Abstract
The advancements in automated diagnostic tools allow researchers to obtain more and more information from medical images. Recently, to obtain more informative medical images, multi-modality images have been used. These images have significantly more information as compared to traditional medical images. However, the construction of multi-modality images is not an easy task. The proposed approach, initially, decomposes the image into sub-bands using a non-subsampled contourlet transform (NSCT) domain. Thereafter, an extreme version of the Inception (Xception) is used for feature extraction of the source images. The multi-objective differential evolution is used to select the optimal features. Thereafter, the coefficient of determination and the energy loss based fusion functions are used to obtain the fused coefficients. Finally, the fused image is computed by applying the inverse NSCT. Extensive experimental results show that the proposed approach outperforms the competitive multi-modality image fusion approaches.
Keywords: Fusion, Diagnosis, CNN, Multi-modality, Differential evolution
Introduction
Biomedical images are extensively utilized for automated diagnosis of various diseases such as COVID-19, pneumonia, tuberculosis, cancer, etc. Biomedical imaging systems play an efficient role to monitor and diagnose the internal body organs without utilizing any kind of surgery. Biomedical images come up with various modalities to understand the internal body organs (Du et al. 2016). These modalities are positron emission tomography (PET), magnetic resonance imaging (MRI), computerized tomography (CT), X-ray, ultrasound, etc. (James and Dasarathy 2014). Every modality comes up with its own significance during its usage in the diagnosis process. Due to a single modality, these images are limited to certain diseases or issues. Therefore, multi-modality biomedical images are desirable. A multi-modality image can be obtained by using the efficient fusion approach (Daniel et al. 2017). Thus, these images have more information as compared to classical images and also more helpful to diagnose various kind of diseases (Du et al. 2016).
Recently, many researchers have designed and implemented fusion approaches to obtain efficient multi-modality biomedical images (Ravi and Krishnan 2018; Hu et al. 2020). However, many researchers have utilized the existing image fusion approaches. Therefore, the obtained multi-modality fused images may suffer from various issues such as gradient and texture distortion, especially for the infected region. To overcome the issues associated with the existing approaches, many researchers have utilized deep learning and dictionary learning kind of approaches. These are dictionary learning (Hu et al. 2020), local-features fuzzy sets (Ullah et al. 2020), deep learning (Algarni 2020; Xia et al. 2019; Zhou et al. 2019), deep learning and NSST (Wang et al. 2019), etc. that found to be the best tool to obtain efficient multi-modality fused images.
The design and development of an efficient multi-modality biomedical image fusion approach is still an open area for research. Deep learning is found to be one of the best fusion approaches to obtain promising results. Additionally, a deep transfer learning-based multi-modality biomedical fusion approach can provide better results.
The main contributions are summarized as:
A multi-objective differential evolution and Xception model based multi-modality biomedical fusion model is proposed.
The proposed approach, initially, decomposes the image into subbands using a non-subsampled contourlet transform (NSCT).
An extreme version of the Inception (Xception) is then used for feature extraction of the source images.
The multi-objective differential evolution is used to select the optimal features.
To obtain the fused coefficients, a coefficient of determination and the energy loss based fusion functions are used.
Finally, a fused image is computed by applying the inverse NSCT.
The proposed and the competitive approaches are compared by considering the benchmark multi-modality image fusion dataset.
The remaining structure of this paper is organized as: Existing literature is presented in Sect. 2. The proposed medical image fusion is illustrated in Sect. 3. Experimental results and comparative analyses are discussed in Sect. 4. Section 5 concludes the proposed work.
Literature review
Shu et al. (Zhu et al. 2019) implemented local laplacian energy and phase congruency based fusion approach in the NSCT domain (LEPN). Local laplacian energy utilized weighted local energy and sum of laplacian coefficients to obtain the regulated details and features of input images. Zhu et al. (2020) designed a diffusion-based approach by using the synchronized-anisotropic operators (DSA). A maximum absolute value constraint was also utilized for base layers fusion. The fusion decision map was computed by considering the sum of the modified anisotropic Laplacian approach by using the similar corrosion sub-bands obtained from the anisotropic diffusion. Kumar et al. (2020) proposed a co-learning based fusion maps for obtained more efficient multi-modality fused biomedical images. A convolutional neural network (CNN) was also used for the prediction and segmentation of potential objects.
Lu et al. (2014) designed an edge-guided dual-modality (EGDM) approach to obtain the multi-modality images. It performs significantly better even on highly under-sampled data. Lifeng et al. (2001) utilized wavelet and quality analysis to obtain the biomedical multi-modality fused images. The pyramid wavelet used for the fusion process. Ma et al. (2020) designed a dual-discriminator conditional generative adversarial network (DDcGAN) to obtain multi-modality fused images. It obtained a real-like fused image by using the content loss to dupe both discriminators. Two discriminators were also considered intention to differentiate the composition variations between the fused and source images, respectively.
Wang et al. (2019) developed a 3D auto-context-based locality adaptive multi-modality GANs (3GANs) to obtain more efficient multi-modality fused images. A non-unified kernel was also used along with the adaptive approach for multimodality fusion. Gai et al. (2019) utilized pulse coupled neural network (PCNN) by considering the edge preservation and enhanced sparse representation in the nonsubsampled shearlet transform (NSST). It completely utilized the features of various modalities that can handle edge details well and improves the results.
Liu et al. (2020) utilized a VGG16 based deep transfer learning approaches for image fusion to improve the classification process. VGG16 can obtain more efficient features as compared to the traditional deep learning approaches. The obtained features were then used to fuse the multi-modality biomedical images. Tavard et al. (2014) designed a multi-modal registration and fusion approach to improve the cardiac re-synchronization. The approach helps in improving therapy optimization.
Zhu et al. (2016) designed a novel dictionary learning-based image fusion approach for multi-modality biomedical images. Due to the use of dictionary learning, this approach achieves higher accuracy. But it is computationally complex in nature. Liu et al. (2018) proposed the biomedical image decomposition approach using NSST to fuse the multi-modality images. This approach has shown significant results over the existing approaches, but, suffer from the edge degradation issue. Wang et al. (2020) utilized CNN and contrast pyramid to fuse the biomedical multi-modality images. CNN can fuse the images efficiently. However, it is a computationally extensive approach, and some time also not provide promising results if the biomedical images are very similar to each other.
From the literature review, it has been observed that multi-modality image fusion is still an open area for research. The deep learning is found to be the best promising techniques to obtain better multi-modality fused biomedical images. But these approaches can provide better results if pre-trained deep transfer learning approaches are used. Additionally, the initial parameter selection of the deep learning and deep transfer learning approach is also a challenging issue (Pannu et al. 2019; Kaur and Singh 2019; Pannu et al. 2018). Therefore, in this paper, a well-known multi-objective differential evolution is used to enhance the results.
Proposed multi-objective differential evolution based deep transfer learning model for multi-modality image fusion
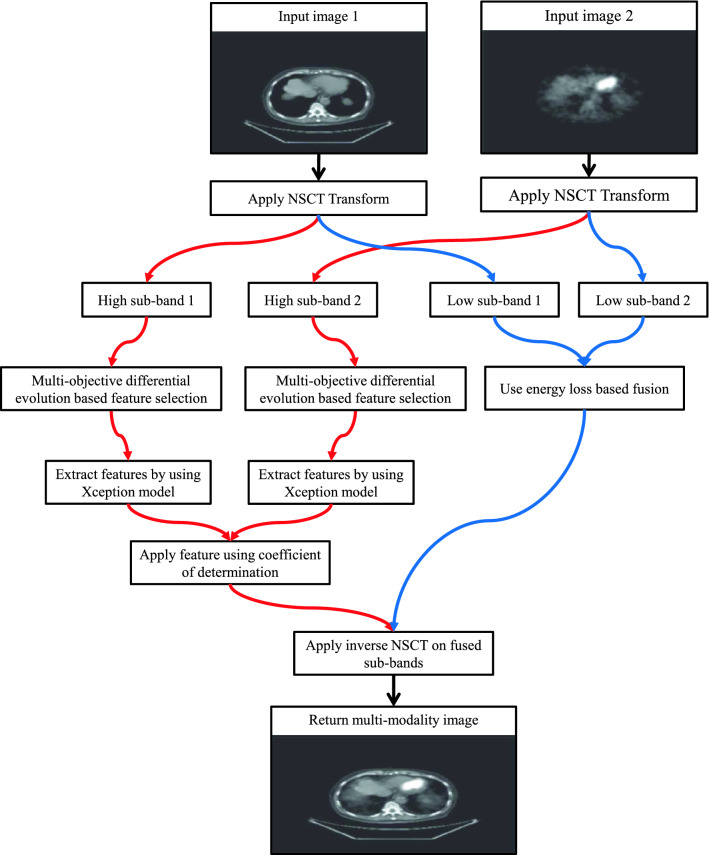
Assume that and show source images. Initially, NSCT is used to decompose and into sub-bands. Our primary objective is to fuse the respective sub-bands of both source images. The fusion of high sub-band is achieved by using the extreme version of the Inception (Xception) model. The coefficient of determination is utilized to evaluate the significance of the computed fused high sub-bands. Low sub-bands fusion is achieved by using the local energy function. Inverse NSCT is utilized to compute the multi-modality fused image. Figure 1 shows the step by step methodology of the proposed model.
Fig. 1.
Diagrammatic flow of the proposed multi-objective differential evolution based deep transfer learning model for multi-modality image fusion
Nonsubsampled contourlet transform
Nonsubsampled Contourlet Transform (NSCT) is a well-known transform used to decompose the images into the wavelet domain. It is a shift-invariant transform which can provide rich directional details. This directionality is effective to convert the transformed images to the actual one with minimum root mean square error (for more details please see Da Cunha et al. 2006).
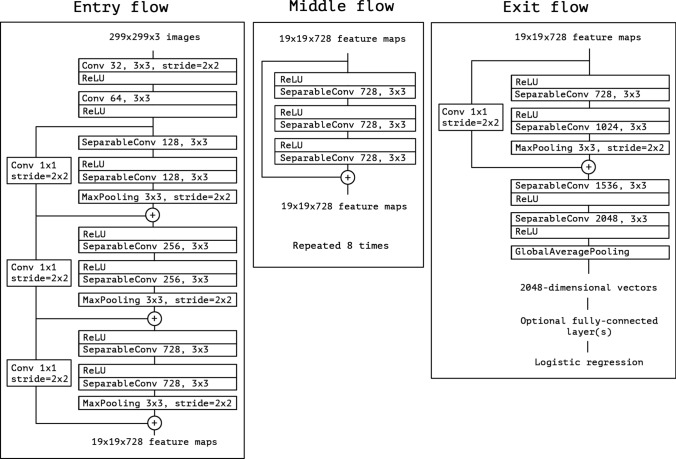
Feature extraction using deep Xception model
CNN may suffer from the under-fitting issue, as many potential features may not be extracted. To overcome this issue, an extreme version of the Inception (Xception) model is used. Figure 2 represents the block diagram of the xception model (for mathematical and other information please see Chollet 2017).
Fig. 2.
Architecture of Xception model (obtained from Chollet 2017)
Both high sub-bands of source images are placed in parallel fashion in the Xception model. Consider and are the obtained features from respective high sub-bands by using the Xception model.
Feature selection using multi-objective differential evolution
In this step, the optimal features are selected from the features obtained from the Xception model. The fusion factor and entropy metrics are used as the fitness function to select the optimal features. A multi-objective differential evolution can solve many computationally complex problems (Babu et al. 2005). It can significantly balance the fast convergence and population diversity. It can be described in the following steps:
I. Initialization: First of all, various parameters related to differential evolution are defined such as population size (), crossover rate (), mutation rate (), etc. Random distribution is used to generate the random solutions . h defines the number of function evaluations. It is used to control the iterative process of differential optimization with maximum function evaluations ().
II. Iterative step Mutation and crossover operators are used to obtain the optimal number of features.
Mutation is implemented on a to evaluate a child vector . In this paper, following mutation is used:
| 1 |
Here, shows index values. . , , and are random numbers selected from .
Crossover is used to obtain the news solutions. A child can be obtained from , as:
| 2 |
where D shows dimensions of the problem. and .
III. Selection: A child vector can be drawn by considering its parent vector as:
| 3 |
IV. Stopping condition: If number of functional evaluations are lesser than the total available evaluation then Steps II and III will be repeated.
Fusion of high sub-bands
The extracted and selected features using the Xception model from high sub-bands are then fused by using the coefficient of determination (R). R between and can be computed as:
| 4 |
Here, and shows the average of high sub-bands, respectively.
The dominated features are preserved in the obtained feature maps as:
| 5 |
Here, and show high sub-bands of and , respectively.
Fusion of low sub-bands
Motivated from (Hermessi et al. 2018), local energy is used to fuse the low sub-bands as:
| 6 |
Here, . represents neighbors of patch placed at (p, q). size of local patch is assigned as . Fused coefficients of low sub-bands can be computed as:
| 7 |
Experimental results
To evaluate the performance of the proposed approach, benchmark multi-modality biomedical images dataset is obtained from Ullah et al. (2020). Fifteen different pairs of modality images are taken for comparative purposes. The main goal is to fuse these images to obtain multi-modality fused images. To draw the comparisons, six competitive multi-modality biomedical fusion approaches such as LEPN (Zhu et al. 2019), DSA (Zhu et al. 2020), CNN (Kumar et al. 2020), EGDM (Lu et al. 2014), DDcGAN (Ma et al. 2020), and 3GANs (Wang et al. 2019) are also implemented on the same set of images. The hyper-parameters of these approaches are assigned as mentioned in their respective papers.
Visual analysis
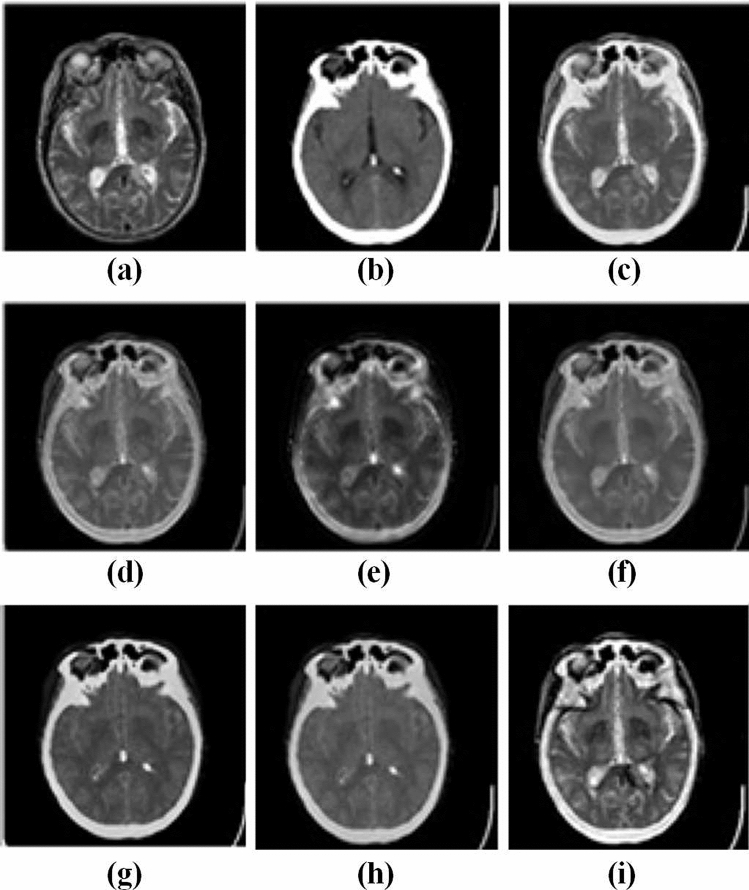
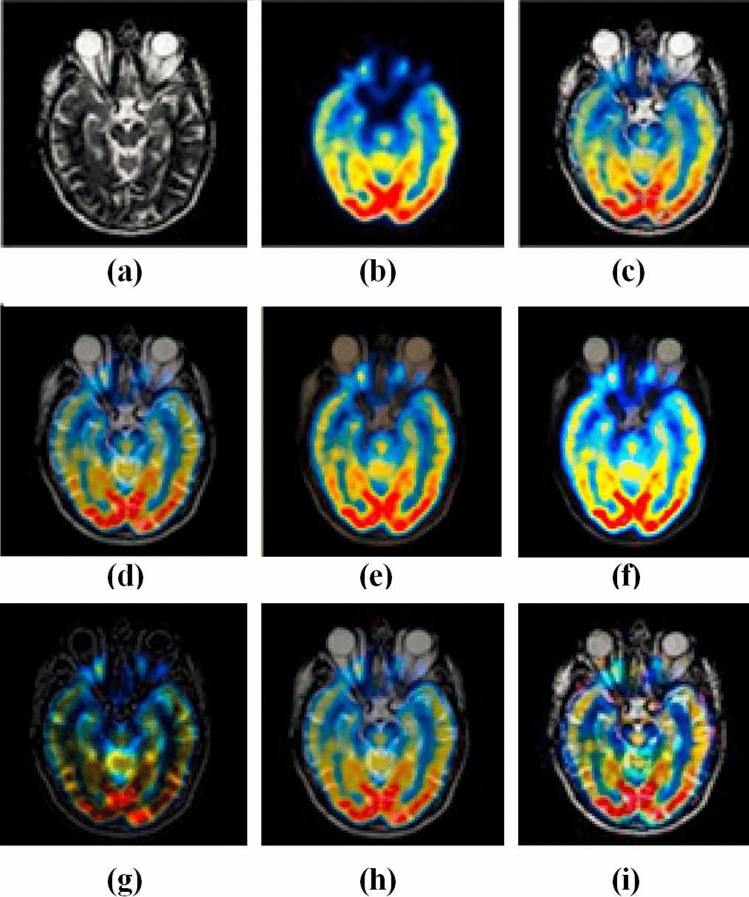
Figures 3 and 4 represent the source images and their respective multi-modality fused biomedical images obtained from the LEPN (Zhu et al. 2019), DSA (Zhu et al. 2020), CNN (Kumar et al. 2020), EGDM (Lu et al. 2014), DDcGAN (Ma et al. 2020), 3GANs (Wang et al. 2019) and the proposed approach. It is clearly shown that the obtained results have better modality as compared to the competitive approaches. Although, the existing approaches such as LEPN (Zhu et al. 2019), DSA (Zhu et al. 2020), CNN (Kumar et al. 2020), EGDM (Lu et al. 2014), DDcGAN (Ma et al. 2020), and 3GANs (Wang et al. 2019) provide significant visual results but have little edge and texture distortion. Figures 3i and 4j show the obtained results from the proposed approach. These images prove that the proposed approach provides a better visual appearance of the obtained multi-modality fused images.
Fig. 3.
Analysis of multi-modality biomedical fusion approaches: a MRI, b CT, c LEPN (Zhu et al. 2019), d DSA (Zhu et al. 2020), e CNN (Kumar et al. 2020), f EGDM (Lu et al. 2014), g DDcGAN (Ma et al. 2020), and h 3GANs (Wang et al. 2019) and i proposed approach
Fig. 4.
Analysis of multi-modality biomedical fusion approaches: a MRI, b CT, c LEPN (Zhu et al. 2019), d DSA (Zhu et al. 2020), e CNN (Kumar et al. 2020), f EGDM (Lu et al. 2014), g DDcGAN (Ma et al. 2020), and h 3GANs (Wang et al. 2019) and i proposed approach
Quantitative analysis
In this section, we have compared the proposed approach with the existing approaches such as LEPN (Zhu et al. 2019), DSA (Zhu et al. 2020), CNN (Kumar et al. 2020), EGDM (Lu et al. 2014), DDcGAN (Ma et al. 2020), and 3GANs (Wang et al. 2019), by considering the some well-known performance metrics. The selected performance measures are as edge strength, fusion symmetry, entropy, and fusion factor (for mathematical information see Prakash et al. 2019).
A multi-modality biomedical image fusion approach generally provides significant entropy values. Table 1 depicts entropy analysis of the proposed deep transfer learning-based multi-modality biomedical image fusion approach. It shows that the proposed approach provides significantly more entropy values than the existing multi-modality biomedical image fusion approaches. It is found that the proposed approach provides improvement over the best available approaches.
Table 1.
Comparative analysis among the proposed deep transfer learning based multi-modality image fusion and the competitive approaches in terms of entropy (maximum is better)
Mutual information represents the preserved details from the sourced image in the fused image. Therefore, it is desirable to be maximum. Table 2 depicts the mutual information analysis of the proposed approach over the competitive approaches. The proposed approach shows an average improvement of .
Table 2.
Comparative analysis among the proposed deep transfer learning based multi-modality image fusion and the competitive approaches in terms of mutual information (maximum is better)
The fusion factor is a well-known performance metric that shows the strength of the fusion process. It is desirable to be maximum. Table 3 shows the fusion factor analysis of the proposed and competitive approaches. The proposed approach shows an average improvement of over the competitive fusion models.
Table 3.
Comparative analysis among the proposed deep transfer learning based multi-modality image fusion and the competitive approaches in terms of fusion factor (maximum is better)
Fusion symmetry evaluates the symmetric details between source and fused images. It is desirable to be maximum. Table 4 shows fusion symmetry analysis of the proposed deep transfer learning model based multi-modality fusion model. It is found that the proposed model achieves an average improvement of over the competitive models.
Table 4.
Comparative analysis among the proposed deep transfer learning based multi-modality image fusion and the competitive approaches in terms of fusion symmetry (maximum is better)
Edge strength evaluates the degree of edge preservation and it is desirable to be maximum (Xydeas and Petrovic 2000). Table 5 shows the edge strength analysis of the proposed deep transfer learning-based multi-modality image fusion model. The proposed model achieves an average improvement of over the competitive approaches.
Table 5.
Comparative analysis among the proposed deep transfer learning based multi-modality image fusion and the competitive approaches in terms of edge strength (maximum is better)
Conclusion
From the literature review, it has been found that multi-modality image fusion is still an open area for research. The deep learning-based fusion approaches are found to be one of the best promising techniques to obtain better multi-modality fused biomedical images. However, these approaches are computationally complex in nature and also still suffer from the under-fitting issue. The proposed approach, initially, decomposes the image into sub-bands using a non-subsampled contourlet transform (NSCT). Thereafter, an extreme version of the Inception (Xception) has been used for feature extraction of the source images. The multi-objective differential evolution has been used to select the optimal features. Thereafter, to obtain the fused coefficients, a coefficient of determination and the energy loss functions are used. Finally, the fused image has been computed by applying the inverse NSCT. Extensive experimental results have shown that the proposed approach outperforms the competitive multi-modality image fusion approaches in terms of various performance metrics. In near future, one may use the proposed model for other applications such as remote sensing images (Singh et al. 2018; Singh and Kumar 2019a), medical images, etc. Additionally, the proposed hyper-parameters tuning approach can be used to tune the hyper parameter of the other approaches such as visibility restoration models (Osterland and Weber 2019; Singh and Kumar 2018, 2019b; Wang et al. 2019; Singh et al. 2019a, 2019b), filtering models (Gupta et al. 2019; Kaur et al. 2020; Wiens 2019), deep learning models (Jaiswal et al. 2020; Basavegowda and Dagnew 2020; Kaur et al. 2019, 2020; Ghosh et al. 2020), etc.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Algarni AD. Automated medical diagnosis system based on multi-modality image fusion and deep learning. Wirel Pers Commun. 2020;111:1033–1058. doi: 10.1007/s11277-019-06899-6. [DOI] [Google Scholar]
- Babu B, Chakole PG, Mubeen JS. Multiobjective differential evolution (mode) for optimization of adiabatic styrene reactor. Chem Eng Sci. 2005;60(17):4822–4837. doi: 10.1016/j.ces.2005.02.073. [DOI] [Google Scholar]
- Basavegowda HS, Dagnew G. Deep learning approach for microarray cancer data classification. CAAI Trans Intell Technol. 2020;5(1):22–33. doi: 10.1049/trit.2019.0028. [DOI] [Google Scholar]
- Chollet F (2017) Xception: deep learning with depthwise separable convolutions. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 1251–1258
- Da Cunha AL, Zhou J, Do MN. The nonsubsampled contourlet transform: theory, design, and applications. IEEE Trans Image Process. 2006;15(10):3089–3101. doi: 10.1109/TIP.2006.877507. [DOI] [PubMed] [Google Scholar]
- Daniel E, Anitha J, Kamaleshwaran K, Rani I. Optimum spectrum mask based medical image fusion using gray wolf optimization. Biomed Signal Process Control. 2017;34:36–43. doi: 10.1016/j.bspc.2017.01.003. [DOI] [Google Scholar]
- Du J, Li W, Lu K, Xiao B. An overview of multi-modal medical image fusion. Neurocomputing. 2016;215:3–20. doi: 10.1016/j.neucom.2015.07.160. [DOI] [Google Scholar]
- Gai D, Shen X, Cheng H, Chen H. Medical image fusion via pcnn based on edge preservation and improved sparse representation in nsst domain. IEEE Access. 2019;7:85413–85429. doi: 10.1109/ACCESS.2019.2925424. [DOI] [Google Scholar]
- Ghosh S, Shivakumara P, Roy P, Pal U, Lu T. Graphology based handwritten character analysis for human behaviour identification. CAAI Trans Intell Technol. 2020;5(1):55–65. doi: 10.1049/trit.2019.0051. [DOI] [Google Scholar]
- Gupta B, Tiwari M, Lamba SS. Visibility improvement and mass segmentation of mammogram images using quantile separated histogram equalisation with local contrast enhancement. CAAI Trans Intell Technol. 2019;4(2):73–79. doi: 10.1049/trit.2018.1006. [DOI] [Google Scholar]
- Hermessi H, Mourali O, Zagrouba E. Convolutional neural network-based multimodal image fusion via similarity learning in the shearlet domain. Neural Comput Appl. 2018;30(7):2029–2045. doi: 10.1007/s00521-018-3441-1. [DOI] [Google Scholar]
- Hu Q, Hu S, Zhang F. Multi-modality medical image fusion based on separable dictionary learning and gabor filtering. Signal Process Image Commun. 2020;83:115758. doi: 10.1016/j.image.2019.115758. [DOI] [Google Scholar]
- Jaiswal A, Gianchandani N, Singh D, Kumar V, Kaur M. Classification of the COVID-19 infected patients using DenseNet201 based deep transfer learning. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1788642. [DOI] [PubMed] [Google Scholar]
- James AP, Dasarathy BV. Medical image fusion: a survey of the state of the art. Inf Fusion. 2014;19:4–19. doi: 10.1016/j.inffus.2013.12.002. [DOI] [Google Scholar]
- Kaur M, Singh D. Fusion of medical images using deep belief networks. Clust Comput. 2019 doi: 10.1007/s10586-019-02999-x. [DOI] [Google Scholar]
- Kaur M, Gianey HK, Singh D, Sabharwal M. Multi-objective differential evolution based random forest for e-health applications. Mod Phys Lett B. 2019;33(05):1950022. doi: 10.1142/S0217984919500222. [DOI] [Google Scholar]
- Kaur M, Singh D, Kumar V, Sun K. Color image dehazing using gradient channel prior and guided l0 filter. Inf Sci. 2020;521:326–342. doi: 10.1016/j.ins.2020.02.048. [DOI] [Google Scholar]
- Kaur M, Singh D, Uppal R Singh. Parallel strength pareto evolutionary algorithm-II based image encryption. IET Image Process. 2020;14(6):1015–1026. doi: 10.1049/iet-ipr.2019.0587. [DOI] [Google Scholar]
- Kumar A, Fulham M, Feng D, Kim J. Co-learning feature fusion maps from PET-CT images of lung cancer. IEEE Trans Med Imaging. 2020;39(1):204–217. doi: 10.1109/TMI.2019.2923601. [DOI] [PubMed] [Google Scholar]
- Lifeng Y, Donglin Z, Weidong W, Shanglian B. Multi-modality medical image fusion based on wavelet analysis and quality evaluation. J Syst Eng Electron. 2001;12(1):42–48. [Google Scholar]
- Liu X, Mei W, Du H. Multi-modality medical image fusion based on image decomposition framework and nonsubsampled shearlet transform. Biomed Signal Process Control. 2018;40:343–350. doi: 10.1016/j.bspc.2017.10.001. [DOI] [Google Scholar]
- Liu Z, Wu J, Fu L, Majeed Y, Feng Y, Li R, Cui Y. Improved kiwifruit detection using pre-trained VGG16 with RGB and NIR information fusion. IEEE Access. 2020;8:2327–2336. doi: 10.1109/ACCESS.2019.2962513. [DOI] [Google Scholar]
- Lu Y, Zhao J, Wang G. Edge-guided dual-modality image reconstruction. IEEE Access. 2014;2:1359–1363. doi: 10.1109/ACCESS.2014.2371994. [DOI] [Google Scholar]
- Ma J, Xu H, Jiang J, Mei X, Zhang X. Ddcgan: a dual-discriminator conditional generative adversarial network for multi-resolution image fusion. IEEE Trans Image Process. 2020;29:4980–4995. doi: 10.1109/TIP.2020.2977573. [DOI] [PubMed] [Google Scholar]
- Osterland S, Weber J. Analytical analysis of single-stage pressure relief valves. Int J Hydromech. 2019;2(1):32–53. doi: 10.1504/IJHM.2019.098951. [DOI] [Google Scholar]
- Pannu HS, Singh D, Malhi AK. Improved particle swarm optimization based adaptive neuro-fuzzy inference system for benzene detection. CLEAN-Soil Air Water. 2018;46(5):1700162. doi: 10.1002/clen.201700162. [DOI] [Google Scholar]
- Pannu HS, Singh D, Malhi AK. Multi-objective particle swarm optimization-based adaptive neuro-fuzzy inference system for benzene monitoring. Neural Comput Appl. 2019;31:2195–2205. doi: 10.1007/s00521-017-3181-7. [DOI] [Google Scholar]
- Prakash O, Park CM, Khare A, Jeon M, Gwak J. Multiscale fusion of multimodal medical images using lifting scheme based biorthogonal wavelet transform. Optik. 2019;182:995–1014. doi: 10.1016/j.ijleo.2018.12.028. [DOI] [Google Scholar]
- Ravi P, Krishnan J (2018) Image enhancement with medical image fusion using multiresolution discrete cosine transform. In: Materials today: proceedings 5 (1, part 1) 1936 – 1942, international conference on processing of materials, minerals and energy (July 29th–30th) 2016, Ongole, Andhra Pradesh, India
- Singh D, Kumar V. Dehazing of outdoor images using notch based integral guided filter. Multimed Tools Appl. 2018;77(20):27363–27386. doi: 10.1007/s11042-018-5924-6. [DOI] [Google Scholar]
- Singh D, Kumar V. A comprehensive review of computational dehazing techniques. Arch Comput Methods Eng. 2019;26(5):1395–1413. doi: 10.1007/s11831-018-9294-z. [DOI] [Google Scholar]
- Singh D, Kumar V. Single image defogging by gain gradient image filter. Sci China Inf Sci. 2019;62(7):79101. doi: 10.1007/s11432-017-9433-4. [DOI] [Google Scholar]
- Singh D, Kaur M, Singh H. Remote sensing image fusion using fuzzy logic and gyrator transform. Remote Sens Lett. 2018;9(10):942–951. doi: 10.1080/2150704X.2018.1500044. [DOI] [Google Scholar]
- Singh D, Kumar V, Kaur M. Single image dehazing using gradient channel prior. Appl Intell. 2019;49(12):4276–4293. doi: 10.1007/s10489-019-01504-6. [DOI] [Google Scholar]
- Singh D, Kumar V, Kaur M. Image dehazing using window-based integrated means filter. Multimed Tools Appl. 2019 doi: 10.1007/s11042-019-08286-6. [DOI] [Google Scholar]
- Tavard F, Simon A, Leclercq C, Donal E, Hernández AI, Garreau M. Multimodal registration and data fusion for cardiac resynchronization therapy optimization. IEEE Trans Med Imaging. 2014;33(6):1363–1372. doi: 10.1109/TMI.2014.2311694. [DOI] [PubMed] [Google Scholar]
- Ullah H, Ullah B, Wu L, Abdalla FY, Ren G, Zhao Y. Multi-modality medical images fusion based on local-features fuzzy sets and novel sum-modified-laplacian in non-subsampled shearlet transform domain. Biomed Signal Process Control. 2020;57:101724. doi: 10.1016/j.bspc.2019.101724. [DOI] [Google Scholar]
- Wang C, Zhao Z, Ren Q, Xu Y, Yu Y. Multi-modality anatomical and functional medical image fusion based on simplified-spatial frequency-pulse coupled neural networks and region energy-weighted average strategy in non-sub sampled contourlet transform domain. J Med Imaging Health Inform. 2019;9(5):1017–1027. doi: 10.1166/jmihi.2019.2669. [DOI] [Google Scholar]
- Wang Y, Zhou L, Yu B, Wang L, Zu C, Lalush DS, Lin W, Wu X, Zhou J, Shen D. 3d auto-context-based locality adaptive multi-modality gans for pet synthesis. IEEE Trans Med Imaging. 2019;38(6):1328–1339. doi: 10.1109/TMI.2018.2884053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Yu H, Wang G, Zhang G, Wang W. Study on the dynamic and static characteristics of gas static thrust bearing with micro-hole restrictors. Int J Hydromech. 2019;2(3):189–202. doi: 10.1504/IJHM.2019.102893. [DOI] [Google Scholar]
- Wang K, Zheng M, Wei H, Qi G, Li Y. Multi-modality medical image fusion using convolutional neural network and contrast pyramid. Sensors. 2020;20(8):2169. doi: 10.3390/s20082169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiens T. Engine speed reduction for hydraulic machinery using predictive algorithms. Int J Hydromech. 2019;2(1):16–31. doi: 10.1504/IJHM.2019.098949. [DOI] [Google Scholar]
- Xia K-J, Yin H-S, Wang J-Q. A novel improved deep convolutional neural network model for medical image fusion. Clust Comput. 2019;22(1):1515–1527. doi: 10.1007/s10586-018-2026-1. [DOI] [Google Scholar]
- Xydeas C, Petrovic V. Objective image fusion performance measure. Electron Lett. 2000;36(4):308–309. doi: 10.1049/el:20000267. [DOI] [Google Scholar]
- Zhou T, Ruan S, Canu S. A review: deep learning for medical image segmentation using multi-modality fusion. Array. 2019;3:100004. doi: 10.1016/j.array.2019.100004. [DOI] [Google Scholar]
- Zhu Z, Chai Y, Yin H, Li Y, Liu Z. A novel dictionary learning approach for multi-modality medical image fusion. Neurocomputing. 2016;214:471–482. doi: 10.1016/j.neucom.2016.06.036. [DOI] [Google Scholar]
- Zhu Z, Zheng M, Qi G, Wang D, Xiang Y. A phase congruency and local laplacian energy based multi-modality medical image fusion method in NSCT domain. IEEE Access. 2019;7:20811–20824. doi: 10.1109/ACCESS.2019.2898111. [DOI] [Google Scholar]
- Zhu R, Li X, Zhang X, Ma M. Mri and ct medical image fusion based on synchronized-anisotropic diffusion model. IEEE Access. 2020;8:91336–91350. doi: 10.1109/ACCESS.2020.2993493. [DOI] [Google Scholar]