Abstract
Abstract
Anthraquinone derivatives are identified for their immune-boosting, anti-inflammatory, and anti-viral efficacy. Hence, the present study aimed to investigate the reported anthraquinone derivatives as immune booster molecules in COVID-19 infection and evaluate their binding affinity with three reported targets of novel coronavirus i.e. 3C-like protease, papain-like protease, and spike protein. The reported anthraquinone derivatives were retrieved from an open-source database and filtered based on a positive druglikeness score. Compounds with positive druglikeness scores were predicted for their targets using DIGEP-Pred and the interaction among modulated proteins was evaluated using STRING. Further, the associated pathways were recorded concerning the Kyoto Encyclopedia of Genes and Genomes pathway database. Finally, the docking was performed using autodock4 to identify the binding efficacy of anthraquinone derivatives with 3C-like protease, papain-like protease, and spike protein. After docking the pose of ligand scoring minimum binding energy was chosen to visualize the ligand–protein interaction. Among 101 bioactives, 36 scored positive druglikeness score and regulated multiple pathways concerned with immune modulation and (non-) infectious diseases. Similarly, docking study revealed torososide B to possess the highest binding affinity with papain-like protease and 3C-like protease and 1,3,6-trihydroxy-2-methyl-9,10-anthraquinone-3-O-(6′-O-acetyl)-β-d-xylopyranosyl-(1 → 2)-β-d-glucopyranoside with spike protein.
Graphic Abstract
Electronic supplementary material
The online version of this article (10.1007/s13659-020-00260-2) contains supplementary material, which is available to authorized users.
Keywords: 3CLpro, Anthroquine derivatives, Coronavirus, COVID-19, Immune boost
Introduction
Presently, CoV Disease (COVID-19) has lead to millions of death throughout the world beginning from the December of 2019 [1]. Further, the risk of getting affected with COVID-19 is supplemented in subjects with lower immunity primarily special subjects like pediatrics/geriatrics and the patients suffering from infectious and non-infectious diseases [2]. Although approaches are being made to prevent this virus to get spread via social distancing and so on, boosting of immunity in subjects could play an important role in inhibiting the transmission of the virus and its invasion into the body. Although investigations are undergoing to develop the vaccine against COVID-19, it may still take time as the drug discovery process is much complicated. Hence, it is important to identify any alternative approach as prophylaxis against COVID-19 which can be implemented via the utilization of immune boosters from natural sources. Three targets of novel coronavirus i.e. 3C-like protease (3CLpro), papain-like protease (PLpro), and spike protein [3–6] are being targeted by multiple investigators to identify the new hit molecule for the management of COVID-19. Further, inflammation and cell necrosis are contributing factors in worsening the COVID-19 pathogenesis which suggests identifying the molecule with anti-viral, anti-inflammatory and immune-boosting properties.
Anthroquinolines are the group of compounds from multiple folk medicines like Senna species which are utilized in Ayurvedic system of medicines and Traditional Chinese Medicines for the management of various infectious and non-infectious diseases [7, 8]. Further, anthraquinone derivatives are also reported for anti-viral property [9], anti-inflammatory efficacy [10], and as immune booster [11]. Hence, in the COVID-19 infection, it may be beneficial if the bioactives are identified with an immune boost, anti-inflammatory, and anti-viral properties which can be demonstrated via the concept of network pharmacology or polypharmacological approach. Hence, based on the above theme, of anti-viral/anti-inflammatory/immune boosting reports of anthraquinones we attempted to screen the multiple anthraquinone derivatives as an immune booster and anti-viral efficacy using in silico molecular docking and other system biology tools.
Materials and methods
Bioactives and their Druglikeness Score
The reported phytoconstituents under the phytochemistry of anthraquinone were retrieved from available literature/Chemical Entities of Biological Interest (ChEBI) records (https://www.ebi.ac.uk/chebi/). All the compounds were then predicted for the druglikeness score by querying the SMILES of each molecule in MolSoft (https://molsoft.com/mprop/).
Target Prediction and their Enrichment Analysis to Assess Immune-boosting Efficacy
Anthraquinone derivatives with positive druglikeness scores were queried in DIGEP-Pred [12] to identify “Proteins based targets” (up-regulated/downregulated proteins) at the probable activity of 0.5. The list of regulated proteins was queried in STRING [13] to identify the biological process, cellular function, and molecular processes of combined gene-set. Further, the probably modulated pathways were also identified concerning the Kyoto Encyclopedia of Genes and Genomes database. Network among the bioactives, their targets, and modulated pathways was constructed using the Cytoscape [14] version 3.5.1. Any duplicates interconnection of two nodes was eliminated to avoid the appearance of a false hit. The whole network was analyzed using the “network analyzer” tool based on node size and count representing the edge count as “low values to small size” and “low values to bright colors” respectively as explained previously [15].
Prediction of Probable Anti-viral Activity
Anti-viral activity of each compound was predicted by querying the SMILES in Prediction of Activity Spectra for Substances [16] at the pharmacological activity (Pa) > pharmacological inactivity (Pi) and retrieving the probable biological spectrum for keyword “anti-viral”. Records were queried for their probable pharmacological spectrum against multiple viruses like Adenovirus, Cytomegalovirus (CMV), Hepatitis B, Hepatitis C, Hepatitis, Herpes, Human immunodeficiency virus (HIV), Influenza A, Influenza, Parainfluenza, Picornavirus, Poxvirus, Rhinovirus, and Trachoma.
In Silico Molecular Docking
Preparation of Ligand Molecules
All the 3D. sdf format of ligand molecules with positive drug-likeness scores was retrieved from PubChem database (https://pubchem.ncbi.nlm.nih.gov/) or structures were drawn in ChemSketch (https://www.acdlabs.com/resources/freeware/chemsketch/) as applicable. The ligand molecules were converted into.pdb format using Discovery studio 2019 [17]. All the bioactives were minimized using MMFF94 forcefield [18] using conjugate gradients as an optimization algorithm. After the minimization of energy, all ligand molecules were converted into.pdbqt format.
Preparation of Macromolecules
Three target proteins of COVID-19 i.e. 3clpro (PDB: 6LU7), PLpro (PDB: 4M0W), and spike proteins (homology modeled target, accession number: AVP78042.1 as query sequence and PDB: 6VSB as a template using SWISS-MODEL [19]) were selected. The retrieved proteins from Research Collaboratory for Structural Bioinformatics database were in complex with other heteroatoms which were removed using Discovery studio 2019 and saved in.pdb format.
Ligand–protein Docking
The ligand molecules were docked with protein molecules using autodock4 [20] by setting 8 exhaustiveness as default to obtain 10 different poses of ligand molecules. After docking the pose of ligand scoring lowest binding energy was selected to visualize the ligand–protein interaction in Discovery Studio 2019 as explained previously [21, 22].
Results
Bioactives and their Druglikeness Score
The complete datasheet of 101 compounds including their name, ChEBI ID, molecular formula/weight, and synonym including phytochemistry for the retrieved compounds were summarized (Table S1). Among 101 different compounds, 36 were identified with positive druglikeness scores. Among them, laccaic acid A scored highest druglikeness score i.e. 0.85 with molecular weight 537.09, 12 hydrogen bond acceptor, 8 hydrogen bond donors, and 2.88 MolLogP. The details of the druglikeness score of each compound are summarized in Table 1.
Table 1.
Druglikeness of anthraquinone derivatives with positive score
| Bioactives | Molecular formula | Molecular weight | NHBA | NHBD | MolLogP | MolPSA (A2) | MolVol (A3) | DLS |
|---|---|---|---|---|---|---|---|---|
| Versicolorone tricyclic form | C20H18O8 | 386.1 | 8 | 5 | 1.67 | 122.82 | 375.84 | 0.09 |
| (1′S,5′S)-5′-hydroxyaverantin | C20H20O8 | 388.12 | 8 | 6 | 1.9 | 125 | 371.14 | 0.17 |
| (1′S,5′R)-5′-hydroxyaverantin | C20H20O8 | 388.12 | 8 | 6 | 1.9 | 125 | 371.14 | 0.17 |
| chrysophanol 8-O-β-d-glucoside | C21H20O9 | 416.11 | 9 | 5 | 1.11 | 122.67 | 379.02 | 0.45 |
| 1,3,6-trihydroxy-2-methyl-9,10-anthraquinone-3-O-(6′-O-acetyl)-α-l-rhamnopyranosyl-(1- > 2)-β-d-glucopyranoside | C29H32O15 | 620.17 | 15 | 7 | 1.46 | 189.78 | 556.02 | 0.6 |
| 1,3,6-trihydroxy-2-methyl-9,10-anthraquinone-3-O-(6′-O-acetyl)-β-d-glucopyranoside | C23H22O11 | 474.12 | 11 | 5 | 2.07 | 143.34 | 436.83 | 0.45 |
| 1,3,6-trihydroxy-2-methyl-9,10-anthraquinone-3-O-α-l-rhamnopyranosyl-(1- > 2)-β-d-glucopyranoside | C27H30O14 | 578.16 | 14 | 8 | 0.85 | 185.66 | 510.27 | 0.5 |
| 1,3,6-trihydroxy-2-methyl-9,10-anthraquinone-3-O-(6′-O-acetyl)-β-d-xylopyranosyl-(1- > 2)-β-d-glucopyranoside | C28H30O15 | 606.16 | 15 | 7 | 1.01 | 190.6 | 540.52 | 0.69 |
| 1,3,6-trihydroxy-2-methyl-9,10-anthraquinone-3-O-(3′-O-acetyl)-α-l-rhamnopyranosyl-(1- > 2)-β-d-glucopyranoside | C29H32O15 | 620.17 | 15 | 7 | 1.42 | 190.3 | 556.1 | 0.76 |
| 1-hydroxy-2-(β-d-glucosyloxy)-9,10-anthraquinone | C20H18O9 | 402.1 | 9 | 5 | 1.1 | 121.6 | 358.41 | 0.04 |
| (2S)-versicolorone | C20H18O8 | 386.1 | 8 | 5 | 1.67 | 122.82 | 375.84 | 0.09 |
| (S)-5′-oxoaverantin | C20H18O8 | 386.1 | 8 | 5 | 1.77 | 121.86 | 374.32 | 0.16 |
| BDA-366 | C24H29N3O4 | 423.22 | 5 | 3 | 2.54 | 74.7 | 440.1 | 0.09 |
| Emodin 8-glucoside | C21H20O10 | 432.11 | 10 | 6 | 0.61 | 140.29 | 389.71 | 0.74 |
| Anthragallol | C21H20O10 | 432.11 | 10 | 6 | 0.61 | 140.29 | 389.71 | 0.74 |
| Nogalonic acid | C20H14O8 | 382.07 | 8 | 3 | 2.33 | 113.17 | 370.61 | 0.44 |
| Mitoxantrone | C22H28N4O6 | 444.2 | 8 | 8 | -0.55 | 135.33 | 432.17 | 0.53 |
| Torososide B | C40H52O25 | 932.28 | 25 | 14 | -4.46 | 320.41 | 789.36 | 0.63 |
| Versicolorone | C20H16O7 | 368.09 | 7 | 3 | 2.68 | 97.9 | 364.43 | 0.14 |
| 4′-O-demethylknipholone-4′-O-β-d-glucopyranoside | C29H26O13 | 582.14 | 13 | 8 | 1.91 | 184.8 | 529.73 | 0.61 |
| Aklanonic acid | C21H16O8 | 396.08 | 8 | 3 | 2.83 | 112.59 | 389.73 | 0.51 |
| Kermesic acid | C16H10O8 | 330.04 | 8 | 5 | 2.1 | 119.71 | 304.77 | 0.06 |
| Gaboroquinone A | C24H18O9 | 450.1 | 9 | 5 | 3.35 | 130.76 | 428.49 | 0.27 |
| Variecolorquinone A | C20H18O9 | 402.1 | 9 | 4 | 1.36 | 121.77 | 383.49 | 0.71 |
| Laccaic acid A | C26H19NO12 | 537.09 | 12 | 8 | 2.88 | 186.68 | 496.66 | 0.85 |
| Laccaic acid B | C24H16O12 | 496.06 | 12 | 8 | 3.19 | 178.91 | 446.68 | 0.65 |
| Laccaic acid C | C25H17NO13 | 539.07 | 14 | 10 | 0.68 | 211.46 | 476.5 | 0.68 |
| Carminic acid | C22H20O13 | 492.09 | 13 | 9 | 0.62 | 189.58 | 434.4 | 0.77 |
| Ekatetrone | C19H13NO7 | 367.07 | 7 | 4 | 2.12 | 112.76 | 348.31 | 0.5 |
| 4-bromo-1-hydroxyanthraquinone-2-carboxylic acid | C15H7BrO5 | 345.95 | 5 | 2 | 4.39 | 70.07 | 269.12 | 0.24 |
| Scutianthraquinone A | C39H32O13 | 708.18 | 13 | 4 | 7.75 | 165.64 | 711.03 | 0.45 |
| Scutianthraquinone B | C38H30O13 | 694.17 | 13 | 4 | 7.41 | 165.64 | 693.71 | 0.26 |
| Scutianthraquinone C | C34H24O12 | 624.13 | 12 | 5 | 5.88 | 161.07 | 618.22 | 0.31 |
| 1,3,6-trihydroxy-2-hydroxymethyl-9,10-anthraquinone-3-O-(6′-O-acetyl)-β-d-glucopyranoside | C23H22O12 | 490.11 | 12 | 6 | 0.93 | 160.45 | 445.01 | 0.41 |
| Rubianthraquinone | C16H12O5 | 284.07 | 5 | 2 | 3.26 | 68.06 | 281.86 | 0.07 |
| 5-hydroxyanthraquinone-1,3-dicarboxylic acid | C16H8O7 | 312.03 | 7 | 3 | 3.47 | 99.54 | 281.87 | 0.28 |
DLS Druglikeness Score NHBA Number of Hydrogen Bond Acceptor NHBD Number of Hydrogen Bond Donor
Target Prediction and their Enrichment Analysis to Assess Immune-Boosting Efficacy
Among the compounds with positive druglikeness score, anthragallol was predicted to modulate the highest number of genes i.e. 25. Similarly, human carbonyl reductase 1 (CBR1) was targeted by the highest number of bioactives i.e. 33. Further, the enrichment analysis identified the modulation of 54 different pathways in which pathways in cancer was majorly modulated by regulating 12 genes (AR, CASP8, CDK4, CTNNB1, EPAS1, HMOX1, KLK3, MMP2, NFE2L2, NOS2, RAC1, and RARA) under the background of 515 proteins at the false discovery rate of 7.71E−08. Table 2 summarizes the gene enrichment analysis of the modulated gene set along with modulated pathways with their respective gene codes. The protein–protein interaction of modulated proteins is presented in Fig. 1. Similarly, the combined bioactives-proteins-pathways also reflected the anthrogallol to target the highest number of proteins. Likewise, TNFRSF1A and pathways in cancer were majorly targeted/modulated protein and pathways respectively (Fig. 2).
Table 2.
Enrichment analysis of modulated proteins by the reported anthraquinone derivatives
| Term ID | Term description | Observed gene count | Background gene count | False discovery rate | Matching proteins in network |
|---|---|---|---|---|---|
| hsa05200 | Pathways in cancer | 12 | 515 | 7.71E−08 | AR, CASP8, CDK4, CTNNB1, EPAS1, HMOX1, KLK3, MMP2, NFE2L2, NOS2, RAC1, RARA |
| hsa05418 | Fluid shear stress and atherosclerosis | 7 | 133 | 1.18E−06 | CTNNB1, HMOX1, MMP2, NFE2L2, PLAT, RAC1, TNFRSF1A |
| hsa05167 | Kaposi's sarcoma-associated herpesvirus infection | 6 | 183 | 0.00012 | CASP8, CD86, CDK4, CTNNB1, RAC1, TNFRSF1A |
| hsa05215 | Prostate cancer | 5 | 97 | 0.00012 | AR, CTNNB1, KLK3, PLAT, PLAU |
| hsa05014 | Amyotrophic lateral sclerosis (ALS) | 4 | 50 | 0.00015 | CAT, GPX1, RAC1, TNFRSF1A |
| hsa04932 | Non-alcoholic fatty liver disease (NAFLD) | 5 | 149 | 0.00044 | ADIPOQ, CASP8, PPARA, RAC1, TNFRSF1A |
| hsa05202 | Transcriptional misregulation in cancer | 5 | 169 | 0.00068 | CD86, FLT1, PLAT, PLAU, RARA |
| hsa04066 | HIF-1 signaling pathway | 4 | 98 | 0.0012 | FLT1, HMOX1, NOS2, TIMP1 |
| hsa00380 | Tryptophan metabolism | 3 | 40 | 0.0017 | CAT, CYP1A1, CYP1A2 |
| hsa04915 | Estrogen signaling pathway | 4 | 133 | 0.003 | FKBP5, MMP2, PGR, RARA |
| hsa05416 | Viral myocarditis | 3 | 56 | 0.0037 | CASP8, CD86, RAC1 |
| hsa00980 | Metabolism of xenobiotics by cytochrome P450 | 3 | 70 | 0.0053 | CBR1, CYP1A1, CYP1A2 |
| hsa04115 | p53 signaling pathway | 3 | 68 | 0.0053 | CASP8, CDK4, CHEK1 |
| hsa04920 | Adipocytokine signaling pathway | 3 | 69 | 0.0053 | ADIPOQ, PPARA, TNFRSF1A |
| hsa05152 | Tuberculosis | 4 | 172 | 0.0053 | CASP8, NOS2, TNFRSF1A, VDR |
| hsa05225 | Hepatocellular carcinoma | 4 | 163 | 0.0053 | CDK4, CTNNB1, HMOX1, NFE2L2 |
| hsa05203 | Viral carcinogenesis | 4 | 183 | 0.0056 | CASP8, CDK4, CHEK1, RAC1 |
| hsa05204 | Chemical carcinogenesis | 3 | 76 | 0.0056 | CBR1, CYP1A1,CYP1A2 |
| hsa05205 | Proteoglycans in cancer | 4 | 195 | 0.0065 | CTNNB1, MMP2, PLAU, RAC1 |
| hsa04933 | AGE-RAGE signaling pathway in diabetic complications | 3 | 98 | 0.0097 | CDK4, MMP2, RAC1 |
| hsa04620 | Toll-like receptor signaling pathway | 3 | 102 | 0.01 | CASP8, CD86, RAC1 |
| hsa05142 | Chagas disease (American trypanosomiasis) | 3 | 101 | 0.01 | CASP8, NOS2, TNFRSF1A |
| hsa05145 | Toxoplasmosis | 3 | 109 | 0.0113 | CASP8, NOS2, TNFRSF1A |
| hsa04670 | Leukocyte transendothelial migration | 3 | 112 | 0.0117 | CTNNB1, MMP2, RAC1 |
| hsa05166 | HTLV-I infection | 4 | 250 | 0.0121 | CDK4, CHEK1, CTNNB1, TNFRSF1A |
| hsa04215 | Apoptosis—multiple species | 2 | 31 | 0.0131 | CASP8, TNFRSF1A |
| hsa04216 | Ferroptosis | 2 | 40 | 0.0204 | GSS, HMOX1 |
| hsa04310 | Wnt signaling pathway | 3 | 143 | 0.0204 | CTNNB1, MMP7, RAC1 |
| hsa05219 | Bladder cancer | 2 | 41 | 0.0204 | CDK4, MMP2 |
| hsa05224 | Breast cancer | 3 | 147 | 0.0204 | CDK4, CTNNB1, PGR |
| hsa05165 | Human papillomavirus infection | 4 | 317 | 0.0226 | CASP8, CDK4, CTNNB1, TNFRSF1A |
| hsa00480 | Glutathione metabolism | 2 | 50 | 0.0261 | GPX1, GSS |
| hsa04978 | Mineral absorption | 2 | 51 | 0.0263 | HMOX1, VDR |
| hsa04151 | PI3K-Akt signaling pathway | 4 | 348 | 0.0285 | CDK4, FLT1, GH1, RAC1 |
| hsa00140 | Steroid hormone biosynthesis | 2 | 58 | 0.0316 | CYP1A1, CYP1A2 |
| hsa00590 | Arachidonic acid metabolism | 2 | 61 | 0.0337 | CBR1, GPX1 |
| hsa00830 | Retinol metabolism | 2 | 62 | 0.0338 | CYP1A1, CYP1A2 |
| hsa04024 | cAMP signaling pathway | 3 | 195 | 0.0339 | PPARA, RAC1, TNNI3 |
| hsa04510 | Focal adhesion | 3 | 197 | 0.034 | CTNNB1, FLT1, RAC1 |
| hsa04015 | Rap1 signaling pathway | 3 | 203 | 0.0359 | CTNNB1, FLT1, RAC1 |
| hsa05211 | Renal cell carcinoma | 2 | 68 | 0.0363 | EPAS1, RAC1 |
| hsa01524 | Platinum drug resistance | 2 | 70 | 0.0374 | CASP8, TOP2A |
| hsa04520 | Adherens junction | 2 | 71 | 0.0375 | CTNNB1, RAC1 |
| hsa03320 | PPAR signaling pathway | 2 | 72 | 0.0376 | ADIPOQ, PPARA |
| hsa05100 | Bacterial invasion of epithelial cells | 2 | 72 | 0.0376 | CTNNB1, RAC1 |
| hsa05212 | Pancreatic cancer | 2 | 74 | 0.0379 | CDK4, RAC1 |
| hsa04610 | Complement and coagulation cascades | 2 | 78 | 0.0409 | PLAT, PLAU |
| hsa04146 | Peroxisome | 2 | 81 | 0.0429 | CAT, NOS2 |
| hsa05132 | Salmonella infection | 2 | 84 | 0.045 | NOS2, RAC1 |
| hsa05210 | Colorectal cancer | 2 | 85 | 0.045 | CTNNB1, RAC1 |
| hsa05323 | Rheumatoid arthritis | 2 | 84 | 0.045 | CD86, FLT1 |
| hsa04211 | Longevity regulating pathway | 2 | 88 | 0.0462 | ADIPOQ, CAT |
| hsa05222 | Small cell lung cancer | 2 | 92 | 0.0492 | CDK4, NOS2 |
| hsa04064 | NF-kappa B signaling pathway | 2 | 93 | 0.0493 | PLAU, TNFRSF1A |
Fig. 1.
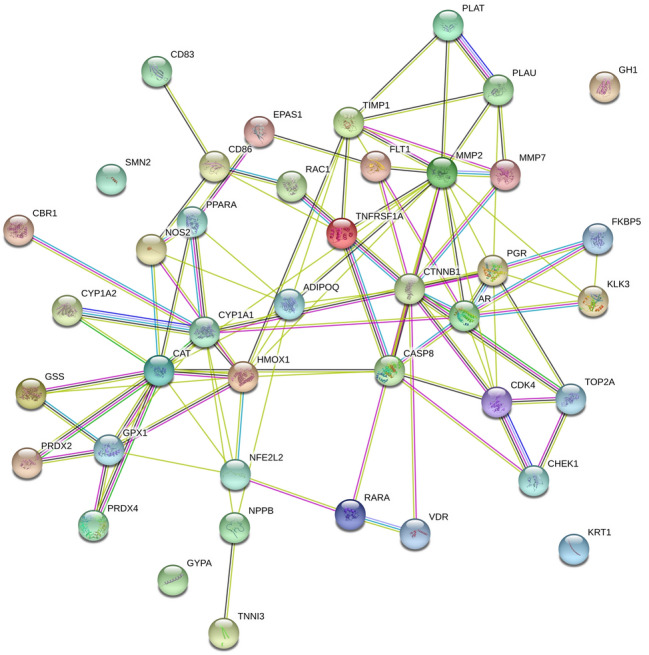
Protein–protein interaction of regulated proteins
Fig. 2.
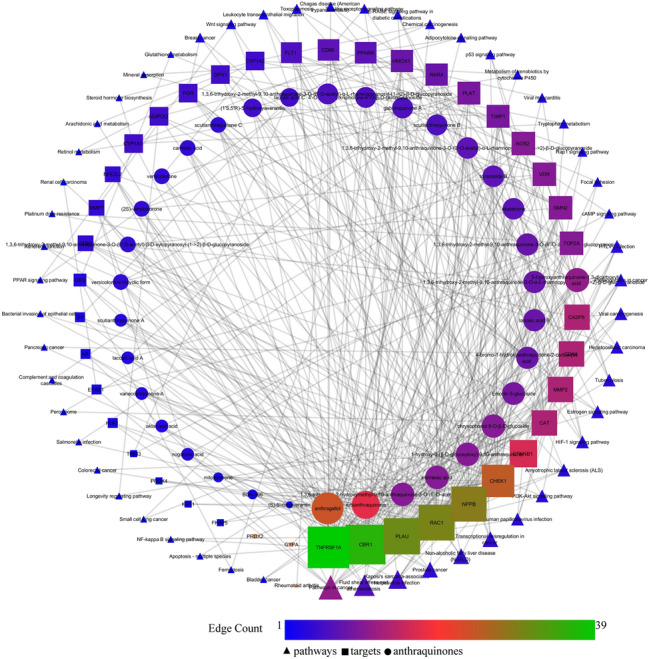
Network interaction of anthraquinone derivatives with their proteins and regulated pathways
Prediction of Probable Anti-viral Activity
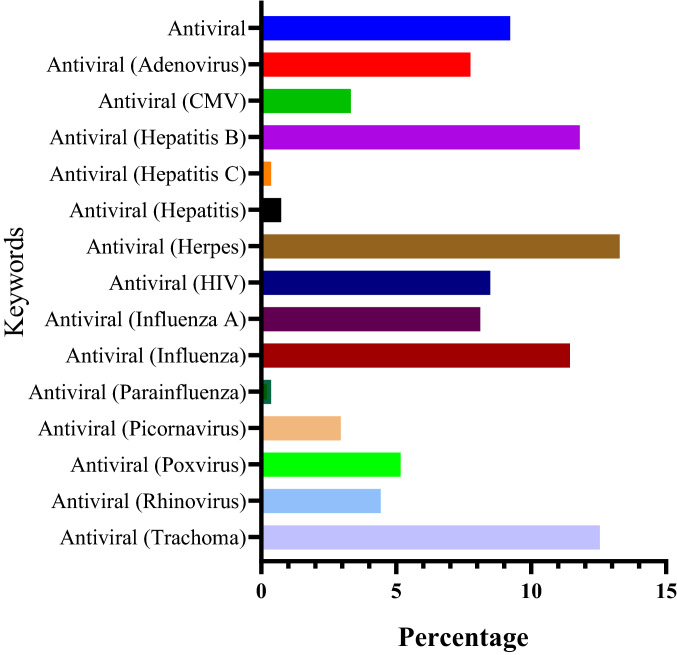
The anthraquinones were found to be anti-viral agents against adenovirus, CMV, hepatitis B and C, hepatitis, herpes, HIV, influenza A, influenza, parainfluenza, picornavirus, poxvirus, rhinovirus, and trachoma. Among them, the majority of the compounds were active against herpes virus i.e. 13.28%. The overall activity of compounds against multiple viruses is summarized in Fig. 3.
Fig. 3.
Predicted anti-viral activity of anthraquinone derivatives against multiple viruses
In Silico Molecular Docking
Torososide B was predicted to have the highest binding affinity (− 8.7 kcal/mol) with PLpro with 9 hydrogen bond interactions via THR302, ASP303, TYR274, TYR265, ARG167, TYR269, ASP165. Further, Torososide B was predicted to possess the highest binding affinity (− 9.3 kcal/mol) with 3CLpro with 14 hydrogen bond interactions with LEU287, TYR237, THR199, ARG131, LYS137, LYS5, GLU290, ILE281, LEU282, and PHE3. Similarly, 1,3,6-trihydroxy-2-methyl-9,10-anthraquinone-3-O-(6′-O-acetyl)-β-d-xylopyranosyl-(1– > 2)-β-d-glucopyranoside was predicted to possess the highest binding affinity (− 8.7 kcal/mol) with spike protein with the highest number of hydrogen bond interactions i.e. 6 with ASP820, ILE816, ASP815, GLN825, and MET703. The binding affinity of each compound with individual targets with the number of hydrogen bond interactions and residues is summarized in Table S2. The interaction of Torososide B with PLpro and 3CLpro and 1,3,6-trihydroxy-2-methyl-9,10-anthraquinone-3-O-(6′-O-acetyl)-β-d-xylopyranosyl-(1– > 2)-β-d-glucopyranoside with spike protein is presented in Fig. 4.
Fig. 4.
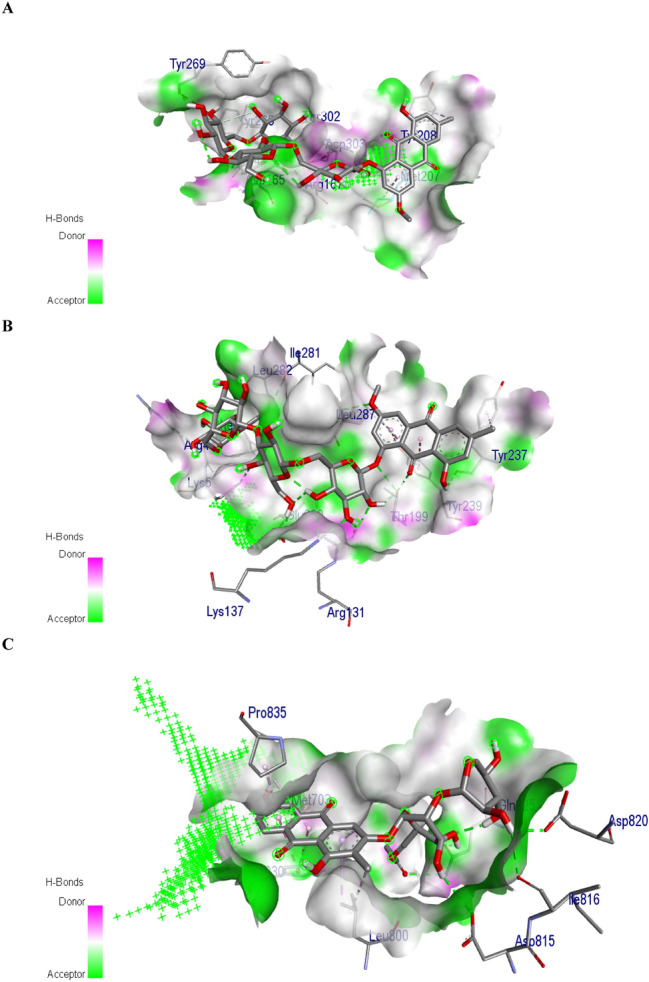
Interaction of torososide B with (a) Papain-like protease and (b) coronavirus main proteinase and 1,3,6-trihydroxy-2-methyl-9,10-anthraquinone-3-O-(6′-O-acetyl)-β-d-xylopyranosyl-(1→ 2)-β-d-glucopyranoside (c) with spike protein
Discussion
During the COVID-19 infection, severe necrosis and inflammation lead to defects in the supply of necessary nutrients and oxygen into the cells which are more terrible in the subjects with compromised immunity. Hence, in the present study, we investigated multiple anthraquinone derivatives from various traditional medicines to act against COVID-19 targets i.e. 3CLpro, PLpro, and spike protein, and their combined immune-boosting efficacy. Initially, we calculated the druglikeness score of each molecule based on the “Lipinksi’s Rule of Five” [23] as the majority of the plant-based medicines are utilized via the oral route which identified 36 different compounds with positive druglikeness score and considered to get absorbed orally (Table 1) which were contemplated for further study.
The conventional drug discovery process utilizes the concept of “single drug-single protein-single disease” [24] which may not be applicable in the management of infectious diseases. This is due to the affinity of the pathogens (viruses/bacteria) to affect the multiple homeostatic functions of protein molecules. It means multiple proteins from the pathogens are involved to generate this effect. Hence, this can be managed via the utilization of modified drug development process “multi compound-multi protein-disease” interaction in which multiple bioactives regulate multiple proteins [25] which can also be taken as a basic key of boosting the immune system. Hence, in the present study, the combined synergistic phenomena of anthraquinones were investigated rather than a single bioactive molecule to identify multiple pathways that are directly or indirectly involved in the immune system.
Gene set enrichment analysis identified multiple pathways like p53 signaling pathway [26], PI3K-Akt signaling pathway [27], Rap1 signaling pathway [28], NF-kappa B signaling pathway [29] which are directly involved in the boosting the immune system. Similarly, some other pathways like pathways in cancer, PPAR signaling pathway, colorectal cancer, chemical carcinogenesis, estrogen signaling pathway were also identified which reflects the potency of anthraquinones to be beneficial in the subjects which are suffering from these pathways associated diseases like cancer. Further, pathways like p53 signaling pathways, PI3K-Akt, Wnt signaling pathways are also associated with diseases like diabetes and obesity where the immune system is compromised. Hence, regulation of these pathways could be beneficial in managing the diseases from which they are suffering, and boosting the immune system will also act as prophylaxis against COVID-19. Additionally, the enrichment analysis also identified the modulation of multiple pathways that are associated with a pathogenic infection like viral myocarditis and tuberculosis which reflects the potency of anthraquinone derivatives to manage the infectious diseases. Further, herbal medicines rich in anthraquinones also possess the anti-viral potency against various viruses. Hence, we attempted to identify the probable anti-viral activity of the anthraquinones with positive druglikeness score which identified their efficacy against multiple viruses like the rhino, influenza, herpes trachoma, pox, and CMV.
3CLpro alters the ubiquitin system to incorporate the viral polypeptides and deregulates the homeostatic task of functional proteins [30] which was majorly targeted by torososide B. Further, PLpro alters the function of protein phosphatase 1A and protein phosphatase 1B into the replicase proteins to adjust viral life cycle [31] which was majorly inhibited by torososide B. Similarly, spike protein utilizes angiotensin-converting enzyme 2 (ACE-2) as a receptor to enter inside the host cell [32, 33] which was chiefly regulated by modulated by 1,3,6-trihydroxy-2-methyl-9,10-anthraquinone-3-O-(6′-O-acetyl)-β-d-xylopyranosyl-(1→ 2)-β-d-glucopyranoside. These results reflect the probability of the anthraquinone derivatives to act as the anti-viral against COVID-19. However, as the time proceeds, it is to be understood that the binding affinity of probable lead hit molecules may get altered due to mutation in the possible protein targets and the inhibitory function may not occur as predicted.
Conclusion
The present study utilized in silico molecular docking tools to identify the binding affinity of previously recorded anthraquinones derivatives against 3clpro, PLpro, and spike protein which identified Torososide B and 1,3,6-trihydroxy-2-methyl-9,10-anthraquinone-3-O-(6′-O-acetyl)-β-d-xylopyranosyl-(1→ 2)-β-d-glucopyranoside as a lead hits. Similarly, the combined synergies of the network identified the modulation of multiple pathways involved in the immune system like p53, chemokine, and PI3K-Akt signaling pathways. Additionally, anthraquinone derivatives were also identified as the modulators of the disease pathways where the immune system is compromised like diabetes and obesity. All these results suggest the probable therapeutic option of the utilization of anthraquinones as an immune booster and anti-viral against novel coronavirus by acting on the three targets as investigated. However, the present findings are only based on the computer simulations which need to be further validated using well designed experimental protocols.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
All the authors are thankful to Principal KLE College of Pharmacy, Belagavi, KAHER Belagavi for his support for this completion of this work.
Author Contributions
Pukar Khanal developed the protocol, performed the work, and drafted the manuscript. Prof. B.M. Patil reviewed and finalized the manuscript draft. Jagdish Chand and Yasmin Naaz had an equal contribution to mining the database and assisting in the work performed.
Funding
Not available.
Compliance with Ethical Standards
Conflict of interest
There is no conflict of interest.
Research Involving Human and Animal Participants
This manuscript doesn’t include any animal or human studies.
Footnotes
Jagdish Chand and Yasmin Naaz have contributed equally to this work.
Contributor Information
Pukar Khanal, Email: pukarkhanal58@gmail.com.
B. M. Patil, Email: drbmpatil@klepharm.edu, Email: bmpatil59@hotmail.com
References
- 1.Miura TA, Holmes KV. J. Leukoc. Biol. 2009;86:1145–1151. doi: 10.1189/jlb.0209078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.University of Melbourne. Science News. https://www.sciencedaily.com/releases/2020/03/200317103815.htm. Accessed 28 May 2020
- 3.Khanal P, Duyu T, Patil BM, Dey YN, Pasha I, Kavalapure RS. Res Sq. 2020 doi: 10.21203/rs.3.rs-32233/v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khanal P, Duyu T, Dey YN, Pasha I, Wanjari M. Res. Sq. 2020 doi: 10.21203/rs.3.rs-31776/v1. [DOI] [Google Scholar]
- 5.Zhang DH, Wu KL, Zhang X, Deng SQ, Peng B. J. Integr. Med. 2020;18:152–158. doi: 10.1016/j.joim.2020.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mahdian S, Ebrahim-Habibi A, Zarrabi M. J. Diabetes Metab. Disord. 2020 doi: 10.1007/s40200-020-00546-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malik EM, Müller CE. Med. Res. Rev. 2016;36:705–748. doi: 10.1002/med.21391. [DOI] [PubMed] [Google Scholar]
- 8.Chien SC, Wu YC, Chen ZW, Yang WC. Evid.-Based Complement. Altern. Med.: eCAM. 2015;2015:357357. doi: 10.1155/2015/357357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andersen DO, Weber ND, Wood SG, Hughes BG, Murray BK, North JA. Antivir. Res. 1991;16:185–196. doi: 10.1016/0166-3542(91)90024-L. [DOI] [PubMed] [Google Scholar]
- 10.Kshirsagar AD, Panchal PV, Harle UN, Nanda RK, Shaikh HM. Int. J. Inflamm. 2014;2014:690596. doi: 10.1155/2014/690596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Panigrahi GK, Yadav A, Mandal P, Tripathi A, Das M. Toxicol. Lett. 2016;245:15–23. doi: 10.1016/j.toxlet.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 12.Lagunin A, Ivanov S, Rudik A, Filimonov D, Poroikov V. Bioinformatics. 2013;29:2062–2063. doi: 10.1093/bioinformatics/btt322. [DOI] [PubMed] [Google Scholar]
- 13.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, et al. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khanal P, Patil BM. Adv. Tradit. Med. 2020 doi: 10.1007/s13596-020-00426-w. [DOI] [Google Scholar]
- 16.Poroikov VV, Filimonov DA, Ihlenfeldt WD, Gloriozova TA, Lagunin AA, Borodina YV, et al. J. Chem. Inf. Comput. Sci. 2003;43:228–236. doi: 10.1021/ci020048r. [DOI] [PubMed] [Google Scholar]
- 17.Biovia DS. Discovery Studio client. San Diego: DassaultSystèmes; 2019. p. 2019. [Google Scholar]
- 18.Halgren TA. J. Comput. Chem. 1996;17:490–519. doi: 10.1002/(SICI)1096-987X(199604)17:5/6<490::AID-JCC1>3.0.CO;2-P. [DOI] [Google Scholar]
- 19.Schwede T, Kopp J, Guex N, Peitsch MC. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, et al. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khanal P, Patil BM, Hullatti KK. Indian J. Pharm. Sci. 2019;81:550–555. [Google Scholar]
- 22.Khanal P, Magadum P, Patil BM, Hullatti KK. Indian J. Pharm. Sci. 2019;81:326–332. [Google Scholar]
- 23.Lipinski CA. Drug. Discov. Today Technol. 2004;1:337–341. doi: 10.1016/j.ddtec.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 24.Khanal P, Patil BM. Asian Pac. J. Trop. Biomed. 2019;9:263. doi: 10.4103/2221-1691.260399. [DOI] [Google Scholar]
- 25.Khanal P, Patil BM. J. Diabetes Metab. Disord. 2020 doi: 10.1007/s40200-020-00554-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muñoz-Fontela C, Mandinova A, Aaronson SA, Lee SW. Nat. Rev. Immunol. 2016;16:741–750. doi: 10.1038/nri.2016.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weichhart T, Säemann MD. Ann. Rheum. Dis. 2008;iii67:iii70–iii74. doi: 10.1136/ard.2008.098459. [DOI] [PubMed] [Google Scholar]
- 28.Johnson DS, Chen YH. Curr. Opin. Pharmacol. 2012;12:458–463. doi: 10.1016/j.coph.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sokol CL, Luster AD. Cold Spring Harb. Perspect. Biol. 2015;7:a016303. doi: 10.1101/cshperspect.a016303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bhoj VG, Chen ZJ. Nature. 2009;458:430–437. doi: 10.1038/nature07959. [DOI] [PubMed] [Google Scholar]
- 31.Lindner HA, Fotouhi-Ardakani N, Lytvyn V, Lachance P, Sulea T, Ménard R. J. Virol. 2005;79:15199–15208. doi: 10.1128/JVI.79.24.15199-15208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC, Choe H, Farzan M. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuhn JH, Li W, Choe H, Farzan M. Cell Mol. Life Sci. 2004;61:2738–2743. doi: 10.1007/s00018-004-4242-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.