Although electric voltage and current have been studied for a long time in terms of their ability to suppress the growth of bacteria and to kill bacteria, increasing interest has been aroused more recently due to the prevalence of antibiotic resistance of microbes in past decades. Toward understanding the antimicrobial mechanism of low electric voltage and current, previous studies showed that treating bacteria with milliampere electric currents (≥5 mA) for ≥72 h led to significant damage of the bacterial membrane, which likely resulted in leakage of cellular contents and influx of toxic substances through the damaged membrane. However, it remains unclear to what extent membrane damage and two-way (i.e., inward and outward) leakage are caused by lower (i.e., microampere) electric current in a shorter time frame. In this work, we set out to answer this question. We observed that the membrane damage was caused by microampere electric current in half an hour, which allowed two-way leakage of ions, small molecules, and proteins.
KEYWORDS: ultralow electric current, leakage, membrane damage, single-molecule localization microscopy, superresolution fluorescence microscopy
ABSTRACT
Physical agents, such as low electric voltage and current, have recently gained attention for antimicrobial treatment due to their bactericidal capability. Although microampere electric current was shown to suppress the growth of bacteria, it remains unclear to what extent the microampere current damaged the bacterial membrane. Here, we investigated the membrane damage and two-way leakage caused by microampere electric current (≤100 μA) with a short exposure time (30 min). Based on MitoTracker staining, propidium iodide staining, filtration assays, and quantitative single-molecule localization microscopy, we observed significant membrane damage, which allowed two-way leakage of ions, small molecules, and proteins. This study paves the way to new development of antimicrobial applications for ultralow electric voltage and current.
IMPORTANCE Although electric voltage and current have been studied for a long time in terms of their ability to suppress the growth of bacteria and to kill bacteria, increasing interest has been aroused more recently due to the prevalence of antibiotic resistance of microbes in past decades. Toward understanding the antimicrobial mechanism of low electric voltage and current, previous studies showed that treating bacteria with milliampere electric currents (≥5 mA) for ≥72 h led to significant damage of the bacterial membrane, which likely resulted in leakage of cellular contents and influx of toxic substances through the damaged membrane. However, it remains unclear to what extent membrane damage and two-way (i.e., inward and outward) leakage are caused by lower (i.e., microampere) electric current in a shorter time frame. In this work, we set out to answer this question. We observed that the membrane damage was caused by microampere electric current in half an hour, which allowed two-way leakage of ions, small molecules, and proteins.
INTRODUCTION
As antibiotic resistance of bacteria has become one of the biggest threats to public health (1), alternatives to antibiotics have been attracting broad interest and attention (2, 3). Physical methods, such as sunlight/UV, fire, drying, high temperature, and high pressure, have played important roles in sterilization and disinfection from very early times of human history (4). Since the 1960s, electric voltage, current, and field have been explored as physical means for suppressing the growth, and/or killing, of bacteria. While most of the early studies focused on the bactericidal effects of high electric voltage and current (5–8), it was found more recently that low electric voltage and current can also effectively kill bacteria and biofilms (9–27). For example, Pareilleux and Sicard investigated the effects of low electric current, ranging from 10 to 200 mA, on the viability of Escherichia coli bacteria and found that current as low as 25 mA could kill the bacteria (27). The lethal activity of milliampere current was confirmed by other reports (9, 17–19, 25, 26). In addition, many studies showed that ultralow electric current at microamperes was also bactericidal (10, 13, 18, 20, 21, 24); even an electric field (without current) can inhibit the growth of planktonic bacteria (16). Furthermore, electric currents at microamperes and/or milliamperes have been shown to be effective for treating biofilms (11, 12, 14, 15, 18, 22–24, 28).
Efforts have been made toward understanding the antimicrobial mechanisms of low electric voltage and current. Possible mechanisms include membrane damage and disruption, reduction in ATP production and enzymatic activities (17, 26), and generation of reactive oxygen species (ROS) (13–15). Membrane damage caused by high electric voltage has been well known for a long time. For example, electroporation is a commonly used microbiological technique to deliver DNA and proteins into bacteria and cells by applying high electric voltage at kilovolts (29–31). More recently, transmission electron microscopy showed that treating bacteria at a low electric current of 5 mA for ≥72 h led to significant damage based on which leakage of cellular contents and influx of toxic substances through the damaged membrane were suggested (17). An interesting follow-up question is to what extent membrane damage and two-way (i.e., inward and outward) leakage are caused by microampere electric current in a shorter time frame. Although previous results showed that microampere electric currents were effective for suppressing the growth of bacteria, this question remains unanswered.
In this work, we set out to answer this question by investigating possible membrane damage of bacteria after subjecting them to microampere electric current of ≤100 μA (corresponding to voltage of ≤2.5 V) for 30 min. The effect of the electric current on the bacterial membrane was first examined by MitoTracker staining and fluorescence microscopy. In addition, propidium iodide (PI) staining and filtration assays were carried out to assess the two-way leakage of ions, small molecules, and proteins due to the membrane damage caused by the microampere electric current. Furthermore, quantitative single-molecule localization microscopy, which allowed us to localize, and count the number of, histone-like nucleoid structuring (H-NS) proteins inside individual bacteria, was performed to evaluate the changes in the organization and clustering of the H-NS proteins and to quantify the leakage of the H-NS proteins out of the bacteria caused by microampere electric current.
RESULTS
Membrane damage caused by microampere electric currents.
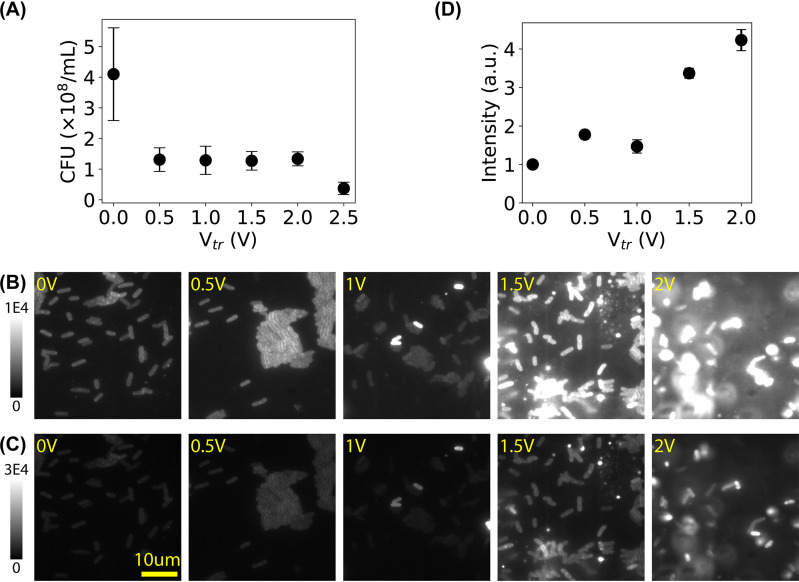
Microampere electric current was applied to the bacterial culture by applying low electric DC voltage (Fig. 1). As the resistance (R) of the bacterial culture was measured to ≈32 kΩ, DC voltage (Vtr) in the range of 0 to 2.5 V resulted in current (I) of 0 to 78 μA (I = Vtr/R). After treatment for 30 min, we performed CFU assays and found that the number of live cells decreased after subjecting the bacteria to low electric voltage/current (Fig. 2A), confirming the antimicrobial effect of low electric voltage/current. In addition, we stained the bacterial membrane by MitoTracker Green FM dye (32). As shown in Fig. 2B and C, the intensity of the bacterial membrane became higher after subjecting the bacteria to the microampere electric current. To quantify this observation, we estimated the mean intensities of the bacteria and examined the dependence of the mean intensity on the voltage (Fig. 2D). The intensity increased slightly at 0.5 V or 16 μA, indicating changes in the bacterial membrane. The average intensity increased ≥4-fold when cells were exposed to a voltage of ≥1.5 V (or current of ≥47 μA), and many bacterial cells showed extremely high brightness (up to 19 times brighter) than the negative control, suggesting that many of the treated cells had significant changes in their membrane. Although controversy exists (33, 34), previous studies showed that the fluorescence of membrane-incorporated MitoTracker Green dye depends on the membrane potential (35, 36); it is possible that the microampere electric current altered the bacterial membrane potential.
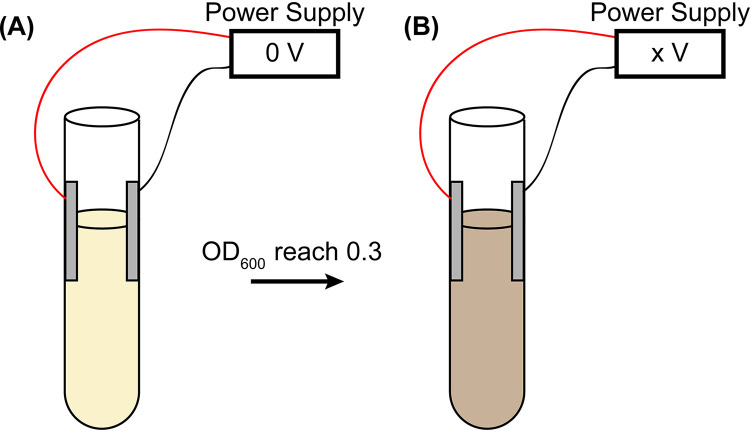
FIG 1.
Treating bacteria with low DC voltage. (A) Overnight bacterial culture was diluted in fresh medium and regrown until the OD600 reached 0.3 without applying DC voltage (0 V). (B) Applying low DC voltage (0.5, 1.0, 1.5, 2.0, and 2.5 V) on the bacterial culture in culture tubes for 30 min, followed by quantifications.
FIG 2.
Membrane damage in bacteria caused by microampere electric current. (A) CFU assay showing the number of live bacteria decreased after treating them with Vtr. (B) Representative fluorescence images of bacterial membrane stained by MitoTracker Green FM dye after treatment of low DC voltages at 0 V (untreated negative control) to 2.0 V (corresponding to 0 to 63 μA). (C) The same images from panel B but with a wider intensity scale. (D) Dependence of the mean fluorescence intensity (rescaled by the negative control) on the applied voltage. a.u., arbitrary units. Error bars in panels A and D represent the standard errors of the means.
Inward leakage due to membrane damage caused by microampere electric currents.
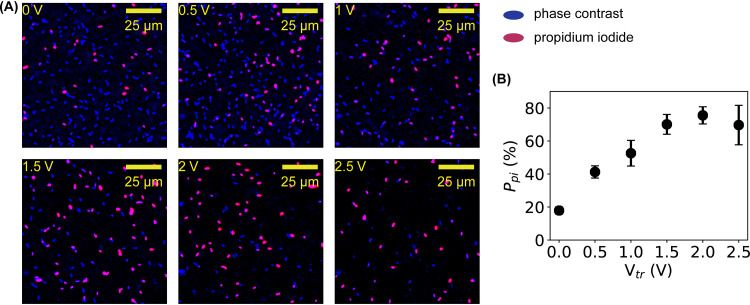
It was previously proposed that membrane damage caused by low electric current allowed the influx of toxicants (17). To test this possibility with microampere electric current, the staining agent PI (37, 38) was used to stain the DNA of the bacteria after treatment by microampere electric current. The rationale was that if electrically induced membrane damage allowed PI to enter the bacteria to stain DNA, then the influx of ions and other small organic molecules would also be possible. Representative images of untreated (0 V or 0 μA) and treated (0.5 to 2.5 V or 16 to 78 μA) bacteria are shown in Fig. 3A, where the fluorescent images from PI staining (red) are superposed on the inverted phase-contrast images (blue). It was observed that more bacteria were stained by PI at higher voltages and currents. Quantifying the percentage of PI-stained bacteria (Ppi) showed that the percentage of membrane-damaged bacteria increased quickly from 20% to ≈ 80% at 1.5 to 2 V or 47 to 63 μA (Fig. 3B). This observation confirmed that the membrane damage caused by microampere electric current was significant enough to enable inward leakage of at least ions and small molecules with a size of ≤1 nm.
FIG 3.
Quantification of inward leakage due to membrane damage by PI staining. (A) Representative fluorescence images (red) of bacteria stained by PI before and after electric treatment superimposed on the inverted phase-contrast images (blue) of the bacteria in the same fields of view. (B) Dependence of the percentage of PI-stainable bacteria on the applied voltage. Error bars represent the standard deviations.
Outward leakage due to membrane damage caused by microampere electric currents.
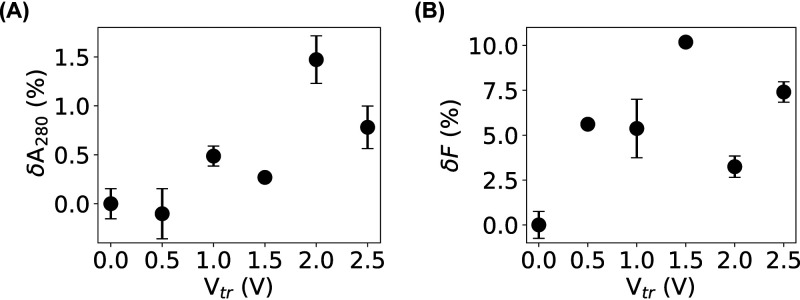
Our PI-staining result showed that influx of ions and small organic molecules (≤1 nm) was possible due to membrane damage caused by microampere electric current, which also implied that small cellular contents, including amino acids, could leak out of the bacteria. Another question is whether biological macromolecules, such as proteins and nucleic acids, also could leak out. To answer this question, we adopted a filtration assay that was used to study the membrane damage caused by carbon nanotubes (39). From the relative changes in the absorbance (δA280) of the filtrates for the bacteria before and after microampere current treatment, we observed a slight increase, ∼1% (Fig. 4A), providing weak evidence of possible outward leakage of macromolecules. Because the culture media could contribute significantly to the absorbance, leading to a high baseline in the measurement, the measured increase in δA280 was likely underestimated.
FIG 4.
Quantification of outward leakage of bacterial content due to membrane damage by the filtration assays. (A and B) Relative changes in the absorbance at 280 nm (A) and fluorescence (B) of the filtrates of bacterial samples before and after electric treatment. Error bars represent the standard deviations.
To lower the contribution of the culture media, a modified filtration assay based on fluorescence was performed using an E. coli strain that expresses histone-like nucleoid structuring (H-NS) proteins fused to mEos3.2 fluorescent proteins (40). By quantifying the relative changes in the fluorescence intensities, δF, of the filtrates, we observed an increase of ∼10% for the bacteria treated at 1.5 V or 47 μA (Fig. 4B). This observation suggested that the membrane damage caused by the microampere electric current was significant enough to allow proteins to leak out of the bacteria.
Quantification of protein leakage by single-molecule localization microscopy.
The filtration assays suggested that proteins leaked out of bacteria after treating the bacteria with ≤100 μA DC. To further confirm such protein leakage, we measured the number of the histone-like nucleoid structuring (H-NS) proteins inside individual bacteria before and after microampere current treatment for 30 min using single-molecule localization microscopy (SMLM) (41–44). SMLM is one type of superresolution fluorescence microscopy that localizes individual molecules of interest with a precision of ≤10 nm (45); therefore, it not only produces superresolved images with high spatial resolution but also provides a convenient way to count the number of molecules. Without applying sophisticated algorithms (46–48), the number of molecules of interest is, on average, proportional to the intensity in the superresolved images or the number of localizations obtained by SMLM (49).
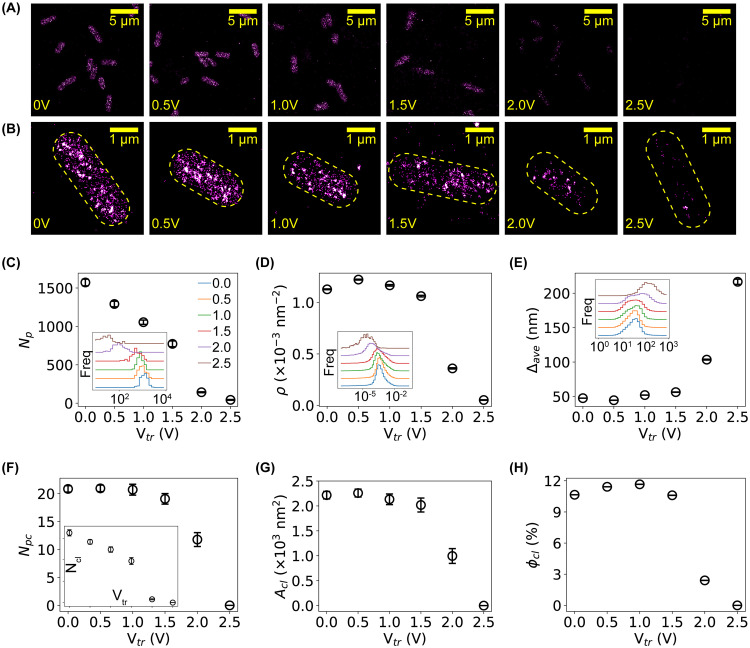
We performed single-molecule localization microscopy on untreated and treated bacteria using the E. coli strain expressing H-NS–mEos3.2 fusion proteins, as mEos3.2 fluorescent proteins are photoactivable and allow superresolution imaging (40, 50). Representative images of H-NS proteins in untreated and treated bacteria are shown in Fig. 5A and B. For the untreated negative control, H-NS proteins were organized as clusters inside the bacteria (Fig. 5A and B, first column), consistent with previously reported results (50–52). After subjecting the bacteria to a DC voltage of 1 to 2.5 V (or current of 31 to 78 μA), the intensities on the superresolved images became significantly dimmer (Fig. 5A and B, columns 3 to 6). As the intensity in superresolved images correlates with the probability of localizing the molecules of interest, this result indicates that fewer H-NS proteins were present in the bacteria after treating the bacteria with microampere electric current.
FIG 5.
Quantification of outward leakage of bacterial content due to membrane damage by single-molecule localization microscopy. (A) Representative superresolved images of H-NS proteins in E. coli bacteria before and after electric treatment (from left to right, Vtr of 0, 0.5, 1.0, 1.5, 2.0, and 2.5 V). Scale bar, 5 μm. (B) Zoom-in view of superresolved images showing individual bacteria. Scale bar, 1 μm. (C to E) Dependence of the number of detected H-NS proteins per cell, Np (C), the molecular density, ρ (D), and the average intermolecular distance, Δave (E), on the applied voltage. Insets are the corresponding distributions of the three quantities. Error bars stand for the standard errors of the means (SEM). (F to H) Dependence of the number of clusters per cell, Ncl (inset), the number of proteins per cluster, Npc (F), the area of clusters, Acl (G), and the fraction of clustering proteins per cell, ϕcl (H), on the applied voltage. Error bars stand for SEM.
To quantify this result, we segmented the bacteria (50) and counted the number of localizations of H-NS proteins in each cell, Np. Due to the stochastic nature in the activation and detection of the mEos3.2 fluorescent proteins, Np is expected to be proportional to, on average, the number of H-NS proteins in the bacteria (49, 50). The histograms of Np are shown in the inset of Fig. 5C, where the baselines of the histograms were vertically shifted for better visualization of the differences. It is clear that the peaks of the Np distributions translated to the left as the applied voltage increased, suggesting that the number of H-NS proteins per bacterial cell decreased when subjecting the bacteria to low DC voltage and current. We also calculated the mean Np and standard error of the mean (SEM) for each sample. Note that as the distributions of Np were bell-shaped in the linear-log scale (Fig. 5C, inset), we estimated the mean log(Np) and then calculated the mean Np from Np = exp(log(Np)). As shown in Fig. 5C, the average number of H-NS proteins per cell decreased nearly linearly. It is also noted that the observed decrease in the number of H-NS proteins per bacterial cell is unlikely due to cellular responses (e.g., lower expression level of H-NS proteins) for several reasons. First, the treatment time (30 min) is relatively short compared to the degradation time of proteins in bacteria (approximately hours) (53). Second, our previous work on H-NS proteins in bacteria treated by antimicrobial silver ions and silver nanoparticles showed that the number of H-NS proteins did not decrease within several hours (50). Therefore, the microscopic data on individual bacteria directly suggested that H-NS proteins leaked out of the bacteria after microampere electric current treatment.
We examined how the spatial organization of the H-NS proteins was affected by the electric treatment based on Voronoi diagram tessellation (50, 54). We computed the molecular density (ρ) of the H-NS proteins and observed that the distribution of ρ shifted to the left, indicating the density decreased as the applied voltage increased (Fig. 5D, inset), which is expected as a result of the decreased number of H-NS proteins per cell. Interestingly, the mean molecular density started to decrease at 1.5 to 2 V (or 47 to 63 μA), as shown in Fig. 5D, which is different from the dependence of the number of H-NS proteins per cell on the applied electric voltage and current (Fig. 5C). We also observed a shift to the right in the distribution of the mean interneighbor distances, Δave (Fig. 5E, inset), suggesting that the intermolecular distances for some H-NS proteins became larger when bacteria were treated by low DC voltage and current, consistent with the result of the molecular density. In addition, we identified the clusters of H-NS proteins based on Voronoi diagrams (50, 54) and counted the number of clusters per bacterial cell (Ncl) and the number of proteins per cluster (Npc). We found that both Ncl and Npc decreased after treating the bacteria with DC voltages and currents (Fig. 5F). Furthermore, by quantifying the area of clusters of H-NS proteins, we observed that the H-NS clusters became smaller (Fig. 5G). Lastly, we examined the fraction of H-NS proteins forming clusters in individual bacteria, ϕc = Ncp/Np, where Np is the number of localizations (i.e., detected proteins) per cell and Npc the number of proteins that form clusters in the cell. We observed that the clustering fraction decreased from 10% for the negative control to 0% at 2.5 V or 78 μA (Fig. 5H).
DISCUSSION
To summarize, we investigated the membrane damage of bacteria and the two-way leakage caused by microampere electric current. We observed that bacteria subjected to ≥47 μA current for 30 min showed much higher intensities with MitoTracker staining, suggesting that the bacterial membrane was altered by the microampere electric current. The membrane damage caused by the microampere current was large enough to allow PI molecules to enter the bacteria, suggesting that inward leakage of ions and small molecules was possible. In addition, we found, based on filtration assays and superresolution fluorescence microscopy, that the membrane damage was significant enough to allow proteins to leak out of the cells. More importantly, using histone-like nucleoid structuring (H-NS) proteins as an example, we quantified the decrease in the number of H-NS proteins per bacterial cell and characterized the changes in the spatial organization of the H-NS proteins caused by the electric treatment. This study highlights that treating bacteria with electric current at ≤100 μA for 30 min caused significant membrane damage and led to two-way leakages of ions, small molecules, and proteins.
It was noted previously that the bactericidal effects of electric voltage and current are complex and involve various interactions between the bacteria, electricity, electrode materials, and medium (17). For example, in addition to the electrical effects, metal ions released from the electrodes into the medium are likely to affect the growth of bacteria, which has been shown in the literature (55). In addition, ROS generated by the electric voltage and current could be another significant contributor (13–15). The current study did not aim to distinguish the contributions of these different effects, while these are interesting questions worth pursuing in future investigations. On the other hand, we argue that heating by electric voltage/current is unlikely to cause damage to the bacteria in this study, because the used electric power is very low. Considering the resistance of the bacterial culture was ≈32 kΩ, the electric power was below 200 μW; at this power, it would take more than 1 day to heat up the bacterial culture (5 ml) by one degree.
It is worthwhile to highlight that the electric power leading to serious membrane damage of bacteria is very low, which is expected to facilitate the use of microampere electric current (and low electric voltage) for antimicrobial applications. For example, commonly used household batteries can provide the needed voltages to kill bacteria. More importantly, solar panels have an output power of ∼10 mW per cm2 (56); therefore, a solar panel of 1 cm2 can easily generate the needed electric power for damaging bacterial membranes, suppressing the growth of bacteria and/or killing bacteria. We anticipate that this study will lead to new development of antibiotic applications for low electric current.
MATERIALS AND METHODS
Bacterial strains and growth.
Two K-12-derived E. coli strains were used in this study: the first strain is MG1655 (from the Yale Coli Genetic Stock Center) (57, 58), and the second strain has the hns gene on the bacterial DNA fused to the gene for mEos3.2 fluorescent protein, which was derived from the EosFP fluorescent protein from the stony coral Lobophyllia hemprichii (40, 59). The second strain expresses fluorescent H-NS–mEos3.2 fusion proteins, allowing us to perform fluorescence-based filtration assays and quantitative single-molecule localization microscopy (40, 50–52, 60), as described below. The bacterial stocks were stored at −80°C as glycerol stocks by following standard protocols (61, 62). The purity of the stocks was regularly monitored by phenotypic characterizations based on colony morphology and growth curve. The bacterial morphology was also regularly monitored under a bright-field or phase-contrast microscope. In addition, for the E. coli strain expressing fluorescent proteins, the fluorescence of the bacteria and the emission of individual fluorescent proteins were regularly monitored.
The bacteria were grown at 37°C overnight with orbital rotation at 250 rpm in defined M9 minimal medium (autoclave sterilized), supplemented with 1% glucose (filter sterilized using a 0.22-μm filter), 0.1% Casamino Acids (filter sterilized using a 0.22-μm filter), 0.01% thiamine (filter sterilized using a 0.22-μm filter), and appropriate antibiotics (none for MG1655; chloramphenicol at a final concentration of 34 μg/ml for the strain expressing H-NS–mEos3.2 proteins) (49–51, 60). On the second day, the overnight culture was diluted 50 to 100 times into fresh medium so that the optical density at 600 nm (OD600) was 0.05 (49–51, 60). The fresh cultures were grown again at 37°C in culture tubes equipped with two sterile aluminum electrodes with orbital rotation at 250 rpm without electric voltage or current (i.e., the wires were shorted) (Fig. 1A). The resistance of the bacterial cultures (≈32 kΩ) was measured using a multimeter. When the fresh bacterial culture reached an OD600 of ≈0.3, low DC voltage was applied to the culture for 30 min (0 [untreated negative control], 0.5, 1, 1.5, 2, and 2.5 V) (Fig. 1B). The corresponding currents were 0, 16, 31, 47, 63, and 78 μA, respectively. After the 30-min treatment, the bacteria were quantified as described below.
CFU assay.
For the CFU assays, the untreated and treated bacteria were diluted by a factor of 4 × 105 or 4 × 106 in fresh LB medium and plated on agar plates. The plates were incubated at 37°C overnight, followed by imaging the plates with a gel documentation system (Analytik Jena US LLC, Upland, CA) and manually counting the number of colonies on each plate. Four replicates were performed for each sample.
MitoTracker staining and quantification.
To stain the bacterial membrane, MitoTracker Green FM dye (Thermo Fisher Scientific, Waltham, MA) was added to the untreated and treated bacteria at a final concentration of 300 nM and incubated in a shaking incubator (250 rpm, 37°C) for 30 min (32). Eight microliters of the stained bacteria was transferred to 5-mm by 5-mm agarose pads (3% in 1× phosphate-buffered saline [PBS], ∼1 mm thick) for mounting. The agarose pads with bacteria were flipped and attached to clean coverslips (cleaned by sonication sequentially in 1 M NaOH, 100% ethanol, and ultrapure water). Chambers were then constructed by sandwiching rubber O rings between the coverslips and clean microscope slides. The chambers were sealed using epoxy glue and then mounted on a microscope for fluorescence imaging (excitation at 488 nm). From the acquired images, the average fluorescence intensities of 100 bacteria treated at each voltage were measured using ImageJ (63–65).
PI staining and quantification.
To measure possible inward leakage due to membrane damage, the untreated and treated bacteria were first fixed by 3.7% formaldehyde (Sigma-Aldrich, St. Louis, MO) and stained with PI (G-Biosciences, St. Louis, MO) (37, 38). The PI-stained bacteria were mounted on agarose pads and prepared for fluorescence imaging (similar to the MitoTracker experiments, except that the excitation wavelength was 532 nm) and phase-contrast imaging. The stained cells were counted, and their percentages were calculated based on the acquired fluorescence and phase-contrast images.
Filtration assays.
Filtration assays to assess the leakage of cellular contents due to membrane damage were performed similarly to that of Chen et al. (39). Briefly, the untreated and treated bacteria (the strain expressing H-NS–mEos3.2 fusion proteins) were filtered by 0.2-μm filters (VWR International LLC, Radnor, PA), resulting in filtrates containing leaked cellular contents if membrane damage occurred, in addition to proteins and molecules from the culture media. For each sample, the filtrate was aliquoted into a 96-well plate (200 μl per well), followed by measuring the absorbance at 280 nm and fluorescence at 525 nm (excitation at 498 nm) on a multimode microplate reader (Synergy H1, BioTek, Winooski, VT). The relative change in the absorbance was estimated by , where and were the absorbances at 280 nm of the filtrates for bacteria before and after microampere current treatment, respectively. Similarly, the relative change in the fluorescence intensities of the filtrates was estimated by , where F− and F+ were the fluorescence of the filtrates for samples before and after microampere current treatment, respectively.
Quantitative single-molecule localization microscopic assay.
Quantitative single-molecule localization microscopy (41) was performed on the bacteria subjected to microampere electric current by following our previous work (49–51, 60). Briefly, the untreated and treated bacteria were first fixed by 3.7% formaldehyde for 30 min at room temperature and harvested by centrifugation (1,000 × g for 10 min). The harvested cells were resuspended in 1× PBS buffer and washed three times in 1× PBS buffer. The prepared bacteria were then mounted on agarose pads for imaging. The single-molecule localization microscope was home-built on an Olympus IX-73 inverted microscope with an Olympus TIRF 100× (numeric aperture, 1.49) oil immersion objective. The microscope and data acquisition were controlled by Micro-Manager (66, 67). A 405-nm laser and a 532-nm laser from a multilaser system (iChrome MLE; TOPTICA Photonics, Farmington, NY) were used to activate and excite the H-NS–mEos3.2 fusion proteins in the bacteria. Emissions from the fluorescent proteins were collected by the objective and imaged on an electron-multiplying charge-coupled device camera (Andor Technology Ltd.–Oxford Instruments, Concord, MA) with an exposure time of 30 ms (the actual time interval between frames was 45 ms). The effective pixel size of acquired images was 160 nm, while the field of view was 256 by 256 pixels.
The resultant movies (20,000 frames) were analyzed with RapidStorm (68), generating x/y positions, x/y widths, intensity, and background for each detected fluorescent spot. If the intensity of a spot was below 4,000 (i.e., too dim) or above 50,000 (i.e., too bright), or if the width of a spot was below 200 nm (i.e., too narrow) or above 600 nm (i.e., too wide), the spot was rejected and excluded from later analysis (42). The positions of the spots surviving the rejection criteria were further corrected for sample drift using a mean cross-correlation algorithm (69). The localizations that appeared in adjacent frames and within 10 nm of each other were regrouped as a single molecule (70). The resultant localizations were used for the reconstruction of superresolved images and for further quantitative analysis.
The localizations of H-NS molecules from each acquisition were first manually segmented into individual bacterial cells using custom MATLAB programs (50). Analysis and quantification of the spatial organization of H-NS proteins were performed on individual cells. Following the segmentation of cells, the number of localizations per bacterium, Np, was counted (50). In addition, Voronoi-based quantification and clustering analysis were performed using custom MATLAB programs by following Levet et al. (54), from which we estimated the molecular density of H-NS proteins, ρ, the mean interneighbor distances, Δave, the number of clusters per bacterium, Ncl, the number of proteins per cluster, Ncp, the area of clusters of H-NS proteins, Acl, and the fraction of proteins forming clusters, ϕcl (50, 54).
ACKNOWLEDGMENTS
We thank Joshua N. Milstein for the generous gift of the E. coli strain expressing mEos3.2–H-NS fusion proteins.
This work was supported by the University of Arkansas, the Arkansas Biosciences Institute (grant no. ABI-0189, no. ABI-0226, and no. ABI-0277), and the National Science Foundation (grant no. 1826642). A.R. and J.P. were supported through the Research Experience for Undergraduates Program, funded by the National Science Foundation (grant no. 1460754). We are also grateful for support from the Arkansas High Performance Computing Center (AHPCC), which is funded in part by the National Science Foundation (grant no. 0722625, 0959124, 0963249, and 0918970) and the Arkansas Science and Technology Authority.
REFERENCES
- 1.World Health Organization. 2014. World health statistics 2014. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.François B, Jafri HS, Bonten M. 2016. Alternatives to antibiotics. Intensive Care Med 42:2034–2036. doi: 10.1007/s00134-016-4339-y. [DOI] [PubMed] [Google Scholar]
- 3.Allen HK, Trachsel J, Looft T, Casey TA. 2014. Finding alternatives to antibiotics. Ann N Y Acad Sci 1323:91–100. doi: 10.1111/nyas.12468. [DOI] [PubMed] [Google Scholar]
- 4.Blancou J. 1995. History of disinfection from early times until the end of the 18th century. Rev Sci Tech 14:21–39. doi: 10.20506/rst.14.1.831. [DOI] [PubMed] [Google Scholar]
- 5.Rosenberg B, Vancamp L, Krigas T. 1965. Inhibition of cell division in escherichia coli by electrolysis products from a platinum electrode. Nature 205:698–699. doi: 10.1038/205698a0. [DOI] [PubMed] [Google Scholar]
- 6.Sale A, Hamilton W. 1967. Effects of high electric fields on microorganismsI. Killing of bacteria and yeasts. Biochim Biophys Acta Gen Subj 148:781–788. doi: 10.1016/0304-4165(67)90052-9. [DOI] [Google Scholar]
- 7.Hülsheger H, Potel J, Niemann EG. 1981. Killing of bacteria with electric pulses of high field strength. Radiat Environ Biophys 20:53–65. doi: 10.1007/BF01323926. [DOI] [PubMed] [Google Scholar]
- 8.Hülsheger H, Potel J, Niemann EG. 1983. Electric field effects on bacteria and yeast cells. Radiat Environ Biophys 22:149–162. doi: 10.1007/BF01338893. [DOI] [PubMed] [Google Scholar]
- 9.Blenkinsopp SA, Khoury AE, Costerton JW. 1992. Electrical enhancement of biocide efficacy against Pseudomonas aeruginosa biofilms. Appl Environ Microbiol 58:3770–3773. doi: 10.1128/AEM.58.11.3770-3773.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu WK, Tebbs SE, Byrne PO, Elliott TS. 1993. The effects of electric current on bacteria colonising intravenous catheters. J Infect 27:261–269. doi: 10.1016/0163-4453(93)92068-8. [DOI] [PubMed] [Google Scholar]
- 11.Costerton JW, Ellis B, Lam K, Johnson F, Khoury AE. 1994. Mechanism of electrical enhancement of efficacy of antibiotics in killing biofilm bacteria. Antimicrob Agents Chemother 38:2803–2809. doi: 10.1128/AAC.38.12.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stoodley P, deBeer D, Lappin-Scott HM. 1997. Influence of electric fields and pH on biofilm structure as related to the bioelectric effect. Antimicrob Agents Chemother 41:1876–1879. doi: 10.1128/AAC.41.9.1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu WK, Brown MR, Elliott TS. 1997. Mechanisms of the bactericidal activity of low amperage electric current (DC). J Antimicrob Chemother 39:687–695. doi: 10.1093/jac/39.6.687. [DOI] [PubMed] [Google Scholar]
- 14.Stewart PS, Wattanakaroon W, Goodrum L, Fortun SM, McLeod BR. 1999. Electrolytic generation of oxygen partially explains electrical enhancement of tobramycin efficacy against Pseudomonas aeruginosa biofilm. Antimicrob Agents Chemother 43:292–296. doi: 10.1128/AAC.43.2.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brinkman CL, Schmidt-Malan SM, Karau MJ, Greenwood-Quaintance K, Hassett DJ, Mandrekar JN, Patel R. 2016. Exposure of bacterial biofilms to electrical current leads to cell death mediated in part by reactive oxygen species. PLoS One 11:e0168595. doi: 10.1371/journal.pone.0168595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giladi M, Porat Y, Blatt A, Wasserman Y, Kirson ED, Dekel E, Palti Y. 2008. Microbial growth inhibition by alternating electric fields. Antimicrob Agents Chemother 52:3517–3522. doi: 10.1128/AAC.00673-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valle A, Zanardini E, Abbruscato P, Argenzio P, Lustrato G, Ranalli G, Sorlini C. 2007. Effects of low electric current (LEC) treatment on pure bacterial cultures. J Appl Microbiol 103:1376–1385. doi: 10.1111/j.1365-2672.2007.03374.x. [DOI] [PubMed] [Google Scholar]
- 18.Schmidt-Malan SM, Karau MJ, Cede J, Greenwood-Quaintance KE, Brinkman CL, Mandrekar JN, Patel R. 2015. Antibiofilm activity of low-amperage continuous and intermittent direct electrical current. Antimicrob Agents Chemother 59:4610–4615. doi: 10.1128/AAC.00483-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.del Pozo JL, Rouse MS, Mandrekar JN, Sampedro MF, Steckelberg JM, Patel R. 2009. Effect of electrical current on the activities of antimicrobial agents against Pseudomonas aeruginosa, Staphylococcus aureus, and Staphylococcus epidermidis biofilms. Antimicrob Agents Chemother 53:35–40. doi: 10.1128/AAC.00237-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Del Pozo JL, Rouse MS, Euba G, Kang C-I, Mandrekar JN, Steckelberg JM, Patel R. 2009. The electricidal effect is active in an experimental model of Staphylococcus epidermidis chronic foreign body osteomyelitis. Antimicrob Agents Chemother 53:4064–4068. doi: 10.1128/AAC.00432-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van der Borden AJ, van der Mei HC, Busscher HJ. 2005. Electric block current induced detachment from surgical stainless steel and decreased viability of Staphylococcus epidermidis. Biomaterials 26:6731–6735. doi: 10.1016/j.biomaterials.2004.04.052. [DOI] [PubMed] [Google Scholar]
- 22.Kim YW, Subramanian S, Gerasopoulos K, Ben-Yoav H, Wu H-C, Quan D, Carter K, Meyer MT, Bentley WE, Ghodssi R. 2015. Effect of electrical energy on the efficacy of biofilm treatment using the bioelectric effect. NPJ Biofilms Microbiomes 1:15016. doi: 10.1038/npjbiofilms.2015.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caubet R, Pedarros-Caubet F, Chu M, Freye E, de Belém Rodrigues M, Moreau JM, Ellison WJ. 2004. A radio frequency electric current enhances antibiotic efficacy against bacterial biofilms. Antimicrob Agents Chemother 48:4662–4664. doi: 10.1128/AAC.48.12.4662-4664.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van der Borden AJ, van der Werf H, van der Mei HC, Busscher HJ. 2004. Electric current-induced detachment of Staphylococcus epidermidis biofilms from surgical stainless steel. Appl Environ Microbiol 70:6871–6874. doi: 10.1128/AEM.70.11.6871-6874.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rabinovitch C, Stewart PS. 2006. Removal and inactivation of Staphylococcus epidermidis biofilms by electrolysis. Appl Environ Microbiol 72:6364–6366. doi: 10.1128/AEM.00442-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ranalli G, Iorizzo M, Lustrato G, Zanardini E, Grazia L. 2002. Effects of low electric treatment on yeast microflora. J Appl Microbiol 93:877–883. doi: 10.1046/j.1365-2672.2002.01758.x. [DOI] [PubMed] [Google Scholar]
- 27.Pareilleux A, Sicard N. 1970. Lethal effects of electric current on Escherichia coli. Appl Microbiol 19:421–424. doi: 10.1128/AEM.19.3.421-424.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davis CP, Weinberg S, Anderson MD, Rao GM, Warren MM. 1989. Effects of microamperage, medium, and bacterial concentration on iontophoretic killing of bacteria in fluid. Antimicrob Agents Chemother 33:442–447. doi: 10.1128/aac.33.4.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neumann E, Schaefer-Ridder M, Wang Y, Hofschneider PH. 1982. Gene transfer into mouse lyoma cells by electroporation in high electric fields. EMBO J 1:841–845. doi: 10.1002/j.1460-2075.1982.tb01257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dower WJ, Miller JF, Ragsdale CW. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res 16:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crawford R, Torella JP, Aigrain L, Plochowietz A, Gryte K, Uphoff S, Kapanidis AN. 2013. Long-lived intracellular single-molecule fluorescence using electroporated molecules. Biophys J 105:2439–2450. doi: 10.1016/j.bpj.2013.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pogliano J, Ho TQ, Zhong Z, Helinski DR. 2001. Multicopy plasmids are clustered and localized in Escherichia coli. Proc Natl Acad Sci U S A 98:4486–4491. doi: 10.1073/pnas.081075798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pendergrass W, Wolf N, Poot M. 2004. Efficacy of MitoTracker Green and CMXrosamine to measure changes in mitochondrial membrane potentials in living cells and tissues. Cytometry A 61:162–169. doi: 10.1002/cyto.a.20033. [DOI] [PubMed] [Google Scholar]
- 34.Poot M, Pierce RH. 1999. Detection of changes in mitochondrial function during apoptosis by simultaneous staining with multiple fluorescent dyes and correlated multiparameter flow cytometry. Cytometry 35:311–317. doi:. [DOI] [PubMed] [Google Scholar]
- 35.Keij JF, Bell-Prince C, Steinkamp JA. 2000. Staining of mitochondrial membranes with 10-nonyl acridine orange, MitoFluor Green, and MitoTracker Green is affected by mitochondrial membrane potential altering drugs. Cytometry 39:203–210. doi:. [DOI] [PubMed] [Google Scholar]
- 36.Buckman JF, Hernández H, Kress GJ, Votyakova TV, Pal S, Reynolds IJ. 2001. MitoTracker labeling in primary neuronal and astrocytic cultures: influence of mitochondrial membrane potential and oxidants. J Neurosci Methods 104:165–176. doi: 10.1016/s0165-0270(00)00340-x. [DOI] [PubMed] [Google Scholar]
- 37.Rosenberg M, Azevedo NF, Ivask A. 2019. Propidium iodide staining underestimates viability of adherent bacterial cells. Sci Rep 9:6483. doi: 10.1038/s41598-019-42906-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boulos L, Prévost M, Barbeau B, Coallier J, Desjardins R. 1999. LIVE/DEAD BacLight: application of a new rapid staining method for direct enumeration of viable and total bacteria in drinking water. J Microbiol Methods 37:77–86. doi: 10.1016/S0167-7012(99)00048-2. [DOI] [PubMed] [Google Scholar]
- 39.Chen H, Wang B, Gao D, Guan M, Zheng L, Ouyang H, Chai Z, Zhao Y, Feng W. 2013. Broad-spectrum antibacterial activity of carbon nanotubes to human gut bacteria. Small 9:2735–2746. doi: 10.1002/smll.201202792. [DOI] [PubMed] [Google Scholar]
- 40.Zhang M, Chang H, Zhang Y, Yu J, Wu L, Ji W, Chen J, Liu B, Lu J, Liu Y, Zhang J, Xu P, Xu T. 2012. Rational design of true monomeric and bright photoactivatable fluorescent proteins. Nat Methods 9:727–729. doi: 10.1038/nmeth.2021. [DOI] [PubMed] [Google Scholar]
- 41.Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. 2006. Imaging intracellular fluorescent proteins at nanometer resolution. Science 313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 42.Huang B, Wang W, Bates M, Zhuang X. 2008. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science 319:810–813. doi: 10.1126/science.1153529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bates M, Huang B, Dempsey GT, Zhuang X. 2007. Multicolor super-resolution imaging with photo-switchable fluorescent probes. Science 317:1749–1753. doi: 10.1126/science.1146598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van de Linde S, Löschberger A, Klein T, Heidbreder M, Wolter S, Heilemann M, Sauer M. 2011. Direct stochastic optical reconstruction microscopy with standard fluorescent probes. Nat Protoc 6:991–1009. doi: 10.1038/nprot.2011.336. [DOI] [PubMed] [Google Scholar]
- 45.Huang B, Bates M, Zhuang X. 2009. Super-resolution fluorescence microscopy. Annu Rev Biochem 78:993–1016. doi: 10.1146/annurev.biochem.77.061906.092014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee S-H, Shin JY, Lee A, Bustamante C. 2012. Counting single photoactivatable fluorescent molecules by photoactivated localization microscopy (PALM). Proc Natl Acad Sci U S A 109:17436–17441. doi: 10.1073/pnas.1215175109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nino D, Rafiei N, Wang Y, Zilman A, Milstein JN. 2017. Molecular counting with localization microscopy: a Bayesian estimate based on fluorophore statistics. Biophys J 112:1777–1785. doi: 10.1016/j.bpj.2017.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nino D, Djayakarsana D, Milstein JN. 2019. Nanoscopic stoichiometry and single‐molecule counting. Small Methods 3:1900082. doi: 10.1002/smtd.201900082. [DOI] [Google Scholar]
- 49.Wang Y, Penkul P, Milstein JN. 2016. Quantitative localization microscopy reveals a novel organization of a high-copy number plasmid. Biophys J 111:467–479. doi: 10.1016/j.bpj.2016.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alqahtany M, Khadka P, Niyonshuti I, Krishnamurthi VR, Sadoon AA, Challapalli SD, Chen J, Wang Y. 2019. Nanoscale reorganizations of histone-like nucleoid structuring proteins in Escherichia coli are caused by silver nanoparticles. Nanotechnology 30:385101. doi: 10.1088/1361-6528/ab2a9f. [DOI] [PubMed] [Google Scholar]
- 51.Sadoon AA, Wang Y. 2018. Anomalous, non-Gaussian, viscoelastic, and age-dependent dynamics of histonelike nucleoid-structuring proteins in live Escherichia coli. Phys Rev E 98:042411. doi: 10.1103/PhysRevE.98.042411. [DOI] [Google Scholar]
- 52.Mazouchi A, Milstein JN. 2016. Fast Optimized Cluster Algorithm for Localizations (FOCAL): a spatial cluster analysis for super-resolved microscopy. Bioinformatics 32:747–754. doi: 10.1093/bioinformatics/btv630. [DOI] [PubMed] [Google Scholar]
- 53.Nath K, Koch AL. 1970. Protein degradation in Escherichia coli. I. Measurement of rapidly and slowly decaying components. J Biol Chem 245:2889–2900. [PubMed] [Google Scholar]
- 54.Levet F, Hosy E, Kechkar A, Butler C, Beghin A, Choquet D, Sibarita J-B. 2015. SR-Tesseler: a method to segment and quantify localization-based super-resolution microscopy data. Nat Methods 12:1065–1071. doi: 10.1038/nmeth.3579. [DOI] [PubMed] [Google Scholar]
- 55.Bal W, Protas AM, Kasprzak KS. 2011. Genotoxicity of metal ions: chemical insights. Met Ions Life Sci 8:319–373. [PubMed] [Google Scholar]
- 56.Haeberlin A, Zurbuchen A, Walpen S, Schaerer J, Niederhauser T, Huber C, Tanner H, Servatius H, Seiler J, Haeberlin H, Fuhrer J, Vogel R. 2015. The first batteryless, solar-powered cardiac pacemaker. Heart Rhythm 12:1317–1323. doi: 10.1016/j.hrthm.2015.02.032. [DOI] [PubMed] [Google Scholar]
- 57.Guyer MS, Reed RR, Steitz JA, Low KB. 1981. Identification of a sex-factor-affinity site in E. coli as gamma delta. Cold Spring Harbor Symp Quant Biol 45(Part 1):135–140. doi: 10.1101/sqb.1981.045.01.022. [DOI] [PubMed] [Google Scholar]
- 58.Jensen KF. 1993. The Escherichia coli K-12 “wild types” W3110 and MG1655 have an rph frameshift mutation that leads to pyrimidine starvation due to low pyrE expression levels. J Bacteriol 175:3401–3407. doi: 10.1128/jb.175.11.3401-3407.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wiedenmann J, Ivanchenko S, Oswald F, Schmitt F, Röcker C, Salih A, Spindler K-D, Nienhaus GU. 2004. EosFP, a fluorescent marker protein with UV-inducible green-to-red fluorescence conversion. Proc Natl Acad Sci U S A 101:15905–15910. doi: 10.1073/pnas.0403668101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sadoon AA, Khadka P, Freeland J, Gundampati RK, Manso RH, Ruiz M, Krishnamurthi VR, Thallapuranam SK, Chen J, Wang Y. 2020. Silver ions caused faster diffusive dynamics of histone-like nucleoid-structuring proteins in live bacteria. Appl Environ Microbiol 86:e02479-19. doi: 10.1128/AEM.02479-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Koh CM. 2013. Storage of bacteria and yeast. Methods Enzymol 533:15–21. doi: 10.1016/B978-0-12-420067-8.00002-7. [DOI] [PubMed] [Google Scholar]
- 62.Gorman R, Adley CC. 2004. An evaluation of five preservation techniques and conventional freezing temperatures of −20 degrees C and −85 degrees C for long-term preservation of Campylobacter jejuni. Lett Appl Microbiol 38:306–310. doi: 10.1111/j.1472-765x.2004.01490.x. [DOI] [PubMed] [Google Scholar]
- 63.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rasband WS. 1997. ImageJ. U. S. National Institutes of Health, Bethesda, MD. [Google Scholar]
- 66.Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N. 2010. Computer control of microscopes using μManager. Curr Protoc Mol Biol Chapter 14:Unit14.20. doi: 10.1002/0471142727.mb1420s92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Edelstein AD, Tsuchida MA, Amodaj N, Pinkard H, Vale RD, Stuurman N. 2014. Advanced methods of microscope control using μManager software. J Biol Methods 1:10. doi: 10.14440/jbm.2014.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wolter S, Löschberger A, Holm T, Aufmkolk S, Dabauvalle M-C, van de Linde S, Sauer M. 2012. rapidSTORM: accurate, fast open-source software for localization microscopy. Nat Methods 9:1040–1041. doi: 10.1038/nmeth.2224. [DOI] [PubMed] [Google Scholar]
- 69.Wang Y, Schnitzbauer J, Hu Z, Li X, Cheng Y, Huang Z-L, Huang B. 2014. Localization events-based sample drift correction for localization microscopy with redundant cross-correlation algorithm. Opt Express 22:15982–15991. doi: 10.1364/OE.22.015982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Coltharp C, Kessler RP, Xiao J. 2012. Accurate construction of photoactivated localization microscopy (PALM) images for quantitative measurements. PLoS One 7:e51725. doi: 10.1371/journal.pone.0051725. [DOI] [PMC free article] [PubMed] [Google Scholar]