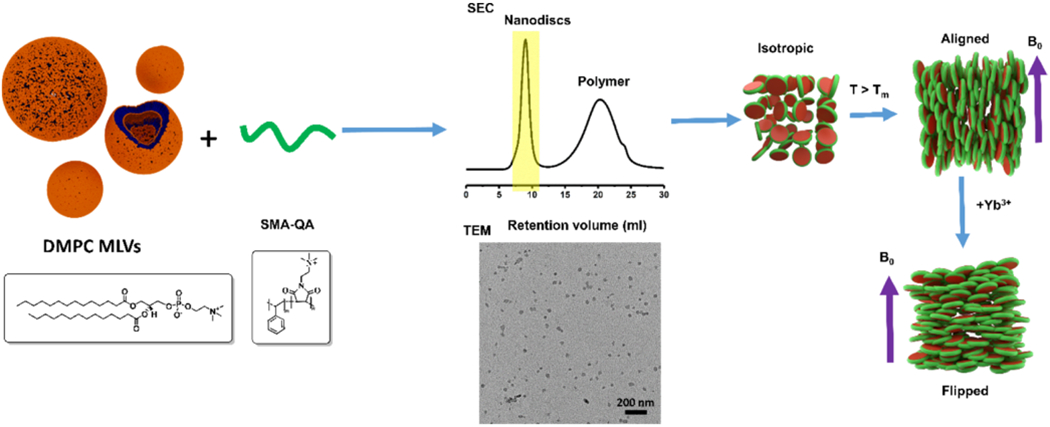
Nanodisc preparation and characterization. (Top row) Schematic representation of the preparation of nanodiscs for solid-state NMR experiments, isotropic nanodiscs, and the alignment of macro–nanodiscs in the presence of an external magnetic field. (Bottom row) DMPC and SMA-QA chemical formulae, SEC chromatogram, TEM, and an illustration of flipped macro–nanodiscs in the presence of lanthanide ions.
NMR Sample Preparation.
To prepare samples for solid-state NMR experiments, 8 mg of DMPC dissolved in CHCl3 was used for each sample preparation. The samples were dried under a stream of nitrogen gas, followed by overnight drying under a vacuum (30 °C) to completely remove any residual solvent. Tris buffer (10 mM Tris, pH 7.4) was added to hydrate the lipid film, vortexed for 2 min above the lipid’s phase transition temperature, and freeze-thawed between liquid nitrogen temperature and 35 °C for at least four times. To obtain a uniform size of large unilamellar vesicles (LUVs) of 1 μm in diameter, the LUVs were extruded through polycarbonate filters (pore size of 1 μm, Nuclepore, Whatman, NJ, USA) mounted on a mini extruder (Avanti Polar Lipids, AL, USA) fitted with two 1.0 mL Hamilton gastight syringes (Hamilton, NV, USA). The samples were typically subjected to 23 passes through the filter. An odd number of passages were used to avoid contamination of the sample by vesicles that have not passed through the first filter. The SMA-QA sample, dissolved in Tris buffer, was added to LUVs to prepare the desired polymer concentration by making a final sample volume of 150 μL. The mixture was freeze-thawed between liquid nitrogen and 35 °C for three to five times to get a transparent solution.
31P NMR Experiments.
Time-dependent NMR experiments were performed on an Agilent NMR spectrometer operating at the resonance frequency of 699.88 MHz for 1H and 283.31 MHz for 31P nuclei. A 4 mm triple-resonance HXY magic-angle spinning (MAS) NMR probe (Agilent) was used under static conditions. 31P NMR spectra were acquired using a 5.5 μs 90° pulse followed by acquisition under 24 kHz two-pulse phase modulation (TPPM) proton decoupling. A total of 128 scans was acquired for each sample with a relaxation/recycle delay time of 2.0 s. Each sample was put in a 4 mm pyrex glass tube, which was cut to fit into the 4 mm MAS probe. A Varian/Agilent temperature control unit was used to maintain the sample temperature. All 31P NMR spectra were processed using 150 Hz line broadening and referenced externally to 85% phosphoric acid (0 ppm). Temperature-dependent experiments were performed on a Bruker NMR spectrometer at a resonance frequency of 400.11 MHz for protons and 161.97 MHz for 31P nuclei. A 5 mm triple-resonance HXY MAS NMR probe was used under static conditions. A 5 mm glass tube was used for the sample. 31P NMR spectra were acquired using a 5 μs 90° pulse followed by 25 kHz TPPM proton decoupling. In total, 512 scans were acquired for each sample with a relaxation/recycle delay of 2.0 s.
14N NMR Experiments.
Nitrogen-14 NMR spectra were acquired using a Bruker 400 MHz solid-state NMR spectrometer and a 5 mm double-resonance probe operating at the 14N resonance frequency of 28.910 MHz. 14N NMR spectra were recorded using the quadrupole echo pulse sequence53 with a 90° pulse length of 8 μs and an echo delay of 80 μs. 14N magnetization was acquired using 25 ms acquisition time, 20,000 scans, and a recycle delay of 0.9 s with no 1H decoupling.
RESULTS AND DISCUSSION
In this study, we used nanodiscs composed of a positively charged SMA-QA polymer50 and DMPC lipids. SMA-QA was synthesized and characterized similar to the procedure as reported in our previous studies.50 Polymer macro–nanodiscs were prepared by the addition of DMPC liposomes to the polymer stock solution of weight ratio 1:0.5 DMPC:SMA-QA to give the final lipid concentration of 100 mg/mL. The resulting solution was subjected to freeze thaw cycles until it became transparent, and the polymer nanodisc solution was transferred to the NMR tube. To monitor the time-dependent changes in the alignment properties of the sample, a series of 31P NMR spectra were acquired as a function of time at 308 K (Figure 2). Initially, we observed a peak around ~−1 ppm and a small shoulder peak at ~−15 ppm. The observed peak at −1 ppm is from the isotropically tumbling nanodiscs, whereas the peak at −15 ppm is from the macro–nanodiscs aligned with the bilayer normal perpendicular to the magnetic field direction. The peak intensity for the aligned macro–nanodiscs increased, whereas the isotropic peak intensity decreased as a function of time, and after 3 h, the aligned peak became predominant. Considering the negative magnetic susceptibility of DMPC,54 these results suggest that macro–nanodiscs align in such a way that the lipid bilayer normal orients perpendicular to the magnetic field direction, as shown in previous studies.40,47,50
Figure 2.
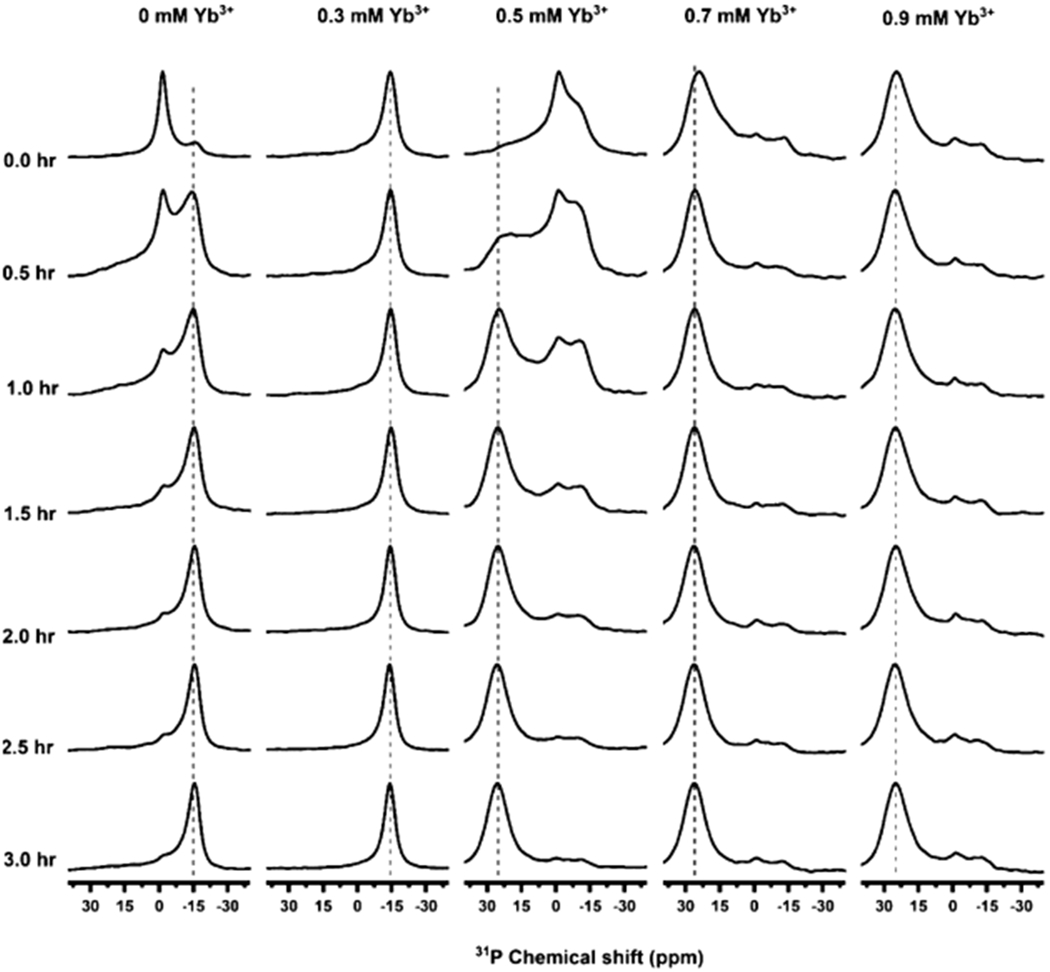
Magnetic alignment and flipping of macro–nanodiscs. 31P NMR spectra of unpurified DMPC:SMA-QA nanodiscs recorded as a function of time at 308 K and at various concentrations of YbCl3. 31P peaks appearing at ~−15, −1, and 25 ppm indicate a perpendicular orientation, isotropic phase, and a parallel orientation, respectively. Here, the perpendicular and parallel orientations mean the orientation of the lipid bilayer normal relative to the direction of an external magnetic field. All of these NMR spectra were acquired using an Agilent 700 MHz NMR spectrometer.
The magnetic susceptibility of these nanodiscs can be changed by the addition of paramagnetic metal ions such as YbCl3 which has a positive magnetic susceptibility.41,55 To study the alignment behavior of macro–nanodiscs, 31P NMR spectra were acquired as a function of time by adding different concentrations of YbCl3 to the aligned macro–nanodiscs. Upon the addition of sufficient Yb3+ ions, we observed a change in the orientation of macro–nanodiscs, resulting in the bilayer normal oriented parallel to the magnetic field direction. At a low concentration of YbCl3 (0.3 mM), no change in the 31P NMR spectrum was observed. However, as we increased the concentration of the YbCl3 salt to 0.5 mM, a powder pattern along with an isotropic peak in the 31P NMR spectrum was observed initially. After 0.5 h, a new peak appeared at 25 ppm whose intensity increased with further time, and it became the predominant peak after 3 h. This observation suggests that the macro–nanodisc alignment is flipped to orient the bilayer normal parallel to the magnetic field direction. A further increase in the concentration of YbCl3 did not show any difference in the 31P peak intensity, indicating no major changes in the alignment of macro–nanodiscs.
Although the simple addition of polymer to lipids result in the formation of nanodiscs, there is always a portion of the added polymer that stays in solution. The free polymer in solution can be removed from the sample by using SEC to yield purified nanodiscs. We observed that the purified nanodiscs aligned much faster (<1 h) as compared to the unpurified nanodiscs (Figure 3). These nanodiscs exhibited a sharp peak at −1 ppm, suggesting the isotropic nature of the macro–nanodiscs. As the temperature is increased above the gel-to-liquid crystalline phase transition temperature of the lipids (T > Tm), an additional peak at −13 ppm in the 31P spectrum appeared, suggesting the spontaneous magnetic alignment of nanodiscs (Figure 3). As shown in Figure 3, at high temperatures (>310 K), the isotropic peak at −1 ppm appeared with far less intensity, suggesting a significantly reduced isotropic population of macro–nanodiscs. In fact, for a range of temperatures (between 295 and 310 K), the 31P NMR spectra exhibited no isotropic behavior indicating an alignment of all macro–nanodiscs in the sample. The 31P NMR spectra were also recorded upon the addition of different concentrations of YbCl3 as a function of temperature (Figure 3). For 0.5 and 1 mM concentrations of YbCl3, the 31P NMR spectra showed a combination of powder pattern, isotropic peak, alignment with the bilayer normal perpendicular to the magnetic field direction, and flipping of nanodiscs with the bilayer normal parallel to the magnetic field direction depending on the temperature of the sample, as shown in Figure 3. Although the presence of 0.5 mM YbCl3 showed flipping of partial macro–nanodisc population, the addition of a 1 mM YbCl3 showed complete flipping of nanodiscs at a temperature of 305 K. A further increase in temperature above 305 K showed the presence of an isotropic peak along with a peak at 21 ppm for samples containing YbCl3. At 1.5 mM YbCl3, the samples exhibited no significant powder pattern and aligned with the bilayer normal parallel to the magnetic field direction at 305 K. Thus, the 31P NMR experimental results presented in Figures 2 and 3 clearly demonstrate the magnetic alignment of macro–nanodiscs, and their direction of alignment can be flipped by the addition of lanthanide ions like YbCl3. The observed chemical shifts are reported in Table 1. In addition, these results show that temperature and the concentration of lanthanide ions need to be optimized to achieve a desirable magnetic alignment of macro–nanodiscs. The sample and experimental conditions also depend on the type of lipids present in the nanodiscs. This behavior of macro–nanodiscs is similar to that of the well-studied bicelle samples.40,41
Figure 3.
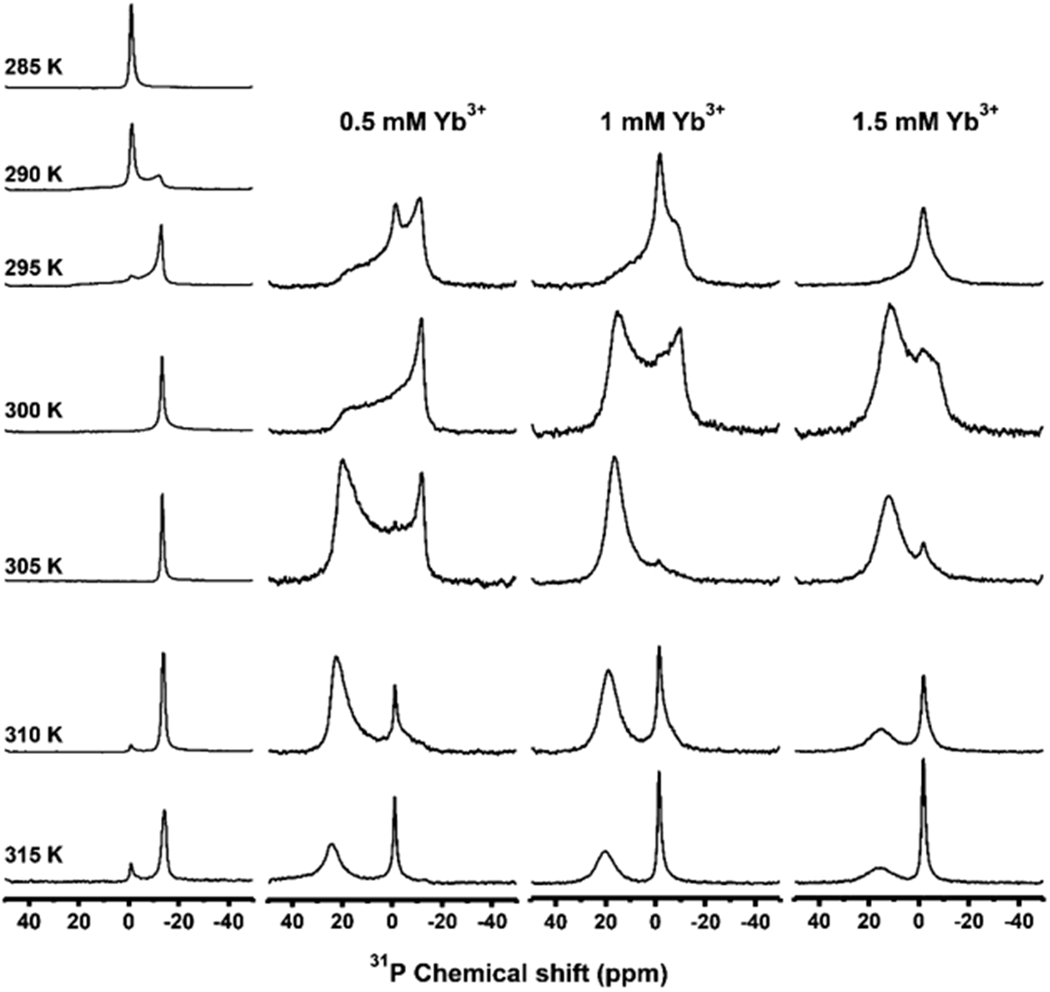
Temperature dependence of magnetic alignment and flipping of macro–nanodiscs. 31P NMR spectra of DMPC:SMA-QA (1:0.5 w/w) macro–nanodiscs in the absence (column 1) and presence of YbCl3 (columns 2–4) at different temperatures. A 100 mg/mL lipid concentration was used in the sample. All of these NMR spectra were acquired using a Bruker 400 MHz NMR spectrometer.
Table 1.
31P Chemical Shifts and Linewidths Measured from the NMR Spectra of DMPC:SMA-QA (1:0.5 w/w) Macro–Nanodiscs in the Absence (Column 1) and Presence of YbCl3 (Columns 2–4) at Different Temperatures, as Shown in Figure 3a,b.
| temperature (K) | 0 mM YbCl3 | 0.5 mM YbCl3 | 1 mM YbCl3 | 1.5 mM YbCl3 |
|---|---|---|---|---|
| 305 | −13.4 ppm (181 Hz) | 19.9 ppm (2741 Hz) | 16.5 ppm (1265 Hz) | 12.0 ppm (1723 Hz) |
| 310 | −13.8 ppm (272 Hz) | 22.6 ppm (1265 Hz) | 18.8 ppm (1355 Hz) | 14.9 ppm (1841 Hz) |
| 315 | −14.3 ppm (365 Hz) | 23.8 ppm (1292 Hz) | 20.1 ppm (1359 Hz) | 15.5 ppm (1933 Hz) |
Unlike the samples used to obtain the spectra given in Figure 2, free polymers were removed using SEC for these macro–nanodisc samples.
Line widths are shown in parentheses.
In addition to 31P NMR, 14N NMR spectroscopy is a powerful technique in sensing the membrane surface charge potential as well as useful to study the interaction of membrane-active biomolecules because of the choline moiety.56–61 14N is a quadrupolar nucleus with the nuclear spin quantum number 1, and the unique axis of the 14N quadrupole coupling tensor is parallel to the C–N bond vector of DMPC’s choline group. Therefore, the quadrupolar coupling constant depends on the orientation of the C–N bond vector of the choline group of DMPC with respect to the applied magnetic field direction, hydration, temperature, ions and other ligands, and mobility.56–58 14N NMR experiments have previously been used to study the magnetic alignment of bicelles.57,59,60 The 14N NMR spectra of macro–nanodiscs at 295 K showed the presence of a narrow peak centered at 0 kHz arising from the isotropic phase. As the temperature of the sample is increased to 310 K, the 14N NMR spectra showed the 0 kHz isotropic peak as well as two additional peaks corresponding to a quadrupole splitting of 9.4 kHz. The quadrupolar splitting of 9.4 ± 0.3 kHz arises from the magnetic alignment of macro–nanodiscs, with the bilayer normal oriented perpendicular to the magnetic field axis. The small peak at 0 kHz may be assigned to the quaternary ammonium group from the SMA-QA polymer. With the addition of YbCl3, the choline group’s 14N quadrupolar splitting has doubled from 9.4 ± 0.3 to 18.5 ± 0.5 kHz (Figure 4), indicating the flip of the nanodisc alignment from perpendicular to parallel orientation of the bilayer normal with respect to the applied magnetic field axis. The 14N NMR spectra acquired at different temperatures revealed that macro–nanodiscs at low temperatures showed the presence of an isotropic phase, whereas the increase in temperature (305 K and above) showed a quadrupolar splitting of ~18 kHz (Figure 4 and Figure 5). A further increase in the sample temperature showed an increase in the intensity of the center peak at 0 kHz, suggesting the coexistence of an isotropic phase and flipped macro–nanodiscs. These results are in excellent agreement with the above-presented 31P NMR results (Figures 2 and 3).
Figure 4.
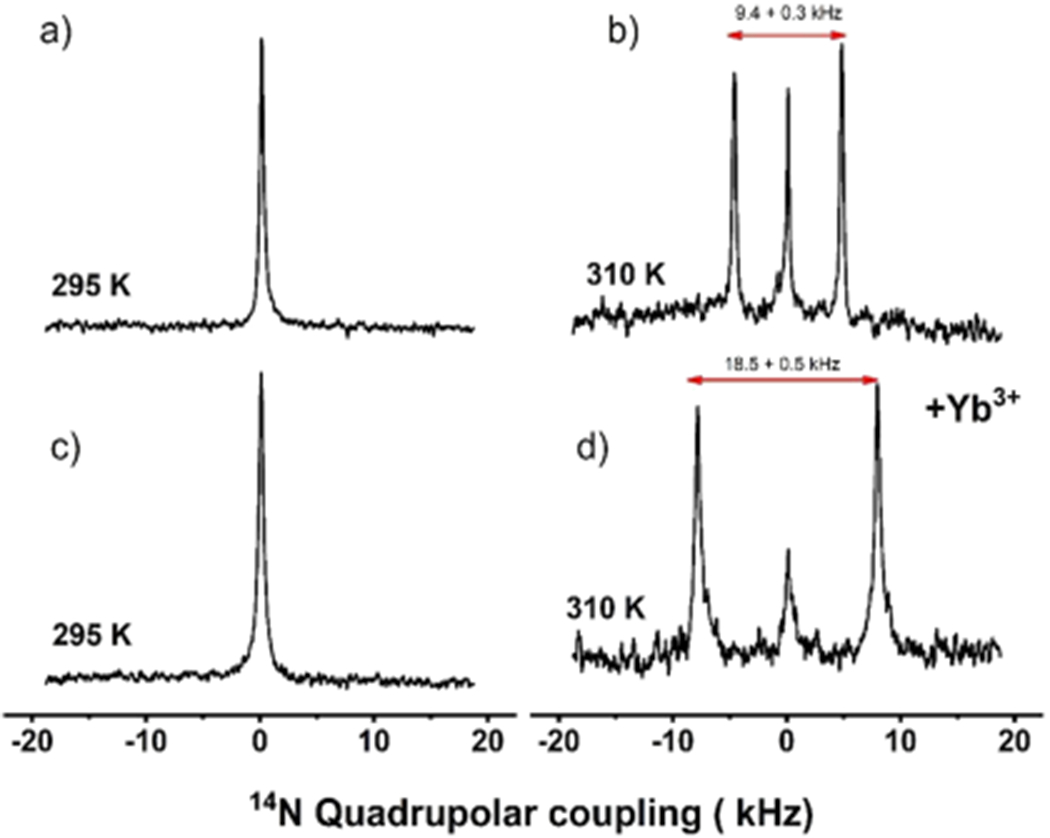
Magnetic alignment and flipping of macro–nanodiscs by 14N NMR. Nitrogen-14 NMR spectra of DMPC:SMA-QA with (c,d) and without (a,b) YbCl3 acquired at 295 K (a,c) and 310 K (b,d). The isotropic 14N NMR spectra (a,c) indicate that the macro–nanodiscs are isotropic in the gel phase, that is, below the gel-to-liquid crystalline phase transition temperature of DMPC lipids. The magnetic alignment (b) and its flipping because of the presence of 1 mM YbCl3 (d) are revealed by the observed 14N quadrupole splitting of 9.4 (b) and 18.8 kHz (d) for the bilayer normal oriented perpendicular (b) and parallel (d) to the applied magnetic field direction, respectively. All of these NMR spectra were acquired using a Bruker 400 MHz NMR spectrometer.
Figure 5.
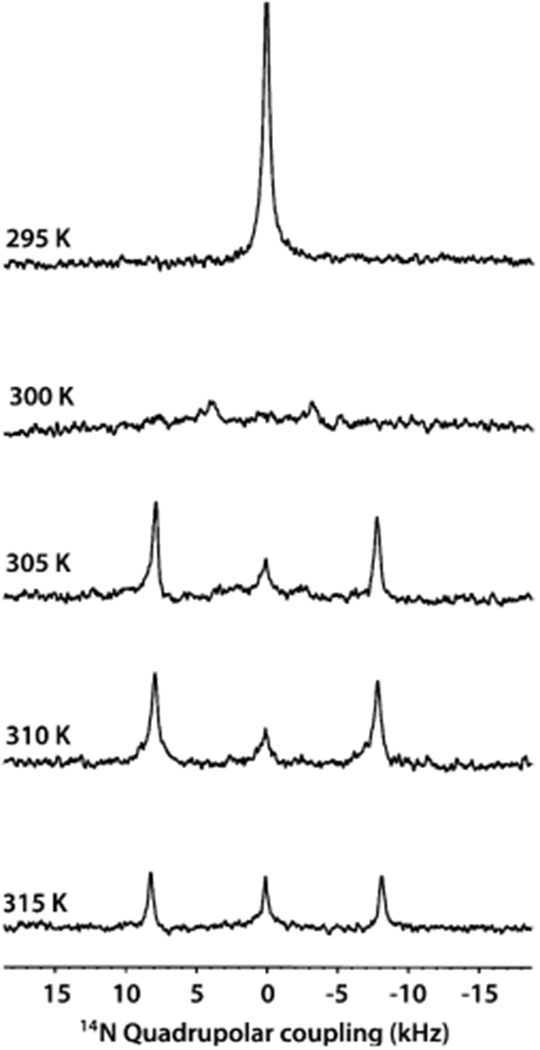
Temperature dependence of the magnetic alignment of macro–nanodiscs by 14N NMR. Nitrogen-14 NMR spectra of flipped DMPC:SMA-QA macro–nanodiscs containing 1 mM YbCl3 as a function of temperature. Macro–nanodiscs are isotropic (and unaligned, indicated by 0 kHz 14N quadrupole coupling) below the gel-to-liquid crystalline phase transition temperature (295 K) of DMPC lipids. Above the phase transition temperature (>300 K), the macro–nanodiscs align with the lipid bilayer normal parallel to the applied magnetic field direction in the presence of 1 mM YbCl3, as revealed by the observed quadrupole splitting of 18.8 kHz. All of these NMR spectra were acquired using a Bruker 400 MHz NMR spectrometer.
CONCLUSIONS
Polymer nanodiscs are a valuable tool used to study membrane proteins. With the ability to align in the presence of a magnetic field such that the bilayer normal is oriented perpendicular to the magnetic field direction, macro–nanodiscs show a promising use in solid-state NMR studies. The 14N NMR spectra reported in this study reveal the spontaneous alignment and the time and temperature required to align in the presence of an external magnetic field. Our results successfully demonstrate the ability of YbCl3 ions to flip the direction of macro–nanodiscs. We believe that this study will be useful in optimizing the conditions that are needed to study the membrane proteins reconstituted in polymer nanodiscs by solid-state NMR spectroscopy.62–66 It is also worth mentioning that pH tolerance and divalent metal ion resistance of SMA-QA polymer nanodiscs further expand the solid-state NMR applications including structural studies on membrane proteins. In addition, as shown in our recent study, magnetically aligned polymer macro–nanodiscs can be used to measure residual dipolar couplings by well-established solution NMR methods to study the structure and dynamics of water-soluble molecules including proteins, peptides, DNA, RNA, and small-molecule compounds.67–69 We expect these unique properties of polymer-based macro–nanodiscs to open avenues to expand the applications of both solution and solid-state NMR techniques and create opportunities for further development of novel NMR approaches.2,70–73
ACKNOWLEDGMENTS
This study was supported by NIH (GM084018 and AG048934 to A.R.). We thank Nathaniel Hardin for his help in preparing the manuscript.
Footnotes
The authors declare no competing financial interest.
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.langmuir.9b03538
Contributor Information
Thirupathi Ravula, Biophysics Program and Department of Chemistry, Macromolecular Science and Engineering, Biomedical Engineering, University of Michigan Ann Arbor, Michigan 48109-1055, United States.
JaeWoong Kim, Department of Fine Chemistry, Seoul National University of Science and Technology, Seoul 01811, Republic of Korea.
Dong-Kuk Lee, Department of Fine Chemistry, Seoul National University of Science and Technology, Seoul 01811, Republic of Korea.
Ayyalusamy Ramamoorthy, Biophysics Program and Department of Chemistry, Macromolecular Science and Engineering, Biomedical Engineering, University of Michigan, Ann Arbor, Michigan 48109-1055, United States.
REFERENCES
- (1).Garavito RM; Ferguson-Miller S Detergents as tools in membrane biochemistry. J. Biol. Chem 2001, 276, 32403–32406. [DOI] [PubMed] [Google Scholar]
- (2).Xiao P; Bolton D; Munro RA; Brown LS; Ladizhansky V Solid-state NMR spectroscopy based atomistic view of a membrane protein unfolding pathway. Nat. Commun 2019, 10, 3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Wylie BJ; Bhate MP; McDermott AE Transmembrane allosteric coupling of the gates in a potassium channel. Proc. Natl. Acad. Sci. U.S.A 2014, 111, 185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Chipot C; Dehez F; Schnell JR; Zitzmann N; Pebay-Peyroula E; Catoire LJ; Miroux B; Kunji ERS; Veglia G; Cross TA; Schanda P Perturbations of Native Membrane Protein Structure in Alkyl Phosphocholine Detergents: A Critical Assessment ofNMRand Biophysical Studies. Chem. Rev 2018, 118, 3559–3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Frey L; Lakomek N-A; Riek R; Bibow S Micelles, Bicelles, and Nanodiscs: Comparing the Impact of Membrane Mimetics on Membrane Protein Backbone Dynamics. Angew. Chem., Int. Ed. Engl 2017, 56, 380–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Lee SC; Pollock NL Membrane proteins: is the future disc shaped? Biochem. Soc. Trans 2016, 44, 1011–1018. [DOI] [PubMed] [Google Scholar]
- (7).Helenius A; Simons K Solubilization of membranes by detergents. Biochim. Biophys. Acta 1975, 415, 29–79. [DOI] [PubMed] [Google Scholar]
- (8).Seddon AM; Curnow P; Booth PJ Membrane proteins, lipids and detergents: not just a soap opera. Biochim. Biophys. Acta 2004, 1666, 105–117. [DOI] [PubMed] [Google Scholar]
- (9).Tanford C; Reynolds JA Characterization of membrane proteins in detergent solutions. Biochim. Biophys. Acta 1976, 457, 133–170. [DOI] [PubMed] [Google Scholar]
- (10).Czerski L; Sanders CR Functionality of a membrane protein in bicelles. Anal. Biochem 2000, 284, 327–333. [DOI] [PubMed] [Google Scholar]
- (11).Parmar MJ; Lousa CDM; Muench SP; Goldman A; Postis VLG Artificial membranes for membrane protein purification, functionality and structure studies. Biochem. Soc. Trans 2016, 44, 877–882. [DOI] [PubMed] [Google Scholar]
- (12).Kalipatnapu S; Chattopadhyay A Membrane protein solubilization: recent advances and challenges in solubilization of serotonin1A receptors. IUBMB Life 2005, 57, 505–512. [DOI] [PubMed] [Google Scholar]
- (13).Rigaud J-L; Lévy D Reconstitution of Membrane Proteins into Liposomes Methods in Enzymology; Academic Press, 2003; Vol. 372, pp 65–86. [DOI] [PubMed] [Google Scholar]
- (14).Tribet C; Audebert R; Popot J-L Amphipols: Polymers that keep membrane proteins soluble in aqueous solutions. Proc. Natl. Acad. Sci. U.S.A 1996, 93, 15047–15050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Sanders CR; Prosser RS Bicelles: a model membrane system for all seasons? Structure 1998, 6, 1227–1234. [DOI] [PubMed] [Google Scholar]
- (16).Morrison EA; DeKoster GT; Dutta S; Vafabakhsh R; Clarkson MW; Bahl A; Kern D; Ha T; Henzler-Wildman KA Antiparallel EmrE exports drugs by exchanging between asymmetric structures. Nature 2011, 481, 45–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Denisov IG; Sligar SG Nanodiscs in Membrane Biochemistry and Biophysics. Chem. Rev 2017, 117, 4669–4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Denisov IG; Grinkova YV; Lazarides AA; Sligar SG Directed self-assembly of monodisperse phospholipid bilayer Nanodiscs with controlled size. J. Am. Chem. Soc 2004, 126, 3477–3487. [DOI] [PubMed] [Google Scholar]
- (19).Denisov IG; Sligar SG Nanodiscs for structural and functional studies of membrane proteins. Nat. Struct. Mol. Biol 2016, 23, 481–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Nath A; Atkins WM; Sligar SG Applications of Phospholipid Bilayer Nanodiscs in the Study of Membranes and Membrane Proteins†. Biochemistry 2007, 46, 2059–2069. [DOI] [PubMed] [Google Scholar]
- (21).Kariyazono H; Nadai R; Miyajima R; Takechi-Haraya Y; Baba T; Shigenaga A; Okuhira K; Otaka A; Saito H Formation of stable nanodiscs by bihelical apolipoprotein A-I mimetic peptide. J. Pept. Sci 2016, 22, 116–122. [DOI] [PubMed] [Google Scholar]
- (22).Mishra VK; Palgunachari MN; Krishna NR; Glushka J; Segrest JP; Anantharamaiah GM Effect of Leucine to Phenylalanine Substitution on the Nonpolar Face of a Class A Amphipathic Helical Peptide on Its Interaction with Lipid. J. Biol. Chem 2008, 283, 34393–34402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Zhang M; Huang R; Ackermann R; Im S-C; Waskell L; Schwendeman A; Ramamoorthy A Reconstitution of the Cytb5-CytP450 Complex in Nanodiscs for Structural Studies Using NMR Spectroscopy. Angew. Chem., Int. Ed 2016, 55, 4497–4499. [DOI] [PubMed] [Google Scholar]
- (24).Knowles TJ; Finka R; Smith C; Lin Y-P; Dafforn T; Overduin M Membrane proteins solubilized intact in lipid containing nanoparticles bounded by styrene maleic acid copolymer. J. Am. Chem. Soc 2009, 131, 7484–7485. [DOI] [PubMed] [Google Scholar]
- (25).Lee SC; Knowles TJ; Postis VLG; Jamshad M; Parslow RA; Lin Y.-p.; Goldman A; Sridhar P; Overduin M; Muench SP; Dafforn TR A method for detergent-free isolation of membrane proteins in their local lipid environment. Nat. Protoc 2016, 11, 1149–1162. [DOI] [PubMed] [Google Scholar]
- (26).Dörr JM; Koorengevel MC; Schäfer M; Prokofyev AV; Scheidelaar S; van der Cruijsen EAW; Dafforn TR; Baldus M; Killian JA Detergent-free isolation, characterization, and functional reconstitution of a tetrameric K+channel: The power of native nanodiscs. Proc. Natl. Acad. Sci. U.S.A 2014, 111, 18607–18612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Hagn F; Etzkorn M; Raschle T; Wagner G Optimized phospholipid bilayer nanodiscs facilitate high-resolution structure determination of membrane proteins. J. Am. Chem. Soc 2013, 135, 1919–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Nasr ML; Wagner G Covalently circularized nanodiscs; challenges and applications. Curr. Opin. Struct. Biol 2018, 51, 129–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Denisov IG; Sligar SG Nanodiscs for structural and functional studies of membrane proteins. Nat. Struct. Mol. Biol 2016, 23, 481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Redhair M; Clouser AF; Atkins WM Hydrogen-deuterium exchange mass spectrometry of membrane proteins in lipid nanodiscs. Chem. Phys. Lipids 2019, 220, 14–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Overduin M; Esmaili M Structures and Interactions of Transmembrane Targets in Native Nanodiscs. SLAS Discov. 2019, 24, 943. [DOI] [PubMed] [Google Scholar]
- (32).Sun C; Benlekbir S; Venkatakrishnan P; Wang Y; Hong S; Hosler J; Tajkhorshid E; Rubinstein JL; Gennis RB Structure of the alternative complex III in a supercomplex with cytochrome oxidase. Nature 2018, 557, 123–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Orwick MC; Judge PJ; Procek J; Lindholm L; Graziadei A; Engel A; Gröbner G; Watts A Detergent-free formation and physicochemical characterization of nanosized lipid-polymer complexes: Lipodisq. Angew. Chem., Int. Ed. Engl 2012, 51, 4653–4657. [DOI] [PubMed] [Google Scholar]
- (34).Kopf AH; Koorengevel MC; van Walree CA; Dafforn TR; Killian JA A simple and convenient method for the hydrolysis of styrene-maleic anhydride copolymers to styrene-maleic acid copolymers. Chem. Phys. Lipids 2019, 218, 85–90. [DOI] [PubMed] [Google Scholar]
- (35).Domínguez Pardo JJ; van Walree CA; Egmond MR; Koorengevel MC; Killian JA Nanodiscs bounded by styrene-maleic acid allow trans-cis isomerization of enclosed photoswitches of azobenzene labeled lipids. Chem. Phys. Lipids 2019, 220, 1–5. [DOI] [PubMed] [Google Scholar]
- (36).Danielczak B; Meister A; Keller S Influence of Mg2+ and Ca2+ on nanodisc formation by diisobutylene/maleic acid (DIBMA) copolymer. Chem. Phys. Lipids 2019, 221, 30–38. [DOI] [PubMed] [Google Scholar]
- (37).Danielczak B; Keller S Collisional lipid exchange among DIBMA-encapsulated nanodiscs (DIBMALPs). Eur. Polym. J 2018, 109, 206–213. [Google Scholar]
- (38).Oluwole AO; Klingler J; Danielczak B; Babalola JO; Vargas C; Pabst G; Keller S Formation of Lipid-Bilayer Nanodiscs by Diisobutylene/Maleic Acid (DIBMA) Copolymer. Langmuir 2017, 33, 14378–14388. [DOI] [PubMed] [Google Scholar]
- (39).Oluwole AO; Danielczak B; Meister A; Babalola JO; Vargas C; Keller S Solubilization of Membrane Proteins into Functional Lipid-Bilayer Nanodiscs Using a Diisobutylene/Maleic Acid Copolymer. Angew. Chem. Int. Ed. Engl 2017, 56, 1919–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Sanders CR; Hare BJ; Howard KP; Prestegard JH Magnetically-Oriented Phospholipid Micelles as a Tool for the Study of Membrane-Associated Molecules. Prog. Nucl Mag. Res. Sp 1994, 26, 421–444. [Google Scholar]
- (41).Prosser RS; Hwang JS; Vold RR Magnetically aligned phospholipid bilayers with positive ordering: a new model membrane system. Biophys. J 1998, 74, 2405–2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Naito A; Nagao T; Obata M; Shindo Y; Okamoto M; Yokoyama S; Tuzi S; Saitô H Dynorphin induced magnetic ordering in lipid bilayers as studied by 31P NMR spectroscopy. Biochim. Biophys. Acta—Biomembranes 2002, 1558, 34–44. [DOI] [PubMed] [Google Scholar]
- (43).Zhang M; Huang R; Ackermann R; Im S-C; Waskell L; Schwendeman A; Ramamoorthy A Reconstitution of the Cytb5-CytP450 Complex in Nanodiscs for Structural Studies using NMR Spectroscopy. Angew. Chem., Int. Ed. Engl 2016, 55, 4497–4499. [DOI] [PubMed] [Google Scholar]
- (44).Ramamoorthy A Beyond NMR spectra of antimicrobial peptides: dynamical images at atomic resolution and functional insights. Solid State Nucl. Magn. Reson 2009, 35, 201–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Ravula T; Barnaba C; Mahajan M; Anantharamaiah GM; Im S-C; Waskell L; Ramamoorthy A Membrane environment drives cytochrome P450’s spin transition and its interaction with cytochrome b5. Chem. Commun 2017, 53, 12798–12801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Park SH; Berkamp S; Cook GA; Chan MK; Viadiu H; Opella SJ Nanodiscs versus macrodiscs for NMR of membrane proteins. Biochemistry 2011, 50, 8983–8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Ravula T; Ramadugu SK; Di Mauro G; Ramamoorthy A Bioinspired, Size-Tunable Self-Assembly of Polymer-Lipid Bilayer Nanodiscs. Angew. Chem., Int. Ed. Engl 2017, 56, 11466–11470. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- (48).Zhang R; Sahu ID; Liu L; Osatuke A; Comer RG; Dabney-Smith C; Lorigan GA Characterizing the structure of lipodisq nanoparticles for membrane protein spectroscopic studies. Biochim. Biophys. Acta 2015, 1848, 329–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Radoicic J; Park SH; Opella SJ Macrodiscs Comprising SMALPs for Oriented Sample Solid-State NMR Spectroscopy of Membrane Proteins. Biophys. J 2018, 115, 22–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Ravula T; Hardin NZ; Ramadugu SK; Cox SJ; Ramamoorthy A Formation of pH-Resistant Monodispersed Polymer-Lipid Nanodiscs. Angew. Chem., Int. Ed. Engl 2018, 57, 1342–1345. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- (51).Ravula T; Hardin NZ; Ramamoorthy A Polymer nanodiscs: Advantages and limitations. Chem. Phys. Lipids 2019, 219, 45–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Ravula T; Ramamoorthy A Magnetic Alignment of Polymer Macro-Nanodiscs Enables Residual-Dipolar-Coupling-Based High-Resolution Structural Studies by NMR Spectroscopy. Angew. Chem., Int. Ed. Engl 2019, 58, 14925–14928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Weisman ID; Bennett LH Quadrupolar Echoes in Solids. Phys. Rev 1969, 181, 1341–1350. [Google Scholar]
- (54).Scholz F; Boroske E; Helfrich W Magnetic anisotropy of lecithin membranes. A new anisotropy susceptometer. Biophys. J 1984, 45, 589–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Prosser RS; Hunt SA; DiNatale JA; Vold RR Magnetically Aligned Membrane Model Systems with Positive Order Parameter: Switching the Sign ofSzzwith Paramagnetic Ions. J. Am. Chem. Soc 1996, 118, 269–270. [Google Scholar]
- (56).Lindström F; Williamson PTF; Gröbner G Molecular Insight into the Electrostatic Membrane Surface Potential by14N/31P MAS NMR Spectroscopy: Nociceptin–Lipid Association. J. Am. Chem. Soc 2005, 127, 6610–6616. [DOI] [PubMed] [Google Scholar]
- (57).Ramamoorthy A; Lee D-K; Santos JS; Henzler-Wildman KA Nitrogen-14 Solid-State NMR Spectroscopy of Aligned Phospholipid Bilayers to Probe Peptide–Lipid Interaction and Oligomerization of Membrane Associated Peptides. J. Am. Chem. Soc 2008, 130, 11023–11029. [DOI] [PubMed] [Google Scholar]
- (58).Ramamoorthy A; Thennarasu S; Lee D-K; Tan A; Maloy L Solid-state NMR investigation of the membrane-disrupting mechanism of antimicrobial peptides MSI-78 and MSI-594 derived from magainin 2 and melittin. Biophys. J 2006, 91, 206–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Smith PES; Brender JR; Ramamoorthy A Induction of negative curvature as a mechanism of cell toxicity by amyloidogenic peptides: the case of islet amyloid polypeptide. J. Am. Chem. Soc 2009, 131, 4470–4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Semchyschyn DJ; Macdonald PM Conformational response of the phosphatidylcholine headgroup to bilayer surface charge: torsion angle constraints from dipolar and quadrupolar couplings in bicelles. Magn. Reson. Chem 2004, 42, 89–104. [DOI] [PubMed] [Google Scholar]
- (61).Scherer PG; Seelig J Electric charge effects on phospholipid headgroups. Phosphatidylcholine in mixtures with cationic and anionic amphiphiles. Biochemistry 1989, 28, 7720–7728. [DOI] [PubMed] [Google Scholar]
- (62).Aisenbrey C; Salnikov ES; Bechinger B Solid-State NMR Investigations of the MHC II Transmembrane Domains: Topological Equilibria and Lipid Interactions. J. Membr. Biol 2019, 252, 371–384. [DOI] [PubMed] [Google Scholar]
- (63).Salnikov ES; De Zotti M; Bobone S; Mazzuca C; Raya J; Siano AS; Peggion C; Toniolo C; Stella L; Bechinger B Trichogin GA IV Alignment and Oligomerization in Phospholipid Bilayers. Chembiochem 2019, 20, 2141–2150. [DOI] [PubMed] [Google Scholar]
- (64).Grage SL; Sani M-A; Cheneval O; Henriques ST; Schalck C; Heinzmann R; Mylne JS; Mykhailiuk PK; Afonin S; Komarov IV; Separovic F; Craik DJ; Ulrich AS Orientation and Location of the Cyclotide Kalata B1 in Lipid Bilayers Revealed by Solid-State NMR. Biophys. J 2017, 112, 630–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Morton CJ; Sani M-A; Parker MW; Separovic F Cholesterol-Dependent Cytolysins: Membrane and Protein Structural Requirements for Pore Formation. Chem. Rev 2019, 119, 7721–7736. [DOI] [PubMed] [Google Scholar]
- (66).Laadhari M; Arnold AA; Gravel AE; Separovic F; Marcotte I Interaction of the antimicrobial peptides caerin 1.1 and aurein 1.2 with intact bacteria by 2 H solid-state NMR. Biochim. Biophys. Acta 2016, 1858, 2959–2964. [DOI] [PubMed] [Google Scholar]
- (67).Tjandra N; Bax A Direct Measurement of Distances and Angles in Biomolecules by NMR in a Dilute Liquid Crystalline Medium. Science 1997, 278, 1111. [DOI] [PubMed] [Google Scholar]
- (68).Peti W; Meiler J; Bruschweiler R; Griesinger C Model-free analysis of protein backbone motion from residual dipolar couplings. J. Am. Chem. Soc 2002, 124, 5822–5833. [DOI] [PubMed] [Google Scholar]
- (69).Liu Y; Navarro-Vázquez A; Gil RR; Griesinger C; Martin GE; Williamson RT Application of anisotropic NMR parameters to the confirmation of molecular structure. Nat. Protoc 2019, 14, 217–247. [DOI] [PubMed] [Google Scholar]
- (70).Wang S; Gopinath T; Veglia G Improving the quality of oriented membrane protein spectra using heat-compensated separated local field experiments. J. Biomol. NMR 2019, 73, 617–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Hong M; Su Y Structure and dynamics of cationic membrane peptides and proteins: insights from solid-state NMR. Protein Sci. 2011, 20, 641–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Leninger M; Sae Her A; Traaseth NJ Inducing conformational preference of the membrane protein transporter EmrE through conservative mutations. elife 2019, 8, 48909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Pinto C; Mance D; Sinnige T; Daniels M; Weingarth M; Baldus M Formation of the beta-barrel assembly machinery complex in lipid bilayers as seen by solid-state NMR. Nat. Commun. 2018, 9, 4135. [DOI] [PMC free article] [PubMed] [Google Scholar]


