The cell wall, composed mainly of peptidoglycan, is key to maintaining the cell shape and protecting the cell from bursting. Peptidoglycan degradation by peptidoglycan hydrolysis and autolysins occurs during growth and cell division. Since peptidoglycan hydrolases are important for virulence, envelope integrity, and regulation of cell division, it is valuable to investigate their function and regulation. Notably, PcsB-like proteins such as Usp45 have been proposed as new targets for antimicrobial drugs and could also be target for the development of food-grade suicide systems. In addition, although various other expression and secretion systems have been developed for use in Lactococcus lactis, the most-used signal peptide for protein secretion in this bacterium is that of the Usp45 protein. Thus, elucidating the biological function of Usp45 and determining the factors affecting its expression would contribute to optimize several applications.
KEYWORDS: Lactococcus lactis, Usp45, cell morphology, cell separation, peptidoglycan synthesis
ABSTRACT
Lactococcus lactis is a Gram-positive bacterium that is widely used as a cell factory for the expression of heterologous proteins that are relevant in the pharmaceutical and nutraceutical fields. The signal peptide of the major secreted protein of L. lactis, Usp45, has been employed extensively in engineering strategies to secrete proteins of interest. However, the biological function of Usp45 has remained obscure despite more than 25 years of research. Studies on Usp45 homologs in other Gram-positive bacteria suggest that Usp45 may play a role in cell wall turnover processes. Here, we show the effect of inactivation and overexpression of the usp45 gene on L. lactis growth, phenotype, and cell division. Our results are in agreement with those obtained in streptococci and demonstrate that the L. lactis Usp45 protein is essential for proper cell division. We also show that the usp45 promoter is highly activated by galactose. Overall, our results indicate that Usp45 mediates cell separation, probably by acting as a peptidoglycan hydrolase.
IMPORTANCE The cell wall, composed mainly of peptidoglycan, is key to maintaining the cell shape and protecting the cell from bursting. Peptidoglycan degradation by peptidoglycan hydrolysis and autolysins occurs during growth and cell division. Since peptidoglycan hydrolases are important for virulence, envelope integrity, and regulation of cell division, it is valuable to investigate their function and regulation. Notably, PcsB-like proteins such as Usp45 have been proposed as new targets for antimicrobial drugs and could also be target for the development of food-grade suicide systems. In addition, although various other expression and secretion systems have been developed for use in Lactococcus lactis, the most-used signal peptide for protein secretion in this bacterium is that of the Usp45 protein. Thus, elucidating the biological function of Usp45 and determining the factors affecting its expression would contribute to optimize several applications.
INTRODUCTION
Lactococcus lactis has proven to be a suitable bacterial host for the expression and secretion of heterologous proteins (1). The most used signal peptide (SP) for protein secretion in L. lactis is that of the native Usp45 protein (2).
Previous studies have attempted to characterize Usp45 and have led to the use of the usp45 promoter (Pusp45) for gene expression and to the use of the Usp45 export signal (SPUsp45) for protein secretion in L. lactis (3–5). A role of Usp45 in the proteolytic system of L. lactis was excluded, and it did not possess any antimicrobial activity against Gram-positive bacteria (3). Thus, its biological function remained elusive (6).
A significant body of research on the PcsB protein in Streptococcus pneumoniae, which is homologous to Usp45 in L. lactis (7), has shown that it is required for normal growth and cell division (8). Importantly, comparative genomic analysis reveals that homologs of PcsB are widely distributed in Gram-positive bacteria (9). Among these proteins, PcsB protein has 41.8% similarity to Usp45 from L. lactis and 28.3% similarity to P45 from Listeria monocytogenes (7, 10). Although muralytic activity of the purified catalytic domain (cysteine, histidine-dependent amidohydrolases/peptidases [CHAP]) of PcsB was recently demonstrated (11), the full-length P45 protein from L. monocytogenes is the only protein exhibiting murein hydrolase activity in vitro (7, 12). P45 from L. monocytogenes does not contain a CHAP domain; instead, it contains an NLPC/P60 domain (13). Moreover, recent studies have shown that PcsB is recruited to the septum during the bacterial cell division process, where its muralytic activity is triggered by an ATP-driven conformational change, which might explain the nondetectable catalytic activity of recombinant PcsB in vitro (11).
Studies in S. pneumoniae and Streptococcus pyogenes show that the two-component system (TCS) WalKR is essential and that it positively regulates the PcsB protein (14, 15). Two-component systems are commonly used by bacteria to sense and respond to environmental signals (16). The WalKR system, originally described in Bacillus subtilis, is highly conserved and specific to low-G+C-content Gram-positive bacteria. It is responsible for the coordination of cell wall metabolism and cell division (17). Interestingly, the WalR orthologue, LlrC, is not essential in L. lactis (18). Considering all of these observations, the differences in the regulation or function of the PcsB-like proteins might indicate subtle differences in cell division in Gram-positive bacteria. In the current report, we investigated the biological function of the Usp45 protein of L. lactis and its contribution to the cell division process. These findings now explain the problem of making knockouts of the usp45 gene in previous studies and its essentiality under certain conditions.
Since usp45 encodes the major extracellular protein from L. lactis and its SP has been extensively used to drive the secretion of proteins and peptides in L. lactis (4), we also studied growth conditions to enhance the usp45 promoter activity. Our results show that the usp45 promoter is induced by galactose.
RESULTS
Bioinformatic analysis identifies mreCE genes upstream of usp45.
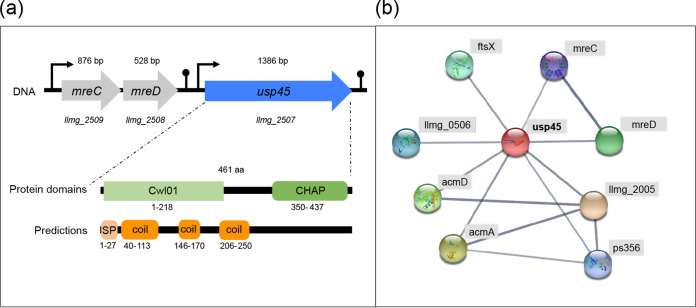
Analysis of the Lactococcus lactis MG1363 genome reveals that the two genes upstream from usp45 encode homologs of the cell shape-determining proteins MreC and MreD found in other Gram-positive bacteria (Fig. 1a). Previous studies identified the mreCD genes in ovococcus species always upstream from an usp45 orthologue (pcsB) (19). The conservation of this genomic organization in several Gram-positive bacteria suggests a relationship between the biological function of these proteins in cell wall metabolism and cell shape. We performed an analysis of the amino acid sequence of Usp45 using the Conserved Domains Database (CDD-NCBI) and InterProScan software to identify protein domains and to structurally predict protein motifs (see Fig. 1a) (20). We identified the Cwl01 and CHAP domains, which suggest that Usp45 has an amidase function (21). Cwl01 is a domain described as uncharacterized N-terminal domain of peptidoglycan hydrolase, and the CHAP domain is present in proteins involved in cell wall metabolism of bacteria (22). The structural prediction identified the Usp45 export signal (SPUsp45) and three coil motifs. Since a previous study of PcsB from S. pneumoniae describes key protein motifs for the protein activity, such as a coiled coil (CC) motif (11), we performed a sequence and structure alignment of the Usp45 protein sequence and determined the three-dimensional (3D) structural information of PcsB using the PROMALS3D software (see Fig. S1 in the supplemental material) (23). This alignment reveals the presence of the CC domain in Usp45 and corroborates the presence of the CHAP domain, including the conservation of three key amino acid residues of the catalytic CHAP domain (C292, H343, and E360). The results of the analysis of Usp45 protein domains and motifs are consistent with the structure of PcsB (11).
FIG 1.
Features of the L. lactis protein Usp45. (a) Genetic organization of the llmg_2508, llmg_2509 and usp45 genes and protein features (domain and predictions) of Usp45 of L. lactis. The products of llmg_2508 and llmg_2509 show homology to MreD and MreC, which were described as cell shape-determining proteins in previous studies (19). The arrows indicate transcription start sites. The domain features and predictions were identified with InterProScan software (EMBL-EBI) (https://www.ebi.ac.uk/interpro/) and the Conserved Domains Database (CDD-NCBI) (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). Cw101, uncharacterized N-terminal domain of peptidoglycan hydrolase (COG3883); CHAP, amidase function (pfam05257). Structural predictions identified a signal peptide (SP; highlighted in light orange color) and three coil motifs (indicated in orange). (b) The interaction network (as displayed by EMBL-STRING) for genetically interacting proteins possibly related in function with L. lactis Usp45 is shown. Protein-protein interaction network analysis obtained from the STRING database (https://string-db.org/cgi/input.pl?sessionId=LiYGlCGsH5Uw&input_page_show_search=on). Usp45 is illustrated as a red node. The thickness of the network edges indicates the strength of data support. Llmg_2005, cell wall-associated hydrolase; AcmA, N-acetylmuramidase; AcmD, N-acetylmuramidase, MreD, cell shape-determining protein; MreC, cell shape-determining protein; FtsX, cell division protein; Ps456, endolysin; Llmg_0505, uncharacterized protein.
To obtain insight into the proteins related to Usp45, a protein-protein interaction network analysis was performed with the STRING database (24) (Fig. 1b). Proteins identified in the network have a role in cell division or cell lysis, except for Llmg_0506, the function of which is unclear. For instance, the enzymes AcmA and AcmD have been extensively characterized in L. lactis (25, 26). AcmA participates in cell division and autolysis, whereas AcmD is also involved in cell separation and contributes to autolysis when AcmA is present (25).
The usp45 gene is essential for growth of L. lactis.
Genes homologous to lactococcal usp45 are essential in serotype 2 S. pneumoniae (pcsB) (8) and Enterococcus faecium (sagA) (27), but not in Staphylococcus aureus (ssa) (28), serotype 4 S. pneumoniae (29), or Streptococcus mutans (gsp-781) (30). None of the strategies employed by van Asseldonk et al. (3) to inactivate the chromosomal usp45 gene of L. lactis by homologous recombination were successful, suggesting that usp45 is essential under the conditions used. We constructed the pCSPusp45-usp45 L. lactis strain and tried several times to delete usp45. Homologous recombination in two steps was attempted, using the pCS1966 double crossover (DCO) strategy (31), by growing L. lactis cells on synthetic amino acid (SA) medium plates (32) supplemented with 30 μg · ml−1 5-fluoroorotic acid hydrate. No usp45 deletion mutant L. lactis strain was obtained. A second strategy entailed the replacement of Pusp45 by PnisA (the native usp45 gene driven by the nisA promoter). The DCO strategy was attempted in the pCS1966-PnisA-usp45 strain by growing this strain on SA medium plates with 30 μg · ml−1 5-fluoroorotic acid hydrate and nisin at different concentrations (10, 20, 30, 40, and 50 ng · ml−1). Again, no L. lactis colonies were obtained when Pusp45 was replaced by PnisA. These results clearly indicate that usp45 is an essential gene in L. lactis.
Use of the CRISPR-Cas9 system to target the usp45 gene.
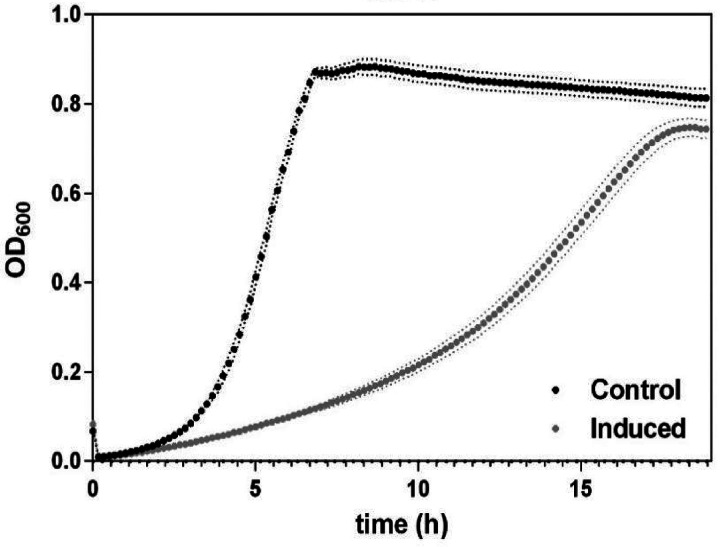
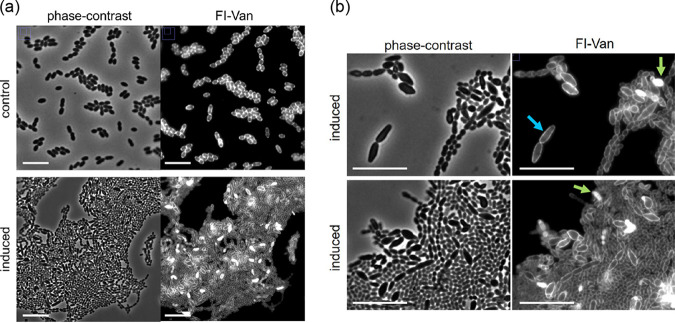
To repress usp45 by CRISPR interference (CRISPRi), the L. lactis sgRNA(usp45) strain was constructed. The production of a single guide RNA (sgRNA) against usp45 is induced with nisin (10 ng · ml−1). Repression of usp45 in L. lactis was performed in rich GM17 medium. Figure 2 shows that induction of sgRNA(usp45) results in a growth defect, i.e., a lower growth rate compared to that under noninduced conditions is observed when the cells are grown in GM17. Remarkably, Fig. 3a shows the effect of knocking down usp45 by the CRISPRi system on the cell phenotype in L. lactis. Cells grown under nisin-induced conditions show a variety of aberrant cell shapes, including small and large cells, clumps, and chains (Fig. 3b). Staining with fluorescent vancomycin (Fl-Van) was performed to visualize accumulation of peptidoglycan precursors during the cell division process (33). Besides the observation of the aberrant phenotype, some cells were stained entirely with Fl-Van. This accumulation of peptidoglycan precursors suggests that the cells failed to divide. Moreover, Fig. 3b highlights a cell division defect (blue arrow), where septum formation was not evident with the fluorescent vancomycin staining. In agreement with the results of inactivation of homologs to usp45 in Streptococcus agalactiae (7), the repression of usp45 in L. lactis cells shows irregular cell division compared to that in noninduced cells.
FIG 2.
Effect of induction of the single guide RNA against the usp45 gene on growth of Lactococcus lactis. Growth curves of the L. lactis sgRNA(usp45) strain in GM17 medium over 20 h, nisin-induced single guide RNA (induced; nisin was added at a concentration of 10 ng · ml−1) and control sample (noninduced). Points are means of 3 replicates for each growth curve. Data are presented as mean ± standard deviation (SD). Error bars represent SD.
FIG 3.
Induction of the single guide RNA against the usp45 gene of Lactococcus lactis results in aberrant cell shape phenotype. Growth of the L. lactis sgRNA(usp45) strain was performed in GM17 medium. (a) Cells were grown in two conditions, nisin-induced single guide RNA (induced; nisin was added at a concentration of 10 ng · ml−1) and control sample (noninduced). (b) Accumulation of fluorescent vancomycin (Fl-Van) is indicated with a green arrow, and disturbed cell separation is indicated with a blue arrow. Samples of each bacterial culture were taken at exponential growth phase (optical density at 600 nm of 0.5). Representative images of fluorescent vancomycin (Fl-Van) and phase-contrast are shown. Bars, 10 μm.
Overexpression of the usp45 gene.
Overexpression of the usp45 gene was performed by nisin induction in the L. lactis pNZ-PnisA-usp45 strain. Figure 4 shows that overexpression of usp45 driven by nisin induction results in a lower growth rate and lower cell density compared to those of the control. Next, we evaluated the effect on the phenotype of cells at exponential growth phase (Fig. 5). The overexpression of usp45 causes no visible effect on cell shape, nor does it have an effect on the localization of cell wall synthesis. Moreover, no irregularities in septum formation during cell division were found.
FIG 4.
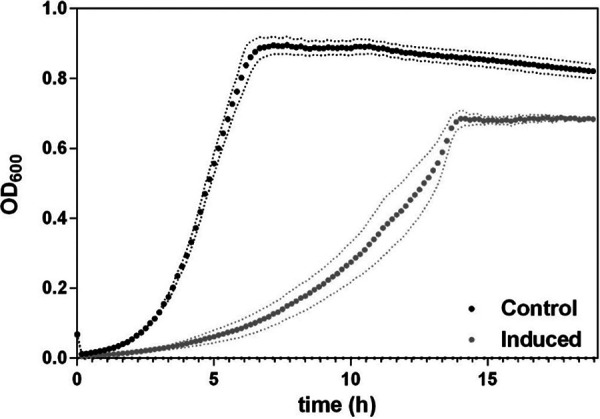
Effect of overexpression of usp45 on the growth of L. lactis. Growth curves of the L. lactis PnisA-usp45 strain in GM17 medium over 20 h, nisin-induced usp45 expression (induced; nisin was added at a concentration of 10 ng · ml−1) and control sample (noninduced). Points are means of 3 replicates for each growth curve. Data are presented as mean ± standard deviation (SD). Error bars represent SD.
FIG 5.
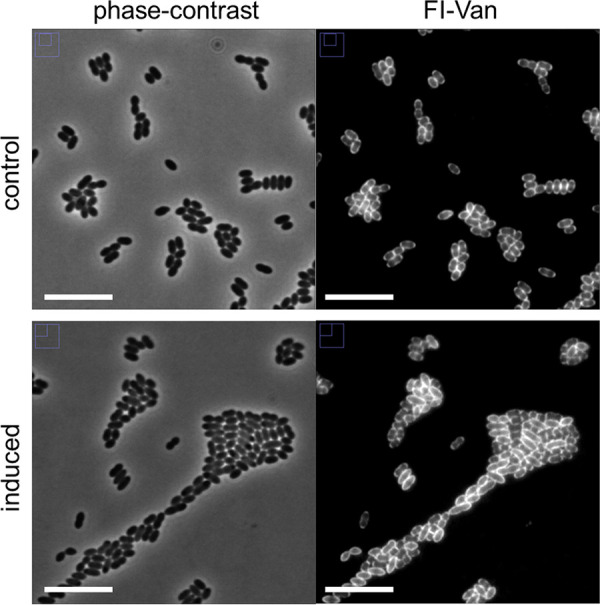
Effect of overexpression of usp45 on phenotype of L. lactis. Growth of the L. lactis PnisA-usp45 strain was performed in GM17 medium. Cells were grown under the following two conditions: nisin-induced single guide RNA (induced; nisin was added at a concentration of 10 ng · ml−1) and control sample (noninduced). Samples of each bacterial culture were taken in the exponential growth phase (optical density at 600 nm [OD600] of 0.5). Peptidoglycan synthesis was observed by staining the cells with fluorescent vancomycin (Fl-Van). Representative fluorescent vancomycin and phase-contrast images are shown. Bars, 10 μm.
The usp45 promoter is induced by galactose.
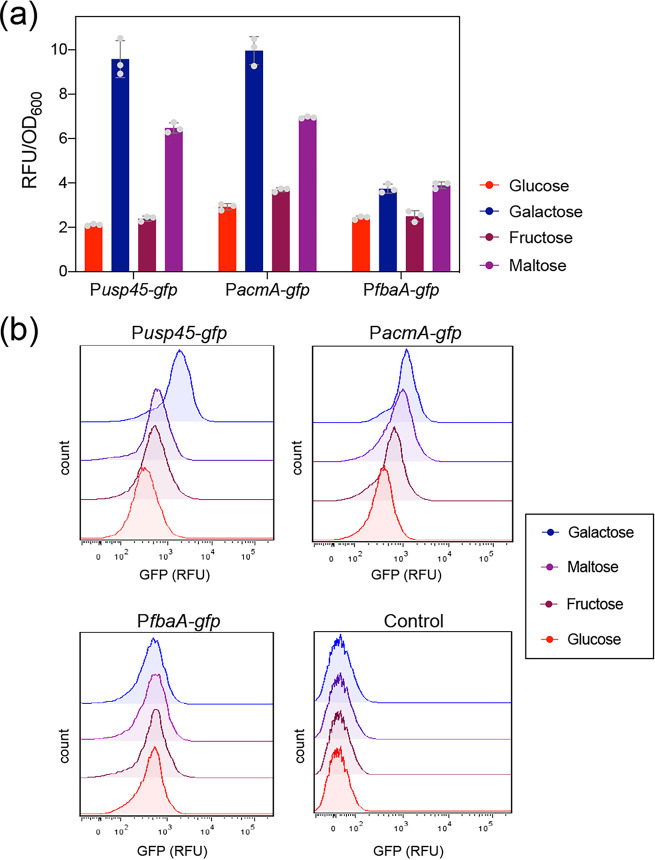
Although the usp45 promoter has been previously described as a strong promoter in L. lactis (34), we investigated whether the activity of the usp45 promoter is affected when L. lactis is grown with a carbon source other than glucose. Previous studies have shown that the binding of the major autolysin AcmA to the peptidoglycan is reduced when L. lactis cells are grown on galactose (26). Therefore, we tested the activity of both the acmA and usp45 promoters in L. lactis Pusp45-gfp and PacmA-gfp strains grown in chemically defined medium (CDM) supplemented with different carbon sources. Figure 6a shows that both promoters are highly induced in the presence of galactose, and to a lesser extent when maltose is present. An ∼4-fold increase of the usp45 promoter activity and an ∼3.5-fold increase of the acmA promoter activity is observed in cells grown on galactose compared to that in cells grown on glucose (Fig. 6a). To validate our results, we aimed to test the activity of a constitutive promoter to the presence of various sugars. Thus, we constructed another transcriptional fusion with the promoter PfbaA. The fbaA gene encodes the fructose-bisphosphate aldolase, a key enzyme in the glycolysis pathway, i.e., it has a housekeeping role in metabolism (35). The green fluorescent protein (GFP) expression values of the PfbaA-gfp strain were more homogenous in the presence of different sugars than those of the Pusp45-gfp and PacmA-gfp strains. Although all of the strains were inoculated with the same dilution (see Materials and Methods), they differed in growth rate when grown in CDM supplemented with different carbon sources. This growth effect was taken into account to correct the GFP expression values at different optical density values (see Materials and Methods and Fig. S2 in the supplemental material). Moreover, we performed single-cell GFP measurements by flow cytometry to corroborate the effects of sugars on the activity of the usp45 and acmA promoters. Accordingly, Fig. 6b shows a shift in the GFP expression levels of the Pusp45-gfp and PacmA-gfp strains grown in the presence of galactose and maltose compared to the GFP expression levels when grown in glucose. We also measured the GFP expression of the PfbaA-gfp strain and observed homogenous GFP expression when it was grown in the presence of different sugars. We added a second control for this experiment by constructing a L. lactis strain (indicated as the control strain in Fig. 6b) bearing the empty vector pSEUDO-gfp (promoterless) to distinguish the background fluorescence values. Together, these findings confirm that both promoters, acmA and usp45, are highly induced in the presence of galactose.
FIG 6.
The usp45 and acmA promoters are induced by sugars. (a) The usp45, acmA, and fbaA promoters were fused to the green fluorescent protein reporter gene (gfp). L. lactis strains bearing each construct (Pusp45-gfp, PacmA-gfp, and PfbaA-gfp) were grown in chemically defined medium (CDM) in the presence of different sugars (glucose, galactose, fructose, and maltose) at concentrations of 0.5% (wt/vol). Population-level normalized green fluorescent protein (GFP) expression (relative fluorescence units [RFU]/OD600) of bacterial cultures. Data are presented as mean ± standard deviation (SD). Dots represent the single values of independent experiments (n = 3). Error bars represent SD. (b) Single-cell fluorescence measurements by flow cytometry in the presence of different sugars at concentrations of 0.5% (wt/vol). Fluorescence measurements were taken at the beginning of the stationary-growth phase. Ten thousand ungated events for each sample are shown. A control sample of a L. lactis strain bearing an empty vector (promoterless pSEUDO-gfp) was used to compare background fluorescence of the bacterial cells.
Similarly to what was seen when the cells are grown on glucose, different cell shapes were observed when usp45 was inactivated in cells grown on galactose (Fig. 7). There was a reduced variation in cell shape, with long-chain formation being the predominant phenotypic change. This observation could be due to the fact that when the galactose–mediated induction results in large amounts of Usp45, the repression of usp45 by the nisin-induced CRISPRi system might not reach the same repression levels as those when cells are grown with glucose.
FIG 7.
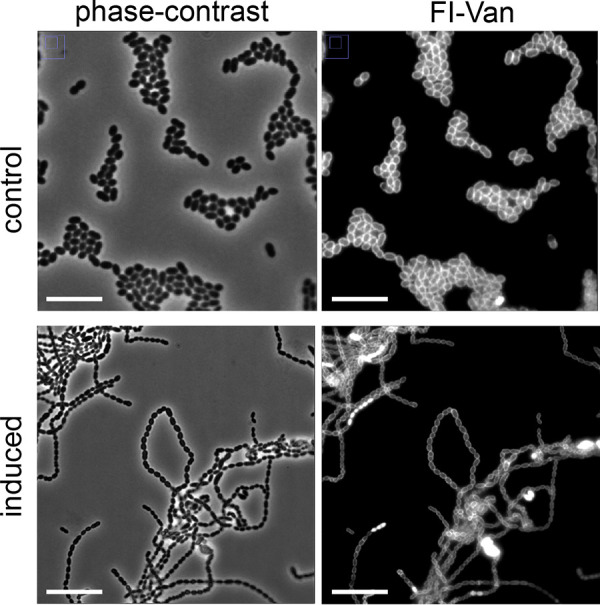
Effect of sugars on cell separation. The nisin-induced single guide RNA against usp45 affects the cell phenotype of L. lactis sgRNA(usp45) when it is grown in M17 containing 0.5% (wt/vol) galactose. Cells were grown under two conditions, nisin-induced single guide RNA (induced; nisin was added at a concentration of 10 ng · ml−1) and control sample (noninduced). Representative images of the control sample (noninduced) are shown. Peptidoglycan synthesis was observed by staining the cells with fluorescent vancomycin (Fl-Van). Representative fluorescent vancomycin (Fl-Van) and phase-contrast images are shown. Bars, 10 μm.
DISCUSSION
We report here that the long-known Usp45 protein of L. lactis is essential for proper cell division. Usp45 from L. lactis has catalytic domains similar to that of PcsB in other streptococcal species (36). Inactivation of the usp45 gene affects cell wall synthesis and cell shape in L. lactis, which is consistent with a role of Usp45 acting as a cell wall hydrolase, and a function that is attributed to the cysteine, histidine-dependent amidohydrolases/peptidases (CHAP) domain (Pfam identifier PF05257). Moreover, sequence and structure alignment of the Usp45 protein sequence and 3D structural information of PcsB reveals the conservation of three key amino acid residues of the catalytic CHAP domain (C292, H343, and E360; see Fig. S1 in the supplemental material). The essentiality of the Usp45 in L. lactis is comparable to that of the PcsB in S. pneumoniae, in which the cysteine and histidine residues from the CHAP domain were shown to be required for viability (22, 37). Since previous studies performed by van Asseldonk et al. reported that the purified Usp45 protein has no function in the proteolytic system of L. lactis, nor antimicrobial activity against other Gram-positive bacteria (3), we speculate that Usp45 requires an ATP-driven conformational change to activate the catalytic domains in vivo, as is predicted for its homolog PcsB in S. pneumoniae (11). The VicRK TCS regulates pcsB in S. pneumoniae, and the VicR regulator is essential for viability in this bacterium (38). The essentiality of VicR is caused by regulation of multiple genes, including genes that mediate wall teichoic acid biosynthesis, virulence, or exopolysaccharide production (17, 39). Conversely, in the homologous TCS in L. lactis (KinC and LlrC), VicR (KinC) is not essential, but its deletion results in a clumping phenotype (18). In addition, L. lactis strains where either the kinC or llrC gene is disrupted show a similar phenotype to that of the wild type (WT) (see Fig. S3 in the supplemental material). These findings suggest that either the lactococcal usp45 may not be completely regulated by the homologous TCS in L. lactis (KinC and LlrC) or that this TCS only indirectly affects Usp45 expression (15).
The CRISPRi-mediated knockdown of usp45 led to aberrantly shaped cells, variation in cell size, and different patterns of septum formation (Fig. 3a). The formation of long chains of cells is a phenotype often described for strains deficient in cell wall hydrolytic activity (40). Thus, inactivation of usp45 causes a cell separation defect and unusual cell wall synthesis. Accordingly, we suggest that Usp45 acts as a cell wall hydrolase that participates in cell separation and cell wall synthesis.
The usp45 gene product is a secreted 45-kDa protein. There is a discrepancy in the literature regarding the description of Usp45 as a surface protein with two possible types of surface display, either as a lipoprotein or anchored by transmembrane helices (TMH). This description is based on a study that combined software prediction and proteomic data, which suggested that the Usp45 protein might exist in an equilibrium between unbound and bound states (41). However, recent studies of the PcsB protein provide experimental evidence of the PcsB anchoring to the cell via its interaction with the membrane-embedded protein FtsX (11, 42). Our study further supports the supposition of Usp45 being a surface protein that plays an important role in cell division.
In a previous study, the usp45 gene was used as an internal standard because it is presumably constitutively expressed (43). Here, we reveal that the usp45 promoter is strongly induced by galactose. Previous studies on the major autolysin AcmA of L. lactis have shown that the nature of the carbon source influences binding of AcmA to peptidoglycan (26). The authors confirmed that growth of L. lactis on galactose affects the carbohydrate composition in the lipoteichoic acids (LTAs) in the cell wall in such a way that AcmA binding is decreased compared to that when cells are grown on glucose. As alanylation or galactosylation of the LTAs affects the function of AcmA, these changes in cell wall composition, when L. lactis is grown on galactose, could diminish efficient peptidoglycan hydrolysis by Usp45. Therefore, large amounts of Usp45 are required for efficient cell division when galactose is present, and thus the usp45 promoter is highly induced. In support of this hypothesis, Fig. 6a shows that repression of usp45, when the cells are grown on galactose, causes the formation of long chains of cells. We speculate that, although cell separation is impaired, the lesser variation in cell shape and cell size compared to those when usp45 is inactivated in cells grown on glucose (Fig. 3) might result from a higher expression of usp45 that counterbalances the inactivation.
The present results are significant in at least two major respects. First, we provide evidence for a role of L. lactis Usp45 in cell shape and division. Second, since the usp45 promoter and Usp45 signal peptide are commonly used for heterologous protein secretion, induction of Pusp45 by galactose might be further developed for novel engineering strategies. Moreover, based on our results, food-grade suicide systems can be developed, which would remove recombinant L. lactis cells from food products after they have performed their function.
Further research should be undertaken to investigate whether one of the TCSs in L. lactis is responsible of the regulation of usp45. Since all known homologs of Usp45 in other Gram-positive bacteria are regulated by the WalKR (VicRK) two-component system, it would be relevant to determine what makes L. lactis an exception to this rule. One of the issues that emerges from previous studies and our findings is that PcsB-like proteins might not only be typical hydrolases but also be enzymes that organize the cell division process (42), a role that would be consistent with the essential nature of the usp45 gene and its homologs (44).
The present study supports a model in which Usp45 mediates cell separation by its catalytic activity as peptidoglycan hydrolase. Gram-positive bacteria produce several enzymes that hydrolyze peptidoglycan (45, 46). Lactococcus lactis produces three different types of peptidoglycan hydrolases (47, 48). According to our data, when Usp45 is expressed at low levels, the balance between cell wall synthesis and cell division is affected. Usp45 may affect the activity of other cell wall hydrolases such as AcmA and AcmD. The great variation in aberrant cell shapes caused by irregular cell wall synthesis is a remarkable sign of the consequences of a perturbation in the peptidoglycan synthesis and turnover harmony.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains used in this study are listed in Table 1. L. lactis was grown as standing cultures at 30°C in M17 broth (Difco BD, NJ, USA) with 0.5% (wt/vol) glucose (GM17) or in chemically defined medium (CDM) (49) supplemented with glucose (GCDM), galactose, fructose, or maltose (Sigma-Aldrich, MO, USA). All sugars were added at concentrations of 0.5% (wt/vol). GM17-agar plates contained 1.5% (wt/vol) agar. When necessary, culture media were supplemented with erythromycin (Sigma-Aldrich, MO, USA) and/or chloramphenicol (Sigma-Aldrich, MO, USA), both at 5 μg · ml−1.
TABLE 1.
Strains and plasmids used in this study
| Strain name/genotype or plasmida | Relevant genotype and/or descriptionb | Source or reference |
|---|---|---|
| L. lactis | ||
| NZ9000 (WT) | MG1363 pepN::nisRK | 50 |
| sgRNA(usp45) | Eryr Cmr, NZ9000 carrying pNZ-PnisA-dcas9 and pTLR-Pusp45-sgRNA(usp45) | This work |
| PnisA-usp45 | Cmr, NZ9000 carrying pNZ-PnisA-usp45 | This work |
| Pusp45-gfp | Eryr, NZ9000 carrying pSEUDO::Pusp45-gfp | 53 |
| PacmA-gfp | Eryr, NZ9000 carrying pSEUDO::PacmA-gfp | This work |
| pCSPusp45-usp45 | Eryr, NZ9000 carrying pCS1966::Pusp45-usp45 | This work |
| pCSPnisA-usp45 | Eryr, NZ9000 carrying pCS1966::PnisA-usp45 | This work |
| PfbaA-gfp | Eryr, NZ9000 carrying pSEUDO::PfbaA-gfp | This work |
| pSEUDO vector | Eryr, NZ9000 carrying pSEUDO::gfp (promoterless) | This work |
| ΔkinC | Eryr derivative of MG1363 with 800-bp disruption in the kinC gene; MGKinC | 18 |
| ΔllrC | Eryr derivative of MG1363 with 500-bp disruption in the llrC gene; MGRrC | 18 |
| E. coli | ||
| DH5α | F− Φ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK+) phoA supE44 λ− thi-1 gyrA96 relA1 | Laboratory stock |
| Plasmids | ||
| pCS1966 | Eryr, oroP integration vector | 31 |
| pSEUDO-sfgfp(Bs) | Eryr, pCS1966 derivative for integration in llmg_pseudo10 locus | 56 |
| pNZ8048 | Cmr, PnisA nisin-inducible gene expression vector | 51 |
| pJWV102-Plac-dcas9sp | Ampr, Plac-dcas9sp | 54 |
| pNZ-PnisA-sgRNA(usp45) | Cmr, pNZ8048 carrying PnisA-sgusp45 | This work |
| pSEUDO::Pusp45-dcas9 | Eryr, pSEUDO carrying Pusp45-dcas9 | This work |
| pNZ-PnisA-usp45 | Cmr, pNZ8048 carrying PnisA-usp45 | This work |
| pSEUDO::Pusp45-gfp | Eryr, pSEUDO carrying Pusp45-sfgfp(Bs) | 53 |
| pSEUDO::PacmA-gfp | Eryr, pSEUDO carrying PacmA-sfgfp(Bs) | This work |
| pSEUDO::PfbaA-gfp | Eryr, pSEUDO carrying PfbaA-sfgfp(Bs) | This work |
| pCS1966::Pusp45-usp45 | Eryr, pSEUDO carrying Pusp45-usp45 | This work |
| pCS1966::PnisA-usp45 | Eryr, pSEUDO carrying PnisA-usp45 | This work |
| pNZ-PnisA-dcas9 | Cmr, pNZ8048 carrying PnisA-dcas9 | This work |
| pTLR-Pusp45-sgRNA(usp45) | Eryr, pTLR carrying Pusp45-sgRNA(usp45) | This work |
WT, wild type; sgRNA, single guide RNA.
Cmr, chloramphenicol resistant; Eryr, erythromycin resistant; Ampr, ampicillin resistant.
Escherichia coli DH5α (Life Technologies, Gaithersburg, MD) was used to perform all recombinant DNA techniques. Cells were grown at 37°C in Luria-Bertani (LB) broth or Luria-Bertani agar 1.5% (wt/vol) (Difco BD). For screening of colonies containing recombinant plasmids, chloramphenicol (25 μg · ml−1) or erythromycin (150 μg · ml−1) was added.
Nisin induction was performed by diluting an overnight culture of L. lactis 1:50 and adding nisin (Sigma-Aldrich, Munich, Germany) to a final concentration of 10 ng · ml−1.
For microscopy experiments and plate reader assays, L. lactis was grown in GCDM. Exponentially growing cells (optical density at 600 nm [OD600] of 0.3) were collected by centrifugation in a Microfuge 16 centrifuge (Beckman Coulter, Woerden, The Netherlands) and washed three times with phosphate-buffered saline (PBS) (pH 7.2) solution containing KH2PO4 at 15.44 μM, NaCl at 1.55 mM, and Na2HPO4 at 27.09 μM.
Recombinant DNA techniques and oligonucleotides.
DNA amplifications by PCR were performed using a PCR mix containing Phusion high-fidelity (HF) buffer (Thermo Fisher Scientific, Inc., MA, USA), 2.5 mM deoxynucleoside triphosphate (dNTP) mix, Phusion HF DNA polymerase (Thermo Fisher Scientific, Inc.), oligonucleotides (0.5 μM each), and 50 ng of L. lactis chromosomal DNA as the template. Oligonucleotides (Table 2) were purchased from Biolegio (Nijmegen, The Netherlands). PCRs were performed in an Eppendorf thermal cycler (Eppendorf, Hamburg, Germany). The DNA target sequence of interest was amplified by 35 cycles of denaturation (98°C for 30 s), annealing (5°C or more, lower than the melting temperature [Tm] for 30 s), and extension (70°C for 1 min per 1 kbp). Amplifications were confirmed by the 1% agarose gel electrophoresis method.
TABLE 2.
Oligonucleotides used in this study
| Name | Sequence (5′–3′) |
|---|---|
| usp45-Rv_NcoI | AGAACAGCCATGGAAAAAAAGATTATCTCAG |
| usp45-Fw_KpnI | TAAGACGGTACCCTAGTTTGGCATCAAGAAAGTAAC |
| PacmA-Fw | GGCATACTCGAGTACCTTTCTAAAGATTACAAA |
| PacmA-Rv | TATCCAGCATGCTCATTTACATCATCTATTCTATC |
| PfbaA-Fw | GGGTCGATCGAATTCGGTCCTCGGGATATG |
| PfbaA-Rv | GACTTTGCAAGCTTGCATGCCTGCAGGTCG |
| FwKI_A_XbaI | GCAATTGTCTAGAGCGGCCGCCAACAACCTTGTA |
| RvKI_A_KpnI | CAATTGGGTACCCAGCTTTTGTTCCCTTTAGTGAGG |
| FwKI_B_KpnI | GTGCTTGGGTACCTTCTTTTTGTTGAGCTTCATAAGCC |
| RvKI_B_EcoRI | ATTAAAGAATTCCAGTTATGAAAAAAAAGATT |
| FwKI_C_BamHI | ACAGCTCGGATCCCTAGTCTTATAACTATACTG |
| RvKI_C_EcoRI | TGCAGTAGAATTCGTGAGTGCCTCCTTATAATT |
| FwKI_D_BamHI | TCCAGCGATTAGTAAATATAGGATCCTGTATA |
| RvKI_D_XbaI | CTAACTCTCTAGATGAGGGTGGAACACCAAGTG |
| KOusp45_1Fw | GCATTCTAGATAAGTAGTGAGTCGATTTAC |
| KOusp45_2Rv | GCATGGATCCTTAAAATAGCTGAGATAATC |
| KOusp45_3Fw | GCATGGATCCGTGCGTCTGGTGTTACTTTC |
| KOusp45_4Rv | CGTACTCGAGAATGATCGCGTGCACCAAAC |
| 0310-sgRNA(usp45)_F | AGTATAATATGTTTAAGCGTAAACACCTGACAACGGTTTAAGAGCTATGC |
| 0311-sgRNA(usp45)_R | GCATAGCTCTTAAACCGTTGTCAGGTGTTTACGCTTAAACATATTATACT |
| 0153-sgRNA_backbone_FW | GTTTAAGAGCTATGCTGGAAACAG |
| 0154-sgRNA_backbone_RV | TAAACATATTATACTATTCCTACCCCAC |
| 0217-pNZ8048_USER_F | AGCTTTATAAGUAATTACAGCACGTGTTGCTTTGATTG |
| 0221-Pnis_pNZ8048_R | ATTTCTTATCCAUTGGTGAGTGCCTCCTTATAATTT |
| 0032-Pnis_dCas9_F | ATGGATAAGAAAUACTCAATAGGCTTAG |
| 0220-dCas9-R | ACTTATAAAGCUCTCGAGGTCGACTTAGTCAC |
For DNA cloning, we used FastDigest restriction enzymes and T4 DNA ligase (Thermo Fisher Scientific, Inc.). Reactions were performed according to the manufacturer’s recommendations. The ligation products were transformed into E. coli DH5α (Life Technologies) competent cells by electroporation. Cells were plated on Luria-Bertani agar plates with appropriate antibiotics and grown overnight at 37°C. Screening of colonies to confirm the genetic construct was performed by colony PCR. Positive colonies with correct constructs were inoculated in Luria-Bertani broth with the appropriate antibiotic. Plasmid DNA and PCR products were isolated and cleaned up with a High Pure plasmid isolation kit (Roche Applied Science, Mannheim, Germany) according to the protocol of the manufacturer. DNA sequences of constructs were always confirmed by DNA sequencing (Macrogen Europe, Amsterdam, The Netherlands).
Construction of L. lactis strains.
Lactococcus lactis NZ9000 was used throughout (50). To overexpress the Usp45 protein, plasmid pNZ-PnisA-usp45 was constructed. It carries the L. lactis usp45 gene under the control of the nisA promoter. The usp45 coding region was amplified by PCR using the oligonucleotides Usp45_Fw and Usp45_Rv and chromosomal DNA of L. lactis NZ9000 as a template. The PCR fragment was cleaved with the enzymes NcoI and KpnI, after which it was inserted into the high-copy-number plasmid pNZ8048 digested with the same enzymes (51). Plasmid pNZ-PnisA-usp45 was introduced into L. lactis NZ9000 by electroporation as described by Holo and Nes (52). Transformants were selected on M17-agar plates supplemented with sucrose, glucose, and chloramphenicol (5 μg · ml−1), yielding L. lactis PnisA-usp45.
To construct the plasmids pSEUDO::PacmA-gfp and pSEUDO::PfbaA-gfp carrying the L. lactis acmA and fbaA promoters upstream from the gfp gene, the acmA and fbaA promoters were amplified by PCR using the oligonucleotides PacmA_fw and PacmA_Rv, PfbaA_fw, and PfbaA_Rv and chromosomal DNA of L. lactis NZ9000 as a template. The PCR fragments were digested with PaeI and XhoI and inserted into pSEUDO-sfgfp(Bs) digested with the same enzymes. Plasmids pSEUDO::PacmA-gfp and pSEUDO::PfbaA-gfp were integrated into the silent llmg_pseudo10 locus of L. lactis NZ9000 by single-crossover integration as described previously (53), yielding L. lactis PacmA-gfp and L. lactis PfbaA-gfp, respectively.
To obtain usp45 double-crossover gene deletion mutants, upstream and downstream regions of usp45 were selected and amplified using the oligonucleotides KOusp45_1Fw plus KOusp45_2Rv and KOusp45_3Fw plus KOusp45_4Rv, respectively. The upstream fragment was ligated into pCS1966 (31) using XbaI and BamHI restriction sites. The plasmid obtained was named pCS1966-A. Downstream fragment B was cloned into pCS1966-A using BamHI and XhoI restriction, and the resulting plasmid was named pCS1966::Pusp45-usp45. All pCS1966 derivatives were initially constructed in E. coli DH5α and then introduced to L. lactis by electroporation. Homologous recombination in two steps was attempted by growing L. lactis cells on SA medium plates (32) supplemented with 30 μg · ml−1 5-fluoroorotic acid hydrate (Sigma-Aldrich, Munich, Germany). A second strategy entailed the replacement of Pusp45 by PnisA. Four PCR products were obtained: fragment A (oligonucleotides FwKI_A_XbaI and RvKI_A_KpnI amplify the backbone pCS1966), fragment B (oligonucleotides FwKI_B_KpnI and RvKI_B_EcoRI amplify the upstream region of the usp45 promoter), fragment C (oligonucleotides FwKI_C_BamHI and RvKI_C_EcoRI amplify the nisA promoter from pNZ8048), and fragment D (oligonucleotides FwKI_D_BamHI and RvKI_EcoRI amplify the downstream region of the usp45 promoter). The PCR products were ligated into pCS1966 via their corresponding restriction sites. The obtained vector was named pCS1966-PnisA-usp45 and was introduced by homologous recombination in L. lactis NZ9000. The strategy was then employed by using SA medium plates with 30 μg · ml−1 5-fluoroorotic acid hydrate and nisin (Sigma-Aldrich, Munich, Germany) at different concentrations (10, 20, 30, 40, and 50 ng · ml−1).
To obtain L. lactis sgRNA(usp45) (pNZ-PnisA::dcas9; pTLR-Pusp45::sgRNA-usp45), the dcas9 gene was amplified from plasmid pJWV102-Plac-dcas9sp with the oligonucleotides 0217-pNZ8048_F and 0221-Pnis_pNZ8048_R. Plasmid pNZ8048 was amplified by PCR with oligonucleotides 0032-Pnis_dCas9_F and 0220-dCas9-R. The two fragments were ligated and used to transform E. coli, yielding pNZ-PnisA-dcas9. This plasmid was introduced into L. lactis NZ9000. The second plasmid carries the sgRNA gene targeting usp45. It was obtained by the infusion cloning method (54). The set of oligonucleotides 0153-sgRNA_backbone_FW and 0154-sgRNA_backbone_RV were designed to obtain the linearized version of plasmid pTLR-Pusp45-sgRNA(acmA) (Chenxi Huang, unpublished data). The 20-nucleotide (nt) guide sequence targeting acmA was replaced by a sequence targeting usp45 (AGCGTAAACACCTGACAACG). To this end, two 50-nt complementary oligonucleotides were designed [0310_sgRNA(usp45)_F and 0311-sgRNA(usp45)_R], with each oligonucleotide containing 15-nt overlaps with the linearized plasmid, one on each side. The plasmid obtained in E. coli pTLR-Pusp45::sgRNA(usp45) was introduced in L. lactis pNZ-PnisA::dcas9.
Plate-reader assays.
Cultures of L. lactis were grown and prepared as described above. For growth curves, L. lactis cells were diluted 1:50 in CDM or M17, both containing either glucose, maltose, fructose, or galactose. All sugars were added at concentrations of 0.5% (wt/vol). Growth was recorded in 0.2-ml cultures in 96-well microtiter plates and monitored using the microtiter plate reader VarioSkan (Thermo Fisher Scientific, Inc.). The OD600 was recorded every 10 min for 24 h. The signal was corrected for background noise of the medium.
The effect of sugars on promoter activities was determined as follows. All sugars were added at concentrations of 0.5% (wt/vol). Growth and GFP expression were monitored using the microtiter plate reader VarioSkan (Thermo Fisher Scientific, Inc.) by measuring the optical density at 600 nm (OD600) and the fluorescence signal (excitation, 485 nm; emission, 535 nm) every 10 min for 24 h. Both signals were corrected for the background values of the medium used for growth. The OD600 values used were corrected for the background value of the corresponding medium used for growth (CDM). The calculation used for resolving the relative GFP measurements (relative fluorescence units [RFU]/OD600) of the cultures is depicted by the following formula:
GFPpromoter and ODpromoter are the fluorescence and optical density values of the L. lactis strain bearing the promoter of interest fused to the gfp gene. GFPmedium and ODmedium are the fluorescence and optical density values of the growth medium. GFPcontrol and ODcontrol are the fluorescence and optical density values of the control L. lactis strain (empty vector; see Table 1).
The maximum value of the fluorescence peak in each sample was considered to be the GFP value in all figures of this work and was corrected with the formula mentioned above, yielding the relative fluorescent values (RFU/OD600).
Flow cytometry.
L. lactis cultures were grown overnight in CDM as described above, washed three times in PBS, and transferred to fresh CDM supplemented with various carbon sources (glucose, galactose, fructose, and maltose). All sugars were added at a concentration of 0.5% (wt/vol). The cultures were incubated at 30°C, and samples were taken at the beginning of the stationary-growth phase. Thresholds for the forward scatter (FSC) and side scatter (SCC) parameters were set (200 in both) in a FACSCanto flow cytometer (BD Biosciences, CA, USA) to remove all events that did not correspond to cells. The GFP signal in all measured cells was recorded in 10,000 events and used for downstream analysis (ungated events shown in Fig. 6b). GFP signal measurements were obtained with a FACS Canto flow cytometer (BD Biosciences) using a 488-nm argon laser. Raw data were collected using FACSDiva Software 5.0.3 (BD Biosciences), and FlowJo software was used for data analysis.
Fluorescence microscopy.
Washed, exponentially growing cells were transferred to a solidified thin layer of CDM with high-resolution agarose 1.5% (wt/vol) (Sigma-Aldrich). A standard microscope slide was prepared with a layer of solidified agar. Bacterial cells were spotted on the agar and covered with a standard microscope coverslip. For Fl-Van experiments, 10 μl of bacterial culture was taken at the end of the stationary phase and stained without fixation with Fl-Van (a 1:1 mixture of vancomycin [Sigma-Aldrich)] and Bodipy FL-conjugated vancomycin (Sigma-Aldrich) at a final concentration of 2 μg · ml−1), followed by 5 min of incubation in the dark at room temperature.
Microscopy was performed with a temperature-controlled (Cube and Box incubation system; Life Imaging Services, Basel, Switzerland) DeltaVision (Applied Precision, WA, USA) IX7I microscope (Olympus, PA, USA) at 30°C. Images were obtained with a CoolSnap HQ2 camera (Princeton Instruments, NJ, USA) at 60-fold or 100-fold magnification using a 300-W xenon light source for bright-field and Fl-Van detection (filter from Chroma; excitation, 470/40 nm, and emission, 525/50 nm). Snapshots in bright-field microscopy and for Fl-Van detection were taken with 10% white light-emitting diode (LED) light (Applied Precision); 0.05-s exposure was used for bright-field detection and a 100% xenon light and 0.8 s of exposure were used for Fl-Van signal detection. The raw data were stored using softWoRx 3.6.0 (Applied Precision) and analyzed using ImageJ software (55).
Statistics.
Statistical analyses were performed using Prism 6.01 (GraphPad Software, Inc.). All experiments were repeated independently at least three times. All micrographs show representative images from three independent replicate experiments.
Bioinformatics.
Protein structural predictions and domain features were identified using InterProScan software (20). Protein domain features were identified using the Conserved Domains Database (CDD-NCBI). Protein-protein interaction network analysis was performed by employing the STRING database (https://string-db.org/) (24). Protein alignments (sequences and structure) were performed by using the PROMALS3D multiple sequence and structure alignment server (23).
Supplementary Material
ACKNOWLEDGMENTS
We thank Saulius Kulakauskas (INRA, France) for helpful discussions. We thank Douwe van Sinderen and Mary O’Connell Motherway (both from APC Microbiome Institute, Ireland) for providing the L. lactis MGKinC and MGRrC strains.
J.A.H.-V. and O.P.K. were financed by the Netherlands Organization for Scientific Research (NWO), research program TTW (grant 13858). C.H. holds a China Scholarship Council (CSC) scholarship (no. 201505990303).
We declare no conflicts of interest.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Villatoro-Hernández J, Kuipers OP, Saucedo-Cárdenas O, Montes-de-Oca-Luna R. 2012. Heterologous protein expression by Lactococcus lactis. Methods Mol Biol 824:155–165. doi: 10.1007/978-1-61779-433-9_8. [DOI] [PubMed] [Google Scholar]
- 2.Ng DTW, Sarkar CA. 2013. Engineering signal peptides for enhanced protein secretion from Lactococcus lactis. Appl Environ Microbiol 79:347–356. doi: 10.1128/AEM.02667-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Asseldonk M, Rutten G, Oteman M, Siezen RJ, de Vos WM, Simons G. 1990. Cloning of usp45, a gene encoding a secreted protein from Lactococcus lactis subsp. lactis MG1363. Gene 95:155–160. doi: 10.1016/0378-1119(90)90428-t. [DOI] [PubMed] [Google Scholar]
- 4.Borrero J, Jiménez JJ, Gútiez L, Herranz C, Cintas LM, Hernández PE. 2011. Use of the usp45 lactococcal secretion signal sequence to drive the secretion and functional expression of enterococcal bacteriocins in Lactococcus lactis. Appl Microbiol Biotechnol 89:131–143. doi: 10.1007/s00253-010-2849-z. [DOI] [PubMed] [Google Scholar]
- 5.van Asseldonk M, de Vos WM, Simons G. 1993. Functional analysis of the Lactococcus lactis usp45 secretion signal in the secretion of a homologous proteinase and a heterologous α-amylase. Mol Gen Genet 240:428–434. doi: 10.1007/BF00280397. [DOI] [PubMed] [Google Scholar]
- 6.Song AAL, In LLA, Lim SHE, Rahim RA. 2017. A review on Lactococcus lactis: from food to factory. Microb Cell Fact 16:55. doi: 10.1186/s12934-017-0669-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reinscheid DJ, Gottschalk B, Schubert A, Eikmanns BJ, Chhatwal GS. 2001. Identification and molecular analysis of PcsB, a protein required for cell wall separation of group B streptococcus. J Bacteriol 183:1175–1183. doi: 10.1128/JB.183.4.1175-1183.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ng WL, Kazmierczak KM, Winkler ME. 2004. Defective cell wall synthesis in Streptococcus pneumoniae R6 depleted for the essential PcsB putative murein hydrolase or the VicR (YycF) response regulator. Mol Microbiol 53:1161–1175. doi: 10.1111/j.1365-2958.2004.04196.x. [DOI] [PubMed] [Google Scholar]
- 9.Mattos-Graner RO, Jin S, King WF, Chen T, Smith DJ, Duncan MJ. 2001. Cloning of the Streptococcus mutans gene encoding glucan binding protein B and analysis of genetic diversity and protein production in clinical isolates. Infect Immun 69:6931–6941. doi: 10.1128/IAI.69.11.6931-6941.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pancholi V, Boël G, Jin H. 2010. Streptococcus pyogenes Ser/Thr kinase-regulated cell wall hydrolase is a cell division plane-recognizing and chain-forming virulence factor. J Biol Chem 285:30861–30874. doi: 10.1074/jbc.M110.153825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartual SG, Straume D, Stamsås GA, Muñoz IG, Alfonso C, Martínez-Ripoll M, Håvarstein LS, Hermoso JA. 2014. Structural basis of PcsB-mediated cell separation in Streptococcus pneumoniae. Nat Commun 5:3842. doi: 10.1038/ncomms4842. [DOI] [PubMed] [Google Scholar]
- 12.Schubert K, Bichlmaier AM, Mager E, Wolff K, Ruhland G, Fiedler F. 2000. P45, an extracellular 45 kDa protein of Listeria monocytogenes with similarity to protein p60 and exhibiting peptidoglycan lyric activity. Arch Microbiol 173:21–28. doi: 10.1007/s002030050003. [DOI] [PubMed] [Google Scholar]
- 13.Layec S, Decaris B, Leblond-Bourget N. 2008. Characterization of proteins belonging to the CHAP-related superfamily within the Firmicutes. J Mol Microbiol Biotechnol 14:31–40. doi: 10.1159/000106080. [DOI] [PubMed] [Google Scholar]
- 14.Liu M, Hanks TS, Zhang J, McClure MJ, Siemsen DW, Elser JL, Quinn MT, Lei B. 2006. Defects in ex vivo and in vivo growth and sensitivity to osmotic stress of group A Streptococcus caused by interruption of response regulator gene vicR. Microbiology 152:967–978. doi: 10.1099/mic.0.28706-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ng WL, Tsui HCT, Winkler ME. 2005. Regulation of the pspA virulence factor and essential pcsB murein biosynthetic genes by the phosphorylated VicR (YycF) response regulator in Streptococcus pneumoniae. J Bacteriol 187:7444–7459. doi: 10.1128/JB.187.21.7444-7459.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitrophanov AY, Groisman EA. 2008. Signal integration in bacterial two-component regulatory systems. Genes Dev 22:2601–2611. doi: 10.1101/gad.1700308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dubrac S, Bisicchia P, Devine KM, Msadek T. 2008. A matter of life and death: cell wall homeostasis and the WalKR (YycGF) essential signal transduction pathway. Mol Microbiol 70:1307–1322. doi: 10.1111/j.1365-2958.2008.06483.x. [DOI] [PubMed] [Google Scholar]
- 18.O’Connell-Motherway M, Van Sinderen D, Morel-Deville F, Fitzgerald GF, Ehrlich SD, Morel P. 2000. Six putative two-component regulatory systems isolated from Lactococcus lactis subsp. cremoris MG1363. Microbiology 146:935–947. doi: 10.1099/00221287-146-4-935. [DOI] [PubMed] [Google Scholar]
- 19.Land AD, Winkler ME. 2011. The requirement for pneumococcal MreC and MreD is relieved by inactivation of the gene encoding PBP1a. J Bacteriol 193:4166–4179. doi: 10.1128/JB.05245-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, Pesseat S, Quinn AF, Sangrador-Vegas A, Scheremetjew M, Yong SY, Lopez R, Hunter S. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Geer LY, Bryant SH. 2017. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res 45:D200–D203. doi: 10.1093/nar/gkw1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bateman A, Rawlings ND. 2003. The CHAP domain: a large family of amidases including GSP amidase and peptidoglycan hydrolases. Trends Biochem Sci 28:234–237. doi: 10.1016/S0968-0004(03)00061-6. [DOI] [PubMed] [Google Scholar]
- 23.Pei J, Kim BH, Grishin NV. 2008. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res 36:2295–2300. doi: 10.1093/nar/gkn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, Bork P, von Mering C. 2009. STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res 37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Visweswaran GRR, Steen A, Leenhouts K, Szeliga M, Ruban B, Hesseling-Meinders A, Dijkstra BW, Kuipers OP, Kok J, Buist G. 2013. AcmD, a homolog of the major autolysin AcmA of Lactococcus lactis, binds to the cell wall and contributes to cell separation and autolysis. PLoS One 8:e72167. doi: 10.1371/journal.pone.0072167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steen A, Buist G, Kramer NE, Jalving R, Benus G, Venema G, Kuipers OP, Kok J. 2008. Reduced lysis upon growth of Lactococcus lactis on galactose is a consequence of decreased binding of the autolysin AcmA. Appl Environ Microbiol 74:4671–4679. doi: 10.1128/AEM.00103-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teng F, Kawalec M, Weinstock GM, Hryniewicz W, Murray BE. 2003. An Enterococcus faecium secreted antigen, SagA, exhibits broad-spectrum binding to extracellular matrix proteins and appears essential for E. faecium growth. Infect Immun 71:5033–5041. doi: 10.1128/IAI.71.9.5033-5041.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin PK, Bao Y, Boyer E, Winterberg KM, McDowell L, Schmid MB, Buysse JM. 2002. Novel locus required for expression of high-level macrolide-lincosamide-streptogramin B resistance in Staphylococcus aureus. J Bacteriol 184:5810–5813. doi: 10.1128/jb.184.20.5810-5813.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giefing C, Meinke AL, Hanner M, Henics T, Minh DB, Gelbmann D, Lundberg U, Senn BM, Schunn M, Habel A, Henriques-Normark B, Örtqvist Å, Kalin M, Von Gabain A, Nagy E. 2008. Discovery of a novel class of highly conserved vaccine antigens using genomic scale antigenic fingerprinting of pneumococcus with human antibodies. J Exp Med 205:117–131. doi: 10.1084/jem.20071168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chia JS, Chang LY, Shun CT, Chang YY, Tsay YG, Chen JY. 2001. A 60-kilodalton immunodominant glycoprotein is essential for cell wall integrity and the maintenance of cell shape in Streptococcus mutans. Infect Immun 69:6987–6998. doi: 10.1128/IAI.69.11.6987-6998.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Solem C, Defoor E, Jensen PR, Martinussen J. 2008. Plasmid pCS1966, a new selection/counterselection tool for lactic acid bacterium strain construction based on the oroP gene, encoding an orotate transporter from Lactococcus lactis, p 4772–4775. Appl Environ Microbiol 74:4772–4775. doi: 10.1128/AEM.00134-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jensen PR, Hammer K. 1993. Minimal requirements for exponential growth of Lactococcus lactis. Appl Environ Microbiol 59:4363–4366. doi: 10.1128/AEM.59.12.4363-4366.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scheffers D-J, Pinho MG. 2005. Bacterial cell wall synthesis: new insights from localization studies. Microbiol Mol Biol Rev 69:585–607. doi: 10.1128/MMBR.69.4.585-607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kobierecka PA, Olech B, Książek M, Derlatka K, Adamska I, Majewski PM, Jagusztyn-Krynicka EK, Wyszyńska AK. 2016. Cell wall anchoring of the Campylobacter antigens to Lactococcus lactis. Front Microbiol 7:165. doi: 10.3389/fmicb.2016.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shams F, Oldfield NJ, Wooldridge KG, Turner D. 2014. Fructose-1,6-bisphosphate aldolase (FBA)—a conserved glycolytic enzyme with virulence functions in bacteria: iIll met by moonlight. Biochem Soc Trans 42:1792–1795. doi: 10.1042/BST20140203. [DOI] [PubMed] [Google Scholar]
- 36.Ng WL, Robertson GT, Kazmierczak KM, Zhao J, Gilmour R, Winkler ME. 2003. Constitutive expression of PcsB suppresses the requirement for the essential VicR (YycF) response regulator in Streptococcus pneumoniae R6. Mol Microbiol 50:1647–1663. doi: 10.1046/j.1365-2958.2003.03806.x. [DOI] [PubMed] [Google Scholar]
- 37.Rigden DJ, Jedrzejas MJ, Galperin MY. 2003. Amidase domains from bacterial and phage autolysins define a family of γ-d,l-glutamate-specific amidohydrolases. Trends Biochem Sci 28:230–234. doi: 10.1016/s0968-0004(03)00062-8. [DOI] [PubMed] [Google Scholar]
- 38.Wayne KJ, Li S, Kazmierczak KM, Tsui HCT, Winkler ME. 2012. Involvement of WalK (VicK) phosphatase activity in setting WalR (VicR) response regulator phosphorylation level and limiting cross-talk in Streptococcus pneumoniae D39 cells. Mol Microbiol 86:645–660. doi: 10.1111/mmi.12006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Delaune A, Poupel O, Mallet A, Coic YM, Msadek T, Dubrac S. 2011. Peptidoglycan crosslinking relaxation plays an important role in Staphylococcus aureus WalKR-dependent cell viability. PLoS One 6:e17054. doi: 10.1371/journal.pone.0017054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Las Rivas B, García JL, López R, García P. 2002. Purification and polar localization of pneumococcal LytB, a putative endo-β-N-acetylglucosaminidase: the chain-dispersing murein hydrolase. J Bacteriol 184:4988–5000. doi: 10.1128/jb.184.18.4988-5000.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berlec A, Zadravec P, Jevnikar Z, Štrukelj B. 2011. Identification of candidate carrier proteins for surface display on Lactococcus lactis by theoretical and experimental analyses of the surface proteome. Appl Environ Microbiol 77:1292–1300. doi: 10.1128/AEM.02102-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sham LT, Barendt SM, Kopecky KE, Winkler ME. 2011. Essential PcsB putative peptidoglycan hydrolase interacts with the essential FtsX Spn cell division protein in Streptococcus pneumoniae D39. Proc Natl Acad Sci U S A 108:E1061–E1069. doi: 10.1073/pnas.1108323108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marugg JD, Van Kranenburg R, Laverman P, Rutten GAM, De Vos WM. 1996. Identical transcriptional control of the divergently transcribed prtP and prtM genes that are required for proteinase production in Lactococcus lactis SK11. J Bacteriol 178:1525–1531. doi: 10.1128/jb.178.6.1525-1531.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sham LT, Jensen KR, Bruce KE, Winkler ME. 2013. Involvement of FtsE ATPase and FtsX extracellular loops 1 and 2 in FtsEX-PcsB complex function in cell division of Streptococcus pneumoniae D39. mBio 4:e00431-13. doi: 10.1128/mBio.00431-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uehara T, Bernhardt TG. 2011. More than just lysins: peptidoglycan hydrolases tailor the cell wall. Curr Opin Microbiol 14:698–703. doi: 10.1016/j.mib.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vermassen A, Leroy S, Talon R, Provot C, Popowska M, Desvaux M. 2019. Cell wall hydrolases in bacteria: insight on the diversity of cell wall amidases, glycosidases and peptidases toward peptidoglycan. Front Microbiol 10:331. doi: 10.3389/fmicb.2019.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huard C, Miranda G, Redko Y, Wessner F, Foster SJ, Chapot-Chartier MP. 2004. Analysis of the peptidoglycan hydrolase complement of Lactococcus lactis: identification of a third N-acetylglucosaminidase, AcmC. Appl Environ Microbiol 70:3493–3499. doi: 10.1128/AEM.70.6.3493-3499.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chapot-Chartier MP, Kulakauskas S. 2014. Cell wall structure and function in lactic acid bacteria. Microb Cell Fact 13:S9. doi: 10.1186/1475-2859-13-S1-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goel A, Santos F, de Vos WM, Teusink B, Molenaar D. 2012. Standardized assay medium to measure Lactococcus lactis enzyme activities while mimicking intracellular conditions. Appl Environ Microbiol 78:134–143. doi: 10.1128/AEM.05276-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kuipers OP, De Ruyter P, Kleerebezem M, De Vos WM. 1998. Quorum sensing-controlled gene expression in lactic acid bacteria. J Biotechnol 64:15–21. doi: 10.1016/S0168-1656(98)00100-X. [DOI] [Google Scholar]
- 51.De Ruyter P, Kuipers OP, Beerthuyzen MM, Van Alen-Boerrigter I, De Vos WM. 1996. Functional analysis of promoters in the nisin gene cluster of Lactococcus lactis. J Bacteriol 178:3434–3439. doi: 10.1128/jb.178.12.3434-3439.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Holo H, Nes IF. 1995. Transformation of Lactococcus by electroporation. Methods Mol Biol 47:195–199. doi: 10.1385/0-89603-310-4:195. [DOI] [PubMed] [Google Scholar]
- 53.Overkamp W, Beilharz K, Weme RDO, Solopova A, Karsens H, Kovács ÁT, Kok J, Kuipers OP, Veening JW. 2013. Benchmarking various green fluorescent protein variants in Bacillus subtilis, Streptococcus pneumoniae, and Lactococcus lactis for live cell imaging. Appl Environ Microbiol 79:6481–6490. doi: 10.1128/AEM.02033-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu X, Gallay C, Kjos M, Domenech A, Slager J, Kessel SP, Knoops K, Sorg RA, Zhang J, Veening J. 2017. High-throughput CRISPRi phenotyping identifies new essential genes in Streptococcus pneumoniae. Mol Syst Biol 13:931. doi: 10.15252/msb.20167449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pinto JPC, Zeyniyev A, Karsens H, Trip H, Lolkema JS, Kuipers OP, Kok J. 2011. pSEUDO, a genetic integration standard for Lactococcus lactis. Appl Environ Microbiol 77:6687–6690. doi: 10.1128/AEM.05196-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.