Abstract
Porcine epidemic diarrhea virus (PEDV) is a fatal epizootic swine coronavirus that presents a financial threat to the global swine industry. Since the discovery of the low-pathogenic genotype 1b (G1b) in 2014, it has been responsible for sporadic outbreaks in South Korea. In this study, we identified novel G1b variants arising from the natural recombination of a major pandemic-like G2b virus and a minor G1b virus currently circulating in the domestic field. The whole-genome sequences of two 2018–19 G1b recombinants, KNU-1808 and KNU-1909, were determined. A genomic comparison showed that these two viruses share the highest nucleotide sequence similarity with the 2017 G1b strain but share less similarity with the 2014 G1b emergent strain KNU-1406. However, the putative recombination breakpoints spanning the first 1,170 nucleotides of the spike (S) gene were almost identical among the emergent and contemporary G1b strains. Recombination detection indicated that the inter-subgroup G1b recombinant first emerged in 2017 by introducing the N-terminal domain of S from KNU-1406 into the backbone of KNU-1703, possibly leading to antigenic shift. It then evolved into KNU-1808 and KNU-1909 through genetic drift, moving toward a more G2b-like genotype. Phylogenetic analysis revealed that the 2018–2019 G1b recombinants belong to a cluster containing other G1b strains but form a new branch. This study provides an important advance warning in regard to the emergence and prevalence of new genotypes or variants that can result from genetic recombination between two different PEDV genotypes circulating in endemic areas and continuous non-lethal mutations essential for viral fitness in the host environment.
Electronic supplementary material
The online version of this article (10.1007/s00705-020-04767-4) contains supplementary material, which is available to authorized users.
Introduction
Porcine epidemic diarrhea virus (PEDV) is a highly contagious and deadly swine coronavirus that belongs to the genus Alphacoronavirus in the family Coronaviridae of the order Nidovirales. PEDV causes a serious enteric disease in pigs that results in severe watery diarrhea, vomiting, and dehydration, with very high mortality rates in newborn piglets. Since the 2013–2014 fatal pandemic, the virus has gained global attention and a reputation as an economic menace to the world pork industry [2]. PEDV can be divided into two genotypes comprising two subgenotypes: low-pathogenic (LP) genotype 1 (historic G1a and recombinant G1b) and highly pathogenic (HP) genotype 2 (local epidemic G2a and global epidemic or pandemic G2b) [2, 3, 7]. Although the HP-G2b genotype has been the dominant epidemic strain in South Korea since the 2013–2014 national disaster, it is not uncommon for the LP-G1b strains of PEDV to cause small-scale local, economically insignificant outbreaks across the mainland [7–9]. This study provides, for the first time, direct evidence of emerging novel recombinant LP-PEDV G1b strains that share the backbone sequence of pandemic G2b and an N-terminal domain (NTD) of the spike (S) gene from G1b circulating in South Korea.
Materials and methods
Clinical sample collection
In December 2018 and January 2019, enteric and diarrheal diseases accompanied by low mortality rates (10–20%) in newborn piglets (< 7 days of age) occurred consecutively in two different farrow-to-finish commercial swine farms with no previous herd history of PED vaccination or PED outbreaks located in Chungcheong Province (Supplementary Fig. S1). Diarrheic piglets (n = 2) from those farms were submitted for laboratory analysis. Intestinal homogenates (n = 2) were prepared as 10% (wt/vol) suspensions in phosphate-buffered saline (PBS) using a MagNA Lyser Instrument (Roche Diagnostics, Mannheim, Germany) with three rounds of 15 s at a force of 8,000 × g. Fecal samples (n = 2) were also diluted with PBS to prepare 10% (wt/vol) suspensions. The suspensions were vortexed and centrifuged for 10 min at 4,500 × g (Hanil Centrifuge FLETA5, Incheon, South Korea). The clarified supernatants were initially subjected to RT-PCR analysis using a LiliF PED RT-PCR Kit (iNtRON Biotechnology, Seongnam, South Korea) according to the manufacturer’s instructions and to quantitative real-time RT-PCR as described previously [9, 10]. To determine if any other diarrhea-causing pathogens, besides PEDV, were present in clinical samples, we performed virus-specific RT-PCR analyses for transmissible gastroenteritis virus, porcine deltacoronavirus, and porcine rotaviruses.
Nucleotide sequence analysis
The S glycoprotein gene sequences of the virus isolates from intestinal or fecal suspensions with the highest viral load (n = 2) representing each farm were determined by traditional Sanger methods. Two overlapping cDNA fragments spanning the entire S gene of each isolate were amplified by RT-PCR as described previously [4]. The individual cDNA amplicons were gel-purified, cloned using the pGEM-T Easy Vector System (Promega, Madison, WI), and sequenced in both directions using two commercial vector-specific T7 and SP6 primers and gene-specific primers. The complete genomes of representative PEDV field strains were also sequenced. Ten overlapping cDNA fragments spanning the entire genome of each virus strain were amplified by RT-PCR as described previously [5, 7–10], and each PCR product was sequenced as described above. The 5′ and 3′ ends of the genomes of the individual isolates were determined by rapid amplification of cDNA ends (RACE) as described previously [11]. The complete genomic sequences of KOR/KNU-1808/2018/G1b and KOR/KNU-1909/2019/G1b in this study were deposited in the GenBank database under accession numbers MN816181 and MN844888, respectively. For sequencing analysis, we selected at least five colonies obtained from pGEM cloning to rule out the possibility of a mixed G1b-G2b PEDV infection in a single sample as well as to exclude any contamination during sample preparation.
Recombination analysis
Recombination events were detected using two different methods. First, whole genome sequences were aligned and analyzed using the Recombination Detection Program (RDP 4 version 4.95) to detect potential recombination events by eight algorithms (RDP, GENECONV, BootScan, MaxChi, Chimaera, SiScan, 3Seq, and LARD) [14]. Recombination breakpoint detection by at least four of these methods was considered confirmation of any putative recombination event. Second, the potential recombination events and breakpoints were further verified by similarity plot analysis using SimPlot version 3.5.1 [12].
Multiple alignments and phylogenetic analysis
The sequences of 72 fully sequenced S genes and 63 complete genomes of global PEDV isolates were used independently in sequence alignments and phylogenetic analysis. Multiple sequence alignments were generated using the ClustalX 2.0 program [16], and the percentage of nucleotide sequence divergence was assessed using the same software. Phylogenetic trees were constructed from the aligned nucleotide or amino acid sequences using the neighbor-joining method and were subjected to bootstrap analysis with 1000 replicates to determine the percentage reliability values for each internal node of the tree [15]. All phylogenetic trees were generated using MEGA X software [1].
Results
Identification of novel LP-PEDV G1b strains
Small-intestine specimens and feces collected from diarrheic piglets were initially subjected to virus-specific RT-PCR assays. All samples were positive for PEDV (Supplementary Fig. S1A) with Ct values ranging from 17.1 to 22.5, and no other viral pathogens that cause diarrhea were detected in these cases. Subsequently, the complete S gene sequences of the isolates were determined. Nucleotide (nt) sequencing analysis revealed that two strains, KNU-1808 and KNU-1909, were genetically cognate, having 99.6% amino acid (aa) sequence identity. Since these isolates showed 98.4–98.5% and 95.3–95.4% aa sequence identity to the Korean prototype G1b KNU-1406 and G2b KNU-141112 strains, respectively, they were classified as the LP-G1b subtype (Supplementary Table S1).
The LP-PEDV G1b strains appear to have originated from a recombination event between a virus G1a as the minor parent and a virus G2b as the major parent (e.g. > 99% sequence identity to G2b for the whole genome but ≤ 89% and 95% sequence identity to G2b and G1a, respectively, from nt 1 to 1,170 of S) [7, 17]. Their S genes contain typical genetic and phylogenetic features: identical length, no INDELs when compared to the historic G1a CV777 strain, and a different phylogenetic classification (G1b or G2) based on the sequence of the S gene or whole genome [2, 6]. All G1b isolates identified in this study possessed the aforementioned characteristics (Supplementary Fig. S2). When comparing the first 1,170 nt of the S1 NTD, the KNU-1808 and KNU-1909 strains have 93% and 88% sequence identity to G1a CV777 and G2b KNU-141112 strains, respectively. The genetic similarity between the full S gene sequences of the query G1a (consensus) strain and the G1b or G2b (consensus) isolates was plotted using SimPlot analysis. As expected, the sequence similarity of the first 1,170 nt of S was distinctly different between the G2b and KNU-1406 strains (Supplementary Fig. S3). Likewise, the same region showed the genetic distance between the G2b virus and the 2017 G1b isolates (KNU-1701, -1702, and -1707) identified in our previous study [8]. However, when the G2b and 2018–19 G1b (KNU-1808 and KNU-1909) strains were compared, their nucleotide sequences varied by approximately 690 nt, suggesting the emergence of new PEDV variants.
Complete genomic characterization of novel LP-PEDV G1b strains
The full genomes of the 2018–19 G1b strains were sequenced and analyzed to clarify the genetic origin of the novel G1b variants with respect to other domestic PEDV strains. Like other G1b viruses, the genomes of the KNU-1808 and KNU-1909 viruses were 28,029 nt in length, excluding the 3′ poly(A) tail, and no INDELs were identified throughout their entire genomes. The 2018–19 variants showed high overall sequence similarity to each other (99.8% identity) and with other global G1b PEDV strains (98.7–99.7% identity) (Supplementary Table S2). The number of nt or aa differences and the percent identity between the contemporary (2017–19) G1b isolates and their emergent (2014) strain KNU-1406 are summarized in Supplementary Table S3. Aligning the genome sequences of all G1b sequences revealed marked variation between the Korean emergent and contemporary G1b strains but clear similarity among the contemporary isolates (Fig. 1). However, the NTD of the S gene, which includes the first 1,170 nt residues and corresponds to nt 20,634 and nt 21,803 of the genome sequence, showed significant sequence homology among the emergent and contemporary G1b strains. These data indicate a potential for the occurrence of a novel recombination event.
Fig. 1.
Schematic diagram of the PEDV genome alignment relative to the overall G2b consensus sequence derived from at least 50% of the genomic sequences of the 60 global G2b strains, using Geneious software version 10.2.4. Genetic subgroups of PEDV are color-coded: G1a (purple), G1b (blue), and G2b (black). The genomic regions are shown above with the green bars representing the identified open reading frames (ORFs) and the black arrows indicating the nonstructural proteins (nsp1–16) that are produced when ORF1a/1b is translated and processed by viral-encoded proteases. The light grey arrows represent 5′ and 3′ untranslated regions (UTRs). Lightly shaded regions are identical to the consensus sequence, and the vertical black bars represent differences from the consensus nucleotide sequence. The thin horizontal dashed lines indicate deleted nucleotides. The Korean G1b strains are indicated by a red line box, and the novel G1b variants identified in this study (KNU-1808 and KNU-1909) are indicated by an asterisk. The nt 1–1,170 region of the Korean G1b spike gene (corresponding to genomic nt positions 20,634–21,803) is shaded blue
Whole-genome recombination analysis
To determine whether the 2017–19 G1b variants were new recombinants from field isolates, the RDP4 program was used to carry out an analysis based on a comparison between G1b strains and 20 Korean G2b isolates. Eight methods in the RDP4 software package were used to detect recombination events and breakpoints, and the results are summarized in Supplementary Table S4. Two putative recombination sites were detected between nt 20,661 and nt 22,597 in the genome of KNU-1702, corresponding to nt 28–1,964 of the S gene. All eight detection methods revealed that the major and minor parental viruses of the recombination event are the PEDV pandemic-like HP-G2b strain KNU-1703 (MH052682) and the LP-G1b strain KNU-1406, respectively (average P-value, 1.76 × 10−25). Similarly, the recombination events in KNU-1808 and KNU-1909 were independently confirmed by all eight modules with a high degree of confidence (average P-values, 7.95 × 10−20 and 3.07 × 10−22, respectively), consistently indicating that KNU-1703 and KNU-1406 represent the backbone and inserted sequence, respectively. However, the genomes of the 2018–19 G1b variants all contained putative recombination points at nt 20,661 and nt 21,318 nt, corresponding to nt 28 and nt 684 in the S gene. Similarity plots showed high overall sequence similarity between the G1b recombinant and the parental G2b KNU-1703 strains, but the sequence similarity significantly decreased in the putative recombinant positions between nt 28 and nt 1,964, or nt 28 and nt 684 of the S gene (Fig. 2). Analogous to the S gene-based SimPlot data (Supplementary Fig. S3), all methods detected the truncated sequence endpoint for the 2018–19 G1b strains, which was predicted at nt 684 in the S gene. Interestingly, the 2018–19 recombinants had four unique substitutions (S232I, D242E, S243P, and L266V) within the truncated segment spanning nt 684–1,964 of the S gene when compared to that of other G1b strains, including KNU-1702. However, the changed amino acids were identical to those found in the G2b field isolates (Supplementary Fig. S2), possibly leading to the alteration of the sequence endpoint from nt 22,597 to nt 21,318 in the genome (or nt 1,964 to 684 in the S gene). Considering that the most recently identified G1b virus (KNU-1702) and the major parental virus (KNU-1703) were isolated in the same year, the inter-subgroup recombination event appears to have occurred in 2017. This event created the novel G1b variant, which in turn underwent point mutations in the S1 region that shifted it toward the more-G2b-like strain.
Fig. 2.
Recombination analyses of 2018–19 Korean PEDV G1b strains. The x-axis indicates the genomic position, and the y-axis represents the pairwise identity between KNU-1406 and KNU-1703, KNU-1406 and KNU-1702, or KNU-1702 and KNU-1703 (top panel); between KNU-1406 and KNU-1703, KNU-1406 and KNU-1808, or KNU-1808 and KNU-1703 (middle panel); and between KNU-1406 and KNU-1703, KNU-1406 and KNU-1909, or KNU-1909 and KNU-1703 (bottom panel), illustrated with yellow, purple, and green lines, respectively. The beginning and end of each recombinant breakpoint is shaded grey and labeled with position numbers. A schematic diagram of the genomes of Korean G1b recombinants and a major parental virus KNU-1703 relative to the G2b consensus is depicted. The major parental region representing the backbone sequence of KNU-1703 is indicated by blue bars, while the minor parental region spanning the S1 NTD of KNU-1406 is indicated by red bars. The novel G1b variants identified in this study (KNU-1808 and KNU-1909) are indicated by an asterisk
Phylogenetic analysis
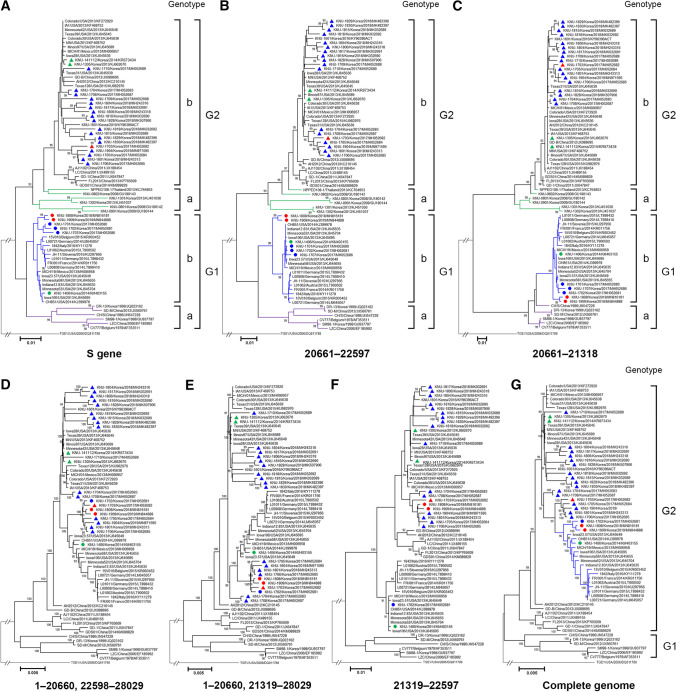
The complete S-gene-based phylogenetic analysis clearly segregated the PEDV strains in two genogroup clusters, G1 and G2, which were further defined into subgroups 1a, 1b, 2a, and 2b (Fig. 3A). The KNU-1808 and KNU-1909 strains still belonged to subgroup G1b, but they formed an independent clade with the 2017 G1b isolates within the same subgroup. However, they were positioned differently based on the phylogenies of their parental regions (Fig. 3B–E). Whereas the minor parental region (the NTD encompassing nt 28–1,964, or nt 28–684 of the S gene) of the 2017–19 G1b strains showed a close relationship to G1b (Fig. 3B and C), the contemporary G1b viruses were grouped with G2b isolates in the phylogenetic tree based on the major parental region (Fig. 3D and E). In addition, a phylogenetic analysis of nt 684–1,964 of S revealed that only the 2018–19 G1b recombinants, excluding the 2017 G1b isolates, clustered together with the domestic G2b strains, including the major parental virus KNU-1703 (Fig. 3F). The whole-genome phylogeny indicated that the novel 2018–19 G1b recombinants were grouped with the global G1b strains within the G2 clade, but they produced a new branch close to the one containing the major parental G2b virus (Fig. 3G).
Fig. 3.
Phylogenetic analysis based on (A) the full-length S genes, (B and C) minor parental regions covering nt 28–1,964 or nt 28–684 of S (corresponding to genomic nt positions 20,661–22,597 or 20,661–21,318, respectively), (D and E) major parental regions (i.e., the whole genome excluding nt 28–1,964 or nt 28–684 of S, respectively), (F) nt 684–1,964 of S (corresponding to genomic nt positions 21,319–22,597), and (G) complete genomes of the PEDV strains. In each case, the corresponding region of the TGEV genome was included as an outgroup. The numbers at each branch indicate bootstrap values greater than 50% based on 1000 replicates. The name of each strain and its country and year of isolation, GenBank accession number, genotype, and subgenotypes proposed in this study are shown. Red dots indicate the 2018–2019 G1b strains identified in this study, blue dots indicate the 2017 G1b strains identified in 2017, a green dot indicates the emergent Korean G1b strain identified in 2014, a red triangle indicates a major parental G2b virus identified in 2017, blue triangles indicate Korean G2b strains identified during 2017–19 outbreaks, and green triangles indicate Korean G2b strains identified during the 2013–14 pandemic. Scale bars indicate nucleotide substitutions per site
Discussion
This is the first report describing the emergence of new PEDV G1b variants resulting from an initial genetic shift (i.e., novel recombination occurring between the Korean emergent G1b strain and the currently circulating pandemic G2b strains as the minor and major parental viruses, respectively) and subsequent genetic drift (i.e., point mutations). Genetic recombination of RNA viruses, including coronaviruses, is a major driving force of rapid viral evolution that leads to the emergence of new strains or viruses. Thus, new traits or phenotypes and novel coding sequences can frequently be acquired by recombination between members of the same or different viral species or families [18]. Our data indicate that the NTD of the S gene is a common target for natural recombination between two different genotypes of PEDV. Since this domain includes one putative neutralizing epitope (NTD/S0) located between aa positions 19 and 220 [19], frequent recombination events may afford some advantages (e.g., antigenic shift) that allow the virus to evade host immune defenses such as neutralizing antibodies (Supplementary Fig. S4). Also, we found ongoing genetic drift in both the major and minor parental portions of the novel G1b recombinants. In particular, the NTD of the S gene, representing the minor parental virus, contained four aa changes outside of the NTD/S0 epitope, which could genetically shift the G1b recombinant to the G2b-like virus and possibly enhance pathogenesis. Indeed, the two PEDV-naïve swine farms in this study experienced mild neonatal mortality rates, which differed somewhat from previous reports describing the low pathogenicity of G1b isolates, which commonly showed minimal to no clinical signs or mortality associated with PED in piglets [8, 17]. To corroborate this field observation, we are currently isolating new PEDV recombinants that can grow in cell culture, which will allow us to conduct a challenge study. Further, reverse genetics studies will likely provide fundamental clues regarding the possible relationship of recombination and additional mutations to PEDV pathogenesis.
Although the Korean emergent G1b KNU-1406 strain was first detected in Gyeongbuk Province and subsequently spread across the country, the contemporary 2017–19 G1b recombinants and the major parental 2017 G2b viruses were identified in Chungcheong Province and the adjacent Gyeonggi Province (Supplementary Fig. S5). It is noteworthy that all of the pig farms where those 2017–19 G1b or G2b viruses were detected belong to the same farmers’ cooperative. Therefore, it is likely that the cooperative shared several farm-to-farm transmission sources, including traffic and humans, and that these transmission sources acted as an intermediate environment that provided favorable conditions for two circulating viruses (i.e. KNU-1406 and KNU-1703) to simultaneously or sequentially contaminate a single farm during the 2017 outbreaks. This generated a new G1b recombinant (i.e., KNU-1702), which continued to undergo non-lethal mutations and was probably dispersed throughout its associated farms (KNU-1808 and KNU-1909), possibly recovering G2b-like virulence. These types of circumstances may favor the emergence of new genotypes or variants in the domestic herd with greater pathogenicity than the prototype G1b and against which the current G2b vaccine may provide incomplete protection. Continuous surveillance for novel variants is paramount for RNA viruses, including PEDV, that have rapid evolutionary rates. Therefore, the present study underlines the importance of performing periodic monitoring and surveillance investigations, which aid in the development of vaccines against variant or new-genotype viruses that may cause future epidemics or pandemics.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We gratefully thank Chang Won Im for providing clinical samples and clinical information about the current status of PEDV on the farms. This research was financially supported by the Ministry of Small and Medium-Sized Enterprises (SMEs) and Startups (MSS), Korea, under the “Regional Enterprise Linked with National-Innovation-Cluster Development Program (R&D, P0010011)” supervised by the Korea Institute for Advancement of Technology (KIAT).
Funding
This research was financially supported by the Ministry of Small and Medium-Sized Enterprises (SMEs) and Startups (MSS), Korea, under the “Regional Enterprise Linked with National-Innovation-Cluster Development Program (R&D, P0010011)” supervised by the Korea Institute for Advancement of Technology (KIAT).
Data availability
The data supporting the findings of this study are available in the supplementary material of this article.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with animals performed by any of the authors.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee C. Porcine epidemic diarrhea virus: an emerging and re-emerging epizootic swine virus. Virol J. 2015;12:193. doi: 10.1186/s12985-015-0421-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee C. Porcine epidemic diarrhoea virus. In: Zakaryan H, editor. Porcine viruses: from pathogenesis to strategies for control. Norfolk, UK: Caister Academic Press; 2019. pp. 107–134. [Google Scholar]
- 4.Lee DK, Park CK, Kim SH, Lee C. Heterogeneity in spike protein genes of porcine epidemic diarrhea viruses isolated in Korea. Virus Res. 2010;149:175–182. doi: 10.1016/j.virusres.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee S, Kim Y, Lee C. Isolation and characterization of a Korean porcine epidemic diarrhea virus strain KNU-141112. Virus Res. 2015;208:215–224. doi: 10.1016/j.virusres.2015.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee S, Ko DH, Kwak WK, Lim CH, Moon SU, Lee DS, Lee C. Reemergence of porcine epidemic diarrhea virus on Jeju Island. Korean J Vet Res. 2014;54:185–188. doi: 10.14405/kjvr.2014.54.3.185. [DOI] [Google Scholar]
- 7.Lee S, Lee C. Outbreak-related porcine epidemic diarrhea virus strains similar to US strains, South Korea, 2013. Emerg Infect Dis. 2014;20:1223–1226. doi: 10.3201/eid2007.140294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee S, Lee C. Genomic and antigenic characterization of porcine epidemic diarrhoea virus strains isolated from South Korea, 2017. Transbound Emerg Dis. 2018;65:949–956. doi: 10.1111/tbed.12904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee S, Lee DU, Noh YH, Lee SC, Choi HW, Yang HS, Seol JH, Mun SH, Kang WM, Yoo H, Lee C. Molecular characteristics and pathogenic assessment of porcine epidemic diarrhea virus isolates from the 2018 endemic outbreaks on Jeju Island, South Korea. Transbound Emerg Dis. 2019;66:1894–1909. doi: 10.1111/tbed.13219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee S, Son KY, Noh YH, Lee SC, Choi HW, Yoon IJ, Lee C. Genetic characteristics, pathogenicity, and immunogenicity associated with cell adaptation of a virulent genotype 2b porcine epidemic diarrhea virus. Vet Microbiol. 2017;207:248–258. doi: 10.1016/j.vetmic.2017.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee YN, Lee C. Complete genome sequence of a novel porcine parainfluenza virus 5 isolate in Korea. Arch Virol. 2013;158:1765–1772. doi: 10.1007/s00705-013-1770-z. [DOI] [PubMed] [Google Scholar]
- 12.Lole KS, Bollinger RC, Paranjae RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol. 1999;73:152–160. doi: 10.1128/JVI.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li C, Li W, de Esesarte EL, Guo H, van den Elzen P, Aarts E, van den Born E, Rottier PJ, Bosch BJ. Cell attachment domains of the porcine epidemic diarrhea virus spike protein are key targets of neutralizing antibodies. J Virol. 2017;12:e00273-17. doi: 10.1128/JVI.00273-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin DP, Murrell B, Khoosal A, Muhire B. Detecting and analyzing genetic recombination using RDP4. Methods Mol Biol. 2017;1525:433–460. doi: 10.1007/978-1-4939-6622-6_17. [DOI] [PubMed] [Google Scholar]
- 15.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 16.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang L, Byrum B, Zhang Y. New variant of porcine epidemic diarrhea virus, United States. Emerg Infect Dis. 2014;20:917. doi: 10.3201/eid2005.140195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiao Y, Rouzine IM, Bianco S, Acevedo A, Goldstein EF, Farkov M, Brodsky L, Andino R. RNA recombination enhances adaptability and is required for virus spread and virulence. Cell Host Microbe. 2016;19:493–503. doi: 10.1016/j.chom.2016.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this study are available in the supplementary material of this article.