Abstract
Sclerotinia head rot (SHR), caused by the necrotrophic fungus Sclerotinia sclerotiorum, is one of the most devastating sunflower crop diseases. Despite its worldwide occurrence, the genetic determinants of plant resistance are still largely unknown. Here, we investigated the Sclerotinia-sunflower pathosystem by analysing temporal changes in gene expression in one susceptible and two tolerant inbred lines (IL) inoculated with the pathogen under field conditions. Differential expression analysis showed little overlapping among ILs, suggesting genotype-specific control of cell defense responses possibly related to differences in disease resistance strategies. Functional enrichment assessments yielded a similar pattern. However, all three ILs altered the expression of genes involved in the cellular redox state and cell wall remodeling, in agreement with current knowledge about the initiation of plant immune responses. Remarkably, the over-representation of long non-coding RNAs (lncRNA) was another common feature among ILs. Our findings highlight the diversity of transcriptional responses to SHR within sunflower breeding lines and provide evidence of lncRNAs playing a significant role at early stages of defense.
Subject terms: Genetics, Plant sciences
Introduction
Sunflower is one of the most important crops for the production of high-quality oil and seeds consumed by both humans and livestock. In recent years, sunflower production showed a steady increase driven by a boost in sunflower oil consumption (FAO, 2017). However, the projected expansion of the sunflower oil market requires appropriate agronomic management and improved genetic resources to cope with abiotic and biotic stresses. Among the latter, special attention should be paid to fungal diseases, as they have the greatest impact on yield and seed quality1.
The necrotrophic fungus Sclerotinia sclerotiorum is the causal agent of Sclerotinia head (SHR) and stalk (SSR) rots in sunflower. In particular, SHR is a recurrent disease in sunflower-growing areas worldwide. It affects oil quality and, under favourable conditions, may lead to total production loss1,2. Chemical fungicides proved to be ineffective and breeding of resistant genotypes has emerged as the most promising control strategy3. So far, there is no evidence of any major gene controlling the resistance to SHR in sunflower. Instead, inbred lines (ILs) show a broad range of responses in accordance with quantitative disease resistance (QDR) patterns depending on the genotype4–8. During the last 20 years, QTL mapping techniques have been used to unravel the complexity of the defense response to both SHR and SSR in sunflower. Biparental mapping has led to the discovery of several main effect loci and epistatic interactions9–11, whereas association mapping has served to identify candidate genes responsible for S. sclerotiorum resistance12–14. Notwithstanding this, little is known about the genetic architecture of quantitative resistance and the functional components of the defense response.
The fungus penetrates the host cuticle through mechanical means and by secreting an arsenal of cell wall-degrading enzymes, small proteins and secondary metabolite toxins, with oxalic acid being the major virulence factor. Diseased plants develop water-soaked lesions, tissue necrosis and finally bear sclerotia15,16, and in response to damage they can activate an array of perception mechanisms and signal transduction pathways that trigger QDR17. The polyphagous nature of S. sclerotiorum, which has been reported to infect over 400 plant species18, has allowed the identification of pathogen-responsive genes in a variety of model and economically important species. In line with current knowledge of the molecular mechanisms underlying plant immune response, transcriptomic studies in Brassica napus, Arabidopsis thaliana and Glycine max have shown that defense against S. sclerotiorum involves members of the WRKY transcription factor family, pathogenesis related (PR) proteins, as well as genes related to signal transduction, cellular redox state, cell wall composition and hormone signaling pathways19–24. However, there are no reports on the transcriptional response of sunflower to SHR.
Although SHR consistently causes water-soaked lesions and necrosis, the histopathology and time from infection to symptom onset differ among species16. In particular, sunflowers show no visible SHR symptoms for at least ten days after inoculation6, but microscopic lesions have been detected after a 24-h incubation period in susceptible and tolerant varieties, with the latter showing a slower progression of the disease25.
Transcriptional profiling at the site of infection provides an efficient means to reflect the natural epidemiologic cycle and constitutes an appealing alternative to discriminate among responses of genotypes with contrasting behaviour against the disease. RNA-sequencing (RNA-seq) has proven to be a powerful method to detect, map and quantify transcripts in several plant-pathogen interactions, regardless of the tissue under analysis or previous genomic knowledge26. A few studies conducted in cultivated sunflower have exploited the advantages and sensitivity of this methodology to identify differentially-expressed genes under biotic or abiotic stress conditions27,28, and none of them addressed the S. sclerotiorum-sunflower interaction. Indeed, only two low-throughput transcriptomic studies have focused on this pathosystem29,30. In addition, most transcriptomic analyses of Sclerotinia infection were made with data obtained from symptomatic plants20–22. However, transcriptional changes taking place during the asymptomatic period can provide insights into initial defense mechanisms and into the diversity of responses to the disease in different crops.
The aim of this study was to investigate the transcriptional response of sunflower during early stages of SHR. To this end, control and inoculated capitula of one susceptible and two tolerant ILs were subjected to RNA-seq at 0, 4 and 8 days post-inoculation (dpi) to analyze their expression patterns and gain a comprehensive view of the mechanisms involved in the defense against S. sclerotiorum infection.
Results
Disease assessment
ILs HA89, HA853 and RK416 were grown under field conditions during season 2010–11 at the experimental station INTA Balcarce (Buenos Aires, Argentina). After sampling for the RNA-seq experiments, additional plants of the different ILs were maintained in the field to evaluate disease evolution and confirm the efficacy of inoculation. Commercial hybrids ACA861 and Dekalb 3820 were used as susceptible and tolerant controls, respectively. The distinct symptoms of SHR were observed in all cases. The three ILs and the hybrids showed an increase in infection levels over time, as evidenced by both disease incidence (DI) and disease severity (DS) scores. DI values exhibited a fast increment in HA89 and ACA 861 and a slower escalation in HA853 and Dekalb 3820, whereas RK416 showed an intermediate behaviour. DS estimates revealed a rapid spread of the fungus in ACA 861, a slower advance in HA89, while RK416, HA853 and Dekalb 3820 showed a delayed expression of symptoms and did not reach the DS values of the first two cultivars (Supplementary Fig. S1).
Overview of RNA-seq analysis
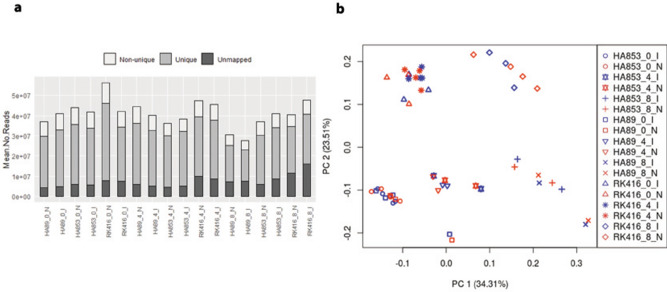
Between 14.1 and 88.04 million reads per sample were mapped to the HanXRQ v1 sunflower genome (mean 42.18 ± 12.62 million reads). An average of 64.18 ± 5.91% and 17.62 ± 1.84% of reads were mapped uniquely and non-uniquely to the reference, respectively (Fig. 1a), whereas 18.19 ± 7.33% of the total reads could not be mapped and were excluded from the analysis (Supplementary Table S1).
Figure 1.
Overview of RNA-seq data. (a) Mean of Unique, Non-unique and Unmapped reads for each treatment and IL-time point combination. (b) Principal component analysis of 53 samples based on the expression levels of 31,673 transcripts.
Transcript expression levels were established at 0, 4 and 8 dpi in inoculated (I) and control (N) capitula of sunflower ILs HA89, HA853 and RK416 (Supplementary Table S2). Transcripts with more than 1 CPM in half of the samples were considered for further analysis, encompassing 31,673 of the 58,050 inferred genes annotated in the HanXRQ v1 sunflower genome31. Of these, 29,329 correspond to protein-coding genes (mRNAs) and 2,344 correspond to non-coding RNAs (ncRNAs).
Overall similarities between ILs and time point conditions were explored using PCA. Three samples from RK416 (4_18_RK416_0_I, 1_5_RK416_0_N and 2_11_RK416_8_I) were identified as outliers and thus removed from the analysis. In the final PCA, the first two principal components accounted for 57.82% of the total variation and grouped samples mainly by IL, separating RK416 samples from those of HA89 and HA853 (Fig. 1b). Within lines, samples were clustered according to the plant developmental stage (0, 4 and 8 dpi), irrespective of the inoculation condition, showing the greatest differences between 8 dpi and the previous time points. These results reveal large transcriptomic differences between RK416 and the other two ILs and imply an increasing time effect, consistent with the progression of disease symptoms.
Analysis of differentially expressed genes
The expression levels of the 31,673 transcripts were estimated for each inoculation treatment and IL-time point combination. To validate these results, we assessed the correlation between RNA-seq transcription ratios (I/N) and those obtained in qPCR assays at 4 and 8 dpi. Between six and twelve genes were compared for each IL and time point. Only three genes were available for comparison in HA853 at 4 dpi, and therefore it was excluded from the analysis. In general, expression results from both methods were highly correlated, with coefficients ranging from 0.90 to 0.98 (p < 0.05). The only exception was HA89 at 8 dpi, which showed a weak and non-significant association (Supplementary Fig. S2).
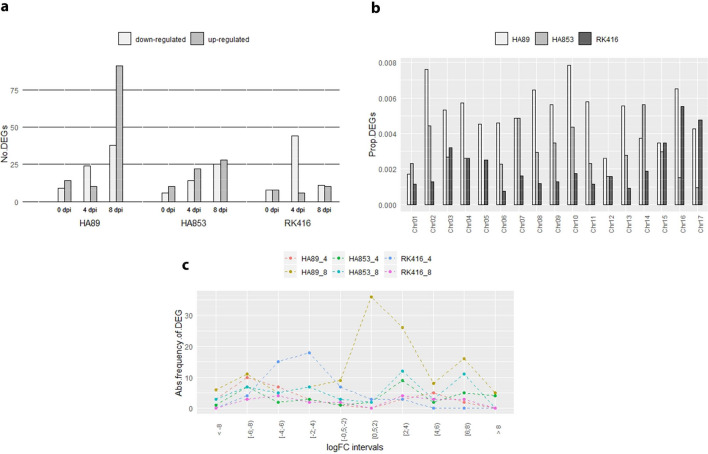
To minimise false positives due to multiple testing, only those genes with FDR < 0.05 were assessed and categorised as differentially expressed (DEGs) at each IL-time point combination. Differences at 0 dpi were also identified to quantify the level of noise generated from sources other than inoculation. A total of 378 DEGs were found, of which 186, 105 and 87 corresponded to HA89, HA853 and RK416, respectively (Supplementary Table S3). The following distribution of DEGs was obtained at 0, 4 and 8 dpi: 23, 34 and 129 for HA89, 16, 36 and 53 for HA853 and 16, 50 and 21 for RK416 (Fig. 2a). As expected, the number of DEGs was larger at 4 and 8 dpi than at 0 dpi. Most IL-time point combinations had similar numbers of up- and down-regulated DEGs. Notably, HA89 showed the largest proportion of up-regulated DEGs at 8 dpi and RK416 the largest proportion of down-regulated ones at 4 dpi.
Figure 2.
Analysis of DEGs. (a) Number of up- and down-regulated DEGs between I and N samples. (b) Proportion of DEGs relative to all expressed transcripts in each IL distributed across chromosomes. (c) Absolute Frequency of DEGs at different intervals of logFC values for each IL-time point combination (0 dpi not shown).
The analysis of the distribution of DEGs across chromosomes indicated that all ILs presented DEGs in the 17 chromosomes, except for HA853 with no representation in chromosome 5. Chromosomes 2 and 10 had the highest number of DEGs, while chromosomes 1 and 12 had the lowest. Since HA89 encompassed the largest number of DEGs, these were the most abundant transcripts in 13 of the 17 chromosomes. Conversely, HA853 and RK416 showed a comparable amount of DEGs across the genome, except for chromosomes 16 and 17 where RK416 DEGs were more abundant (Fig. 2b).
In addition, to uncover distinct patterns of expression in the different IL-time point combinations, the distribution of the log2 fold change (logFC) for down- and up-regulated genes was analysed at 4 and 8 dpi (Fig. 2c). Overall values varied from |0.73| to |10.19|, irrespective of gene regulation. Inspection of Fig. 2c shows a large number of values within the lower intervals for HA89 at 8 dpi and more homogeneous patterns for the remaining IL-time point combinations.
The analysis of the DEG intersections among IL-time point combinations showed that there were only four DEGs in common between HA89 and HA853, albeit with opposite expression behaviour. Genes HanXRQChr04g0102281 and HanXRQChr13g0407331 were down-regulated in the susceptible line and up-regulated in the tolerant HA853 line. Conversely, genes HanXRQChr03g0066301 and HanXRQChr10g0282011 were down-regulated in HA853 and up-regulated in HA89. RK416 shared no DEGs with the other two lines. In addition, the intersection between time points within ILs included two DEGs for HA89, four for HA853 and none for RK416. A summary of these results is shown in Table 1.
Table 1.
Functional description and logFC values of the DEGs shared by the different IL-time point combinations and/or time points within ILs.
| Gene ID | Functional description | HA89_4 | HA89_8 | HA853_4 | HA853_8 |
|---|---|---|---|---|---|
| HanXRQChr04g0102281 | Spliced ncRNA | – | − 7.45 | – | 2.97 |
| HanXRQChr03g0066301 | Transcription factor IBH1 | – | 2.35 | – | − 1.69 |
| HanXRQChr10g0282011 | NA | – | 7.38 | – | − 6.79 |
| HanXRQChr13g0407331 | NA | – | − 1.42 | 7.55 | – |
| HanXRQChr11g0350011 | Retrovirus-related Pol poly from transposon TNT 1–94 | − 6.91 | 7.89 | – | – |
| HanXRQChr13g0418051 | Mitochondrial import inner membrane translocase subunit TIM14-1 | − 7.66 | − 6.91 | – | – |
| HanXRQChr10g0287891 | NA | – | – | − 3.55 | 3.02 |
| HanXRQChr14g0449141 | Spliced ncRNA | – | – | 6.37 | − 6.46 |
| HanXRQChr14g0435741 | NA | – | – | − 7.00 | − 6.27 |
| HanXRQChr11g0331821 | Alcohol dehydrogenase | – | – | 9.32 | − 9.64 |
Altogether, the ensemble of genes responding to the infection was IL-specific, suggesting the coordination of idiosyncratic reactions at early stages of the infection process.
To determine if DEGs were located close to previously reported QTLs11,12, and to reinforce the results obtained by both methodologies, we searched for DEGs in the vicinity of the molecular markers (MM) associated with SHR tolerance (Supplementary Table S4). Examination of these regions allowed the detection of five DEGs within the established 1 Mbp windows (Table 2). In detail, genes HanXRQChr03g0080031 and HanXRQChr03g0080021 were close to each other and within the range of MM HeAn_R_283.1, gene HanXRQChr10g0296411 was the closest to MM ORS437, gene HanXRQChr14g0449141 was in the vicinity of MM G34, while gene HanXRQChr14g0460321 was found near MM HaCOI1-1.
Table 2.
DEGs located in the vicinity of QTLs associated with SHR resistance by biparental (BM) or association (AM) mapping.
| DEG | IL-time point | Chr | Molecular marker (MM) | QTL source | Distance to MM (bp) | Functional description of DEG |
|---|---|---|---|---|---|---|
| HanXRQChr03g0080031 | HA89_4 | 3 | HeAn_R_283.1 | BM | 106,548 | NA |
| HanXRQChr03g0080021 | HA89_4 | 3 | HeAn_R_283.1 | BM | 108,115 | ncRNA |
| HanXRQChr10g0296411 | HA853_8 | 10 | ORS437 | BM | 7,475 | DEAD-box ATP-dependent RNA helicase 40 |
| HanXRQChr14g0449141 | HA853_4-8 | 14 | G34 | AM | 469,108 | ncRNA |
| HanXRQChr14g0460321 | HA89_8 | 14 | HaCOI | AM | 615,017 | Hypoxia-responsive family |
Functional analysis
The functional annotation of DEGs was compiled from the HanXRQ v1 sunflower transcriptome31 (Supplementary Table S5). Gene Ontology (GO) Enrichment Analysis showed that no GO terms were significantly over-represented when strictly considering the DEG list. Despite the lack of enriched GO terms among DEGs in the different IL-time point combinations, at least one gene from each group was associated with defense processes. These include glutathione S-transferase DHAR3, chloroplastic-like (HanXRQChr13g0424161, HA89_4 dpi), probable leucine rich repeat (LRR) receptor-like serine threonine-kinase At1g07650 (HanXRQChr04g0123251, HA89_8 dpi), resistance RGC2, partial (HanXRQChr02g0057101, HA853_4 dpi), S-norcoclaurine synthase-like (HanXRQChr13g0397881, HA853_8 dpi), pathogenesis-related 1 (HanXRQChr04g0109991, RK416_4 dpi) and major allergen Pru ar 1-like (HanXRQChr03g0090261, RK416_8 dpi), among others32–37. Additionally, the MapMan ontology was used to organise the DEGs into main functional categories and relate them to possible processes occurring during the defense response to the infection (Supplementary Table S6 and Supplementary Fig. S3). Of the 34 specific functional categories (BINs) of the ontology, 23 were represented by at least one DEG. However, most DEGs fell within the “not assigned” category (BIN 35). The remaining DEGs were most frequently related to “protein” (BIN 29), “RNA” (BIN 27), “signalling” (BIN 30) and “transport” (BIN 34), in which the most common processes altered were protein degradation, regulation of transcription, as well as the transcriptional variation of receptor kinases. Relevant categories for the defense response included “cell wall” (BIN 10), “hormone metabolism” (BIN 17) and “stress” (BIN 20), which were represented in all ILs. Overall, our results revealed that although there was modulation of similar processes among ILs, they were mediated by different biological molecules.
To extend the exploration of enriched GO terms, a logistic regression was applied on the whole gene set, indexed according to logFC values and the adjusted p-value. This Gene Set Enrichment Analysis (GSEA) approach yielded a total of 1,128 over-represented GO terms (biological process (BP): 704; cellular component (CC): 227; molecular function (MF): 197) (Supplementary Table S7). RK416 presented the largest number of enriched GO terms, followed by HA853 and HA89. The latter two lines showed only a few enriched GO terms at 4 dpi (55 and 26, respectively), but these increased three to ten times at 8 dpi. In comparison, RK416 maintained a similar number of enriched GO terms at both time points.
There was little overlapping of enriched GO terms among IL-time point combinations. Only 15% of GO terms were shared by at least half of the combinations, and just a few were shared by five or six of them (i.e., photosynthesis, BP; photosynthesis, light reaction, BP; ribosome biogenesis, BP; cytosolic part, CC; ribosomal subunit, CC). After checking for redundancy, 699 unique GO terms were identified (BP: 455; CC: 114; MF: 130).
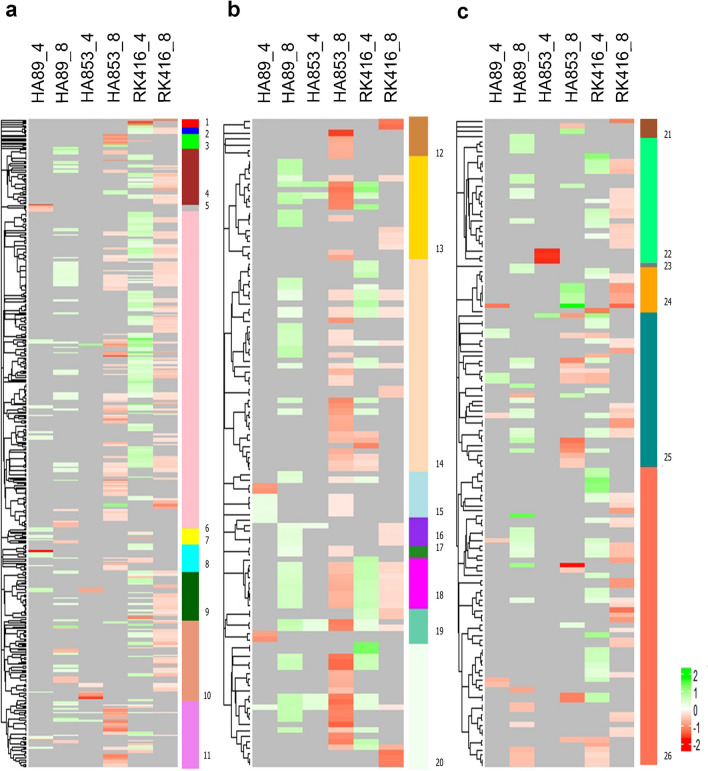
Figure 3 shows the dendrogram derived from the GO term similarity matrix obtained with GoSemSim, with the corresponding LOR plotted as a heatmap for each IL-time point combination. The LORs of the different GO terms were assigned by Multidimensional Gene Set Analysis38 based on the expression levels of the genes included within each category. It is noteworthy that the functional categories over-represented in HA89 at 8 dpi and RK416 at 4 dpi were mainly derived from up-regulated genes, while HA853 and RK416 at 8 dpi showed the opposite pattern.
Figure 3.
Heatmaps based on the LOR values of GO terms grouped by semantic similarities. Functional terms over-represented in up-regulated genes (green) are differentiated from those over-represented in down-regulated genes (red) upon inoculation. (a) BP ontology, (b) CC ontology and (c) MF ontology. Vertical coloured bars represent functional modules in which GO terms were categorised. 1: miscellaneous processes; 2: immune system process; 3: miscellaneous organisation; 4: transport and localisation; 5: reproductive development; 6: metabolic process; 7: developmental process; 8: cell integrity; 9: biological regulation; 10: response to stimuli; 11: cellular organisation and biogenesis; 12: miscellaneous CC ; 13: membrane and membrane-related complexes; 14: organelles; 15: extracellular region and cell junction; 16: Golgi and ER; 17: encapsulating structure; 18: thylakoid; 19: lytic organelles; 20: protein-containing complexes; 21: miscellaneous MF; 22: transporter activity; 23: antioxidant activity; 24: molecular function regulation; 25: binding; 26: catalytic activity.
The classification of GO terms according to their GoSemSim similarities allowed the delimitation of functional modules (Fig. 3, Supplementary Table S8). In this way, apparently unconnected GO terms were grouped into more general processes, thus becoming comparable between the different IL-time point combinations. In regard to BP, “metabolic processes” was the main functional module, i.e. the one including the largest number of related GO terms, and, to a lesser extent, “response to stimuli” and “cellular organisation and biogenesis” were also frequently represented. In the CC category, the main functional module was “organelles”, closely followed by “protein-containing complexes” and “membrane and membrane-related complexes”. As for MF, the main functional module was “catalytic activity”. Secondary modules included “binding” and “transporter activity”. In general, functional modules consisted of GO terms from all the IL-time point combinations, indicating a modulation of similar general processes but with different actors being involved in each condition.
A gene set enrichment analysis based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database revealed two enzyme KEGG identifiers (EC) over-represented in HA89 at 8 dpi, both belonging to Class “Oxidoreductases”. One EC from the class “Hydrolases” was identified in HA853 at 8 dpi, whereas eight EC were identified in RK416 at 4 dpi, half of them being “Transferases” and the other half “Hydrolases”. Finally, one EC classified within “Oxidoreductases” was identified in RK416 at 8 dpi (Supplementary Table S9). Although all of them can be related to defense processes, the enzymatic reactions identified in this analysis were not completely concordant among ILs.
Study of differentially expressed ncRNAs in response to S. sclerotiorum infection
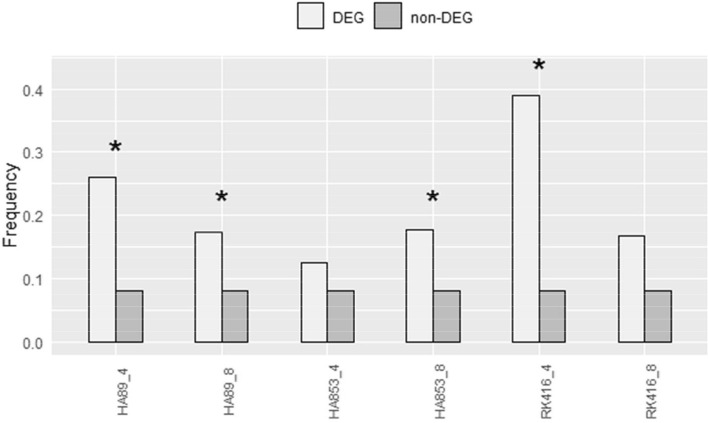
Taking into account that ncRNA genes are not part of GO terms, but that they were frequently represented in the IL-time point combinations, their enrichment in the different groups was examined through Irwin-Fisher tests (Supplementary Table S10). The proportion of differentially expressed ncRNAs compared to that of the remaining ncRNAs was significantly higher in HA89 at both time points, in HA853 at 8 dpi and in RK416 at 4 dpi (p-value < 0.05) (Fig. 4). According to the HanXRQ v1 annotation, all ncRNAs corresponded to the splice ncRNA category. They were considered as long ncRNAs (lncRNAs) based on the length of the identified transcripts (> 200 bp).
Figure 4.
Analysis of ncRNA enrichment for the different treatments; *: p < 0.05.
Because lncRNAs are involved in transcriptional and post-transcriptional regulation of protein-coding RNAs, we used the LncTar algorithm to predict putative mRNA targets among DEGs. This tool computes the normalised binding free energy (ndG) between molecules, thereby estimating a putative interaction. Here, differentially expressed protein-coding genes and lncRNAs of the same IL at 4 and 8 dpi were used as input to predict their binding capability. We identified one possible mRNA-lncRNA interaction for RK416, nine for HA89 and none for HA853 (Table 3). Of these, six pairs were differentially expressed at different time points and four at 8 dpi. Additionally, only four pairs showed the same expression behaviour.
Table 3.
Putative lncRNA-mRNA interactions within DEGs.
| Inbred line | lncRNA | mRNA/target | ndG | Functional description of mRNA | ||||
|---|---|---|---|---|---|---|---|---|
| Gene ID | dpi | Fold-change | Gene ID | dpi | Fold-change | |||
| RK416 | HanXRQChr01g0005231 | 8 | − 2.68 | HanXRQChr14g0427881 | 4 | − 4.46 | − 0.4193 | NA |
| HA89 | HanXRQChr08g0221721 | 4 | 2.44 | HanXRQChr02g0038901 | 8 | − 4.74 | − 0.7460 | NA |
| HA89 | HanXRQChr08g0226611 | 4 | − 8.97 | HanXRQChr08g0235441 | 8 | − 6.13 | − 0.2542 | NA |
| HA89 | HanXRQChr08g0229491 | 4 | 5.07 | HanXRQChr02g0038901 | 8 | − 4.74 | − 0.5924 | NA |
| HA89 | HanXRQChr16g0506901 | 4 | 4.72 | HanXRQChr17g0559391 | 8 | − 6.99 | − 0.2190 | Quinone oxidoreductase 2 homolog |
| HA89 | HanXRQChr02g0033591 | 8 | − 2.00 | HanXRQChr02g0046121 | 4 | − 1.58 | − 0.6558 | NA |
| HA89 | HanXRQChr08g0226611 | 8 | − 8.97 | HanXRQChr10g0314321 | 8 | 3.06 | − 0.1667 | E3 UFM1- ligase 1 homolog |
| HA89 | HanXRQChr13g0406311 | 8 | 3.64 | HanXRQChr02g0041971 | 8 | − 7.42 | − 0.1620 | Mitogen-activated kinase 16, partial |
| HA89 | HanXRQChr13g0406311 | 8 | 3.64 | HanXRQChr02g0046121 | 8 | − 1.58 | − 0.2095 | NA |
| HA89 | HanXRQChr13g0406311 | 8 | 3.64 | HanXRQChr10g0308951 | 8 | 2.28 | − 0.1504 | PREDICTED: uncharacterised protein LOC103323638 |
ndG normalised free energy, NA not available.
Although such interactions need further experimental confirmation, these results provide a first approach to assess the involvement of lncRNAs in sunflower defense processes.
Discussion
The identification of the genes underlying quantitatively inherited traits is one of the most challenging tasks for molecular breeders. Nonetheless, gene pyramiding strategies are considered the most effective way to achieve durable resistance39–41. Large-scale transcriptional studies are a fruitful source of novel candidate genes, as well as a means of validating previous results obtained by other approaches. By conducting a genome-wide transcriptomic analysis of one susceptible and two tolerant sunflower ILs to SHR, we were able to uncover new molecular determinants of defense under conditions that mimic the natural infection process.
In this study, we established the expression profiles of more than half of the annotated genes in the sunflower genome across ILs, inoculation treatments and time points. However, the number of DEGs identified here was relatively low compared to those generally reported in RNA-seq analyses conducted for other pathosystems (e.g. Gao et al., Kamber et al., Song et al., Wu et al.42–45). At early stages, SHR is restricted to a small portion of the inflorescence. Therefore, RNA extraction from the whole set of florets in each capitulum may have diluted the biological signal, limiting our ability to detect the full range of transcriptional changes triggered by the pathogen. Although our approach may have prevented the detection of many of the small expression changes expected at the onset of infection, when no symptoms are visible46,47, it is likely to have detected more reliable and stronger expression variations. Indeed, several DEGs were readily identified at 4 dpi in all ILs, indicating a rapid activation and amplification of plant defense responses.
A comparison of gene activity elicited upon infection revealed limited commonalities among the three ILs and between time points within ILs. HA89 and HA853 were the only ILs that shared DEGs and the fact that these had opposite expression suggests a converging pathway leading to different responses. Of the four genes, one is described as a transcription factor (IBH1) involved in the modulation between growth and immunity in A. thaliana48, another one is inferred to be a ncRNA, while the remaining two have no known function. The singular behaviour of these genes may be indicative of their relevance in the defense pathway, making them promising candidates for improving resistance24,49. On the other hand, although HA853 and RK416 have been characterized as tolerant to SHR6, they had no DEGs in common. The comparison of time points within ILs revealed two and six DEGs at 4 and at 8 dpi in HA89 and in HA853, respectively. The sustained differential expression of these genes suggests a modulation over time, in contrast to the time-specific response observed in most DEGs. Against expectations, more DEGs were detected at 4 dpi than at 8 dpi in RK416, which may indicate an earlier activation of defense mechanisms. Altogether, tolerant lines appear to deploy different responses to fungal attack. This differential pattern of defense is concordant with the diverse origins of the ILs used in this study and with the dynamics of the sunflower domestication process, which included a larger number of genes with a small phenotypic effect than in other domesticated plant species50. Although the three ILs examined here are maintainer lines, HA89 and HA853 originated in USA, whereas RK416 originated in Argentina. HA89 is derived from the Russian domestic oilseed variety “Vniimk 8931”51, and HA853 from the “1975 High Yield Composite”, which in turn was generated from 11 high-oil Russian cultivars52. RK416 was developed by crossing the Composite Ruso, derived from Smena, Arnavirsk, Peredovik and Vniimk open pollinated populations, with the variety Klein, an Argentinian population based on Russian varieties, which were brought by Jewish immigrants as confectionery seeds (Julio Gonzalez, pers. comm.).
It is known that Helianthus species possess a rich spectrum of non-redundant defense mechanisms, many of which were introduced into cultivated sunflower from their wild relatives53,54. QTL mapping has served to confirm the diversity of SHR tolerance sources within breeding germplasm4,11,13,55, whereas combining transcriptomic and QTL mapping data has recently emerged as a powerful tool to identify candidate genes24,56–58. Our transcriptomic study showed that DEGs were homogeneously distributed throughout chromosomes, as expected for a complex quantitative trait. Interestingly, the absence of a cluster of DEGs also suggests a staggered introgression of defense responses, instead of large wild-relative introgression blocks derived from modern breeding. In addition, the results presented here allowed to refine the mapping resolution of the recent QTL analysis by identifying five DEGs that co-localised with QTL regions. Functional annotations of these DEGs include one DEAD-box ATP-dependent RNA helicase 40, one hypoxia-responsive family protein and two ncRNAs, one of which was differentially expressed at 4 and 8 dpi in HA853, and the other has no known function. DEAD-box helicases have been identified as key factors of abiotic stress responses in plants59, and hypoxia-responsive proteins have been related to group VII Ethylene Response Factors, which in turn comprises elements playing a central role in defense against necrotrophic pathogens60–62. These putative functions, together with their relationship to previously identified QTLs, highlight the potential of these genes as candidates to improve resistance.
In this work, different functional approaches were undertaken to explore the molecular responses to SHR. Regardless of the method employed, each IL-time point combination showed a unique array of over-represented GO terms and catalytic enzymes. This suggests a differential contribution of IL-specific features to tolerance and susceptibility, and the existence of alternative control strategies involved in cell defense response. Further grouping of GO terms into broad functional categories revealed that ILs shared common mechanisms (i.e. responses to stimuli, catalytic processes or altered transport activities).
Two types of resistance have been described in sunflower: resistance to penetration and to mycelial spread in tissues63. The type of resistance is likely modulated by the transcriptomic responses elicited upon infection, as they represent the first line of defense and determine the outcome. The functional annotations of the DEGs identified here, as well as their enriched GO terms, may be related to both resistance processes. In tems of QDR, many causal genes are responsible for the defense response and act beyond pathogen recognition64. The perception of the pathogen frequently involves LRR receptor-like kinases (RLKs), which propagate external signals through their kinase domain32,65. Signal transduction pathways are then mediated by kinase/phosphatase activity and the alteration of ion fluxes, such as Ca2+66,67. To counteract pathogen infection, these signaling networks regulate the activity of transcription factors, transcriptional machinery, enzymes and antimicrobial compounds, including pathogenesis-related (PR) proteins68–71. In accordance with this general mechanism, a putative LRR RLK was found in HA89, while candidates involved in signaling networks or transcription factors were detected in HA89 and HA853. In addition, putative PR1 and PR10 proteins were found down-regulated in RK416 at 4 and 8 dpi. As suggested by Cregeen et al. and Upadhyay et al.72,73, the down-regulation of PR proteins may be responsible for the delay in the appearance of symptoms observed in tolerant plants, by blocking fungal spread. Since expression of PR1 is related to a salicylic acid (SA) response, it can also be inferred that the SA-pathway is suppressed upon infection in the tolerant IL RK41674.
In agreement with current knowledge about the initiation of plant immune responses, all three ILs altered the expression of genes involved in the cellular redox state75,76. In addition, the studied ILs appear to reorganise their cell wall composition and use extracellular compounds during defense. We also found genes and GO terms directly associated with defense responses, particularly in RK416 and, to a lesser extent, in HA853 (e.g. a resistance gene candidate 234; a S-norcoclaurine synthase-like gene35; or a Universal stress protein A77). Overall, the functional analysis reinforces the idea of genotype-specific activation of defense pathways. Based on the DEGs and the GO terms enriched in the different ILs, RK416 exhibits the widest arsenal of resources to resist fungal penetration, at least in the environment in which our experiment was performed.
Noteworthy, the most consistent signal in the defense response of sunflower to SHR seems to be related to ncRNAs. Two out of the five DEGs found in the vicinity of previously identified QTLs are ncRNAs, two out of ten DEGs shared by the different IL-time point combinations are ncRNAs, and the ncRNA category is significantly enriched in four out of the six IL-time point combinations. A survey of ncRNA studies has shown that genes encoding RNAs, rather than proteins, have both structural and regulatory functions78,79. In this study, we could only assess spliced ncRNAs over 200 nt (i.e. long ncRNAs) due to a bias towards the sequencing of polyadenylated RNAs. LncRNAs are poorly conserved and display diverse synthesis, processing and regulatory functions. In plants, lncRNAs can function as gene80–82, transcription83–85, or epigenetic regulators86. Moreover, they are known to participate in basal defense against stresses, including response to pathogenic fungi, where they act as precursors of sRNAs, microRNAs (miRNAs) and/or small interfering RNAs (siRNAs)87–91. Recently, it was found that A. thaliana cells secrete exosome-like extracellular vesicles to deliver sRNAs into Botrytis cinerea, a fungal necrotrophic pathogen affiliated to S. sclerotiorum. The sRNAs transferred by the host induce silencing of fungal genes essential for pathogenicity92.
Although ncRNAs appear to play a significant role as a first line of defense to SHR in sunflower, the specific mechanisms by which they operate are yet to be determined. Analysis of RNA–RNA interactions revealed possible mRNA targets within DEGs, but RNA–DNA or RNA–protein interactions cannot be ruled out. In particular, the genes HanXRQChr03g0080021 and HanXRQChr03g0080031, coding a lncRNA and a mRNA, respectively, could represent an example of a RNA–DNA interaction, as their vicinity may facilitate a cis-acting regulatory function of transcription79,93.
The relationship between tolerance to SHR and the expression patterns observed in this study is still difficult to determine. In a recent multi-environment trial, RK416 and HA853 ranked among the top 20 of 137 ILs, with RK416 showing lower DI and DS scores than HA8536. These findings were only partially replicated in our field trial, where HA853 outperformed RK416, although both exhibited a similar behaviour to that of the tolerant control Dekalb 3820. In line with these observations, sunflower response to SHR is known to be greatly affected by environmental variables such as temperature, humidity, and rainfall6. In this context, our results suggest that the ensemble of DEGs identified here may only partially represent the full array of sunflower responses to SHR, with differences in environmental conditions triggering additional reactions, even for the same IL.
In sum, this research represents the first report on the transcriptional changes occurring in sunflower upon infection with S. sclerotiorum. Our results suggest genotype-specific defense responses, in line with the diversity of tolerance levels observed under field conditions6. The combined analysis of transcriptomics and QTL mapping reported here provides a promising approach to identify relevant genes and genomic regions associated with QDR. Finally, the involvement of ncRNAs adds an unexpected complexity to the discovery of the genetic determinants of SHR resistance.
Methods
Plant material and field trial
Three sunflower public ILs were used in this study: HA89, HA853 and RK416. Plants of each IL were grown under field conditions during season 2010–11 at the experimental station INTA Balcarce (Buenos Aires, Argentina). A randomised complete block design comprising three blocks with a furrow per IL was used. Furrows were 6 m long and spaced 75 cm apart. Commercial sunflower lines ACA861 and Dekalb3820 were included in each block as susceptible and tolerant controls, respectively.
Plants that reached the R5.2 flowering stage of the Schneiter and Miller94 scale on the same day were selected in pairs and capitula were ascospore- (2,500 ascospores/ml) (I) or mock- (water) (N) inoculated with 1 ml of inoculum using a portable hand sprayer95. Ascospores were obtained by inducing carpogenic germination of sclerotia, as described by Escande et al.95. Briefly, apothecia were produced from sclerotia collected at the end of the 2009/2010 field trials from naturally and experimentally infected plants at agricultural station INTA Balcarce. Mature apothecia were hold in Petri dishes, incubated for 4 h to favor ascospore release and finally stored at − 18 °C until use. Inoculated capitula were collected at 0 (i.e. immediately after spraying), 4 and 8 dpi, resulting in a total of 56 samples. Four biological replicates at 0 dpi and two biological replicates at 4 and 8 dpi were analysed for HA853 and HA89, whereas four biological replicates were used for RK416 at all time points (Supplementary Table S1). Time 0 after inoculation was included to assess the incidence of false positives in the statistical analysis.
Given that SHR symptoms appear at least after 10 dpi6, 14 to 21 plants per block and IL were maintained in the field to evaluate disease progression and confirm the effect of the inoculation protocol.
Disease assessment
Disease progression was quantified by estimating the phenotypic variables DI, i.e., the number of plants infected over the number of plants inoculated in each furrow, and DS, i.e., the average proportion of capitulum rotted area of plants inoculated in each furrow. These variables have been described as suitable for appraising the resistance to fungal penetration in the first case, and the resistance to the spread of the fungus in the second case96.
For graphical visualization of the results, average DI and DS values per day post inoculation and IL were plotted using GraphPad Prism, v 5.01 for Windows, GraphPad Software, La Jolla California USA, https://www.graphpad.com (Supplementary Fig. S1).
RNA isolation and sequencing
Disc florets and bracts were scraped off each capitulum and grinded in liquid nitrogen. Total RNA of 100 mg grinded material was extracted using the RNAqueous Kit with addition of the Plant RNA Isolation Aid (Ambion, Applied Biosystems, USA). Samples were treated with DNase I (Invitrogen, USA) for 20 min to remove remaining DNA. Sample purity, integrity and concentration were assessed using the RNA 6000 Nano Reagent kit in a 2100 Bioanalyzer (Agilent Technologies, Palo Alto, California, USA).
Sequencing libraries were prepared using an Illumina TruSeq RNA Sample Preparation Kit following the manufacturer’s instructions. Libraries were sequenced using the Illumina HiSeq 2000 system at the Centre Nacional d'Anàlisi Genòmica (CNAG: https://www.cnag.crg.eu/, Barcelona, Spain) as unstranded single-end reads of 50 bp in length. The original data set was deposited at the NCBI Sequence Read Archive (Submission ID: SUB5575431, BioProject ID: PRJNA561716).
Analysis of RNA-seq data
Low-quality bases (average Q-score below 20) and adaptor sequences were trimmed and sequences with less than 36 bases long were removed with Trimmomatic97. The remaining reads were mapped to the Helianthus annuus XRQ (HanXRQ v1) genome-derived transcriptome (https://www.heliagene.org/HanXRQ-SUNRISE/)31 with Bowtie2, using the global sensitive parameter and allowing all multimapping reads to be mapped98. The EM algorithm of eXpress was used for transcriptome quantification99. Differential gene expression analysis was performed with the program edgeR100. In summary, transcripts with < 1 CPM in half of the samples were filtered out and then normalised using the trimmed mean of M-values (TMM) method. Read counts for each gene were fit to a negative binomial generalised log-linear model using the glmFit function, defined as:
which includes the IL (G) i, the day post inoculation (D) j, the inoculation treatment (T) k and the sequencing lane (L) l as factors, and the interaction among G, D and T.
To identify DEGs, genewise likelihood ratio tests between I and N samples were conducted using the glmLRT method for each IL-time point combination. Transcripts were considered as differentially expressed between ascospore- and mock-inoculated treatments if the absolute value of logFC was ≥ 0.6 and FDR was < 0.05.
Validation of RNA-seq analysis by qPCR
qPCR assays, which were previously performed to validate a microarray experiment conducted on the same samples as our RNA-seq analyses, were used to confirm the transcript levels estimated here.
Briefly, cDNA for qPCR was synthesised using Superscript III first strand synthesis system (Invitrogen, USA) and random hexamer primers according to the manufacturer's instructions. For amplification, a 25-µl reaction mix containing 200 nM of each primer, 1 µl of cDNA sample and FastStart Universal SYBR Green Master (Roche Applied Science, Germany) was run in a 96-well plate thermocycler (ABI Prism 7000 Sequence Detection System and software, PE Applied Biosystems, USA). Amplification efficiencies and Ct values were determined for each transcript using LinRegPCR101. The relative expression of the genes was determined using the “RT-PCR comparison of relative gene expressions analysis” included in the InfoStat software102. Actin was established as internal standard after comparison with Elongation Factor-1α and Tubulin using the program Bestkeeper103. Primer pairs used for the qPCR assays are listed in Supplementary Table S11.
Co-localisation of DEGs to previously identified QTLs
To investigate the putative correspondence between previously reported QTLs and the DEGs identified here, 39 MM associated with SHR resistance11,12 were mapped to the HanXRQ v1 genome31 using primersearch v6.6.0.0 for in silico PCR104. DEGs falling within a window size of ± 1 Mbp around the MM were considered to co-localise with the QTLs (Supplementary Table S4).
Functional profiling of genes
To assess the over-representation of GO categories, Single Enrichment analyses were performed on DEGs from each IL and time point using FatiGO105.
Mapman classification106 was used to interpret DEGs in the context of hierarchically organised cellular pathways and processes focused on plant metabolism107,108. To enable mapping, the genes predicted in the HanXRQ v1 sunflower genome annotation were re-annotated through the Mercator pipeline using A. thaliana’s functional annotation109, following Moschen et al.110.
In addition, a GO GSEA based on logistic regression was also applied to the whole gene set using the uvgsa function of the mdgsa R package38, with transcripts indexed by the logFC and the adjusted p-value. Enrichment was considered significant for p < 0.05, after Benjamini & Yekutieli111 correction for multiple testing. To evaluate the affiliations among the over-represented GO terms, semantic similarities were computed using the goSim function of the GOSemSim R package112 based on the genome-wide annotation for Arabidopsis113. Terms not identified in this database were given zero similarity relative to the other terms. GO terms were plotted on heatmaps according to their LOR values and grouped by semantic similarities using the complete-linkage hierarchical clustering method implemented in the R package ComplexHeatmaps.
Finally, to explore systems and chemical information collected in the KEGG database, a KEGG gene set analysis was also performed using the uvgsa function as described previously.
All analyses were performed based on the HanXRQ v1 gene annotation from Badouin et al.31.
Analysis of differentially expressed non-coding RNAs
To include ncRNAs into the functional enrichment analysis, the number of differentially expressed ncRNAs of each IL-time point combination was compared to the total number of expressed ncRNAs by bilateral Irwin-Fisher tests using InfoStat102.
The LncTar software, version of November 10, 2015114 was used to predict the interaction between the ncRNAs and the mRNAs differentially expressed in each IL. Type 1 command line was used to perform all ncRNAs- vs. all mRNAs predictions. Following the developers’ recommendations, a ndG cutoff of − 0.15 was used to obtain high confidence predictions.
Supplementary information
Acknowledgements
This research was supported by Instituto Nacional de Tecnología Agropecuaria (PNBIO 1131022, 1131043), Agencia Nacional de Promoción Científica y Técnica (PICT 2011 1365), Agencia Española de Cooperación Internacional para el Desarrollo (A1/041041/11) and the DEANN project. We thank Silvio Giuliano and Carlos Antonelli for their field assistance at INTA Balcarce and the breeders Julio Gonzalez and Daniel Alvarez for providing the pedigree of the ILs. We are also grateful to Dr. Silvia Pietrokovsky, who kindly revised the English of the manuscript.
Author contributions
M.I.F., M.R. and F.G.G. analyzed the RNA-Seq data. M.I.F., M.R. and J.F.M. prepared the figures. G.F.E. performed the field trial, evaluated disease progression, genotyped the ILs and isolated RNA. C.A.M. and F.Q. assisted with the field trial. R.A.H., N.B.P. and V.V.L. conceived and designed the study. M.I.F., M.R. and V.V.L. wrote the main manuscript. H.E.H. and J.D.B. contributed to the work by the interpretation and discussion of the data. All authors reviewed and approved the final manuscript.
Data availability
All data generated or analysed during this study are included in this published article, its Supplementary Information files or deposited at the NCBI Sequence Read Archive (Submission ID: SUB5575431, BioProject ID: PRJNA561716).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Mónica I. Fass, Máximo Rivarola, Norma B. Paniego and Verónica V. Lia.
Supplementary information
is available for this paper at 10.1038/s41598-020-70315-4.
References
- 1.Pereyra, V. R. & Escande, A. Enfermedades del Girasol en la Argentina : Manual de Reconocimiento. (Instituto Nacional de Tecnología Agropecuaria, 1994).
- 2.Gulya, T., Rashid, K. Y. & Masirevic, S. M. Sunflower diseases. in Sunflower Technology and Production (ed. Schneiter, A. A.) 263–379 (1997).
- 3.Mantecon JD, Pereyra V. Integrated control methods for managing sunflower head rot in Argentina. Int. J. Pest Manag. 1997;43:143–144. [Google Scholar]
- 4.Bert, P. F. et al. Comparative genetic analysis of quantitative traits in sunflower (Helianthus annuus L.) 1. QTL involved in resistance to Sclerotinia sclerotiorum and Diaporthe helianthi. Theor. Appl. Genet.105, 985–993 (2002). [DOI] [PubMed]
- 5.Castaño F, Giussani MA. Effectivness of components of partial resistance in assessing white rot of sunflower head. Helia. 2009;32:59–68. [Google Scholar]
- 6.Filippi CV, et al. Phenotyping sunflower genetic resources for Sclerotinia head rot response: Assessing variability for disease resistance breeding. Plant Dis. 2017;101:1941–1948. doi: 10.1094/PDIS-12-16-1784-RE. [DOI] [PubMed] [Google Scholar]
- 7.Hahn V. Genetic variation for resistance to Sclerotinia head rot in sunflower inbred lines. F. Crop. Res. 2002;77:153–159. [Google Scholar]
- 8.Leclerq P. Influence de facteurs héréditaires sur la résistance apparente du tournesol á Sclerotinia sclerotiorum. Ann. Amélior. Plantes. 1973;23:279–286. [Google Scholar]
- 9.Talukder, Z. I., Seiler, G. J., Song, Q., Ma, G. & Qi, L. SNP discovery and QTL mapping of Sclerotinia basal stalk rot resistance in sunflower using genotyping-by-sequencing. Plant Genome9 (2016). [DOI] [PubMed]
- 10.Yue B, et al. Identifying quantitative trait loci for resistance to Sclerotinia head rot in two USDA sunflower germplasms. Phytopathology. 2008;98:926–931. doi: 10.1094/PHYTO-98-8-0926. [DOI] [PubMed] [Google Scholar]
- 11.Zubrzycki JE, et al. Main and epistatic QTL analyses for Sclerotinia head rot resistance in sunflower. PLoS ONE. 2017;12:e0189859. doi: 10.1371/journal.pone.0189859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Filippi, C. Diversidad Genómica y Mapeo por Asociación Para la Resistencia a la Podredumbre Húmeda del Capítulo Causada por Sclerotinia sclerotiorum en Girasol. (Universidad de Buenos Aires, 2015).
- 13.Fusari CM, et al. Association mapping in sunflower for Sclerotinia head rot resistance. BMC Plant Biol. 2012;12:93. doi: 10.1186/1471-2229-12-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Talukder ZI, et al. Candidate gene association mapping of Sclerotinia stalk rot resistance in sunflower (Helianthus annuus L.) uncovers the importance of COI1 homologs. Theor. Appl. Genet. 2014;127:193–209. doi: 10.1007/s00122-013-2210-x. [DOI] [PubMed] [Google Scholar]
- 15.Amselem J, et al. Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet. 2011;7:e1002230. doi: 10.1371/journal.pgen.1002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bolton MD, Thomma BPHJ, Nelson BD. Sclerotinia sclerotiorum (Lib) de Bary: Biology and molecular traits of a cosmopolitan pathogen. Mol. Plant Pathol. 2006;7:1–16. doi: 10.1111/j.1364-3703.2005.00316.x. [DOI] [PubMed] [Google Scholar]
- 17.Mbengue M, et al. Emerging trends in molecular interactions between plants and the broad host range fungal pathogens Botrytis cinerea and Sclerotinia sclerotiorum. Front. Plant Sci. 2016;7:1–9. doi: 10.3389/fpls.2016.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boland GJ, Hall R. Index of plant hosts of Sclerotinia sclerotiorum. Can. J. Plant Pathol. 1994;16:93–108. [Google Scholar]
- 19.Dai F-M, Xu T, Wolf GA, He Z-H. Physiological and molecular features of the pathosystem Arabidopsis thaliana L.-Sclerotinia sclerotiorum Libert. J. Integr. Plant Biol. 2006;48:44–52. [Google Scholar]
- 20.Girard IJ, et al. RNA sequencing of Brassica napus reveals cellular redox control of Sclerotinia infection. J. Exp. Bot. 2017;68:5079–5091. doi: 10.1093/jxb/erx338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joshi RK, Megha S, Rahman MH, Basu U, Kav NNV. A global study of transcriptome dynamics in canola (Brassica napus L.) responsive to Sclerotinia sclerotiorum infection using RNA-Seq. Gene. 2016;590:57–67. doi: 10.1016/j.gene.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 22.Wu J, et al. Genome-wide association study identifies new loci for resistance to Sclerotinia stem rot in Brassica napus. Front. Plant Sci. 2016;7:1418. doi: 10.3389/fpls.2016.01418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao J, et al. Analysis of gene expression profiles in response to Sclerotinia sclerotiorum in Brassica napus. Planta. 2007;227:13–24. doi: 10.1007/s00425-007-0586-z. [DOI] [PubMed] [Google Scholar]
- 24.Wen Z, et al. Integrating GWAS and gene expression data for functional characterization of resistance to white mould in soya bean. Plant Biotechnol. J. 2018;16:1825–1835. doi: 10.1111/pbi.12918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodriguez MA, Venedikian N, Bazzalo ME, Godeas A. Histopathology of Sclerotinia sclerotiorum attack on flower parts of Helianthus annuus heads in tolerant and susceptible varieties. Mycopathologia. 2004;157:291–302. doi: 10.1023/b:myco.0000024177.82916.b7. [DOI] [PubMed] [Google Scholar]
- 26.Han Y, Gao S, Muegge K, Zhang W, Zhou B. Advanced applications of RNA sequencing and challenges. Bioinform. Biol. Insights. 2015;9:29–46. doi: 10.4137/BBI.S28991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo S, et al. Large-scale transcriptome comparison of sunflower genes responsive to Verticillium dahliae. BMC Genomics. 2017;18:42. doi: 10.1186/s12864-016-3386-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang C, et al. Identification of differentially expressed genes in sunflower (Helianthus annuus) leaves and roots under drought stress by RNA sequencing. Bot. Stud. 2017;58:42. doi: 10.1186/s40529-017-0197-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Monazzah, M., Tahmasebi Enferadi, S. & Rabiei, Z. Enzymatic activities and pathogenesis-related genes expression in sunflower inbred lines affected by Sclerotinia sclerotiorum culture filtrate. J. Appl. Microbiol.125, 227–242 (2018). [DOI] [PubMed]
- 30.Muellenborn C, Krause J-H, Cerboncini C. Analysis of differential transcript expression reveals time-dependent leaf responses to Sclerotinia sclerotiorum in wild and cultivated sunflower. Plant Mol. Biol. Report. 2011;29:597–608. [Google Scholar]
- 31.Badouin H, et al. The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution. Nat. Publ. Gr. 2017;546:148–152. doi: 10.1038/nature22380. [DOI] [PubMed] [Google Scholar]
- 32.Afzal AJ, Wood AJ, Lightfoot DA. Plant receptor-like serine threonine kinases: Roles in signaling and plant defense. Mol. Plant-Microbe Interact. 2008;21:507–517. doi: 10.1094/MPMI-21-5-0507. [DOI] [PubMed] [Google Scholar]
- 33.Breen S, Williams SJ, Outram M, Kobe B, Solomon PS. Emerging insights into the functions of pathogenesis-related protein 1. Trends Plant Sci. 2017;22:871–879. doi: 10.1016/j.tplants.2017.06.013. [DOI] [PubMed] [Google Scholar]
- 34.Christopoulou M, et al. Genome-wide architecture of disease resistance genes in lettuce. G3 (Bethesda). 2015;5:2655–2669. doi: 10.1534/g3.115.020818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Desgagné-Penix I, et al. Integration of deep transcriptome and proteome analyses reveals the components of alkaloid metabolism in opium poppy cell cultures. BMC Plant Biol. 2010;10:252. doi: 10.1186/1471-2229-10-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fan, S. et al. A novel pathogenesis-related class 10 protein Gly m 4l, increases resistance upon Phytophthora sojae infection in soybean (Glycine max [L.] Merr.). PLoS One10, e0140364 (2015). [DOI] [PMC free article] [PubMed]
- 37.Gullner G, Komives T, Király L, Schröder P. Glutathione S-transferase enzymes in plant-pathogen interactions. Front. Plant Sci. 2018;9:1836. doi: 10.3389/fpls.2018.01836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Montaner D, Dopazo J. Multidimensional gene set analysis of genomic data. PLoS ONE. 2010;5:e10348. doi: 10.1371/journal.pone.0010348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fukuoka S, et al. Gene pyramiding enhances durable blast disease resistance in rice. Sci. Rep. 2015;5:7773. doi: 10.1038/srep07773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hittalmani S, Parco A, Mew TV, Zeigler RS, Huang N. Fine mapping and DNA marker-assisted pyramiding of the three major genes for blast resistance in rice. Theor. Appl. Genet. 2000;100:1121–1128. [Google Scholar]
- 41.Joshi RK, Nayak S. Gene pyramiding-A broad spectrum technique for developing durable stress resistance in crops. Biotechnol. Mol. Biol. Rev. 2010;5:51–60. [Google Scholar]
- 42.Gao Y, et al. Time-course transcriptome analysis reveals resistance genes of Panax ginseng induced by Cylindrocarpon destructans infection using RNA-Seq. PLoS ONE. 2016;11:e0149408. doi: 10.1371/journal.pone.0149408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kamber T, et al. Fire blight disease reactome: RNA-seq transcriptional profile of apple host plant defense responses to Erwinia amylovora pathogen infection. Sci. Rep. 2016;6:21600. doi: 10.1038/srep21600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Song Y, et al. Comparative transcriptome analysis of resistant and susceptible kiwifruits in response to Pseudomonas syringae pv. Actinidiae during early infection. PLoS ONE. 2019;14:e0211913. doi: 10.1371/journal.pone.0211913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu J, et al. Comparative transcriptomic analysis uncovers the complex genetic network for resistance to Sclerotinia sclerotiorum in Brassica napus. Sci. Rep. 2016;6:19007. doi: 10.1038/srep19007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Haro LA, et al. Mal de Río Cuarto virus infection causes hormone imbalance and sugar accumulation in wheat leaves. BMC Plant Biol. 2019;19:112. doi: 10.1186/s12870-019-1709-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rezzonico F, Rupp O, Fahrentrapp J. Pathogen recognition in compatible plant-microbe interactions. Sci. Rep. 2017;7:6383. doi: 10.1038/s41598-017-04792-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fan M, et al. The bHLH transcription factor HBI1 mediates the trade-off between growth and pathogen-associated molecular pattern-triggered immunity in Arabidopsis. Plant Cell. 2014;26:828–841. doi: 10.1105/tpc.113.121111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Calla B, Vuong T, Radwan O, Hartman GL, Clough SJ. Gene expression profiling soybean stem tissue early response to Sclerotinia sclerotiorum and in silico mapping in relation to resistance markers. Plant Genome J. 2009;2:149. [Google Scholar]
- 50.Radanović, A., Miladinović, D., Cvejić, S., Jocković, M. & Jocić, S. Sunflower genetics from ancestors to modern hybrids—A review. Genes9 (2018). [DOI] [PMC free article] [PubMed]
- 51.Gavrilova VA, Anisimova IN. Genealogy of the sunflower lines created on the basis of Russian varieties. Helia. 2017;40:133–146. [Google Scholar]
- 52.Korell M, Mosges GFW. Construction of a sunflower pedigree map. Helia. 1992;15:7–16. [Google Scholar]
- 53.Seiler GJ, Qi LL, Marek LF. Utilization of sunflower crop wild relatives for cultivated sunflower improvement. Crop Sci. 2017;57:1083. [Google Scholar]
- 54.Mason CM, et al. Macroevolution of leaf defenses and secondary metabolites across the genus Helianthus. New Phytol. 2016;209:1720–1733. doi: 10.1111/nph.13749. [DOI] [PubMed] [Google Scholar]
- 55.Filippi C, et al. Population structure and genetic diversity characterization of a sunflower association mapping population using SSR and SNP markers. BMC Plant Biol. 2015;15:52. doi: 10.1186/s12870-014-0360-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pandit A, et al. Combining QTL mapping and transcriptome profiling of bulked RILs for identification of functional polymorphism for salt tolerance genes in rice (Oryza sativa L.) Mol. Genet. Genomics. 2010;284:121–136. doi: 10.1007/s00438-010-0551-6. [DOI] [PubMed] [Google Scholar]
- 57.Gelli M, et al. Validation of QTL mapping and transcriptome profiling for identification of candidate genes associated with nitrogen stress tolerance in sorghum. BMC Plant Biol. 2017;17:123. doi: 10.1186/s12870-017-1064-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang S, et al. Integrated RNA sequencing and QTL mapping to identify candidate genes from Oryza rufipogon associated with salt tolerance at the seedling stage. Front. Plant Sci. 2017;8:1427. doi: 10.3389/fpls.2017.01427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nidumukkala S, Tayi L, Chittela RK, Vudem DR, Khareedu VR. DEAD box helicases as promising molecular tools for engineering abiotic stress tolerance in plants. Crit. Rev. Biotechnol. 2019;39:395–407. doi: 10.1080/07388551.2019.1566204. [DOI] [PubMed] [Google Scholar]
- 60.Giuntoli B, Perata P. Group VII ethylene response factors in Arabidopsis: Regulation and physiological roles. Plant Physiol. 2018;176:1143–1155. doi: 10.1104/pp.17.01225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gibbs DJ, et al. Group VII ethylene response factors coordinate oxygen and nitric oxide signal transduction and stress responses in plants. Plant Physiol. 2015;169:23–31. doi: 10.1104/pp.15.00338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schmidt RR, Weits DA, Feulner CFJ, van Dongen JT. Oxygen sensing and integrative stress signaling in plants. Plant Physiol. 2018;176:1131–1142. doi: 10.1104/pp.17.01394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Castaño F, Vear F, de Labrouhe DT. Resistance of sunflower inbred lines to various forms of attack by Sclerotinia sclerotiorum and relations with some morphological characters. Euphytica. 1993;68:85–98. [Google Scholar]
- 64.Corwin JA, Kliebenstein DJ. Quantitative resistance: More than just perception of a pathogen. Plant Cell. 2017;29:655–665. doi: 10.1105/tpc.16.00915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goff KE, Ramonell KM. The role and regulation of receptor-like kinases in plant defense. Gene Regul. Syst. Bio. 2007;1:167–175. [PMC free article] [PubMed] [Google Scholar]
- 66.Blumwald E, Aharon GS, Lam C-HB. Early signal transduction pathways in plant–pathogen interactions. Trends Plant Sci. 1998;3:342–346. [Google Scholar]
- 67.Ranf S, et al. Defense-related calcium signaling mutants uncovered via a quantitative high-throughput screen in Arabidopsis thaliana. Mol. Plant. 2012;5:115–130. doi: 10.1093/mp/ssr064. [DOI] [PubMed] [Google Scholar]
- 68.van Loon LC, Rep M, Pieterse CMJ. Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 2006;44:135–162. doi: 10.1146/annurev.phyto.44.070505.143425. [DOI] [PubMed] [Google Scholar]
- 69.Singh R, Tiwari JK, Sharma V, Singh BP, Rawat S. Role of pathogen related protein families in defence mechanism with potential role in applied biotechnology. Int. J. Adv. Res. 2014;2:210–226. [Google Scholar]
- 70.Tena G, Boudsocq M, Sheen J. Protein kinase signaling networks in plant innate immunity. Curr. Opin. Plant Biol. 2011;14:519–529. doi: 10.1016/j.pbi.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang C, Du X, Mou Z. The mediator complex subunits MED14, MED15, and MED16 are involved in defense signaling crosstalk in Arabidopsis. Front. Plant Sci. 2016;7:1–7. doi: 10.3389/fpls.2016.01947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cregeen S, et al. Different gene expressions of resistant and susceptible hop cultivars in response to infection with a highly aggressive strain of Verticillium albo-atrum. Plant Mol. Biol. Rep. 2015;33:689–704. doi: 10.1007/s11105-014-0767-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Upadhyay P, Rai A, Kumar R, Singh M, Sinha B. Differential expression of pathogenesis related protein genes in tomato during inoculation with A. Solani. J. Plant Pathol. Microbiol. 2014;05:1–7. [Google Scholar]
- 74.El Rahman AT, El Oirdi M, Gonzalez-Lamothe R, Bouarab K. Necrotrophic pathogens use the salicylic acid signaling pathway to promote disease development in tomato. Mol. Plant-Microbe Interact. 2012;25:1584–1593. doi: 10.1094/MPMI-07-12-0187-R. [DOI] [PubMed] [Google Scholar]
- 75.Frederickson Matika DE, Loake GJ. Redox regulation in plant immune function. Antioxid. Redox Signal. 2014;21:1373–1388. doi: 10.1089/ars.2013.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.O’Brien JA, Daudi A, Butt VS, Paul Bolwell G. Reactive oxygen species and their role in plant defence and cell wall metabolism. Planta. 2012;236:765–779. doi: 10.1007/s00425-012-1696-9. [DOI] [PubMed] [Google Scholar]
- 77.Gutiérrez-Beltrán E, Personat JM, de la Torre F, del Pozo O. A universal stress protein involved in oxidative stress is a phosphorylation target for protein kinase CIPK6. Plant Physiol. 2017;173:836–852. doi: 10.1104/pp.16.00949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cech TR, Steitz JA. The noncoding RNA revolution—Trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 79.Fu X-D. Non-coding RNA: A new frontier in regulatory biology. Natl Sci Rev. 2014;1:190–204. doi: 10.1093/nsr/nwu008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bardou F, et al. Long noncoding RNA modulates alternative splicing regulators in Arabidopsis. Dev. Cell. 2014;30:166–176. doi: 10.1016/j.devcel.2014.06.017. [DOI] [PubMed] [Google Scholar]
- 81.Xie Z, et al. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:e104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Franco-Zorrilla JM, et al. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 2007;39:1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- 83.Campalans A. Enod40, a short open reading frame-containing mRNA, induces cytoplasmic localization of a nuclear RNA binding protein in Medicago truncatula. Plant Cell Online. 2004;16:1047–1059. doi: 10.1105/tpc.019406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang Z-W, Wu Z, Raitskin O, Sun Q, Dean C. Antisense-mediated FLC transcriptional repression requires the P-TEFb transcription elongation factor. Proc. Natl. Acad. Sci. 2014;111:7468–7473. doi: 10.1073/pnas.1406635111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wierzbicki AT, Ream TS, Haag JR, Pikaard CS. RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat. Genet. 2009;41:630–634. doi: 10.1038/ng.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhao Y, Chen X. Noncoding RNAs and DNA methylation in plants. Natl. Sci. Rev. 2014;1:219. doi: 10.1093/nsr/nwu003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Di C, et al. Characterization of stress-responsive lncRNAs in Arabidopsis thaliana by integrating expression, epigenetic and structural features. Plant J. 2014;80:848–861. doi: 10.1111/tpj.12679. [DOI] [PubMed] [Google Scholar]
- 88.Wang H, et al. Analysis of non-coding transcriptome in rice and maize uncovers roles of conserved lncRNAs associated with agriculture traits. Plant J. 2015;84:404–416. doi: 10.1111/tpj.13018. [DOI] [PubMed] [Google Scholar]
- 89.Xin M, et al. Identification and characterization of wheat long non-protein coding RNAs responsive to powdery mildew infection and heat stress by using microarray analysis and SBS sequencing. BMC Plant Biol. 2011;11:61. doi: 10.1186/1471-2229-11-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang H, et al. Long non-coding genes implicated in response to stripe rust pathogen stress in wheat (Triticum aestivum L.) Mol. Biol. Rep. 2013;40:6245–6253. doi: 10.1007/s11033-013-2736-7. [DOI] [PubMed] [Google Scholar]
- 91.Zhu Q-H, Stephen S, Taylor J, Helliwell CA, Wang M-B. Long noncoding RNAs responsive to Fusarium oxysporum infection in Arabidopsis thaliana. New Phytol. 2014;201:574–584. doi: 10.1111/nph.12537. [DOI] [PubMed] [Google Scholar]
- 92.Cai Q, et al. Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science. 2018;360:1126–1129. doi: 10.1126/science.aar4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kornienko AE, Guenzl PM, Barlow DP, Pauler FM. Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 2013;11:59. doi: 10.1186/1741-7007-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schneiter A, Miller J. Description of sunflower growth stages. Crop Sci. 1981;21:3–5. [Google Scholar]
- 95.Escande AR, Laich FS, Pedraza MV. Field testing of honeybee-dispersed Trichoderma spp. to manage sunflower head rot (Sclerotinia sclerotiorum) Plant Pathol. 2002;51:346–351. [Google Scholar]
- 96.Gentzbittel L, et al. Cloning of molecular markers for disease resistance in sunflower Helianthus annuus L. Theor. Appl. Genet. 1998;96:519–525. doi: 10.1007/s001220050769. [DOI] [PubMed] [Google Scholar]
- 97.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Roberts A, Pachter L. Streaming fragment assignment for real-time analysis of sequencing experiments. Nat. Methods. 2013;10:71–73. doi: 10.1038/nmeth.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ramakers C, Ruijter JM, Deprez RHL, Moorman AF. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 2003;339:62–66. doi: 10.1016/s0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- 102.Di Rienzo, J. A. et al. InfoStat. Version 2019. Grupo InfoStat, FCA, Universidad Nacional de Córdoba, Argentina. https://www.infostat.com.ar.
- 103.Pfaffl, M. W. Quantification strategies in real-time PCR. in A-Z of Quantitative PCR (ed. Bustin, S. A.) 87–112 (International University Line, 2004).
- 104.Rice P, Longden I, Bleasby A. EMBOSS: The European molecular biology open software suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 105.Al-Shahrour F, Diaz-Uriarte R, Dopazo J. FatiGO: A web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics. 2004;20:578–580. doi: 10.1093/bioinformatics/btg455. [DOI] [PubMed] [Google Scholar]
- 106.Thimm O, et al. mapman: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004;37:914–939. doi: 10.1111/j.1365-313x.2004.02016.x. [DOI] [PubMed] [Google Scholar]
- 107.Usadel B, et al. A guide to using MapMan to visualize and compare Omics data in plants: A case study in the crop species, Maize. Plant. Cell Environ. 2009;32:1211–1229. doi: 10.1111/j.1365-3040.2009.01978.x. [DOI] [PubMed] [Google Scholar]
- 108.Klie S, Nikoloski Z. The choice between MapMan and Gene Ontology for automated gene function prediction in plant science. Front. Genet. 2012;3:115. doi: 10.3389/fgene.2012.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lohse M, et al. Mercator: a fast and simple web server for genome scale functional annotation of plant sequence data. Plant. Cell Environ. 2014;37:1250–1258. doi: 10.1111/pce.12231. [DOI] [PubMed] [Google Scholar]
- 110.Moschen S, et al. Integrating transcriptomic and metabolomic analysis to understand natural leaf senescence in sunflower. Plant Biotechnol. J. 2016;14:719–734. doi: 10.1111/pbi.12422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 2001;29:1165–1188. [Google Scholar]
- 112.Yu G, et al. GOSemSim: An R package for measuring semantic similarity among GO terms and gene products. Bioinformatics. 2010;26:976–978. doi: 10.1093/bioinformatics/btq064. [DOI] [PubMed] [Google Scholar]
- 113.Carlson, M. org.At.tair.db: Genome wide annotation for Arabidopsis. R package version 3.7.0. (2018).
- 114.Li J, et al. LncTar: A tool for predicting the RNA targets of long noncoding RNAs. Brief. Bioinform. 2015;16:806–812. doi: 10.1093/bib/bbu048. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article, its Supplementary Information files or deposited at the NCBI Sequence Read Archive (Submission ID: SUB5575431, BioProject ID: PRJNA561716).